This study examines the activation of a conserved mitogen-activated protein kinase (MAPK) cascade that is essential for fungal pathogenicity in plants. A highly glycosylated mucin-like transmembrane protein is shown to contribute to phosphorylation of the MAPK that promotes invasive growth and virulence in the soilborne vascular wilt fungus Fusarium oxysporum.
Abstract
Fungal pathogenicity in plants requires a conserved mitogen-activated protein kinase (MAPK) cascade homologous to the yeast filamentous growth pathway. How this signaling cascade is activated during infection remains poorly understood. In the soil-borne vascular wilt fungus Fusarium oxysporum, the orthologous MAPK Fmk1 (Fusarium MAPK1) is essential for root penetration and pathogenicity in tomato (Solanum lycopersicum) plants. Here, we show that Msb2, a highly glycosylated transmembrane protein, is required for surface-induced phosphorylation of Fmk1 and contributes to a subset of Fmk1-regulated functions related to invasive growth and virulence. Mutants lacking Msb2 share characteristic phenotypes with the Δfmk1 mutant, including defects in cellophane invasion, penetration of the root surface, and induction of vascular wilt symptoms in tomato plants. In contrast with Δfmk1, Δmsb2 mutants were hypersensitive to cell wall targeting compounds, a phenotype that was exacerbated in a Δmsb2 Δfmk1 double mutant. These results suggest that the membrane mucin Msb2 promotes invasive growth and plant infection upstream of Fmk1 while contributing to cell integrity through a distinct pathway.
INTRODUCTION
Plant pathogenic fungi have evolved sophisticated mechanisms to invade their hosts, overcome their defenses, and colonize their living tissues, thereby causing disease. One of the most broadly conserved pathogenicity mechanisms involves a mitogen-activated protein kinase (MAPK) homologous to the yeast mating/filamentation MAPKs Fus3/Kss1 (Qi and Elion, 2005). The essential role during infection of this so-called pathogenicity MAPK was first reported in the rice blast fungus Magnaporthe oryzae. Mutants lacking the Pmk1 gene were not only defective in formation of appressoria but failed to colonize host plant tissue when inoculated through wound sites, a process known as invasive or infectious growth (Xu and Hamer, 1996). Pmk1 orthologs are essential for infection in a wide range of biologically and taxonomically diverse plant pathogens, suggesting an evolutionarily conserved role of this MAPK cascade in fungal pathogenicity in plants (Zhao et al., 2007).
The soilborne fungus Fusarium oxysporum causes devastating vascular wilt in more than 100 different plant species (Armstrong and Armstrong, 1981). The Pmk1 ortholog of F. oxysporum f. sp lycopersici, Fmk1, is essential for infection of tomato (Solanum lycopersicum) plants (Di Pietro et al., 2001). Mutants lacking Fmk1 are deficient in multiple virulence-related functions, such as adhesion to tomato roots, root penetration, secretion of pectinolytic enzymes, and invasive growth on living plant tissue (Di Pietro et al., 2001; Delgado-Jarana et al., 2005). Fmk1 is also required for vegetative hyphal fusion, a ubiquitous process in filamentous fungi whose biological role remains poorly understood (Prados Rosales and Di Pietro, 2008). Recent work has established that invasive growth, the most critical of these Fmk1-regulated functions for plant infection, is mediated by the homeodomain transcription factor Ste12 (Rispail and Di Pietro, 2009). At present, the signals and receptors that activate this conserved MAPK cascade during plant infection remain largely unknown (Zhao et al., 2007).
In the model fungus Saccharomyces cerevisiae, the orthologous MAPK Kss1 regulates filamentous growth (FG) and agar invasion in response to nutrient limitation (Truckses et al., 2004; Qi and Elion, 2005). Two transmembrane proteins, Msb2 and Sho1, are required for activation of FG through Kss1 (Cullen et al., 2004; Vadaie et al., 2008). Msb2 has a short cytoplasmic tail and a large, highly O-glycosylated extracellular domain that is reminiscent of the mammalian family of signaling mucins (Cullen et al., 2004). Under nutrient-limiting conditions, Msb2 is activated by cleavage of the extracellular inhibitory domain and interacts with Sho1 to activate the FG pathway (Vadaie et al., 2008). Msb2 also participates in a second MAPK pathway in S. cerevisiae, the high osmolarity glycerol (HOG) MAPK pathway (Tatebayashi et al., 2007). Putative orthologs of Sho1 and Msb2 in two plant pathogenic fungi, Ustilago maydis and M. oryzae, have been associated with signaling upstream of the pathogenicity MAPK cascade, recognition of surface signals, and appressorium differentiation (Lanver et al., 2010; Liu et al., 2011).
Here, we report the identification and characterization of F. oxysporum Msb2, a highly glycosylated mucin-type membrane protein with a domain structure similar to S. cerevisiae Msb2. We provide evidence for a dual function of Msb2 in F. oxysporum. It promotes invasive growth and plant infection via the Fmk1 MAPK cascade and contributes to maintenance of cell integrity through a distinct pathway.
RESULTS
F. oxysporum Msb2 Encodes a Predicted Transmembrane Protein with a Large Extracellular Mucin Homology Domain and a Short Cytoplasmic Region
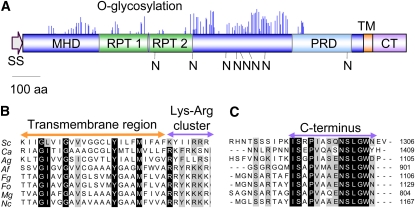
A BLAST search of the complete genome database of F. oxysporum with the amino acid sequence of S. cerevisiae Msb2 identified a single putative ortholog, FOXG_09254, encoding a hypothetical protein of 1129 amino acids with a molecular mass of 117.5 kD and a pI of 4.48. The predicted F. oxysporum Msb2 protein has a domain architecture similar to that of S. cerevisiae Msb2 (Figure 1A), including an N-terminal signal sequence (20 amino acids), a large extracellular domain (amino acids 21 to 991) with a Ser/Thr/Pro-rich region predicted to be highly O-glycosylated (mucin homology domain [MHD], amino acids 106 to 836), a positive regulatory domain (PRD; 176 amino acids), a single transmembrane domain (TM; 22 amino acids), and a short cytoplasmic tail (CT; 95 amino acids). The overall sequence identity between F. oxysporum and S. cerevisiae Msb2 proteins was rather low (20.2%), particularly in the MHD, defined as the extracellular region extending from the first to the last amino acid predicted to be O-glycosylated by the NetOGlyc algorithm (19%). In contrast with S. cerevisiae Msb2, no exact repeats were present in the MHD domain of the F. oxsyporum protein. However, two regions containing nonexact repeats were detected using the Prospero algorithm implemented in the SMART program (Mott, 2000). Somewhat higher identity values were found in the cytoplasmic tail (25.7%) and in a region of ~100 amino acids located upstream of the transmembrane domain (25.8%), named PRD for positive regulatory domain (Cullen et al., 2004). Putative Msb2 orthologs are also present in the genome sequences of other ascomycetes, including plant and human pathogens (Rispail et al., 2009), displaying a similarly conserved domain architecture as F. oxysporum Msb2 (see Supplemental Figure 1 online). While S. cerevisiae and Ashbya gossypii have two paralogs, Msb2 and Hkr1, the other ascomycete species surveyed in this study, contain a single Msb2 ortholog. Sequence identity scores among orthologs were highest in the TM, PRD, and CT regions, with values of above 50% for the TM and CT regions between most filamentous ascomycetes, including F. oxysporum (Figure 1B; see Supplemental Figure 1 online). Collectively, these results suggest that FOXG_09254 encodes a structural ortholog of S. cerevisiae Msb2 and that Msb2 is conserved in ascomycetes.
Figure 1.
F. oxysporum Msb2 Is a Structural Ortholog of the S. cerevisiae Msb2 Mucin.
(A) Schematic representation of the F. oxysporum Msb2 protein. Shown are the N-terminal signal sequence (SS), the extracellular Ser/Thr/Pro-rich MHD with imperfect repeats (RPT), the PRD, the TM, and the CT. O-glycosylation sites (blue peaks) and N-glycosylation sites (N) are represented as predicted by NetOGlyc 3.1 and NetNGlyc 1.0, respectively. Each peak represents the score value calculated for a Ser or Thr residue in the sequence above the limiting threshold. aa, amino acids.
(B) and (C) Amino acid sequence alignment of the TM (B) and C-terminal intracellular regions (C) of the putative Msb2 orthologs of S. cerevisiae (Sc), C. albicans (Ca), A. gossypii (Ag), A. fumigatus (Af), F. graminearum (Fg), F. oxysporum (Fo), M. oryzae (Mg), and N. crassa (Nc). Highly conserved residues are shaded in black; moderately conserved residues are shaded in gray.
We next tested whether F. oxysporum Msb2 could confer signaling function in S. cerevisiae. The open reading frame (ORF) and terminator of F. oxysporum Msb2 was inserted at the genomic MSB2 locus downstream of the native promoter (see Supplemental Figure 2A online) in the yeast strain PC538 carrying a FUS1:HIS3 reporter whose expression depends on the FG pathway (Cullen et al., 2004). Correct gene replacement and the presence of the F. oxysporum Msb2 transcript in the yeast transformants was confirmed by genomic PCR and RT-PCR analysis, respectively (see Supplemental Figures 2B and 2C online). Replacement of native MSB2 with F. oxysporum Msb2 resulted in impaired growth on medium lacking His, similar to the msb2 deletion strain PC948 and in contrast with the ectopic transformants and the untransformed PC538 strain (see Supplemental Figure 2D online). Lack of complementation by F. oxysporum Msb2 was also observed in the agar invasion assay. These results suggest that F. oxysporum Msb2 does not complement the signaling function of S. cerevisiae Msb2.
Msb2 Contributes to Hyphal Growth on Solid Surfaces under Conditions of Nitrogen Limitation and Cell Wall Stress
To explore the biological role of Msb2 in F. oxysporum, we generated a Δmsb2 allele by replacing most of the ORF with the hygromycin resistance cassette (see Supplemental Figure 3A online). The construct was introduced into the wild-type strain or the Δfmk1 mutant to study possible epistatic effects between the two genes. DNA gel blot analysis identified several transformants showing replacement of the 9.7-kb EcoRI fragment corresponding to the wild-type Msb2 allele by a fragment of 5.5 kb (see Supplemental Figure 3B online), demonstrating homologous insertion in these transformants, which were named Δmsb2 and Δfmk1 Δmsb2, respectively. Complementation of the Δmsb2 mutation was performed by introducing a 5.3-kb DNA fragment encompassing the complete Msb2 gene into the Δmsb2#62 mutant, using cotransformation with the phleomycin resistance marker. Several phleomycin-resistant transformants produced a PCR amplification product identical to that obtained from the wild-type strain but absent from the Δmsb2#62 mutant (see Supplemental Figure 3C online). We concluded that these transformants, named Δmsb2+msb2, had integrated an intact copy of the Msb2 gene into their genome.
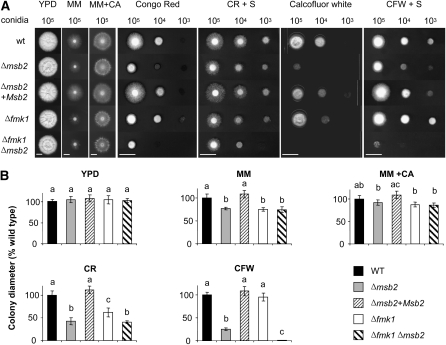
Colonies of the Δmsb2 mutants displayed significantly slower growth than those of the wild-type strain on solid minimal medium (MM) containing the poor nitrogen source NO3 but not on MM containing casaminoacids or on nutrient-rich medium (YPD) (Figure 2). We noted that the Δfmk1 and Δfmk1 Δmsb2 strains displayed a similar decrease in hyphal growth rate as Δmsb2, suggesting the absence of an additive effect in the double mutant. The Δmsb2, Δfmk1, and Δfmk1 Δmsb2 mutants also showed a decrease in colony hydrophobicity on MM + NO3 compared with the wild-type strain (see Supplemental Figure 4 online). By contrast, no significant differences in growth were detected between the strains in submerged culture, either on YPD or on MM + NO3 (see Supplemental Figure 5 online).
Figure 2.
Msb2 Contributes to Hyphal Growth under Conditions of Nutrient Limitation and Cell Integrity Stress.
(A) Colony phenotype of the indicated strains grown on yeast peptone Glc (YPD), minimal medium (MM), MM supplemented with 1% (w/v) casaminoacids (MM+CA), YPD supplemented with 50 μg/mL Congo Red (CR), or 40 μg/mL Calcofluor White (CFW) in the absence or presence of 1 M sorbitol (S). Plates were spot-inoculated with the indicated amount of microconidia, incubated for five (YPD, MM, and MM+CA) or three (CR and CFW) days at 28°C and scanned. Bar = 1 cm. wt, wild type.
(B) Colony diameter on the indicated media was measured after 5 d and plotted relative to the wild-type (WT) strain (100%). Error bars represent standard deviations of colony diameters calculated from five plates. Values with the same letter are not significantly different according to the Mann-Whitney test (P ≤ 0.05).
The Δmsb2, but not the Δfmk1, strains were more sensitive to the cell wall targeting compounds Congo Red (CR) and Calcofluor White (CFW) than the wild-type strain (Figure 2). Addition of 1 M sorbitol partially rescued the growth defect on CR and CFW. Strikingly, a Δfmk1 Δmsb2 double mutant was significantly more sensitive to these compounds than either of the single mutants, indicating that Msb2 and Fmk1 have additive functions in the cell wall stress response. By contrast, no significant differences in sensitivity between strains were detected on menadione (oxidative stress) or sodium chloride (osmotic and salt stress) (see Supplemental Figure 6 online). We conclude that F. oxysporum Msb2 has specific functions in vegetative hyphal growth under conditions of nitrogen limitation and in response to cell wall stress.
Msb2 Is a Highly Glycosylated Membrane Protein
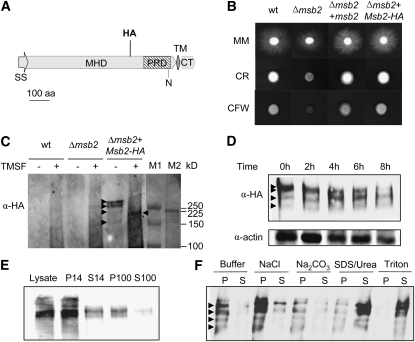
An epitope-tagged allele of Msb2 was generated, by inserting the hemagglutinin (HA) epitope downstream of amino acid residue 722 located in the extracellular MHD domain (Figure 3A). Introduction of the Msb2-HA allele into the Δmsb2#62 mutant fully restored wild-type growth on MM as well as on CR and CFW, suggesting that Msb2-HA is functional in F. oxysporum (Figure 3B). Immunoblot analysis of crude cell extracts of the Δmsb2+Msb2-HA strain with an α-HA antibody detected two major hybridizing bands with an apparent molecular mass of >250 kD as well as two minor bands of ~225 and 170 kD (Figure 3C). These hybridizing bands were lacking in the wild-type and the Δmsb2 controls. A time-course analysis revealed that Msb2-HA was continuously expressed during growth of F. oxysporum on solid MM (Figure 3D).
Figure 3.
Msb2 Is a Glycosylated Membrane Protein.
(A) Schematic representation of the Msb2-HA protein carrying the HA epitope at amino acid (aa) residue 722 located in the MHD region. Symbols and abbreviations are as in Figure 1A.
(B) Colony phenotype of the indicated strains grown on YPD medium in the absence or presence of CR or CFW. Experimental conditions are as in Figure 2. wt, wild type.
(C) Deglycosylation of Msb2. Cell lysates from the indicated strains germinated for 15 h in potato dextrose broth (PDB) and subsequently transferred for 8 h onto MM plates were incubated in the absence or presence of TMSF, separated by SDS-PAGE, and subjected to immunoblot analysis with monoclonal α-HA antibody. Hybridizing bands are marked by arrowheads. Molecular masses of prestained markers from Bio-Rad and Amersham (M1 and M2, respectively) are indicated to the right.
(D) Immunoblot analysis of cell lysates from the Δmsb2+Msb2-HA strain germinated as described in (C) and transferred onto MM plates for the indicated time periods.
(E) Subcellular localization of Msb2-HA. Immunoblot analysis of cell lysates of the Δmsb2+Msb2-HA strain obtained as described in (C) and separated by centrifugation. Crude lysate, supernatant (S), and pellet (P) fractions are shown. 14, 14,000g; 100, 100,000g.
(F) P14 fraction analysis. Treatments were as follows: lysis buffer alone (Buffer) or with 0.5 M NaCl, 100 mM Na2CO3 at pH 11, 5% SDS + 8 M urea, or 1% Triton.
Yeast Msb2 as well as mammalian mucins are highly glycosylated (Silverman et al., 2001; Cullen et al., 2004). The MHD domain of F. oxysporum Msb2 contains seven putative sites for N-linked glycosylation and many predicted sites for O-linked glycosylation (Figure 1A). We noted that the apparent molecular mass of Msb2-HA (>250 kD) was substantially higher than predicted (117.5 kD), suggesting that F. oxysporum Msb2 may be glycosylated. Consistent with this hypothesis, treatment of crude cell extracts of the Δmsb2+Msb2-HA strain with trifluoromethanesulfonic acid (TMSF), which removes both O- and N-linked glycosyl side chains, resulted in increased electrophoretic mobility of the two major hybridizing bands, giving rise to a single band with an apparent molecular mass of ~200 kD (Figure 3C).
To study subcellular localization of Msb2, we performed immunoblot analysis of different subcellular fractions. Almost all of the HA-tagged protein was detected in the P14 fraction, a location consistent with the plasma membrane and associated proteins (Figure 3E). Treatments that disrupt the membrane lipid layer, such as SDS/urea and Triton, completely released the Msb2-HA protein from the P14 to the soluble fraction, confirming the bioinformatic prediction that F. oxysporum Msb2 is an integral membrane protein (Figure 3F).
Msb2 Is Shed from the Cell Surface
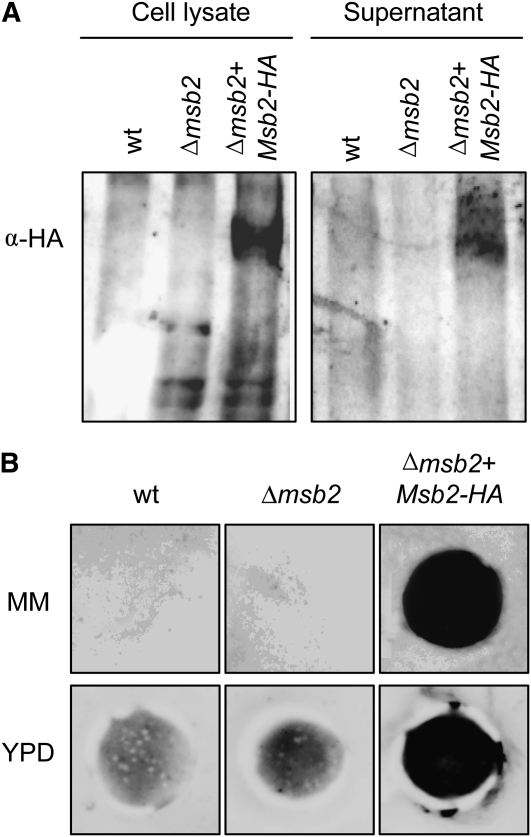
While studying subcellular localization of Msb2, we noted that treatments that solubilize peripheral membrane proteins, such as high salt, urea, or sodium bicarbonate, consistently released part of the membrane-bound protein into the supernatant (Figure 3F). This result suggested that a proportion of Msb2 may be peripherally associated rather than integral to the membrane. Recent work in S. cerevisiae suggested that Msb2 is processed through proteolytic cleavage and that a part of the extracellular domain is shed from the cells (Vadaie et al., 2008). Using immunoblot analysis, we detected a hybridizing band of the expected size in culture supernatants of the Δmsb2+Msb2-HA strain but not in those of the wild type and the Δmsb2 strain (Figure 4A). Shedding of F. oxysporum Msb2 was confirmed by colony blot analysis (Pitoniak et al., 2009), revealing a strong hybridizing signal underneath the colonies of the Δmsb2+Msb2-HA strain grown on MM or YPD medium but not those of the negative controls (Figure 4B). These results suggest that the highly glycosylated extracellular part of F. oxysporum Msb2 is shed from the cell surface into the surrounding medium.
Figure 4.
Msb2 Is Shed from the Cell Surface.
(A) Immunoblot with α-HA antibody of cell lysates and culture supernatants of the indicated strains. wt, wild type.
(B) Colony inmunoblot assay. Microconidia of the indicated strains were germinated for 15 h in PDB, harvested and washed twice in water, transferred onto 0.2-μm pore size filters placed over a plate of MM or YPD overlaid with a nitrocellulose filter, and incubated for 8 h at 28°C. The 0.2-μm filters with the colonies were removed, and nitrocellulose membranes were washed with running water and subjected to immunoblot with α-HA antibody.
Msb2 Regulates Phosphorylation of the MAPK Fmk1 and Expression of Fmk1-Regulated Effector Genes
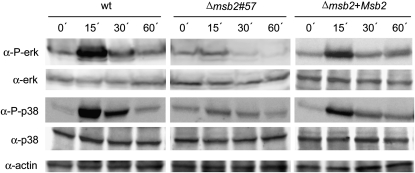
We previously noted that the Δfmk1 and Δmsb2 mutants shared similar hyphal growth defects on solid medium containing the poor nitrogen source nitrate (Figure 2). We therefore used this culture condition to investigate the hypothesis that Msb2 functions upstream of the Fmk1 MAPK cascade. Immunoblot analysis of the wild-type strain with α-phospho-p44/42 MAPK antibody detected a rapid and transient increase in Fmk1 phosphorylation levels upon transfer from submerged culture to solid MM, with a peak in phosphorylation after 15 min (Figure 5). Two independent Δmsb2 mutants failed to show an increase in Fmk1 phosphorylation after transfer to solid MM, while the phosphorylation peak was restored in the complemented strain (Figure 5; see Supplemental Figure 7A online). This suggests that Msb2 is required for transient phosphorylation of the Fmk1 MAPK upon contact with a solid surface.
Figure 5.
Msb2 Contributes to Phosphorylation of the Fmk1 and Hog1 MAPKs.
Transfer to solid medium induces a transient Msb2-dependent increase in phosphorylation of Fmk1 and Hog1. Total protein extracts from the indicated strains germinated for 15 h in PDB and transferred onto MM plates for the indicated time periods (min) were subjected to immunoblot analysis with anti-phospho-p44/42 MAPK antibody (α-P-erk) that only detects the phosphorylated form of Fmk1and anti-p44/p42 MAPK antibody (α-ERK) or with anti-phospho-p38 MAPK antibody (α-P-p38) that only detects the phosphorylated form of Hog1 and anti-p38 MAPK antibody (α-p38). A monoclonal α-actin antibody was used as loading control. wt, wild type.
Besides Fmk1, F. oxysporum has two additional MAPKs, the cell integrity MAPK, Mpk1, and the hyperosmotic response MAPK, Hog1 (Rispail et al., 2009). Phosphorylation of Hog1 was investigated using a commercial p38 MAPK antibody that specifically detects the phosphorylated form of Hog1. Immunoblot with α-phospho-p38 detected a major hybridizing band migrating at a predicted molecular mass of 38 kD, whose intensity increased rapidly and transiently upon transfer of the wild-type strain from submerged culture to solid MM with a peak in phosphorylation at 15 min (Figure 5). Phosphorylation of Hog1 was strongly attenuated in the Δmsb2 mutant and largely restored in the complemented strain.
Phospohorylation status of Mpk1 was monitored with the α-phospho-p44/42 MAPK antibody employed for detection of Fmk1 using prolonged exposure times. As expected from the predicted molecular mass, phospho-Mpk1 migrated at a higher molecular mass position than phospho-Fmk1 (see Supplemental Figure 7B online). In contrast with Fmk1, phosphorylation of Mpk1 did not vary significantly after transfer to solid MM. However, the amount of phospho-Mpk1 appeared slightly higher in the Δmsb2, Δfmk1, and Δfmk1 Δmsb2 mutants compared with the wild-type strain.
Transcript levels of the three MAPK genes were measured using quantitative real-time PCR analysis (qPCR) of cDNA obtained from mycelia 6 h after transfer to MM. Under these conditions, levels of the Fmk1 transcript were reduced by 30% in the Δmsb2 mutant compared with the wild-type strain (see Supplemental Figure 8 online). Levels of the Hog1 transcript decreased between 60 and 80% in the Δmsb2, Δfmk1, and Δfmk1 Δmsb2 mutants compared with wild-type levels. By contrast, no significant differences in transcript levels of Mpk1 were detected between the strains.
qPCR analysis revealed a reduction of Msb2 expression in the Δfmk1 mutant versus the wild-type strain, suggesting that Msb2 may itself be a transcriptional target of the Fmk1 MAPK cascade (Figure 6). We also noted that expression of Msb2 in the complemented strain was significantly higher than in the wild type, possibly due to multiple copies of the complementing allele or to positional effects at the ectopic integration site.
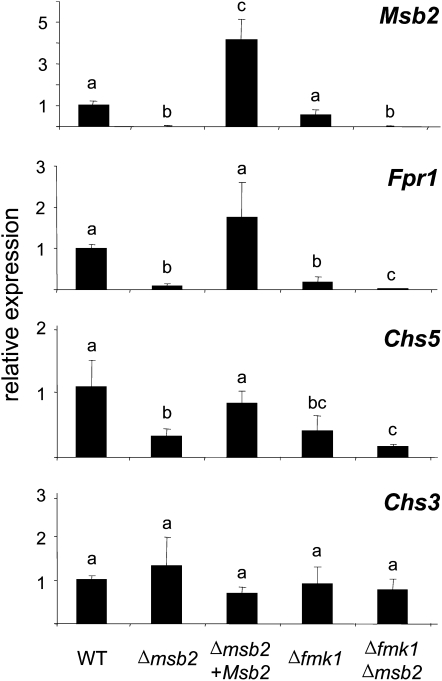
Figure 6.
Msb2 Modulates the Expression of Fmk1-Regulated Effector Genes.
mRNA abundance of the indicated genes was measured 6 h after transfer of the strains to MM plates using real-time qPCR. Relative expression levels represent mean cycle threshold values normalized to actin gene expression levels and relative to expression values in the wild-type (WT) strain. Bars represent standard errors calculated from three biological replicates. Values with the same letter are not significantly different according to the Mann-Whitney test (P ≤ 0.05).
To further investigate the role of Msb2 as an upstream component of Fmk1, we examined expression of Fpr1, a gene encoding a secreted protein with an SCP-PR-1–like domain that was previously shown by RNA gel blot analysis to be transcriptionally activated by the Fmk1 MAPK cascade (R.C. Prados-Rosales and A. Di Pietro, unpublished data). In agreement with the earlier results, Fpr1 transcript levels in the Δfmk1 mutant were 5 times lower than in the wild-type strain. Interestingly, expression of Fpr1 was 10-fold reduced in the Δmsb2 mutant and 100-fold downregulated in the Δfmk1 Δmsb2 double mutant (Figure 6). A similar trend was detected in ChsV, which encodes a class V chitin synthase essential for pathogenicity of F. oxysporum (Madrid et al., 2003). By contrast, Chs3, which codes for a distinct class of chitin synthase, did not display significant differences in transcript levels between the strains (Figure 6). Thus, Msb2 regulates the expression of two Fmk1-regulated effectors that are either secreted (Fpr1) or located at the cell surface (ChsV).
Msb2 Contributes to Fmk1-Dependent Invasive Growth Functions
To investigate the role of Msb2 in regulating virulence-associated functions modulated by Fmk1, we systematically compared the phenotypes of the Δfmk1, Δmsb2, and Δfmk1 Δmsb2 mutants. One of these functions is vegetative hyphal fusion, which can be monitored in submerged culture by the presence of interconnected mycelial networks that are macroscopically visible as hyphal aggregates (see Supplemental Figure 9A online). In contrast with the wild-type strain, Δfmk1 mutants are defective in hyphal fusion and aggregation (Prados Rosales and Di Pietro, 2008). Two independent Δmsb2 mutants produced hyphal aggregates to a similar extent as the wild-type strain, indicating that Msb2 is dispensable for vegetative hyphal fusion. This was further corroborated by microscopy examination of germinated microconidia, which revealed the presence of hyphal fusion events at similar frequencies in the wild type and the Δmsb2 strains, while no such events were observed in the Δfmk1 and Δfmk1 Δmsb2 mutants (see Supplemental Figure 9A online).
Impaired hyphal fusion was previously suggested as a likely cause for the lack of hyphal adhesion of the Δfmk1 mutant to tomato roots (Prados Rosales and Di Pietro, 2008). Consistent with this idea, the Δmsb2 mutants still showed robust root adhesion in contrast with the Δfmk1 and Δfmk1 Δmsb2 strains (see Supplemental Figure 9A online).
Besides hyphal fusion, Fmk1 is required for multiple functions associated with invasive growth on plant tissue. First, Δfmk1 mutants have reduced extracellular pectinolytic activity during growth on plates containing polygalacturonic acid, as shown by a lack of clear halo production (Delgado-Jarana et al., 2005). Two independent Δmsb2 strains showed an intermediate phenotype for pectinolytic activity, which was less extreme than that of the Δfmk1 mutants but clearly reduced compared with the wild type (see Supplemental Figure 9B online). Second, Δfmk1 mutants are defective in penetration of cellophane membranes (Prados Rosales and Di Pietro, 2008; López-Berges et al., 2010). Interestingly, the Δmsb2 strains had a reduced ability to penetrate cellophane, although they retained some invasive capacity in contrast with the Δfmk1 and Δfmk1 Δmsb2 mutants (see Supplemental Figure 9B online). Third, Δfmk1 mutants fail to invade and colonize living fruit tissue (Di Pietro et al., 2001; Rispail and Di Pietro, 2009). The Δmsb2 mutants showed reduced invasive growth on tomato fruits or apple (Malus domestica) slices (see Supplemental Figure 10 online). All the invasive growth phenotypes of the Δmsb2 mutants were fully restored in the complemented strain.
Msb2 Regulates Root Penetration and Virulence on Tomato Plants
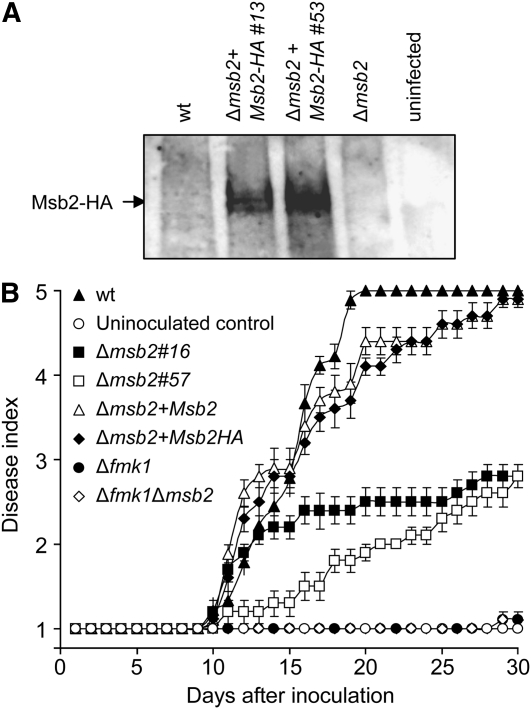
Immunoblot analysis with α-HA antibody detected a robust hybridization signal in protein extracts from tomato roots inoculated with two independent F. oxysporum strains carrying the Msb2-HA allele but not from roots inoculated with the wild type or the Δmsb2 mutant or from uninoculated roots (Figure 7A). This result demonstrates that Msb2 is expressed by F. oxysporum during the early stages of infection. Plants inoculated with the wild-type strain showed a progressive increase in vascular wilt symptoms, and most were dead 20 d after inoculation (Figure 7B). As reported previously (Di Pietro et al., 2001), tomato plants inoculated with the Δfmk1 mutant failed to develop any disease symptoms. Three independent Δmsb2 mutants showed reduced virulence (two representative mutants are shown in Figure 7B) but still produced a low level of disease symptoms in contrast with the Δfmk1 and the Δfmk1 Δmsb2 strains. Complementation of the Δmsb2 mutants with wild-type Msb2 or the Msb2-HA allele restored virulence.
Figure 7.
Msb2 Is Expressed during the Early Stages of Infection and Contributes to Virulence on Tomato Plants.
(A) Total protein extracts were obtained from tomato roots 48 h after inoculation with microconidia of the indicated strains or from uninoculated roots and submitted to immunoblot analysis with α-HA antibody. wt, wild type.
(B) Incidence of Fusarium wilt on tomato plants inoculated with the indicated strains. Severity of disease symptoms was recorded using an index ranging from 1 (healthy plant) to 5 (dead plant). Error bars represent standard errors calculated from 20 plants.
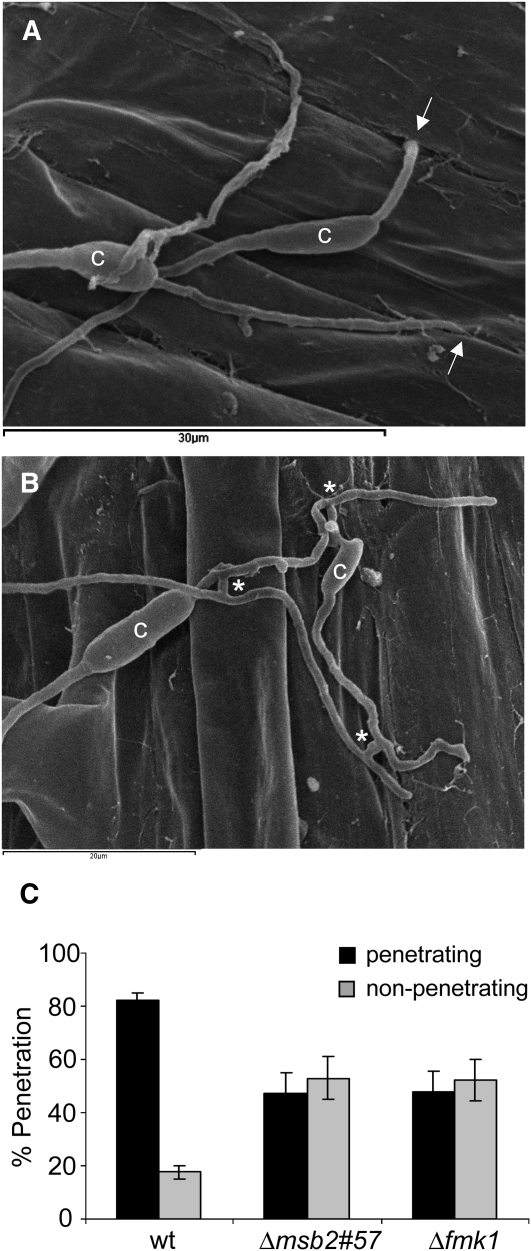
To examine penetration of the tomato root surface by F. oxysporum, roots inoculated with microconidia of the different strains were analyzed after 24 h by scanning electron microscopy. This analysis revealed germ tubes and hyphae entering the root through openings at the junctions between epidermal cells (Figure 8A; see Supplemental Figure 11 online). In the wild-type strain, 83% of the germinated conidia surveyed (n = 80) showed at least one penetration event (Figure 8C). In the Δmsb2 and the Δfmk1 mutants, the frequency of penetration was significantly lower (44 and 50%, respectively), while the proportion of hyphae that failed to enter the root increased (see Supplemental Figure 11C online). The presence of hyphal fusions on the root surface was observed both in the wild type and the Δmsb2 strain (Figure 8B). Collectively, these results demonstrate that Msb2 contributes to root penetration, invasive growth, and virulence of F. oxysporum on tomato plants.
Figure 8.
Msb2 Contributes to Penetration of Tomato Roots.
(A) and (B) Scanning electron microscopy analysis of tomato roots, 24 h after inoculation with microconidia of the F. oxysporum f. sp lycopersici wild-type (A) or Δmsb2 (B) strain. (A) shows fungal hyphae entering the root through openings at intercellular junctions. Arrows point to penetration events. (B) shows multiple hyphal fusion events, indicated by asterisks. c, conidium.
(C) Tomato roots inoculated with the indicated strains and incubated for 24 h were analyzed by scanning electron microscopy. Bars indicate the percentage of fungal germlings showing at least one hypha penetrating the root. In each experiment, 20 germinated conidia per strain were surveyed. Error bars represent standard errors calculated from four independent experiments. Values with the same letter are not significantly different according to the Mann-Whitney test (P ≤ 0.05). wt, wild type.
DISCUSSION
In this work, we describe the genetic and biochemical characterization of Msb2, a mucin-type protein required for invasive growth and virulence of F. oxysporum. We provide evidence that Msb2 functions upstream of Fmk1, a MAPK essential for infection that is conserved in a wide range of plant pathogenic fungi. Our results are in agreement with those of two recent reports on putative Msb2 orthologs in the maize smut pathogen U. maydis (Lanver et al., 2010) and the rice blast fungus M. oryzae (Liu et al., 2011). Collectively, these studies suggest a broadly conserved role of mucin-like proteins in fungal pathogenicity on plants.
F. oxysporum Msb2 Is a Transmembrane Mucin
F. oxysporum Msb2 was identified by its homology to the Msb2 protein from S. cerevisiae. Although the amino acid sequence identity between the two proteins is low, several lines of evidence support the idea that they are structural orthologs. F. oxysporum and S. cerevisiae Msb2 share a common domain structure, including a signal peptide, a large extracellular MHD, a so-called PRD, a TM region, and an intracellular cytoplasmic tail at the C terminus. The presence of an N-terminal signal sequence and a single transmembrane domain suggests that F. oxysporum Msb2 is an integral membrane protein. This hypothesis was confirmed by subcellular fractionation using an HA-tagged version of Msb2, corroborating the results from S. cerevisiae (Cullen et al., 2004). Nevertheless, a fraction of the protein was consistently associated with the P100 fraction corresponding to organelle membranes. Interestingly, in U. maydis and M. oryzae, fluorescent Msb2 fusion proteins localized both to vacuoles and to the cell membrane (Lanver et al., 2010; Liu et al., 2011).
As previously reported in S. cerevisiae, Msb2 in F. oxysporum is shed from the cell surface into the surrounding medium. Shedding was particularly evident during growth on solid surfaces, as detected by a robust hybridizing signal from the secreted Msb2 protein after removal of the fungal colony. The exact mechanism of shedding remains to be determined. In S. cerevisiae, it was suggested that this process involves cleavage of the extracellular Msb2 domain by the glycosylphosphatidyl-inositol–anchored aspartyl protease Yps1p, a member of the yapsin family (Vadaie et al., 2008). A genome-wide inventory of the predicted glycosylphosphatidyl-inositol–anchored proteins in F. oxysporum detected several aspartic proteases, one of which, FOXG_09428, is a putative ortholog of Yps1p (Prados-Rosales et al., 2009). However, immunoblot analysis of F. oxysporum Msb2 proteins obtained from cellular extracts and from supernatants failed to detect an expected difference in electrophoretic mobility, even after prolonged electrophoresis. In S. cerevisiae, immunoblot analysis of an Msb2 version in which the extracellular and cytoplasmic domains were differentially tagged with HA and green fluorescent protein (GFP), respectively, also failed to detect a size difference between Msb2 proteins in the cellular extract and the supernatant (Vadaie et al., 2008). Moreover, hybridization of the cellular extract with α-GFP failed to detect the high molecular mass form of Msb2 but instead detected a 25-kD band (Vadaie et al., 2008). Collectively, these findings would rather argue against the presence of an uncleaved transmembrane form and a cleaved shed form that lacks the transmembrane domain and cytoplasmic tail. Rather, they suggest an alternative mechanism of processing similar to that reported in the human transmembrane mucin MUC1. This protein undergoes autoproteolytic cleavage in the endoplasmic reticulum to generate two subunits, a large extracellular α-subunit and a membrane-tethered β-subunit, that bind together in a strong noncovalent interaction (Ligtenberg et al., 1992; Levitin et al., 2005). A model in which the extracellular domain of Msb2 is peripherally associated rather than integral to the membrane would explain our observation that treatment with high concentrations of salt, urea, or sodium bicarbonate consistently released part of the membrane-bound Msb2 protein into the supernatant. Further studies are needed to clarify the mechanisms involved in processing and shedding of fungal Msb2 proteins.
F. oxysporum and S. cerevisiae Msb2 proteins share a large extracellular MHD (730 residues in F. oxysporum). In S. cerevisiae, the MHD contains several exact tandem repeats rich in Ser, Thr, and Pro residues and is highly glycosylated (Cullen et al., 2004; Yang et al., 2009), two classical hallmarks of mammalian mucins (Hollingsworth and Swanson, 2004). The MHD of F. oxysporum Msb2 is rich in Ser, Thr, and Pro but lacks exact tandem repeats, similar to most of the Msb2 orthologs from filamentous ascomycetes surveyed in this study. The absence of exact repeats in the MHD was previously described in mucins of the protozoan parasite Trypanosoma cruzi, suggesting that exact repeats may not be essential for mucin function but rather represent an evolutionary mechanism for rapid expansion of Ser and Thr residues serving as O-glycosylation sites (Almeida et al., 1994; Di Noia et al., 1996). F. oxysporum Msb2 has multiple predicted sites for O- and N-glycosylation, its apparent molecular mass estimated from immunoblot analysis (>250 kD) was more than double that predicted (117.5 kD), and treatment with the deglycosylating agent TMSF resulted in a decrease in the molecular mass of the protein. Collectively, these results suggest that F. oxysporum Msb2 is a highly glycosylated membrane mucin.
Evidence for a Role of Msb2 in Surface-Induced MAPK Activation
Infection-related development of fungal pathogens is often triggered by contact with the host surface (Kumamoto, 2008). In aerial plant pathogens such as M. oryzae or U. maydis, contact with the leaf surface induces differentiation of an appressorium that builds up turgor pressure to promote entry into the host plant (Wilson and Talbot, 2009). In the non-appressorium-forming soil-borne pathogen F. oxysporum, presence of the host roots induces adhesion and differentiation of infection hyphae that directly penetrate the root (this work; Bishop and Cooper, 1983; Rodríguez-Gálvez and Mendgen, 1995; Lagopodi et al., 2002). The MAPK Fmk1 was previously shown to be essential for these early infection processes (Di Pietro et al., 2001). However, the mechanisms involved in surface sensing upstream of Fmk1 have so far remained elusive.
Here, we provide evidence that contact of F. oxysporum with a solid surface triggers a rapid and transient increase in Fmk1 phosphorylation levels and that this response requires Msb2. These results place Msb2 upstream of the Fmk1 MAPK. Two additional lines of evidence support a role for Msb2 in MAPK activation. Besides Fmk1 phosphorylation, Msb2 also regulates the expression of two Fmk1-regulated genes, Fpr1, which encodes a secreted PR-1-like protein, and ChsV, which encodes a class V chitin synthase with a myosin domain that is essential for plant infection (Madrid et al., 2003). More importantly, Δmsb2 and Δfmk1 mutants share characteristic phenotypes, such as defects in hyphal growth under poor nitrogen conditions, penetration of cellophane membranes, colonization of living fruit tissue, root penetration, and virulence on tomato plants.
It is important to note that, except for hyphal growth and root penetration, the defects of the Δmsb2 strains were consistently less severe than those observed in the Δfmk1 mutant. While this could be explained by the presence of multiple, partially redundant activating components, it cannot be ruled out that Msb2 may not function in a direct hierarchy upstream of Fmk1. In U. maydis, expression of a constitutively active allele of the MAPKK Fuz7 or deletion of the dual specificity phosphatase Rok1, a negative regulator of the downstream MAPKs Kpp2 and Kpp6, restored virulence in the msb2 sho1 double mutant, reinforcing the idea that Msb2 functions upstream in the same pathway (Lanver et al., 2010). Similarly, a dominant active allele of the MAPK kinase MST7 could rescue appressorium formation of the msb2 mutant in M. oryzae (Liu et al., 2011). Further genetic evidence in F. oxysporum will be required to confirm the suggested function of Msb2 upstream of the Fmk1 MAPK.
Additional evidence for a role of Msb2 and Fmk1 in surface sensing comes from the observation that Δmsb2 and Δfmk1 mutants are specifically affected in hyphal growth on solid media but not in submerged culture (this work; Prados Rosales and Di Pietro, 2008). Intriguingly, the hyphal phenotype in the mutants is more severe on a poor nitrogen source, such as nitrate, than on casaminoacids, suggesting that the role of Msb2 and Fmk1 may be linked to nutrient status. A mechanism linking nutrient status to Msb2 activation was recently proposed in S. cerevisiae, whereby the YPS1 gene, which encodes the aspartyl protease involved in processing of Msb2, is induced in response to nutrient limitation (Vadaie et al., 2008).
A key question is how Msb2 mediates surface sensing. The glycosylated extracellular domain of transmembrane mucins has been proposed to function as a sensor of environmental cues (de Nadal et al., 2007; Cullen, 2007). In support of this idea, deletion of the MHD of Msb2 resulted in constitutive activation of the FG pathway (Cullen et al., 2004). Moreover, a combination of O- and N-glycosylation defects induced by tunicamycin and deletion of the O-mannosyltransferase, Pmt4, stimulated FG signaling in an Msb2-dependent manner (Yang et al., 2009). A recent study on in vivo measurement of the mechanical behavior of the glycosylated transmembrane sensor Wsc1, which functions upstream of the yeast cell wall integrity MAPK pathway, suggested that it behaves like a linear nanospring in response to cell surface stress (Dupres et al., 2009). Interestingly, underglycosylation of Wsc1 by pmt4 deletion caused dramatic alterations in protein spring properties (Dupres et al., 2009). Collectively, these studies suggest an important role of extracellular domain glycosylation in the signaling properties of mucin-type transmembrane sensors.
Msb2 and Fmk1 Contribute to Cell Wall Integrity of F. oxysporum through Separate Pathways
In addition to its key role in the FG pathway, S. cerevisiae Msb2 also functions as an osmosensor upstream of the Sho1 branch of the HOG pathway (Tatebayashi et al., 2007). We noted that F. oxysporum Δmsb2 mutants failed to induce a transient peak in phosphorylation of the Hog1 MAPK that was detected in the wild-type strain upon transfer from liquid to solid medium. Intriguingly, the Δmsb2 mutants had no growth defect in the presence of osmotic or oxidative stress but showed increased sensitivity to CR and CFW, two compounds affecting cell wall biosynthesis and composition (Roncero and Durán, 1985). Moreover, the Δfmk1 Δmsb2 double mutant was significantly more sensitive to these compounds than either of the single mutants, revealing a genetic interaction between Msb2 and Fmk1. Based on the mutant phenotypes, we consider it likely that Msb2 and Fmk1 promote cell wall integrity of F. oxysporum through independent pathways. In support of this hypothesis, expression of the chitin synthase gene, chsV, which is required for resistance of F. oxysporum to CR and CFW (Madrid et al., 2003), was decreased in Δmsb2 and Δfmk1 mutants but even more so in the Δfmk1 Δmsb2 double mutant. Deletion of msb2 in the human pathogen Candida albicans also resulted in increased sensitivity to CR and reduced phosphorylation levels of the Fmk1 MAPK ortholog Cek1 in response to cell wall stress (Román et al., 2009). A recent study in S. cerevisiae suggested a cooperative role of the HOG pathway in ensuring survival to cell wall insult (Bermejo et al., 2008). Transcriptional profiling revealed distinct groups of genes whose activation in response to cell wall stress depended on elements of the Sho1 branch of the HOG pathway (García et al., 2009). We speculate that, besides its role upstream of Fmk1, Msb2 functions in the cell wall stress response of F. oxysporum through a separate pathway that may involve components of the Hog1 MAPK cascade.
Msb2 Promotes Fmk1-Dependent Invasive Growth and Plant Pathogenicity
The Δmsb2 strains share several but not all phenotypes of the Δfmk1 mutant. For example, Msb2 is not essential for vegetative hyphal fusion, a process that requires Fmk1 (Prados Rosales and Di Pietro, 2008). On the other hand, all known Fmk1-regulated functions related to invasive growth were affected to some extent in the Δmsb2 mutants, including extracellular pectinolytic activity, penetration of cellophane membranes, and colonization of tomato or apple fruit tissue. Interestingly, a similar subset of Fmk1-dependent invasive growth functions is also impaired in F. oxysporum mutants lacking the homeodomain transcription factor Ste12 (Rispail and Di Pietro, 2009). Moreover, virulence on tomato plants is severely reduced in Δmsb2 and Δste12 mutants, albeit less dramatically than in the Δfmk1 mutant. This suggests that Msb2 contributes to invasive growth, the major virulence function regulated by Fmk1 (Rispail and Di Pietro, 2009), although it cannot be excluded that the reduced virulence of the Δmsb2 mutants could be at least partly linked to the defect in cell wall stress response or to additional yet unknown pathways.
Mutants of U. maydis and M. oryzae lacking Msb2 showed a significant reduction in differentiation of appressoria on inductive surfaces (Lanver et al., 2010; Liu et al., 2011). F. oxysporum does not produce classical appressoria. Here, we investigated the role of Msb2 in penetration of tomato roots using scanning electron microscopy analysis and report a novel penetration mechanism whereby hyphae enter the root through openings located at the junctions of epidermal cells. A previous study using a GFP-marked strain of F. oxysporum f. sp radicis-lycopersici reported preferential colonization of the root surface along the grooves of epidermal cell junctions in the absence of specific infection structures such as appressoria (Lagopodi et al., 2002). The openings between epidermal cells were also present in uninoculated tomato roots, suggesting that they are not caused by the fungus but rather specifically located by the pathogen hyphae. This idea is supported by the remarkably high frequency of penetration (>80% of germinated wild-type spores), suggesting that penetration through preformed openings represents a key mechanism by which F. oxysporum enters the host plant. The Δmsb2 and Δfmk1 mutants showed significantly lower penetration efficiencies, arguing for a role of these two components in the detection and invasion of penetration sites. An intriguing question is through which physical and chemical mechanisms F. oxysporum hyphae successfully locate these preexisting openings on the tomato root surface.
In summary, this study adds to a growing body of evidence implicating the Msb2 transmembrane mucin in early events of surface recognition and plant infection. Because the Δmsb2 mutant has a less severe phenotype in most invasive growth-related functions than the Δfmk1 strain, we speculate that additional upstream components, which are partially redundant with Msb2, also contribute to MAPK activation. One likely candidate is Sho1, a tetraspan membrane protein that is broadly conserved in filamentous ascomycetes (Román et al., 2005; Ma et al., 2008; Rispail et al., 2009), including F. oxysporum (E. Pérez-Nadales and A. Di Pietro, unpublished data). Msb2 and Sho1 were shown to interact physically both in S. cerevisiae and U. maydis (Cullen et al., 2004; Lanver et al., 2010), and a dominant activated version of Sho1 could partially activate the yeast FG MAPK pathway in the absence of Msb2 (Vadaie et al., 2008). In C. albicans, Sho1 cooperates with Msb2 to regulate phosphorylation of the Fmk1 MAPK ortholog Cek1 in response to cell wall stress (Román et al., 2009). Interestingly, individual msb2 or sho1 deletion mutants of U. maydis and M. oryzae showed a partial reduction in appressorium formation and plant infection, while the double mutants were almost completely defective in these functions (Lanver et al., 2010; Liu et al., 2011). Further research will be required to dissect the individual signaling inputs of Msb2 and its possible partner components upstream of the Fmk1 MAPK and to define the downstream factors that regulate pathogenicity in F. oxysporum.
METHODS
Fungal Isolates and Culture Conditions
Fusarium oxysporum f. sp lycopersici race 2 wild-type strain 4287 (FGSC 9935) was used in all experiments. Generation and molecular characterization of the F. oxysporum MAPK mutant Δfmk1 was described previously (Di Pietro et al., 2001). All fungal strains were stored as microconidial suspensions at −80°C with 30% glycerol. For extraction of DNA, microconidium production, and fungal development, cultures were grown in liquid potato dextrose broth (PDB; Difco) at 28°C with shaking at 170 rpm (Di Pietro and Roncero, 1998). For RNA and protein extraction, 5 × 108 freshly obtained microconidia from each strain were inoculated into 200 mL of PDB. After 15 h of incubation at 28°C and 170 rpm, mycelia were harvested, washed twice with sterile minimal medium without trace elements (MM) (Puhalla, 1985), resuspended in 10 mL of MM, and transferred onto three MM agar plates. Plates were incubated for 6 h at 28°C, and mycelia were harvested, frozen in liquid nitrogen, and stored at −80°C. For determination of N-glycosylation, 25 μg/mL tunicamycin (Sigma-Aldrich Chemicals) was added to germlings in PDB obtained as described above, and cultures were incubated for an additional 2 h before harvesting.
Colony Growth Assays
For phenotypic analysis of colony growth, drops of water with serial dilutions (2 × 105, 2 × 104, and 2 × 103) of freshly obtained microconidia were spotted onto agar plates with complete rich medium (YPD: 0.3% yeast extract, 1% peptone, and 2% Glc), MM, or MM supplemented with 10 g/L casaminoacids (Difco). For cell wall stress assays, 50 μg/mL Congo Red or 40 μg/mL Calcofluor white (Sigma-Aldrich) was added to 50 mM MES-buffered YPD plates, pH 6.5 (Ram and Klis, 2006), with or without 1 M sorbitol. For osmotic and oxidative stress assays, YPD plates were supplemented with 0.4, 0.8, or 1 M NaCl, or 10 μg/mL menadione, respectively. Plates were incubated at 28°C for 3 d before the plate was scanned and the colony diameter was measured. Data from four independent experiments, each with two replicates, were analyzed with the software SPSS 15.0 for Windows (LEAD Technologies). Kruskal-Wallis analysis of variance and the Mann-Whitney test were used to assess statistically relevant differences among strains at P ≤ 0.05.
Nucleic Acid Manipulations, Plasmid Vectors, and Fungal Transformations
Total RNA and genomic DNA were extracted from F. oxysporum mycelium as previously reported (López-Berges et al., 2010). All PCRs were routinely performed with the Expand High Fidelity PCR system (Roche).
The F. oxysporum Msb2 gene disruption construct was generated by fusion PCR (Yang et al., 2004). A 1559-bp upstream fragment and a 1716-bp downstream fragment from the F. oxysporum Msb2 locus were amplified from genomic DNA using PCR with primer pairs msb2-for2 and msb2-knock1, and msb2-knock2 and msb2-comp2, respectively (all primers are listed in Supplemental Table 1 online). The hygromycin B resistance gene, under the control of the Aspergillus nidulans gpdA promoter and trpC terminator (Punt et al., 1987) and cloned into the pGEM-T vector (Promega), was amplified with the universal primers M13-For and M13-Rev. The three obtained PCR fragments were used for a final fusion PCR using primers msb2-nest3 and msb2-nest4.
The HA sequence was inserted into the Msb2 ORF at amino acid 722 located in the extracellular domain. A 3707-bp upstream and a 1954-bp downstream PCR fragment were generated by amplification from genomic DNA with primers msb2-for2 and msb2-5′HA-as, and msb2-3′HA-s and msb2-comp2, respectively. Primers msb2-for2 and msb2-comp2 were used for fusion PCR of the two fragments. The 5.6-kb fusion fragment was cloned into pGemT to generate plasmid Msb2-HA-pGemT and sequenced to confirm in-frame insertion of the HA sequence at the correct site.
For targeted gene knockout, the F. oxysporum Msb2-hph fusion construct was used to transform protoplasts of F. oxysporum wild-type strain 4287 and of the Δfmk1 mutant. Hygromycin-resistant transformants were selected and purified by monoconidial isolation as described (Di Pietro et al., 2001). Gene knockout was confirmed by DNA gel blot analysis as previously described (Di Pietro and Roncero, 1998) using the nonisotopic digoxigenin labeling kit (Roche Diagnostics), and, as a probe, an amplification product of the msb2 gene with primers msb2knock2 and msb2rev.
For complementation experiments, a 5.3-kb PCR fragment encompassing either the entire Msb2 gene or the Msb2-HA allele was obtained by PCR amplification from genomic DNA or plasmid Msb2-HA-pGem-T, respectively, using primers msb2-nest3 and msb2-nest4. The obtained fragments were introduced into protoplasts of the Δmsb2#62 mutant by cotransformation with the phleomycin resistance cassette amplified from plasmid pAN8-1, and phleomycin-resistant transformants were isolated as described (Di Pietro et al., 2001). The presence of the wild-type Msb2 allele in the transformants was detected by PCR with gene-specific primers msb2-nest3 and msb2-ORF-as. The presence of the Msb2-HA allele was confirmed by PCR with the epitope-specific primers msb2-HA-s and msb2-3′HA-as.
Real-Time qPCR
Total RNA was isolated from mycelia using the Tripure isolation reagent (Invitrogen) and treated with DNase I (Fermentas). First-strand cDNA was synthesized with Moloney murine leukemia virus reverse transcriptase following the instructions of the manufacturer (Invitrogen) using 1 μg of total RNA.
qPCR reactions were performed in an iCycler apparatus (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad), 400 ng cDNA template, and 300 nM of each gene-specific primer in a final reaction volume of 12.5 μL. All primer pairs amplified products of 160 to 200 bp. The following PCR program was used for all reactions: an initial step of denaturation (5 min, 94°C) followed by 40 cycles of 30 s at 94°C, 30 s at 60°C, 30 s at 72°C, and 20 s at 80°C for measurement of fluorescence emission. A melting curve program was run for which measurements were made at 0.5°C temperature increments every 5 s within a range of 55 to 95°C. Primer pairs act-2 and act-q6, msb2ORF-s and msb2ORF-as, 09795-for and 09795-rev, ChsV-20 and ChsV-36B, Chs3-12 and Chs3-18, Fmk1-F1 and Fmk1-R1, FoHog1-F1 and FoHog1-R1, and FoMpk1-F1 and FoMpk1-R1 were used to amplify Actin, Msb2, Fpr1, ChsV, Chs3, Fmk1, Hog1, and Mpk1 transcripts, respectively (see Supplemental Table 1 online). Real-time PCR primer efficiencies were calculated using LinRegPCR (Ramakers et al., 2003). Actin was used to calculate relative expression levels of the different genes according to the comparative cycle threshold method (Livak and Schmittgen, 2001; Pfaffl, 2001). Data from three independent experiments, each with two technical replicates, were analyzed with the software SPSS 15.0. Kruskal-Wallis analysis of variance and the Mann-Whitney test were used to assess statistically relevant differences among strains for each gene at P ≤ 0.05.
Protein Purification and Immunoblot Analysis
For analysis of the phosphorylation state of the Fmk1 MAPK, germlings from PDB were obtained as described above, transferred onto MM agar plates, and incubated at 28°C for the indicated time periods. Mycelia were harvested, frozen in liquid nitrogen, ground in a mortar, and resuspended in protein extraction buffer (10% glycerol, 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% SDS, 1% Triton, 5 mM EDTA, 1 mM PMSF, Protease inhibitor cocktail P8215 [Sigma-Aldrich], phosphatase inhibitor [50 mM NaF], 5 mM sodium orthovanadate, 50 mM β-glycerophosphate, 1 mM sodium orthovanadate, and PhosSTOP Phosphatase Inhibitor Cocktail tablets [Roche]). Samples were centrifuged to pellet cell debris, and the protein concentration of the supernatant was determined with the Bio-Rad protein assay reagent using BSA as standard. One hundred micrograms of total protein was separated in 4 to 20% Mini-PROTEAN TGX Precast Gels (Bio-Rad) using standard protocols (Sambrook and Russell, 2001). Membranes were blocked using 5% nonfat skimmed milk for 1 h. p44/42 MAP kinases Fmk1 and Mpk1 were detected using the PhosphoPlus p42/p44 MAP kinase (Thr-202/Tyr-204) Antibody kit #9100 (Cell Signaling Technology) according to the manufacturer’s instructions, except that ECL Plus immunoblotting reagent (GE Healthcare) was used for secondary detection of unphosphorylated p44/42 MAPK (Erk1/2). The Hog1 MAP kinase was detected using Phospho-p38 MAPK (Thr180/Tyr182) antibody #9211 and p38 MAPK antibody #9212 (Cell Signaling). Monoclonal α-actin antibody was obtained from Sigma-Aldrich (A3853). Time-course phosphorylation experiments were performed four times independently with similar results.
Immunoblot analysis of Msb2-HA was performed with α-HA-Peroxidase High Affinity antibody 3F10 (Roche) according to the manufacturer’s instructions, followed by detection with ECL Advance reagent. For analysis of Msb2-HA expression during infection of tomato (Solanum lycopersicum) plants, roots of 2-week-old plants of the susceptible cultivar Monika (Syngenta Seeds) were immersed into a microconidial suspension of the different strains in sterile water (2.5 × 106 mL−1) for 48 h at 28°C. Roots with adhering mycelium were collected, frozen in liquid nitrogen, and processed for immunoblot detection of Msb2-HA as described above. Msb2-HA subcellular fractionation experiments were performed as described (Horazdovsky and Emr, 1993; Rieder and Emr, 2001) using Buffer S (10% glycerol, 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1 mM PMSF, and Protease inhibitor cocktail P8215 [Sigma-Aldrich]) for sample homogenization.
For examination of N-glycosylation, cell extracts were treated with EndoHf (New England Biolabs) following the manufacturer’s instructions. Treatment with TMSF to remove N-linked and O-linked carbohydrates from glycoproteins was performed as described (Edge et al., 1981) with some modifications. Briefly, 500 μg of total cell extract protein was dialyzed against water, freeze-dried in a glass vial, resuspended in 150 μL TMSF containing 10% toluene (Sigma-Aldrich), and left at −20°C for 4 h. Neutralization of excess TMSF was performed by adding dropwise 450 μL of pyridine (Sigma-Aldrich) solution (pyridine:methanol:water at a ratio of 3:1:1), followed by the addition of 1200 μL of neutralization solution (0.5% ammonium bicarbonate). Deglycosylated proteins were recovered by dialysis and lyophilization and analyzed by immunoblots.
Analysis of Msb2-HA Secretion
For detection of Msb2-HA in culture supernantants, germlings from PDB obtained as described above were transferred to liquid MM and incubated for 8 h at 28°C at 170 rpm. Cultures were centrifuged to remove mycelia, and supernatants were sterile filtered (0.2 μm pore size), dialyzed against various changes of water for 3 d at 4°C, and lyophilized. Samples were resuspended in water and subjected to immunoblot analysis with α-HA-Peroxidase antibody.
For colony immunoblot assays (Pitoniak et al., 2009), germlings from PDB were obtained as described above, washed twice in sterile water, resuspended in 1 mL of MM, transferred as a colony onto 0.2-μm pore filters placed over an MM agar plate overlaid with a nitrocellulose filter, and incubated for 8 h at 28°C. The 0.2-μm pore filters with the colonies were carefully removed, and the nitrocellulose membranes were washed with running water and submitted to immunoblotting with α-HA-Peroxidase antibody.
Virulence-Related Assays
Assays for polygalacturonase production on polygalacturonic acid plates (Di Pietro and Roncero, 1998), penetration of cellophane membranes (Prados Rosales and Di Pietro, 2008), or adhesion to tomato roots (López-Berges et al., 2010) were performed as described. The presence of vegetative hyphal fusion was determined as described (Prados Rosales and Di Pietro, 2008) using a Leica DMR microscope and the Nomarski technique. Photographs were recorded with a Leica DC 300F digital camera. For determination of colony surface hydrophobicity, a 100-μL drop of water containing 0.01% bromophenol blue (Sigma-Aldrich) was placed on the edge of the colonies grown on YPD or MM and plates were left at room temperature for 4 h before taking pictures. All experiments were performed at least three times with similar results.
Plant Infection Assays
Tomato root infection assays were performed as described (Di Pietro et al., 2001) using the susceptible cultivar Monika (Syngenta Seeds). Briefly, 2-week-old tomato seedlings were inoculated with F. oxysporum strains by immersing the roots in a microconidial suspension, planted in vermiculite, and maintained in a growth chamber. At different times after inoculation, the severity of disease symptoms was recorded using an index ranging from 1 (healthy plant) to 5 (dead plant). Twenty plants were used for each treatment. Assays for invasive growth on tomato fruits (cultivar Daniela) and apple fruit slices (variety Golden Delicious) were performed as previously described (López-Berges et al., 2009). All plant infection experiments were performed at least three times with similar results.
For quantitative analysis of root penetration, tomato roots were immersed in Erlenmeyer flasks containing a suspension of 5 × 106 microconidia/mL and maintained for 24 h at 80 rpm in a growth chamber. Sections of 0.5 cm were excised from the roots at a distance of 3 cm from the root tip and processed for analysis with scanning electron microscopy as described (Pareja-Jaime et al., 2010). In each experiment, 20 germinated conidia per strain were surveyed, whose germ tubes or hyphae were entirely visible (requiring an average of six root sections per strain). Mean values and standard deviations were calculated from four independent experiments.
Complementation of the Saccharomyces cerevisiae MSB2 Gene
Functional complementation of the F. oxysporum Msb2 gene in S. cerevisiae was tested by replacing the MSB2 ORF of S. cerevisiae strain PC538 (Cullen et al., 2004) with the F. oxysporum msb2 ORF and terminator sequence, using PCR targeting and the kanMX4 selectable marker following the method described by Wach et al. (1994). Briefly, F. oxysporum Msb2, including the ORF (3390 bp) and 641-bp upstream and 696-bp downstream regions, were amplified from F. oxysporum genomic DNA with primers Msb2-PstI-F1 and Msb2-Sac2-R1 and cloned into the PstI/SacII sites of plasmid pRS415LEU to generate FoMsb2::pRS415LEU. Next, primer pair Fomsb2-S1 and Fomsb2-S2, containing 5′-45-bp sequences homologous to nucleotides +385 to +430 and +564 to +609, respectively, of the F. oxysporum Msb2 terminator sequence were used for amplification of the kanMX4 module from plasmid pFA6a-kanMX4 (Wach et al., 1994). The kanMX4 PCR fragment (1596 bp) was cotransformed with FoMsb2::pRS415-LEU into S. cerevisiae strain DHD5, selecting for resistance to G418. Plasmid FoMsb2::kanMX4::pRS415-LEU, obtained by homologous recombination, was isolated from geneticin-resistant colonies and used to obtain the final FoMsb2::kanMX4 construct (5404 bp) by amplification with primer pair Fomsb2-C1 and Fomsb2-C2, containing 5′-45-bp sequences homologous to nucleotides −45 to −1 of the S. cerevisiae MSB2 promoter and +26 to +71 of the MSB2 terminator regions, respectively. This construct was used to transform S. cerevisiae strain PC538. Geneticin-resistant colonies were examined by PCR analysis with primers Scmsb2-C3 and Msb2-ORF-as to discriminate between homologous recombination and ectopic transformants. RNA extraction and cDNA synthesis were performed as described above, using cell cultures grown in liquid synthetic complete dextrose (SD) medium (Rose et al., 1990) to an OD600 of 0.8. RT-PCR analysis was done using primer pair msb2-ORF-s and msb2-ORF-as. For phenotypic analysis, strains PC538, PC948 (Δmsb2 deletion mutant in a PC538 background) (Cullen et al., 2004) and the PC538 + Fomsb2::kanMX4 transformants were grown in liquid SD medium to an OD600 of 0.8 and concentrated in water to an OD of 1.0, and 5 μL of this dilution was spotted on solid SD medium lacking His to test for the ability to activate the FUS1:HIS3 reporter or on YPD to test for agar invasion using the plate washing assay, performed as described (Roberts and Fink, 1994; Cullen and Sprague, 2000), with 3 d incubation at 30°C.
Bioinformatic Prediction and Phylogenetic Analysis of Mucins
The F. oxysporum Msb2 protein was identified by a BLASTp search in the Fusarium Comparative Database of the Broad Institute (http://www.broadinstitute.org/annotation/genome/fusarium_group/MultiHome.html) with the S. cerevisiae Msb2 protein sequence. Identification of putative Msb2 orthologs from other fungi was performed as described (Rispail et al., 2009). Protein alignments were made using ClustalW (Thompson et al., 1994), and protein domain predictions were made using the Prosite database (ExPASy; Swiss Institute of Bioinformatics) and SMART (http://smart.embl-heidelberg.de/). The presence of a signal peptide was determined with SignalP version 3.0 (Bendtsen et al., 2004) using a standardized threshold value of 0.5. Putative N-glycosylation and O-glycosylation sites were identified with NetNGlyc 1.0 and NetOGlyc 3.1 (Julenius et al., 2005), respectively. Prediction of transmembrane helices in proteins was done with TMHMM 2.0 (Krogh et al., 2001).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: YGR014W (S. cerevisiae Msb2), AGR019C (A. gossypii Msb2), Afu4g04070 (A. fumigatus Msb2), NCU04373 (N. crassa Msb2), MGG06033 (M. oryzae Msb2), UM00480 (U. maydis Msb2). They also can be found in the Fusarium Comparative Database of the Broad Institute under accession numbers FOXG_09254 (F. oxysporum Msb2) and FGSG_05633 (F. graminearum Msb2) and in the Candida Genome Database under accession number orf19.1490 (C. albicans Msb2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Alignment of Fungal Msb2 Proteins.
Supplemental Figure 2. F. oxysporum Msb2 Does Not Complement Signaling Function in S. cerevisiae.
Supplemental Figure 3. Targeted Disruption of the F. oxysporum Msb2 Gene.
Supplemental Figure 4. Msb2 Contributes to Growth of Aerial Mycelium and Colony Hydrophobicity under Conditions of Nutrient Limitation.
Supplemental Figure 5. Growth of Different Strains in Submerged Culture.
Supplemental Figure 6. Msb2 Is Not Required for Oxidative and Osmotic (Salt) Stress Response.
Supplemental Figure 7. Phosphorylation of the Fmk1 and Mpk1 MAPKs in Different Mutants.
Supplemental Figure 8. Expression of MAPK Genes in Different Fungal Strains.
Supplemental Figure 9. Msb2 Regulates a Subset of Fmk1-Controlled Functions Related to Invasive Growth.
Supplemental Figure 10. Msb2 Contributes to Invasive Growth on Living Fruit Tissue.
Supplemental Figure 11. F. oxysporum Penetrates Roots Preferentially through Openings at the Junctions of Epidermal Cells.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Esther Martinez Aguilera (University of Córdoba) for valuable technical assistance. We thank Paul J. Cullen (State University of New York at Buffalo) for kindly providing S. cerevisiae strains and Peter Philippsen and Doris Albers (University of Basel) for help and support with complementation experiments. This research was supported by the SIGNALPATH Marie Curie Research Training Network (MRTN-CT-2005-019277) and by grants BIO2008-04479-E, EUI2009-03942, and BIO2010-15505 from the Spanish Ministerio de Ciencia e Innovación.
References
- Almeida I.C., Ferguson M.A., Schenkman S., Travassos L.R. (1994). Lytic anti-alpha-galactosyl antibodies from patients with chronic Chagas’ disease recognize novel O-linked oligosaccharides on mucin-like glycosyl-phosphatidylinositol-anchored glycoproteins of Trypanosoma cruzi. Biochem. J. 304: 793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong G.M., Armstrong J.K. (1981). Formae speciales and races of Fusarium oxysporum causing wilt diseases. Fusarium: Diseases, Biology and Taxonomy, Cook R., (University Park, PA: Penn State University Press; ), pp. 391–399 [Google Scholar]
- Bendtsen J.D., Nielsen H., von Heijne G., Brunak S. (2004). Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340: 783–795 [DOI] [PubMed] [Google Scholar]
- Bermejo C., Rodríguez E., García R., Rodríguez-Peña J.M., Rodríguez de la Concepción M.L., Rivas C., Arias P., Nombela C., Posas F., Arroyo J. (2008). The sequential activation of the yeast HOG and SLT2 pathways is required for cell survival to cell wall stress. Mol. Biol. Cell 19: 1113–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop C.D., Cooper R.M. (1983). An ultrastructural study of root invasion of three vascular wilt diseases. Physiol. Mol. Plant Pathol. 22: 15–27 [Google Scholar]
- Cullen P.J. (2007). Signaling mucins: The new kids on the MAPK block. Crit. Rev. Eukaryot. Gene Expr. 17: 241–257 [DOI] [PubMed] [Google Scholar]
- Cullen P.J., Sabbagh W., Jr, Graham E., Irick M.M., van Olden E.K., Neal C., Delrow J., Bardwell L., Sprague G.F., Jr (2004). A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 18: 1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P.J., Sprague G.F., Jr (2000). Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA 97: 13619–13624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Jarana J., Martínez-Rocha A.L., Roldán-Rodriguez R., Roncero M.I., Di Pietro A. (2005). Fusarium oxysporum G-protein beta subunit Fgb1 regulates hyphal growth, development, and virulence through multiple signalling pathways. Fungal Genet. Biol. 42: 61–72 [DOI] [PubMed] [Google Scholar]
- de Nadal E., Real F.X., Posas F. (2007). Mucins, osmosensors in eukaryotic cells? Trends Cell Biol. 17: 571–574 [DOI] [PubMed] [Google Scholar]
- Di Noia J.M., Pollevick G.D., Xavier M.T., Previato J.O., Mendoça-Previato L., Sánchez D.O., Frasch A.C. (1996). High diversity in mucin genes and mucin molecules in Trypanosoma cruzi. J. Biol. Chem. 271: 32078–32083 [DOI] [PubMed] [Google Scholar]
- Di Pietro A., García-MacEira F.I., Méglecz E., Roncero M.I. (2001). A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39: 1140–1152 [PubMed] [Google Scholar]
- Di Pietro A., Roncero M.I. (1998). Cloning, expression, and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum. Mol. Plant Microbe Interact. 11: 91–98 [DOI] [PubMed] [Google Scholar]
- Dupres V., Alsteens D., Wilk S., Hansen B., Heinisch J.J., Dufrêne Y.F. (2009). The yeast Wsc1 cell surface sensor behaves like a nanospring in vivo. Nat. Chem. Biol. 5: 857–862 [DOI] [PubMed] [Google Scholar]
- Edge A.S., Faltynek C.R., Hof L., Reichert L.E., Jr, Weber P. (1981). Deglycosylation of glycoproteins by trifluoromethanesulfonic acid. Anal. Biochem. 118: 131–137 [DOI] [PubMed] [Google Scholar]
- García R., Rodríguez-Peña J.M., Bermejo C., Nombela C., Arroyo J. (2009). The high osmotic response and cell wall integrity pathways cooperate to regulate transcriptional responses to zymolyase-induced cell wall stress in Saccharomyces cerevisiae. J. Biol. Chem. 284: 10901–10911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth M.A., Swanson B.J. (2004). Mucins in cancer: Protection and control of the cell surface. Nat. Rev. Cancer 4: 45–60 [DOI] [PubMed] [Google Scholar]
- Horazdovsky B.F., Emr S.D. (1993). The VPS16 gene product associates with a sedimentable protein complex and is essential for vacuolar protein sorting in yeast. J. Biol. Chem. 268: 4953–4962 [PubMed] [Google Scholar]
- Julenius K., Mølgaard A., Gupta R., Brunak S. (2005). Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15: 153–164 [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Kumamoto C.A. (2008). Molecular mechanisms of mechanosensing and their roles in fungal contact sensing. Nat. Rev. Microbiol. 6: 667–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagopodi A.L., Ram A.F., Lamers G.E., Punt P.J., Van den Hondel C.A., Lugtenberg B.J., Bloemberg G.V. (2002). Novel aspects of tomato root colonization and infection by Fusarium oxysporum f. sp. radicis-lycopersici revealed by confocal laser scanning microscopic analysis using the green fluorescent protein as a marker. Mol. Plant Microbe Interact. 15: 172–179 [DOI] [PubMed] [Google Scholar]
- Lanver D., Mendoza-Mendoza A., Brachmann A., Kahmann R. (2010). Sho1 and Msb2-related proteins regulate appressorium development in the smut fungus Ustilago maydis. Plant Cell 22: 2085–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitin F., Stern O., Weiss M., Gil-Henn C., Ziv R., Prokocimer Z., Smorodinsky N.I., Rubinstein D.B., Wreschner D.H. (2005). The MUC1 SEA module is a self-cleaving domain. J. Biol. Chem. 280: 33374–33386 [DOI] [PubMed] [Google Scholar]
- Ligtenberg M.J., Kruijshaar L., Buijs F., van Meijer M., Litvinov S.V., Hilkens J. (1992). Cell-associated episialin is a complex containing two proteins derived from a common precursor. J. Biol. Chem. 267: 6171–6177 [PubMed] [Google Scholar]
- Liu W., Zhou X., Li G., Li L., Kong L., Wang C., Zhang H., Xu J.R. (2011). Multiple plant surface signals are sensed by different mechanisms in the rice blast fungus for appressorium formation. PLoS Pathog. 7: e1001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- López-Berges M.S., DI Pietro A., Daboussi M.J., Wahab H.A., Vasnier C., Roncero M.I., Dufresne M., Hera C. (2009). Identification of virulence genes in Fusarium oxysporum f. sp. lycopersici by large-scale transposon tagging. Mol. Plant Pathol. 10: 95–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Berges M.S., Rispail N., Prados-Rosales R.C., Di Pietro A. (2010). A nitrogen response pathway regulates virulence functions in Fusarium oxysporum via the protein kinase TOR and the bZIP protein MeaB. Plant Cell 22: 2459–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Qiao J., Liu W., Wan Z., Wang X., Calderone R., Li R. (2008). The sho1 sensor regulates growth, morphology, and oxidant adaptation in Aspergillus fumigatus but is not essential for development of invasive pulmonary aspergillosis. Infect. Immun. 76: 1695–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid M.P., Di Pietro A., Roncero M.I. (2003). Class V chitin synthase determines pathogenesis in the vascular wilt fungus Fusarium oxysporum and mediates resistance to plant defence compounds. Mol. Microbiol. 47: 257–266 [DOI] [PubMed] [Google Scholar]
- Mott R. (2000). Accurate formula for P-values of gapped local sequence and profile alignments. J. Mol. Biol. 300: 649–659 [DOI] [PubMed] [Google Scholar]
- Pareja-Jaime Y., Martín-Urdíroz M., Roncero M.I., González-Reyes J.A., Roldán Mdel.C. (2010). Chitin synthase-deficient mutant of Fusarium oxysporum elicits tomato plant defence response and protects against wild-type infection. Mol. Plant Pathol. 11: 479–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitoniak A., Birkaya B., Dionne H.M., Vadaie N., Cullen P.J. (2009). The signaling mucins Msb2 and Hkr1 differentially regulate the filamentation mitogen-activated protein kinase pathway and contribute to a multimodal response. Mol. Biol. Cell 20: 3101–3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prados-Rosales R., Luque-Garcia J.L., Martínez-López R., Gil C., Di Pietro A. (2009). The Fusarium oxysporum cell wall proteome under adhesion-inducing conditions. Proteomics 9: 4755–4769 [DOI] [PubMed] [Google Scholar]
- Prados Rosales R.C., Di Pietro A. (2008). Vegetative hyphal fusion is not essential for plant infection by Fusarium oxysporum. Eukaryot. Cell 7: 162–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhalla J.E. (1985). Classification of strains of Fusarium oxysporum on the basis of vegetative incompatibility. Can. J. Bot. 63: 179–183 [Google Scholar]
- Punt P.J., Oliver R.P., Dingemanse M.A., Pouwels P.H., van den Hondel C.A. (1987). Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 56: 117–124 [DOI] [PubMed] [Google Scholar]
- Qi M., Elion E.A. (2005). MAP kinase pathways. J. Cell Sci. 118: 3569–3572 [DOI] [PubMed] [Google Scholar]
- Ram A.F., Klis F.M. (2006). Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat. Protoc. 1: 2253–2256 [DOI] [PubMed] [Google Scholar]
- Ramakers C., Ruijter J.M., Deprez R.H., Moorman A.F. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Rieder S.E., Emr S.D. (2001). Overview of subcellular fractionation procedures for the yeast Saccharomyces cerevisiae. Curr. Protoc. Cell Biol. Chapter 3: Unit 3.7. [DOI] [PubMed] [Google Scholar]
- Rispail N., Di Pietro A. (2009). Fusarium oxysporum Ste12 controls invasive growth and virulence downstream of the Fmk1 MAPK cascade. Mol. Plant Microbe Interact. 22: 830–839 [DOI] [PubMed] [Google Scholar]
- Rispail N., et al. (2009). Comparative genomics of MAP kinase and calcium-calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet. Biol. 46: 287–298 [DOI] [PubMed] [Google Scholar]
- Roberts R.L., Fink G.R. (1994). Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: Mating and invasive growth. Genes Dev. 8: 2974–2985 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Gálvez E., Mendgen K. (1995). Cell wall synthesis in cotton roots after infection with Fusarium oxysporum. The deposition of callose, arabinogalactans, xyloglucans, and pectic components into walls, wall appositions, cell plates and plasmodesmata. Planta 197: 535–545 [DOI] [PubMed] [Google Scholar]
- Román E., Cottier F., Ernst J.F., Pla J. (2009). Msb2 signaling mucin controls activation of Cek1 mitogen-activated protein kinase in Candida albicans. Eukaryot. Cell 8: 1235–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Román E., Nombela C., Pla J. (2005). The Sho1 adaptor protein links oxidative stress to morphogenesis and cell wall biosynthesis in the fungal pathogen Candida albicans. Mol. Cell. Biol. 25: 10611–10627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncero C., Durán A. (1985). Effect of Calcofluor white and Congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J. Bacteriol. 163: 1180–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M.D., Winston F., Hieter P. (1990). Methods in Yeast Genetics. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Sambrook J., Russell D. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Silverman H.S., Parry S., Sutton-Smith M., Burdick M.D., McDermott K., Reid C.J., Batra S.K., Morris H.R., Hollingsworth M.A., Dell A., Harris A. (2001). In vivo glycosylation of mucin tandem repeats. Glycobiology 11: 459–471 [DOI] [PubMed] [Google Scholar]
- Tatebayashi K., Tanaka K., Yang H.Y., Yamamoto K., Matsushita Y., Tomida T., Imai M., Saito H. (2007). Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. EMBO J. 26: 3521–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truckses D.M., Garrenton L.S., Thorner J. (2004). Jekyll and Hyde in the microbial world. Science 306: 1509–1511 [DOI] [PubMed] [Google Scholar]
- Vadaie N., Dionne H., Akajagbor D.S., Nickerson S.R., Krysan D.J., Cullen P.J. (2008). Cleavage of the signaling mucin Msb2 by the aspartyl protease Yps1 is required for MAPK activation in yeast. J. Cell Biol. 181: 1073–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pöhlmann R., Philippsen P. (1994). New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808 [DOI] [PubMed] [Google Scholar]
- Wilson R.A., Talbot N.J. (2009). Under pressure: Investigating the biology of plant infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 7: 185–195 [DOI] [PubMed] [Google Scholar]
- Xu J.R., Hamer J.E. (1996). MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 10: 2696–2706 [DOI] [PubMed] [Google Scholar]
- Yang H.Y., Tatebayashi K., Yamamoto K., Saito H. (2009). Glycosylation defects activate filamentous growth Kss1 MAPK and inhibit osmoregulatory Hog1 MAPK. EMBO J. 28: 1380–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Ukil L., Osmani A., Nahm F., Davies J., De Souza C.P., Dou X., Perez-Balaguer A., Osmani S.A. (2004). Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell 3: 1359–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Mehrabi R., Xu J.R. (2007). Mitogen-activated protein kinase pathways and fungal pathogenesis. Eukaryot. Cell 6: 1701–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.