Abstract
Sequence comparisons, biochemical experiments, and studies with mutants in transgenic plants show that the Arabidopsis protein CORYNE, currently thought to be a kinase that acts as part of a receptor kinase complex, is likely to be a pseudokinase and not a kinase.
The apical stem cell niche in plants is restricted by a ligand and receptor-activated feedback loop that regulates accumulation of the transcription factor WUSCHEL (WUS) (Ceresa and Schmid, 2000). Binding of the secreted peptide ligand CLAVATA3 (CLV3) to the CLV1 leucine-rich repeat Ser-Thr kinase is central to this process. CLV2, a transmembrane leucine-rich repeat protein lacking an internal kinase domain, is also important for perception of CLV3 and other CLV3-like ligands throughout the plant (Wang and Fiers, 2010). In Arabidopsis thaliana, CORYNE (CRN), which encodes a predicted transmembrane Ser-Thr kinase with a short extracellular domain, has been shown to act with CLV2 in this process and may regulate CLV2 localization (Müller et al., 2008; Bleckmann et al., 2010). Because mutations in CRN and CLV2 are additive with CLV1 mutants, it has been proposed that CLV2 and CRN functionally assemble into a signaling complex that functions as a receptor kinase in parallel to CLV1 (Müller et al., 2008). The CLV/CRN-WUS circuit is conserved across diverse taxa, and determining how its signaling is activated is necessary for the understanding of plant stem cell regulation. Here, we demonstrate that CRN is unlikely to be a kinase and is likely a pseudokinase, indicating that the existing model needs revision.
CRN CONTAINS FEATURES OF A PSEUDOKINASE
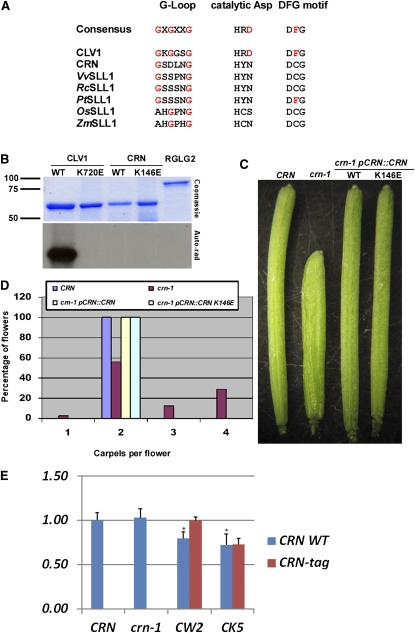
CRN contains several features suggestive of nonfunctional kinases as described by Boudeau et al. (2006). CRN contains a His-Tyr-Asn (HYN) motif in the presumed catalytic loop instead of the His-Arg-Asp (HRD) motif typical of functional kinases and therefore lacks the critical active-site Asp (Figure 1A). The G-loop, GXGXXG, which binds and positions ATP, has diverged in CRN to GXDXXG. Similar mutations in VRK3 family members render the G-loop acidic, inhibiting ATP binding (Scheeff et al., 2009). The activation segment of CRN is shorter (16 amino acids between the DCG and APE motifs, compared with 20 to 35 in most active kinases in the National Center for Biotechnology Information conserved domain database), which should lead to a truncation of either the activation loop or the P+1 loop. Lastly, DCG replaces the DFG motif of the Mg2+ binding loop. Mutation of Phe is correlated with alterations in the positioning of the activation segment relative to the catalytic loop, affecting positioning of the upper and lower lobes (Nolen et al., 2004).
Figure 1.
CRN Encodes an Apparent Pseudokinase.
(A) CRN lacks consensus residues necessary for kinase activity in known kinases. Alignment of critical CRN residues deviating from the consensus kinase sequences. CLV1 is included as an example of an active kinase. Other sequences refer to CRN homologs (referred to as SOL2-like1 [SLL1]) as identified by Miwa et al. (2009). ZmSLL1 represents Zea mays sequence NP_001142196.
(B) CRN lacks autophosphorylation activity. Coomassie blue–stained gel of purified CLV1 and CRN kinase domains. GST-tagged CLV1 or CRN were purified using standard purification techniques, eluted, and allowed to autophosphorylate on beads in the kinase assay buffer (see Methods). Purified protein was then eluted and subjected to SDS-PAGE, followed by staining with Coomassie blue (top panel), and processed and exposed to film for autoradiography (bottom panel). GST-RGLG2, a ubiquitin ligase (Yin et al., 2007), was used as a negative activity control. WT, wild type.
(C) Kinase activity apparently is not required for CRN function in planta. crn-1 plants (Ler background) were transformed with either wild-type CRN or the K146E active-site mutant of CRN driven by pCRN (see Methods for details).
(D) Kinase activity apparently is not required for CRN function in planta. Quantification of carpel number in flowers from the wild type (Ler), crn-1, crn-1 pCRN:CRN-WT, or pCRN:CRN-K146E. Flowers 3 to 21 were counted for individual plants or T1s, and the total number of flowers was combined. Graph represents percentage of flowers displaying different carpel numbers. Six plants were counted for Ler and crn-1, totaling 120 flowers each. Nine T1s were counted for pCRN:CRN-WT, totaling 180 flowers. All pCRN:CRN-WT plants displayed full complementation. Fifteen T1s were counted for pCRN:CRN-K146E, totaling 300 flowers. All pCRN:CRN-K146E lines displayed full complementation. Similar results were obtained with pCRN:CRN-WT and -K146E lines tagged with TAG-RFP (data not shown).
(E) Endogenous crn-1 and CRN transgenes are coexpressed in transgenic lines. Quantitative real-time PCR of endogenous CRN transcripts (CRN WT) and transgenic CRN (CRN-tag). CW2, complementing pCRN:CRN WT-TagRFP line; CK5, complementing pCRN:CRN K146E-TagRFP line. See Methods for RNA extraction and transcript quantitation. No CRN-tag transcripts were detected after 40 cycles in CRN or crn-1 plants. The asterisk denotes a P value < 0.05.
CRN LACKS KINASE ACTIVITY
To test experimentally the prediction that CRN is a kinase, we performed in vitro kinase assays with CRN and CLV1, a known Ser-Thr kinase, using purified wild-type CRN and CLV1 and mutants in the active site Lys of each (K146E and K720E, respectively). Of three CRN splice variants in the database, we chose the variant AT5G13290.2 as this represents the full kinase domain (see below). As expected, CLV1 autophosphorylated, while the CLV1K720E mutant did not (Figure 1B). No activity was seen for either CRN protein, suggesting that wild-type CRN lacks autokinase activity under standard conditions.
CRN DOES NOT REQUIRE KINASE ACTIVITY FOR WILD-TYPE FUNCTION IN VIVO
We next tested the ability of CRN and CRNK146E to complement the crn-1 mutant. There are no known crn null mutants. The crn-1 allele is fully recessive, and overexpression of crn-1 has no effect in wild-type plants, indicating the crn-1 allele is a loss-of-function allele (Müller et al., 2008). We generated in-frame translational fusions of the CRN variants fused at the C terminus to mTFP1 by a 9-Ala linker. When expressed from the endogenous pCRN promoter (2.6 kb of upstream and 0.9 kb downstream), both pCRN:CRN-mTFP1 and pCRN:CRN1K146E-mTFP1 complemented the crn-1 mutant fully, indicating that CRN does not require kinase activity for wild-type function (Figures 1C and 1D). Similar results were obtained with fusions to TAG-red fluorescent protein (TAG-RFP), although no fluorescence signal was visible in any fusion class, consistent with previous results (Bleckmann et al., 2010). In total, more than 30 independent pCRN:CRN1K146E T1s displayed complete complementation of the crn-1 mutant. Expression of endogenous CRN and the CRN-TagRFP transgene were confirmed by quantitative RT-PCR in reference lines (Figure 1E).
CRN HOMOLOGS ENCODE APPARENT PSEUDOKINASES
CRN homologs have been identified in taxonomically diverse species, although functional data exist for Arabidopsis CRN only (Miwa et al., 2009). All CRN homologs examined encoded apparent pseudokinases, arguing that CRN function has been conserved independent of kinase activity for at least 150 million years, perhaps predating the monocot/dicot divergence (Figure 1A). It is therefore implausible that the CRN/CLV2 complex transmits CLV3 signals using a mechanism analogous to that of CLV1. CRN gives rise to two additional splice variants (AT5G13290.1 and AT5G13290.3) that contain large deletions of the kinase catalytic domain, including the entire ATP binding G-loop and a significant portion of the VAVK motif of subdomain II. Thus, none of the three CRN splice variants would appear to give rise to active enzymes with kinase activity.
CLUES TO CRN FUNCTION
Evolutionarily conserved pseudokinases play a role in several Tyr ligand-receptor signaling systems in animals. Pseudokinase roles include steric inhibition of active kinases and scaffolding functions (Boudeau et al., 2006). The additive nature of crn and clv1 alleles suggests that CRN may play a scaffolding role, perhaps to aid export of CLV2 to the plasma membrane and/or to assemble higher-order CLV1 or CLV1 substrate complexes. Consistent with our findings, the CRN kinase-related domain is not necessary for plasma membrane accumulation of CLV2 in transient expression assays (Bleckmann et al., 2010). Previous genomic analysis has suggested that up to 20% of all Arabidopsis receptor kinases, including CRN and members of the SUB family, encode kinase-defective variants (Chevalier et al., 2005; Castells and Casacuberta, 2007). Elucidating the role of CRN in CLV1/CLV2 perception of CLV3 perception should provide a model for understanding the function of this large family of kinase homologs in plants.
METHODS
Vector Construction
The CRN cDNA open reading frame (AT5G13290.2) was amplified from Columbia-0 RNA using CRN forward (Fwd) primer (5′-GGCCATGGGGATCCATGAAGCAAAGAAGAAGAAGAAATGG-3′) and CRN reverse (Rvs) primer (5′-GCCATGGATCGGGCTGCCGCAGCGGCAGCAGCCGCAGCAGGAAAGCTGTGCAGTTGTGTAAGCATG-3′) that generates a NcoI site at either end of the PCR product, deletes the endogenous stop codon, and adds a linker encoding a Pro and nine Ala residues. PFU1 II Ultra was used to minimize mutations (Stratagene). The K146E mutation was generated by recombinant PCR using the CRN Fwd and CRN K-E Rvs primer (5′-CAAGTGAGCCTAGTCTTTCGACTGCAACCACTAG-3′) and the CRN Rvs primer and CRN K-E Fwd primer (5′-CTAGTGGTTGCAGTCGAAAGACTAGGCTCACTTG-3′) in the first round of amplification, followed by PCR amplification of the first round products with the CRN Fwd and CRN Rvs primers using the first round products to generate the CRN K146E full-length mutant. The resulting PCR product was subcloned into pCR2.1 (Invitrogen) and sequenced. These products were cloned into a pBJ based shuttle vector containing either mTFP1 or TAG-RFP vector modified to contain a unique NcoI site to allow in-frame fusions at the N terminus to generate a C-terminal tagged CRN BamHI fragment. The CRN promoter (Pro) was generated by recombinant PCR using CRN Pro Fwd (5′-GGGCGGCCGCGGAGATAAATGAAGCTATTTTTCTTCTCG-3′) and CRN BamHI Rvs (5′-AGTACGTTGGGGGATCCTGCTGCTTCTACGAATAAAAG-3′) and CRN Pro Rvs (5′-GGGCGGCCGCGTAAGTTCTTGTAGAATCCCCAATACGTG-3′) and CRN BamHI Fwd (5′-CTTTTATTCGTAGAAGCAGCAGGATCCCCCAACGTACT-3′) in the first round, followed by amplification using CRN Pro Fwd and CRN Pro Rvs using the first-round products. This fragment was chosen as it has been shown to provide complementation of the crn-1 mutant (Müller et al., 2008). This was cloned into pCR2.1, sequenced, and cloned as a NotI fragment into a modified pMOA33 (Barrell and Conner, 2006) in which the BamHI site was destroyed, thus generating pCRN. The CRN fusions were then cloned into pCRN as BamHI products.
Generation of Transgenic Plants and Plant Growth
pCRN constructs were transformed into Landsberg erecta (Ler) or the crn-1 (Müller et al., 2008) mutant using the GV3101 Agrobacterium tumefaciens strain and the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on B5 media plates containing 30 μg/mL kanamycin and transferred to soil and grown as described before (Clark et al., 1993).
In Vitro Kinase Assays
The regions corresponding to amino acids Met-662 to Phe-980 for CLV1 and Val-85 to Phe-401 for CRN were PCR amplified. For the kinase dead version of these proteins, the catalytic Lys of CLV1 (Lys-720) was mutated to Glu (K720E) and for CRN Lys (Lys-146) was mutated to Glu (K146E) using a recombinant PCR strategy. These products were TOPO cloned into pENTRD/TOPO and moved into pDEST15 by LR recombination to generate N-terminal glutathione S-transferase (GST) fusions. Proteins were expressed in BL21-AI at room temperature for 4 h following the manufacturer's recommendations (Invitrogen). GST-CLV1, GST-CRN, and GST-RGLG proteins were purified with the Glutathione Sepharose 4B affinity matrix.
For the kinase assay, proteins still bound to the GST beads (GE Life Sciences) were quantified, and the appropriate bead volume was used to give a total of 1 μg of bound protein for each kinase reaction. For each reaction, 0.5 mCi of [γ-32P]ATP (MP Biomedicals) was added to the kinase reaction buffer (20 mM Tris-HCl, 10 mM MgCl2, 10 mM MnCl2, 1 mM DTT, and 10 mM cold ATP) and incubated at room temperature or 1 h. The beads were washed three times in cold kinase reaction buffer (lacking ATP) and resolved by SDS-PAGE electrophoresis. After electrophoresis, the gel was stained with Coomassie Brilliant Blue R 250 (Bio-Rad) and dried onto Whatman 3-mm chromatography paper. The dried gel was exposed to film for 18 h.
Gene Expression Analysis
RNA was harvested from inflorescence tissue using the RNeasy Mini Kit (Qiagen). cDNA was synthesized from 1 mg DNase1-treated (Invitrogen) total RNA using SuperScript II (Invitrogen). Quantitative real-time PCR was done using SYBR green (Quantance; SensiMix). Data were analyzed using the ΔΔCt method and normalized with the expression of the reference genes tubulin and NM_128399. Wild-type CRN levels were calculated in reference to wild-type CRN (Ler), and CRN-TAG levels were calculated in reference to transgenic line CW2. For the quantification of wild-type CRN transcript, the following primers were used: CRN-WT Fwd, 5′-AGACCGGCCTTCAAGTGATGA-3′; and CRN-WT 3′UTR, 5′-GAATATATTGATGCAACTGCAGATG-3′. For the quantification of CRN-TAG transcript, the CRN-WT Fwd and CRN-TAG Rvs (5′-GTTGTTCACGGTGCCCTCCA-3′) primer pair was used. Reference genes were quantified using the primers Tubulin F primer, 5′-AAACTCACTACCCCCAGCTTTG-3′; Tubulin R primer, 5′-CACCAGACATAGTAGCAGAAATCAAGT-3′; NM_128399 Fwd, 5′-GGATTTTCAGCTACTCTTCAAGCTA-3′; and NM_128399 Rvs, 5′-TGCCTTGACTAAGTTGACACG-3′.
Acknowledgments
This work was funded by National Institutes of Health (NIH) National Research Service Award F32 GM080843 to Z.L.N., NIH National Research Service Award F32 GM090534 to P.T.T., and NIH Grant 1R01 GM086639 to E.M.M.
References
- Barrell P.J., Conner A.J. (2006). Minimal T-DNA vectors suitable for agricultural deployment of transgenic plants. Biotechniques 41: 708–710 [DOI] [PubMed] [Google Scholar]
- Bleckmann A., Weidtkamp-Peters S., Seidel C.A., Simon R. (2010). Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 152: 166–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudeau J., Miranda-Saavedra D., Barton G.J., Alessi D.R. (2006). Emerging roles of pseudokinases. Trends Cell Biol. 16: 443–452 [DOI] [PubMed] [Google Scholar]
- Castells E., Casacuberta J.M. (2007). Signalling through kinase-defective domains: The prevalence of atypical receptor-like kinases in plants. J. Exp. Bot. 58: 3503–3511 [DOI] [PubMed] [Google Scholar]
- Ceresa B.P., Schmid S.L. (2000). Regulation of signal transduction by endocytosis. Curr. Opin. Cell Biol. 12: 204–210 [DOI] [PubMed] [Google Scholar]
- Chevalier D., Batoux M., Fulton L., Pfister K., Yadav R.K., Schellenberg M., Schneitz K. (2005). STRUBBELIG defines a receptor kinase-mediated signaling pathway regulating organ development in Arabidopsis. Proc. Natl. Acad. Sci. USA 102: 9074–9079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.E., Running M.P., Meyerowitz E.M. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119: 397–418 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Miwa H., Tamaki T., Fukuda H., Sawa S. (2009). Evolution of CLE signaling: Origins of the CLV1 and SOL2/CRN receptor diversity. Plant Signal. Behav. 4: 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R., Bleckmann A., Simon R. (2008). The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen B., Taylor S., Ghosh G. (2004). Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell 15: 661–675 [DOI] [PubMed] [Google Scholar]
- Scheeff E.D., Eswaran J., Bunkoczi G., Knapp S., Manning G. (2009). Structure of the pseudokinase VRK3 reveals a degraded catalytic site, a highly conserved kinase fold, and a putative regulatory binding site. Structure 17: 128–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Fiers M. (2010). CLE peptide signaling during plant development. Protoplasma 240: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X.J., et al. (2007). Ubiquitin lysine 63 chain forming ligases regulate apical dominance in Arabidopsis. Plant Cell 19: 1898–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]