Abstract
Based on their evolutionary origin, MADS box transcription factor genes have been divided into two classes, namely, type I and II. The plant-specific type II MIKC MADS box genes have been most intensively studied and shown to be key regulators of developmental processes, such as meristem identity, flowering time, and fruit and seed development. By contrast, very little is known about type I MADS domain transcription factors, and they have not attracted interest for a long time. A number of recent studies have now indicated a key regulatory role for type I MADS box factors in plant reproduction, in particular in specifying female gametophyte, embryo, and endosperm development. These analyses have also suggested that type I MADS box factors are decisive for setting reproductive boundaries between species.
MADS DOMAIN ENCODING GENES
MADS box genes are of ancient origin and are found in animals, fungi, and plants. All identified MADS box genes encode a highly conserved N-terminal DNA binding domain 55 to 60 amino acids in length named the MADS domain (Figure 1; Tröbner et al., 1992). Homology searches in the nonredundant microbial database using a Hidden Markov Model for seed alignment of the MADS domain suggested that the MADS domain originates from the DNA binding subunit A of topoisomerases IIA subunit A (Gramzow et al., 2010).
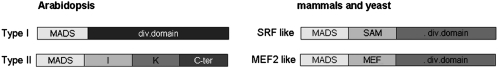
Figure 1.
Schematic Representation of Type I and Type II MADS Box Transcription Factors.
C-ter, the divergent C region; I, intervening region; K, keratin K domain; M, MADS box domain.
The acronym MADS (Schwarz-Sommer et al., 1990) is derived from the initials of MINICHROMOSOME MAINTENANCE1 (MCM1, Saccharomyces cerevisiae; Passmore et al., 1988), AGAMOUS (Arabidopsis thaliana; Yanofsky et al., 1990), DEFICIENS (Antirrhinum majus; Sommer et al., 1990), and SERUM RESPONSE FACTOR (SRF, Homo sapiens; Norman et al., 1988). These members of the MADS box gene family play important biological roles; for example, the human SRF coordinates the transcription of the proto-oncogene c-fos (Masutani et al., 1997; Mo et al., 2001), while MCM1 is central to the transcriptional control of cell type–specific genes and the pheromone response in the yeast S. cerevisiae (Shore and Sharrocks, 1995; Mead et al., 2002).
Plant MADS box genes were first identified as regulators of floral organ identity and have since been reported to control additional developmental processes, such as the determination of meristem identity of vegetative, inflorescence, and floral meristems, root growth, ovule and female gametophyte development, flowering time, fruit ripening, and dehiscence (Zhang and Forde, 1998; Ng and Yanofsky, 2001; Giovannoni, 2004; Whipple et al., 2004; L. Colombo et al., 2008; Liu et al., 2009). Studies using several model species, including Arabidopsis, A. majus, Petunia hybrida, Zea mays, and Oryza sativa, have revealed that many of these functions are conserved among angiosperms (Schwarz-Sommer et al., 2003; Vandenbussche et al., 2003; Kater et al., 2006).
ARABIDOPSIS MADS BOX TYPE I AND TYPE II: AN EVOLUTIONARY OVERVIEW
Based on sequence conservation in the MADS domain, these transcription factors can be grouped into two main lineages, named type I (SRF-like) and type II (MEF2-like; Alvarez-Buylla et al., 2000). In animals, type I genes are involved in response to growth factors, while type II genes play important roles in muscle development (Norman et al., 1988; Yu et al., 1992). Animal type I and II genes can be separated into monophyletic groups, whereas plant type I genes do not group into a monophyletic cluster (Nam et al., 2004).
Type II genes include MEF2-like genes from animals and yeast as well as the plant-specific MIKC-type genes. MIKC MADS box genes were named after the four conserved domains that can be recognized in these proteins: the MADS (M) domain, the intervening (I) domain, the coiled-coil keratin-like (K) domain, and the C-terminal (C) domain (Theissen et al.,1996; Kaufmann et al., 2005) (Figure 1). Studies of MADS box proteins have shown that these domains can have different functions. For instance, the I domain provides specificity in the formation of DNA binding dimers (Masiero et al., 2002), and the K domain mediates dimerization of MADS box proteins and has been shown to be involved in the formation of higher-order complexes (Egea-Cortines et al., 1999). The C domain functions in some MADS box proteins as a transcriptional activation domain and in the formation of higher-order protein complexes (Egea-Cortines et al., 1999; Yang et al., 2003). The C domain also seems to contribute to MADS box protein interaction specificity (van Dijk et al., 2010).
Type I MADS domain transcription factors are grouped together based on their high similarity with the MADS domain of SRF. Furthermore, type I and II proteins differ in the domain immediately C-terminal to the MADS box. Parenicová et al. (2003) divided Arabidopsis type I MADS box genes in three subfamilies: Mα (25 genes), Mβ (20 genes), and Mγ (16 genes), although AGAMOUS LIKE33 (AGL33; At2g26320) was not assigned to any of these. The gene structures of Mα, Mβ, and Mγ clearly differ from those of the MIKC genes; for instance, most of the type I genes are short and encoded by a single exon, whereas the MIKC genes are much longer and contain five to eight exons. The high level of conservation of the MADS box and K domain suggests that these domains evolved under stronger structural/functional constraints than the more divergent I and C domains (De Bodt et al., 2003b). Moreover, MIKC-type genes are distributed over all five Arabidopsis chromosomes in contrast with type I members that are mainly located on chromosomes I and V. The different patterns of distribution may result from different modes of duplication that caused the formation of these two lineages (Martinez-Castilla and Alvarez-Buylla, 2003; Parenicová et al., 2003). While MIKC genes were duplicated during the whole-genome duplication that occurred early in angiosperm evolution, many type I genes in Arabidopsis show signs of intrachromosomal duplication events happening gradually and more recent in evolution (Martinez-Castilla and Alvarez-Buylla, 2003).
An extensive matrix-based yeast two-hybrid screen covering almost the entire Arabidopsis MADS domain transcription factor family showed that MIKC proteins interact preferentially with other type II proteins and barely form dimers with type I proteins (de Folter et al., 2005). Correspondingly, type I MADS domain factors mainly interact with other type I members. Among the type I proteins, most heterodimers are formed between members of the different subclades, Mα, Mβ, and Mγ. Interactions among Mα proteins are rare, and they dimerize preferentially with Mβ and Mγ. Similarly, only a few interactions among members of the Mβ and Mγ clades were observed, and they interact preferentially with Mα proteins. These data suggest that Mα transcription factors might stabilize type I protein complexes, and they might be proposed as the type I factors that establish higher-order complexes (Immink et al., 2009). Nevertheless, it should be kept in mind that the majority of type I proteins were not found to interact with other MADS domain proteins; most of the interactions observed concerned only a few type I factors. This indicates that type I MADS box factors likely do not bind DNA as MADS box dimers.
TYPE I MADS BOX GENES HAVE IMPORTANT FUNCTIONS DURING REPRODUCTIVE DEVELOPMENT
Plant type I MADS box gene sequences have undergone a faster rate of birth and death during evolution compared with animal type I (SRF) and plant type II (MIKC) genes; thus, early reports concluded that type I genes were probably of minor functional importance for plants in comparison to type II genes (De Bodt et al., 2003a; Kofuji et al., 2003).
A first difficulty encountered in the characterization of MADS box type I members was their very low expression level. Preliminary analyses indicated that Mα and Mγ subclasses are predominantly expressed in the inflorescences and siliques, and more meticulous studies using Affymetrix arrays showed that type I MADS box genes are expressed in male and female gametophytes as well as during early stage of endosperm and embryo development (Day et al., 2008; Walia et al., 2009; Tiwari et al., 2010; Wuest et al., 2010).
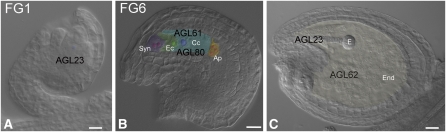
The first Arabidopsis type I gene to be characterized functionally was AGL80/FEM111 (Portereiko et al., 2006), which belongs to the Mγ subclade. AGL80/FEM111 is expressed in the central cell just before polar nuclei fusion, and in accordance with this observation, female gametophytes are severely affected in agl80/fem111 mutants (Schneitz et al., 1995; Christensen et al., 1997; Yadegari and Drews, 2004; Portereiko et al., 2006) (Figure 2,3). The phenotype of agl80/fem111 mutants becomes apparent soon after fusion of the polar nuclei and includes defects in nuclear maturation and maintenance of vacuole size of the central cell (Figure 2). After pollination, agl80/fem111 endosperm fails to develop, the central cell degenerates, and its cavity is filled with highly fluorescent material, indicating that AGL80/FEM111 is important for the initiation of endosperm development (Portereiko et al., 2006). This event appears to be stimulated by fertilization since degeneration does not occur in the absence of pollination. AGL80/FEM111 is also expressed in endosperm from the 1 to 16 nucleate stage (Portereiko et al., 2006), but its function in endosperm development is still unclear. Furthermore, it is expressed in microspores (Parenicová et al., 2003), but reciprocal crosses excluded a vital role for male gametophyte development. By RT-PCR experiments, AGL80 transcripts were detected in most organs (roots, stems, leaves, flowers, and anthers); therefore, weak alleles or inducible silencing strategies might be useful to shed light on other functions that AGL80 might have during development.
Figure 2.
Type I MADS Box Genes Control Female Gametophyte and Seed Development.
Differential interference contrast microscopy images of wild-type Arabidopsis developing ovules and seeds: female gametophyte at stage1 (A), female gametophyte at stage 6 (B), and embryo at globular stage (C). Type I MADS domain proteins are indicated related to their function in the corresponding developmental stages and their function in a specific process. AGL23 (A) is involved in the early phase of gametogenesis (M. Colombo et al., 2008). The agl23 embryo sac arrests at FG1. AGL23 also regulates chloroplast biogenesis, which occurs in the embryo at the globular stage (C) (M. Colombo et al., 2008). AGL80 ([B] and [C]) disruption affects central cell differentiation (Portereiko et al., 2006). AGL80 interacts with AGL61, and genetic evidence supports the yeast two-hybrid assays, as agl61 embryo sacs develop defective central cells (Bemer et al., 2008; Steffen et al., 2008). AGL62 (C) suppresses cellularization and promotes nuclear proliferation during early endosperm development (Kang et al., 2008). The role of AGL80 during endosperm development has yet to be clarified. Ap, antipodal cells; Cc, central cell; Ec, egg cell; E, embryo; End, endosperm; Syn, synergid cells. Bars = 20 μm.
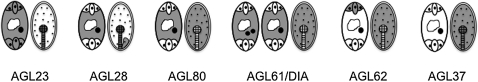
Figure 3.
Overview of Type I MADS Box Gene Expression in the Embryo Sac and Seed.
Diagrammatic representation of expression profiles (in blue) of type I MADS box genes that have a function in female gametophyte and/or seed development.
Yeast two-hybrid assays showed that AGL80 interacts with the Mα-type MADS domain protein AGL61, named DIANA (DIA; de Folter et al., 2005; Bemer et al., 2008; Steffen et al., 2008). DIA/AGL61 contains a distinct N-terminal region in front of the MADS domain, thus being different from most of the Mα proteins, where the MADS domain is located close to the N terminus. AGL61 disruption causes female gametophytic lethality and predominantly affects the differentiation of the central cell, including an overall reduction in size caused by a smaller or absent vacuole (Figure 2; Steffen et al., 2008). Steffen et al. (2008) concluded that synergid, egg, and antipodal cells develop normally, whereas Bemer et al. (2008) described synergid and egg cell defects, although the same SALK line was used in both studies. However, both reports agreed that central cell markers are not expressed in the agl61-1/dia1-1 mature embryo sacs, indicating that central cell fate is disturbed. Moreover, neither endosperm development nor zygote formation occurs, although pollen tubes are attracted and perceived by the mutant ovules. A PRODIA:DIA-GFP-GUS fusion protein is present exclusively in the polar nuclei and the secondary nucleus of the central cell, in agreement with DIA/AGL61 function. Interestingly, nuclear DIA-GFP-GUS localization is abolished when introduced into the agl80/fem111 mutant background, suggesting that AGL61 is translocated to the nucleus only when its partner AGL80 is present, which could also explain the similar phenotypes (Bemer et al., 2008).
In yeast two-hybrid assays, AGL61 and AGL80 heterodimerize with AGL62 (de Folter et al., 2005; Kang et al., 2008). However, AGL62 is only expressed in antipodal cells of mature embryo sacs after cellularization (Figure 3). Although absent from the central cell of unfertilized ovules, AGL62 is expressed in the syncytial endosperm of developing seeds from fertilization until cellularization. Confocal laser microscopy analysis showed that AGL62 suppresses cellularization and promotes nuclear proliferation during early endosperm development. Maternal and paternal alleles seem to be expressed equally, and AGL62 disruption causes recessive seed lethality while embryo sac development and function appear normal (Figure 2; Kang et al., 2008).
FERTILISATION INDEPENDENT SEED (FIS) PcG complex mutants also suppress endosperm cellularization and promote nuclear proliferation, and, interestingly, AGL62 expression is upregulated in PcG mutants (Kang et al., 2008; Tiwari et al., 2010). The core FIS complex is comprised of at least four components, the presumptive HMTase MEDEA, the WD40 domain protein FERTILIZATION INDEPENDENT ENDOSPERM, the Zn-finger protein FIS2, and the homolog of the nucleosome-remodeling factor 55 MULTICOPY SUPPRESSOR OF IRA1 (Chaudhury et al., 1997; Spillane et al., 2000; Köhler et al., 2003b; Gehring et al., 2004; Schönrock et al., 2006; Jullien et al., 2006). Like in animals, the plant PRC2 complex catalyzes histone H3 lysine 27 trimethylation (H3K27me3), a repressive chromatin mark; indeed, many MADS box type I genes have been found to be decorated by this mark that becomes lost in PRC2 null mutants (Zhang et al., 2007; Bouyer et al., 2011). For instance, the expression of the maternal allele of the type I MADS box gene AGL37/PHE1 is silenced by the FIS complex, while the paternal copy was found to be active resulting in a parent-of-origin-dependent expression of PHE1 (i.e., imprinting) in seeds (Köhler et al., 2005). Similar to AGL62, AGL37/PHE1 is transiently expressed during the syncytial phase of endosperm development, and expression is temporally extended in fis mutant seeds (Köhler et al., 2003a; Ingouff et al., 2005). Currently, the pathway by which the FIS PcG complex suppresses AGL62 expression is unknown, especially since AGL62, in contrast with PHE1 and AGL61, was not found to be decorated by H3K27me3 marks (Zhang et al., 2007; Bouyer et al., 2011).
The functional characterization of AGL23, an Mα-type MADS box gene that is subject to PRC2 regulation (Zhang et al., 2007; Bouyer et al., 2011), revealed a role for MADS box transcription factors in embryo sac development (M. Colombo et al., 2008). The agl23 mutant shows, with incomplete penetrance, an arrest at the one-nucleate stage of embryo sac development. The functional megaspore expresses a megaspore marker but is arrested and a one-nucleate female gametophyte persists during subsequent stages of ovule development. Due to incomplete penetrance of the embryo sac phenotype, homozygous agl23 embryos were obtained but were not viable. These embryos can easily be recognized since chloroplasts are absent and only proplastids or etioplasts are formed. However, how AGL23 regulates chloroplast biogenesis is not clear since several pathways (e.g., carothenoid or lipid biosynthesis) could be under the control of AGL23. Expression studies showed that AGL23 expression correlates with the observed phenotypes since the putative AGL23 promoter drives GUS expression in the developing embryos at late globular stage when chloroplast biogenesis takes place and during embryo sac development as soon as the functional megaspore can be detected (M. Colombo et al., 2008).
AGL23 shares high sequence similarity with AGL28, which is also expressed in developing embryos (Bemer et al., 2010). Plants overexpressing AGL28 (PRO35S:AGL28) were reported to have an early flowering phenotype under long-day conditions caused by FLOWERING LOCUS T (Koornneef et al., 1998) and SUPPRESSOR OF OVEREXPRESSION OF CO1 (Lee et al., 2000) upregulation, giving rise to the hypothesis that AGL28 plays a role in the autonomous flowering pathway (Yoo et al., 2006). However, homozygous agl28 mutant plants are viable, and no obvious aberrant phenotypes were observed (Yoo et al., 2006), making the suggested role for AGL28 in the regulation of flowering time premature. Furthermore, the agl28 mutant was combined with agl23 without any enhancement of the agl23 mutant phenotype (S. Masiero, unpublished data). Based on detailed phylogenetic analysis of the type I MADS box gene family, it is expected that significant redundancy will exist within this family, as already shown for MIKC MADS box genes. Therefore, the generation of double or even multiple gene mutants will probably be necessary to unravel their function.
Recently, Wuest et al. (2010) presented the expression profiles of all cell types in the mature Arabidopsis female gametophyte by combining laser-assisted microdissection of individual cells with Affymetrix ATH1 GeneChip expression analysis. The low number of type I MADS box genes present on the ATH1 GeneChip (only 28 out of 61) (Grennan, 2007) is a strong limitation to a global characterization of this class of genes. Nevertheless, 26 of these were exclusively enriched in male or female gametophytes or in the embryo/endosperm. Notably, Wuest et al. observed that AGL62 mRNA was detected exclusively in the central cell, strongly supporting the validity of their laser-assisted microdissection approach. Moreover, seven type I MADS box genes are highly expressed in Arabidopsis embryo sac cells, including AGL23, AGL61, and AGL80, consistent with previously published data (Portereiko et al., 2006; Bemer et al., 2008; M. Colombo et al., 2008; Steffen et al., 2008).
Complementing transcriptomic investigations, the expression pattern of 60 type I MADS box genes in Arabidopsis by translational reporter fusions was recently reported (Bemer et al., 2010). A total of 42 genes were detected in the female gametophytes or developing seeds, confirming their predominant involvement in plant reproduction. Twelve of them (mainly of Mα and Mγ subclades) are expressed in the antipodal cells and 15 in the central cell. Several genes expressed in the central cell are also expressed in the endosperm as previously discussed (Day et al., 2008). These analyses, supplemented with real-time PCR, suggest that in addition to AGL37/PHE1 and AGL62 mentioned above, AGL23, AGL33, AGL35, AGL36, AGL38/PHERES2, AGL40, AGL78, AGL86, AGL91, and AGL102 are expressed in triploid endosperm.
TYPE I MADS BOX TRANSCRIPTION FACTORS REGULATE GENOME DOSAGE AND CONTROL POSTZYGOTIC COMPATIBILITY
A variety of mechanisms have been described that minimize gene flow between species and contribute to their reproductive isolation: the prezygotic mechanisms reduce the frequency at which gametes combine to form a zygote, while the postzygotic ones reduce the viability or reproductive potential of the hybrid. Crosses between diploid A. thaliana (At) and Arabidopsis arenosa (Aa) result in postzygotic incompatibility (Bushell et al., 2003). The observed seed developmental arrest is associated with endosperm hyperproliferation and delayed development similar to paternal-excess interploidy crosses and PRC2 mutants (Scott et al., 1998; Gutiérrez-Marcos et al., 2004). Endosperm size is also strictly controlled by genome dosage, and an increased ratio of paternally to maternally contributed genomes in the seed (paternal excess, resulting from crossing a diploid female with a tetraploid male) generates an increase in endosperm growth, while an increased ratio of maternal to paternal genomes (maternal excess) reduces endosperm growth. Postzygotic incompatibility in such interspecies crosses can be partially overcome by changing ploidy, and in maternal 4n At × paternal 2n Aa semicompatible crosses, partial restoration of seed viability can be observed. Comparison of transcript profiles of semicompatible and compatible crosses revealed that the endosperm expressed PHE1, PHE2, AGL35, AGL36, AGL40, AGL62, and AGL90 are downregulated in 5-d-old semicompatible siliques (Walia et al., 2009). The relative differences between compatible and incompatible crosses increased further by 6 d after pollination in agreement with the abnormal endosperm proliferation that begins 4 to 5 d after fertilization. Walia et al. (2009) propose that the repression of these genes is necessary to restrict endosperm proliferation in hybrids and the transition to the endosperm cellularization stage; moreover, these AGL genes seem to act in a dosage-dependent manner. In accordance, Walia et al. could show that preventing the formation of the AGL62-90 heterodimer, using mutants of AGL62 or AGL90 as female cross partners in maternal 2n At × paternal 2n Aa crosses, enhanced seed survival. This finding was further accompanied by selective transmission of the agl62 or agl90 mutant alleles to the progeny, supporting the interpretation that seed survival in the hybrids is directly related to the loss of AGL62/AGL90 (Walia et al., 2009).
In the offspring of these interploidy crosses, endosperm-expressed type I MADS box genes are affected in their expression level (Tiwari et al., 2010). PHE1 and PHE2 are upregulated in seeds with paternal excess, as well as AGL28 and AGL40 that encode for PHE1 and PHE2 interacting partners. Also, AGL62 and AGL45, which protein products interact with AGL40, show higher expression level in seeds with an increased paternal contribution. In addition, AGL36, AGL62, AGL90, and PHE1 were commonly upregulated in transcriptional profiles of At paternal excess crosses using tetraploid or unreduced jason pollen (Erilova et al., 2009). Overexpression of individual genes (e.g., AGL28, PHE1, and AGL40) under the endosperm-specific promoter of At5g46950 (Tiwari et al., 2006) did not result in the formation of larger seeds; however, seed size is indeed affected by PHE2 overexpression (Tiwari et al., 2010). Concurrent overexpression of two (or more) genes forming dimers or higher-order complexes might be necessary to mimic paternal excess phenotypes.
In line with these findings, overrepresentation of downregulated type I MADS box transcription factors, especially the Mγ subclade, was observed in seeds developing without a paternal contribution to the endosperm (Shirzadi et al., 2011). These authors compared genome-wide transcription profiles of seeds fertilized with the wild type or a mutant in the cell cycle regulator CYCLIN DEPENDENT KINASE A;1 (CKDA;1) (Iwakawa et al., 2006; Nowack et al., 2006). In cdka;1 mutant pollen, the second mitosis is missing or severely delayed, and these pollen can successfully fertilize the egg cell, whereas karyogamy does not take place in the central cell (Nowack et al., 2006; Aw et al., 2010). Although not properly fertilized, the majority of the central cells in cdka;1 fertilized ovules are triggered to initiate endosperm proliferation. Thus, fertilization by cdka;1 sperm cells creates a unique situation where endosperm develops without any paternal contribution (Nowack et al., 2007; Aw et al., 2010). Among the downregulated transcripts in the absence of a paternal genome, type I MADS box transcription factors were significantly overrepresented (PHE1, PHE2, AGL34, AGL35, AGL36, AGL62, AGL90, AGL96, and AGL102) (Shirzadi et al., 2011). From this array, AGL36 was chosen for an in-depth study and was shown to have a parent-of-origin-dependent expression, controlled by the activity of METHYLTRANSFERASE1 maintenance DNA methyltransferase and DEMETER DNA glycosylase (Shirzadi et al., 2011). Interestingly, the active maternal allele of AGL36 was shown to be under the repressive control of the FIS complex, thus highlighting previous reports that genes encoding type I MADS box transcription factors are potent direct or downstream targets of the Polycomb complex (Köhler et al., 2005; Erilova et al., 2009; Walia et al., 2009; Tiwari et al., 2010; Weinhofer et al., 2010).
TYPE I MADS BOX GENES OF OTHER SPECIES
Whereas the plant type II MADS box genes have been well documented and extensively studied through both the functional characterization and moderate to large-scale cDNA sequencing projects in diverse plants, the existence of plant type I MADS box genes was not reported before the completion of the Arabidopsis genome sequence.
In the last 10 years, several other genomes have been sequenced, including rice, poplar (Populus trichocarpa), Glycine max, Physcomitrella patens, cucumber (Cucumis sativus), and apple (Malus domestica). MADS box genome-wide phylogenetic reconstructions using information from those genomes confirmed that this gene family is divided into two main lineages and that the type I members do not have a monophyletic origin, although genes from Arabidopsis, poplar, rice, and apple can be grouped into well-supported sublineages (Goff et al., 2002; Yu et al., 2002; Leseberg et al., 2006; Tuskan et al., 2006; Arora et al., 2007; Rensing et al., 2008; Huang, et al., 2009; Velasco et al., 2010).
In rice, 75 MADS box genes have been identified and classified into type I and II, based on a thorough annotation exercise (Arora et al., 2007; Table 1). Rice Mα (13 members), Mβ (nine members), and Mγ (10 members) genes usually have zero or occasionally up to four introns. The discrepancy between the number of rice and Arabidopsis MADS box genes (75 versus 107) might be explained by the higher number of genome duplications in Arabidopsis in respect to rice. However, the fact that the number of rice and Arabidopsis type II MADS box genes are more or less similar, 38 and 39, respectively, and the observation that none of the Arabidopsis type I genes has a distinct orthologous gene in rice indicate that the type I genes may have increased their number in Arabidopsis due to specific gene duplication events.
Table 1.
Numbers of Annotated Type I MADS Box Genes Present in Fully Sequenced Genomes
| Type I | Arabidopsis | O. sativa | P. trichocarpa | M. domestica |
| Mα | 25 | 13 | 23 | 27 |
| Mβ | 20 | 9 | 12 | 7 |
| Mγ | 16 | 10 | 6 | 20 |
| Total | 61 | 32 | 41 | 54 |
The poplar type I MADS box genes similarly can be classified into three groups: Mα, Mβ, and Mγ (Table 1). As in rice, there are fewer putative functional type I MADS box genes in poplar than in Arabidopsis (i.e., roughly 40 versus the 68 of Arabidopsis). Type I MADS box genes appear to recruit new members from genus-specific duplication events; indeed, few Arabidopsis type I genes had two or more close poplar homologous.
The apple genome has been released recently, and MADS box genome-wide analysis identified 91 type II genes and 27 Mα, seven Mβ, and 20 Mγ type I MADS box genes spread all over the genome (Velasco et al., 2010). As in other species, the apple type I MADS box genes experienced a relatively fast birth and death rate. Moreover, many duplication events have occurred, leading to the formation of apple-specific clusters of very recent origin, which might conceivably be associated with species adaptation and domestication.
The observation that type I MADS box genes seem to be highly variable and genus specific, and also, at least in Arabidopsis, control reproductive boundaries, suggests that type I MADS box genes might be part of the evolutionary tool kit for speciation. Future functional studies of type I MADS box genes in a variety of species are needed to provide more solid evidence for this exciting idea.
PERSPECTIVE
Recent advances in genomics and more powerful molecular tools such as laser dissection microscopy have allowed a first glimpse at the function of MADS box type I transcription factors. Extrapolating from recent insights and the repeated appearance of type I factors in many different studies investigating plant reproduction, we suggest that type I factors might be as important for plant development as their well-studied bigger brother, the type II factors. Our understanding of this class of transcription factors has changed dramatically in just a few years and will presumably continue to change in the future.
In addition, many type I genes have yet to be characterized, not only in Arabidopsis but also other important model systems and crop species. Furthermore, little is known about these factors at the protein level: What types of complexes do they form and which downsteam target genes do they regulate? These questions are technically very challenging, especially for proteins acting in a highly restricted number of cells (such as the embryo sac). Further insight into these important questions awaits the development of highly sensitive proteomic equipment and optimized chromatin immunoprecipitation sequencing approaches. In the near future, most data will come from mutant analysis complemented with large-scale approaches currently available. We are confident that these studies will contribute to the discovery of exciting new functions for this interesting class of MADS box transcription factors.
Acknowledgments
This review is in memory of Zsuzsanna Schwarz-Sommer (1946–2009), the founder of MADS, an unforgettable guide and mentor, but above all, a unique friend. The team that wrote this review was supported by a European Research Area-Net Plant Genomics Grant to M.K., P.E.G., and A.S. and a European Research Council starting grant from the European Union to A.S.
References
- Alvarez-Buylla E.R., Pelaz S., Liljegren S.J., Gold S.E., Burgeff C., Ditta G.S., Ribas de Pouplana L., Martínez-Castilla L., Yanofsky M.F. (2000). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 97: 5328–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R., Agarwal P., Ray S., Singh A.K., Singh V.P., Tyagi A.K., Kapoor S. (2007). MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aw S.J., Hamamura Y., Chen Z., Schnittger A., Berger F. (2010). Sperm entry is sufficient to trigger division of the central cell but the paternal genome is required for endosperm development in Arabidopsis. Development 137: 2683–2690 [DOI] [PubMed] [Google Scholar]
- Bemer M., Heijmans K., Airoldi C., Davies B., Angenent G.C. (2010). An atlas of type I MADS box gene expression during female gametophyte and seed development in Arabidopsis. Plant Physiol. 154: 287–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemer M., Wolters-Arts M., Grossniklaus U., Angenent G.C. (2008). The MADS domain protein DIANA acts together with AGAMOUS-LIKE80 to specify the central cell in Arabidopsis ovules. Plant Cell 20: 2088–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyer D., Roudier F., Heese M., Andersen E.D., Gey D., Nowack M.K., Goodrich J., Renou J.-P., Grini P.E., Colot V., Schnittger A. (2011). Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet 7: e1002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell C., Spielman M., Scott R.J. (2003). The basis of natural and artificial postzygotic hybridization barriers in Arabidopsis species. Plant Cell 15: 1430–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury A.M., Ming L., Miller C., Craig S., Dennis E.S., Peacock W.J. (1997). Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94: 4223–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen C.A., King E.J., Jordan J.R., Drews G.N. (1997). Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex. Plant Reprod. 10: 49–64 [Google Scholar]
- Colombo L., Battaglia R., Kater M.M. (2008). Arabidopsis ovule development and its evolutionary conservation. Trends Plant Sci. 13: 444–450 [DOI] [PubMed] [Google Scholar]
- Colombo M., Masiero S., Vanzulli S., Lardelli P., Kater M.M., Colombo L. (2008). AGL23, a type I MADS-box gene that controls female gametophyte and embryo development in Arabidopsis. Plant J. 54: 1037–1048 [DOI] [PubMed] [Google Scholar]
- Day R.C., Herridge R.P., Ambrose B.A., Macknight R.C. (2008). Transcriptome analysis of proliferating Arabidopsis endosperm reveals biological implications for the control of syncytial division, cytokinin signaling, and gene expression regulation. Plant Physiol. 148: 1964–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S., Raes J., Florquin K., Rombauts S., Rouzé P., Theissen G., Van de Peer Y. (2003a). Genomewide structural annotation and evolutionary analysis of the type I MADS-box genes in plants. J. Mol. Evol. 56: 573–586 [DOI] [PubMed] [Google Scholar]
- De Bodt S., Raes J., Van de Peer Y., Theissen G. (2003b). And then there were many: MADS goes genomic. Trends Plant Sci. 8: 475–483 [DOI] [PubMed] [Google Scholar]
- de Folter S., Immink R.G., Kieffer M., Parenicová L., Henz S.R., Weigel D., Busscher M., Kooiker M., Colombo L., Kater M.M., Davies B., Angenent G.C. (2005). Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell 17: 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea-Cortines M., Saedler H., Sommer H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18: 5370–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erilova A., Brownfield L., Exner V., Rosa M., Twell D., Mittelsten Scheid O., Hennig L., Köhler C. (2009). Imprinting of the polycomb group gene MEDEA serves as a ploidy sensor in Arabidopsis. PLoS Genet. 5: e1000663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M., Choi Y., Fischer R.L. (2004). Imprinting and seed development. Plant Cell 16(suppl.): S203–S213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni J.J. (2004). Genetic regulation of fruit development and ripening. Plant Cell 16(suppl.): S170–S180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S.A., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Gramzow L., Ritz M.S., Theissen G. (2010). On the origin of MADS-domain transcription factors. Trends Genet. 26: 149–153 [DOI] [PubMed] [Google Scholar]
- Grennan A.K. (2007). An analysis of the Arabidopsis pollen transcriptome. Plant Physiol. 145: 3–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Marcos J.F., Costa L.M., Biderre-Petit C., Khbaya B., O’Sullivan D.M., Wormald M., Perez P., Dickinson H.G. (2004). maternally expressed gene1 is a novel maize endosperm transfer cell-specific gene with a maternal parent-of-origin pattern of expression. Plant Cell 16: 1288–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., et al. (2009). The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41: 1275–1281 [DOI] [PubMed] [Google Scholar]
- Immink R.G., Tonaco I.A., de Folter S., Shchennikova A., van Dijk A.D., Busscher-Lange J., Borst J.W., Angenent G.C. (2009). SEPALLATA3: The ‘glue’ for MADS box transcription factor complex formation. Genome Biol. 10: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M., Haseloff J., Berger F. (2005). Polycomb group genes control developmental timing of endosperm. Plant J. 42: 663–674 [DOI] [PubMed] [Google Scholar]
- Iwakawa H., Shinmyo A., Sekine M. (2006). Arabidopsis CDKA;1, a cdc2 homologue, controls proliferation of generative cells in male gametogenesis. Plant J. 45: 819–831 [DOI] [PubMed] [Google Scholar]
- Jullien P.E., Katz A., Oliva M., Ohad N., Berger F. (2006). Polycomb group complexes self-regulate imprinting of the Polycomb group gene MEDEA in Arabidopsis. Curr. Biol. 16: 486–492 [DOI] [PubMed] [Google Scholar]
- Kang I.H., Steffen J.G., Portereiko M.F., Lloyd A., Drews G.N. (2008). The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20: 635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kater M.M., Dreni L., Colombo L. (2006). Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J. Exp. Bot. 57: 3433–3444 [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Melzer R., Theissen G. (2005). MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 347: 183–198 [DOI] [PubMed] [Google Scholar]
- Kofuji R., Sumikawa N., Yamasaki M., Kondo K., Ueda K., Ito M., Hasebe M. (2003). Evolution and divergence of the MADS-box gene family based on genome-wide expression analyses. Mol. Biol. Evol. 20: 1963–1977 [DOI] [PubMed] [Google Scholar]
- Köhler C., Hennig L., Bouveret R., Gheyselinck J., Grossniklaus U., Gruissem W. (2003a). Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 22: 4804–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C., Hennig L., Spillane C., Pien S., Gruissem W., Grossniklaus U. (2003b). The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 17: 1540–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C., Page D.R., Gagliardini V., Grossniklaus U. (2005). The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat. Genet. 37: 28–30 [DOI] [PubMed] [Google Scholar]
- Koornneef M., Alonso-Blanco C., Blankestijn-de Vries H., Hanhart C.J., Peeters A.J. (1998). Genetic interactions among late-flowering mutants of Arabidopsis. Genetics 148: 885–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Suh S.S., Park E., Cho E., Ahn J.H., Kim S.G., Lee J.S., Kwon Y.M., Lee I. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14: 2366–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leseberg C.H., Li A., Kang H., Duvall M., Mao L. (2006). Genome-wide analysis of the MADS-box gene family in Populus trichocarpa. Gene 378: 84–94 [DOI] [PubMed] [Google Scholar]
- Liu C., Thong Z., Yu H. (2009). Coming into bloom: The specification of floral meristems. Development 136: 3379–3391 [DOI] [PubMed] [Google Scholar]
- Martinez-Castilla L.P., Alvarez-Buylla E.R. (2003). Adaptive evolution in the Arabidopsis MADS-box gene family inferred from its complete resolved phylogeny. Proc. Natl. Acad. Sci. USA 100: 13407–13412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiero S., Imbriano C., Ravasio F., Favaro R., Pelucchi N., Gorla M.S., Mantovani R., Colombo L., Kater M.M. (2002). Ternary complex formation between MADS-box transcription factors and the histone fold protein NF-YB. J. Biol. Chem. 277: 26429–26435 [DOI] [PubMed] [Google Scholar]
- Masutani H., Magnaghi-Jaulin L., Ait-Si-Ali S., Groisman R., Robin P., Harel-Bellan A. (1997). Activation of the c-fos SRE through SAP-1a. Oncogene 15: 1661–1669 Erratum. Oncogene 18: 5246 [DOI] [PubMed] [Google Scholar]
- Mead J., Bruning A.R., Gill M.K., Steiner A.M., Acton T.B., Vershon A.K. (2002). Interactions of the Mcm1 MADS box protein with cofactors that regulate mating in yeast. Mol. Cell. Biol. 22: 4607–4621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Y., Ho W., Johnston K., Marmorstein R. (2001). Crystal structure of a ternary SAP-1/SRF/c-fos SRE DNA complex. J. Mol. Biol. 314: 495–506 [DOI] [PubMed] [Google Scholar]
- Nam J., Kim J., Lee S., An G., Ma H., Nei M. (2004). Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc. Natl. Acad. Sci. USA 101: 1910–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M., Yanofsky M.F. (2001). Function and evolution of the plant MADS-box gene family. Nat. Rev. Genet. 2: 186–195 [DOI] [PubMed] [Google Scholar]
- Norman C., Runswick M., Pollock R., Treisman R. (1988). Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 55: 989–1003 [DOI] [PubMed] [Google Scholar]
- Nowack M.K., Grini P.E., Jakoby M.J., Lafos M., Koncz C., Schnittger A. (2006). A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat. Genet. 38: 63–67 [DOI] [PubMed] [Google Scholar]
- Nowack M.K., Shirzadi R., Dissmeyer N., Dolf A., Endl E., Grini P.E., Schnittger A. (2007). Bypassing genomic imprinting allows seed development. Nature 447: 312–315 [DOI] [PubMed] [Google Scholar]
- Parenicová L., de Folter S., Kieffer M., Horner D.S., Favalli C., Busscher J., Cook H.E., Ingram R.M., Kater M.M., Davies B., Angenent G.C., Colombo L. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 15: 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore S., Maine G.T., Elble R., Christ C., Tye B.K. (1988). Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MAT alpha cells. J. Mol. Biol. 204: 593–606 [DOI] [PubMed] [Google Scholar]
- Portereiko M.F., Lloyd A., Steffen J.G., Punwani J.A., Otsuga D., Drews G.N. (2006). AGL80 is required for central cell and endosperm development in Arabidopsis. Plant Cell 18: 1862–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing S.A., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Schneitz K., Hulskamp M., Pruitt R.E. (1995). Wild-type ovule development in Arabidopsis thaliana: A light microscope study of cleared whole-mount tissue. Plant J. 7: 731–749 [Google Scholar]
- Schönrock N., Exner V., Probst A., Gruissem W., Hennig L. (2006). Functional genomic analysis of CAF-1 mutants in Arabidopsis thaliana. J. Biol. Chem. 281: 9560–9568 [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Davies B., Hudson A. (2003). An everlasting pioneer: The story of Antirrhinum research. Nat. Rev. Genet. 4: 657–666 [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Huijser P., Nacken W., Saedler H., Sommer H. (1990). Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 250: 931–936 [DOI] [PubMed] [Google Scholar]
- Scott R.J., Spielman M., Bailey J., Dickinson H.G. (1998). Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125: 3329–3341 [DOI] [PubMed] [Google Scholar]
- Shirzadi R., Andersen E.D., Bjerkan K.N., Gloeckle B.M., Heese M., Ungru A., Winge P., Koncz C., Aalen R.B., Schnittger A., Grini P.E. (2011). Genome-wide transcript profiling of endosperm without paternal contribution identifies parent-of-origin-dependent regulation of AGAMOUS-LIKE36. PLoS Genet. 7: e1001303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore P., Sharrocks A.D. (1995). The ETS-domain transcription factors Elk-1 and SAP-1 exhibit differential DNA binding specificities. Nucleic Acids Res. 23: 4698–4706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer H., Beltrán J.P., Huijser P., Pape H., Lönnig W.E., Saedler H., Schwarz-Sommer Z. (1990). Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. EMBO J. 9: 605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillane C., MacDougall C., Stock C., Köhler C., Vielle-Calzada J.P., Nunes S.M., Grossniklaus U., Goodrich J. (2000). Interaction of the Arabidopsis polycomb group proteins FIE and MEA mediates their common phenotypes. Curr. Biol. 10: 1535–1538 [DOI] [PubMed] [Google Scholar]
- Steffen J.G., Kang I.H., Portereiko M.F., Lloyd A., Drews G.N. (2008). AGL61 interacts with AGL80 and is required for central cell development in Arabidopsis. Plant Physiol. 148: 259–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G., Kim J.T., Saedler H. (1996). Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 43: 484–516 [DOI] [PubMed] [Google Scholar]
- Tiwari S., Spielman M., Day R.C., Scott R.J. (2006). Proliferative phase endosperm promoters from Arabidopsis thaliana. Plant Biotechnol. J. 4: 393–407 [DOI] [PubMed] [Google Scholar]
- Tiwari S., Spielman M., Schulz R., Oakey R.J., Kelsey G., Salazar A., Zhang K., Pennell R., Scott R.J. (2010). Transcriptional profiles underlying parent-of-origin effects in seeds of Arabidopsis thaliana. BMC Plant Biol. 10: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tröbner W., Ramirez L., Motte P., Hue I., Huijser P., Lönnig W.-E., Saedler H., Sommer H., Schwarz-Sommer Z. (1992). GLOBOSA: A homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 11: 4693–4704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan G.A., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Vandenbussche M., Zethof J., Souer E., Koes R., Tornielli G.B., Pezzotti M., Ferrario S., Angenent G.C., Gerats T. (2003). Toward the analysis of the petunia MADS box gene family by reverse and forward transposon insertion mutagenesis approaches: B, C, and D floral organ identity functions require SEPALLATA-like MADS box genes in petunia. Plant Cell 15: 2680–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk A.D.J., Morabito G., Fiers M., van Ham R.C.H.J., Angenent G.C., Immink R.G.H. (2010). Sequence motifs in MADS transcription factors responsible for specificity and diversification of protein-protein interaction. PLoS Comput. Biol. 6: e1001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R., et al. (2010). The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 42: 833–839 [DOI] [PubMed] [Google Scholar]
- Walia H., Josefsson C., Dilkes B., Kirkbride R., Harada J., Comai L. (2009). Dosage-dependent deregulation of an AGAMOUS-LIKE gene cluster contributes to interspecific incompatibility. Curr. Biol. 19: 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhofer I., Hehenberger E., Roszak P., Hennig L., Köhler C. (2010). H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet. 6: e1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple C.J., Ciceri P., Padilla C.M., Ambrose B.A., Bandong S.L., Schmidt R.J. (2004). Conservation of B-class floral homeotic gene function between maize and Arabidopsis. Development 131: 6083–6091 [DOI] [PubMed] [Google Scholar]
- Wuest S.E., Vijverberg K., Schmidt A., Weiss M., Gheyselinck J., Lohr M., Wellmer F., Rahnenführer J., von Mering C., Grossniklaus U. (2010). Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr. Biol. 20: 506–512 [DOI] [PubMed] [Google Scholar]
- Yadegari R., Drews G.N. (2004). Female gametophyte development. Plant Cell 16(suppl.): S133–S141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Fanning L., Jack T. (2003). The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. Plant J. 33: 47–59 [DOI] [PubMed] [Google Scholar]
- Yanofsky M.F., Ma H., Bowman J.L., Drews G.N., Feldmann K.A., Meyerowitz E.M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346: 35–39 [DOI] [PubMed] [Google Scholar]
- Yoo S.K., Lee J.S., Ahn J.H. (2006). Overexpression of AGAMOUS-LIKE 28 (AGL28) promotes flowering by upregulating expression of floral promoters within the autonomous pathway. Biochem. Biophys. Res. Commun. 348: 929–936 [DOI] [PubMed] [Google Scholar]
- Yu J., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]
- Yu Y.T., Breitbart R.E., Smoot L.B., Lee Y., Mahdavi V., Nadal-Ginard B. (1992). Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev. 6: 1783–1798 [DOI] [PubMed] [Google Scholar]
- Zhang H., Forde B.G. (1998). An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409 [DOI] [PubMed] [Google Scholar]
- Zhang X., Clarenz O., Cokus S., Bernatavichute Y.V., Pellegrini M., Goodrich J., Jacobsen S.E. (2007). Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]