Genes that are similarly expressed, or coexpressed, are often involved in related biological processes. Such coexpressed relationships also appear to be conserved across species. The PlaNet platform enables comparative analysis of genome-wide coexpression networks across seven plant species, thus enabling prediction of gene function and elucidation of the identity of functional homologs.
Abstract
The model organism Arabidopsis thaliana is readily used in basic research due to resource availability and relative speed of data acquisition. A major goal is to transfer acquired knowledge from Arabidopsis to crop species. However, the identification of functional equivalents of well-characterized Arabidopsis genes in other plants is a nontrivial task. It is well documented that transcriptionally coordinated genes tend to be functionally related and that such relationships may be conserved across different species and even kingdoms. To exploit such relationships, we constructed whole-genome coexpression networks for Arabidopsis and six important plant crop species. The interactive networks, clustered using the HCCA algorithm, are provided under the banner PlaNet (http://aranet.mpimp-golm.mpg.de). We implemented a comparative network algorithm that estimates similarities between network structures. Thus, the platform can be used to swiftly infer similar coexpressed network vicinities within and across species and can predict the identity of functional homologs. We exemplify this using the PSA-D and chalcone synthase-related gene networks. Finally, we assessed how ontology terms are transcriptionally connected in the seven species and provide the corresponding MapMan term coexpression networks. The data support the contention that this platform will considerably improve transfer of knowledge generated in Arabidopsis to valuable crop species.
INTRODUCTION
Various rapidly evolving genomic and postgenomic technologies, including genome sequences and gene expression data, have greatly enhanced our understanding of how biological systems function. As of June 2010, >1500 genomes from prokaryotic, eukaryotic, and archae organisms have been fully sequenced, and >5500 sequencing projects are in progress (Liolios et al., 2010). In parallel, transcriptional studies via DNA microarrays and deep sequencing methods have generated vast amounts of publicly available expression data for various organisms, with >7000 microarray samples available for Arabidopsis thaliana alone (Gene Expression Omnibus database, as of January 2011).
Now that gene expression data have been generated, they are being mined for hypothesis-driven gene discovery, for example, to reveal transcriptional responses to certain genotypes or external stimuli and to uncover coordinate expression of different genes (Usadel et al., 2009). Data from these types of analyses support the hypothesis that functionally related genes tend to be transcriptionally coordinated (i.e., coexpressed) (Stuart et al., 2003; Persson et al., 2005). Consequently, using guilt-by-association approaches, coexpression analyses have proved valuable for rapid inference of gene function, subcellular localization of gene products, and biological pathway discovery (Wei et al., 2006; Yonekura-Sakakibara et al., 2008; San Clemente et al., 2009; Usadel et al., 2009; Klie et al., 2010).
While coexpression relationships in many cases can provide insight into biological processes and predict genes for functional testing, the representation of genomic content on the microarrays is incomplete (Table 1). For example, the widely used Arabidopsis Affymetrix ATH1 GeneChip and the Affymetrix rice (Oryza sativa) GeneChip cover ~63 and 60% of the genes in the Arabidopsis and rice genomes, respectively (Table 1). It follows that certain transcriptional relationships are not revealed using microarrays. In addition, low spatio-temporal resolution of gene expression contributes both to false negatives (e.g., expression of genes may be rendered as noise due to activity in only specific cell types or stimuli) and to false positives (e.g., difficulties in distinguishing pollen- and ovule-specific genes if only flowers are measured). These caveats should prompt caution by biologists in overreliance, or at least overinterpretation, of whole-genome expression analyses.
Table 1.
Microarray Data Sets Used in This Study
| Organism | Affymetrix GeneChip | Probe Sets | No. of Chipsa | No. of HCCA Obtained Clusters | Source Database of Coding Sequences | Percentage of Represented Genes |
| Arabidopsis | ATH1 | 22,810 | 279 | 229 | TAIR9, http://www.Arabidopsis.org/ | ~63% |
| Barley | Barley1 | 22,840 | 181 | 215 | harvEST Hv35, http://www.harvest-web.org/ | NAb |
| Medicago truncatula, Medicago sativa | Medicago | 61,263 | 163 | 362 | IMGAG 27-02-2008, http://www.medicago.org/genome/IMGAG/ | NAb |
| Poplar | Poplar | 61,413 | 83 | 456 | Poptr 1.1 Jamboree, http://genome.jgi-psf.org/poplar/poplar.home.html | ~65% |
| Rice | Rice | 57,380 | 156 | 565 | Rice Genome annotation v 6.0, http://rice.plantbiology.msu.edu/ | ~60% |
| Soybean | Soybean | 61,170 | 171 | 422 | harvEST Gm 10-12-2009, http://www.harvest-web.org/ | NAb |
| Wheat | Wheat | 61,290 | 165 | 484 | Triticum aestivum, http://www.harvest-web.org/ | NAb |
Microarray data sets used in this study and clustering algorithm are available at http://aranet.mpimp-golm.mpg.de/downloads.
Due to lack of complete genome sequence, this estimation is not possible.
Arabidopsis, as the most studied plant species, has ~50% of its genes functionally annotated by sequence homology, and ~11% of the total genes are associated with distinct biological functions that have been experimentally verified (Saito et al., 2008). Still, a major goal is to transfer the knowledge obtained in a model organism (donor), such as Arabidopsis, to other species (acceptors), which have greater economic and nutritional importance. Once the function of a gene product in the knowledge donor has been proven experimentally, uncovering the identity of the functional equivalent in an acceptor species is, however, not trivial. As plant genomes characteristically contain large gene families, sequence comparison of a gene from the knowledge donor to the genome of the acceptor can return a large list of possible candidate genes. While several of those candidates may perform the same molecular function, they are not necessarily equivalent participants in the biological process of interest. Intuitively, a functional homolog should be present when the relevant biological process occurs. Thus, functional homologs from different species should be reflected in conserved coexpression patterns. Indeed, several studies have shown that coexpressed relationships are conserved across species and even kingdoms (Stuart et al., 2003; Bergmann et al., 2004). Hence, a functional homolog may be identified by combined sequence and coexpression approaches.
Several internet-resident tools that combine coexpression analyses with sequence, protein–protein interaction, cis-element, and subcellular localization prediction have been created for individual plant species (Steinhauser et al., 2004; Manfield et al., 2006; Mutwil et al., 2008; Srinivasasainagendra et al., 2008; Obayashi et al., 2009; Mutwil et al., 2009). The representation of coexpressed relationships as networks has transcended standard single gene analyses, since this enables the biologist to more readily contextualize their genes or proteins of interest (Mao et al., 2009; Mutwil et al., 2010). Several other tools that mediate comparative analyses of coexpression networks across different species have been published (Starnet: Jupiter et al., 2009; CoP: Ogata et al., 2010), but the implemented algorithms permit pairwise comparison of species only.
Here, we present PlaNet (Plant Network), a platform that integrates genomics, transcriptomics, phenomics, and ontology analyses across seven plant species important both for research and human circumstances (http://aranet.mpimp-golm.mpg.de). For comparative analyses, we implemented NetworkComparer, a novel pipeline that compares and displays commonalities and differences between the coexpressed node vicinity networks (NVNs; see Supplemental Figure 1 online) simultaneously across selected species. Importantly, considering the incomplete gene coverage of the microarray probes, comparative analysis between species provided insight into the association of a gene with certain processes despite the absence of corresponding microarray probe. We demonstrate the features of the platform by two examples, the photosynthesis-related PSA-D1 gene and several chalcone synthase (CHS)-related genes in Arabidopsis. These examples illustrate that the utility of the platform can be extended to the plant biology community at large; therefore, we are making it available as a community resource.
Data Sources, Construction, and Structure of PlaNet
Network-based approaches that integrate high-throughput omics data with structured biological knowledge (e.g., sequence information and gene annotation) offer the possibility for transferring biochemical knowledge, including function, localization, and involvement of genes or gene families in (various) cellular processes from model organisms to other investigated species. To guarantee statistical soundness and validity of any prediction, an approach for knowledge transfer should include methods and techniques that necessarily take into consideration only statistically significant observations under a suitably chosen null model. In addition, such an approach should provide the means for discerning which of the available data are most suitable for drawing biologically meaningful conclusions. Finally, with the help of the carefully chosen subset of data, the approach should employ those values for the method/model parameters that maximize the extraction of biologically relevant information.
The three outlined requirements for an approach for knowledge transfer, statistical significance, data quality/selection, and biological relevance, are fully addressed in PlaNet, resulting in a network and a set of parameter values that can be used for statistically significant and robust knowledge transfer. In addition, the implementation allows for exploration of the network not only with the set of optimal parameter values, but also with user-specified values.
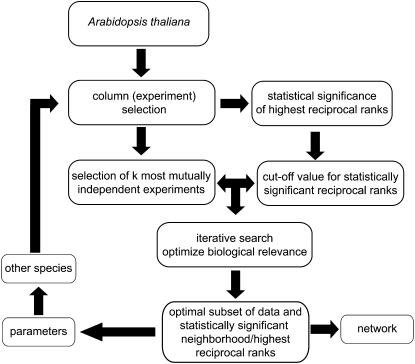
Our approach relies on the following framework, depicted in Figure 1: (1) We provide an extension to a mathematically sound technique for finding k most mutually independent columns from a given set of data; this method resolves the issue of selecting most informative set of experiments. (2) We apply sound statistical tests to determine the optimal cutoff (range) for the reciprocal ranks that then translate into establishing edges that are fully supported by the used data. (3) By providing an iterative search on the allowable ranges for the reciprocal ranks that maximize the similarity of gene function in the vicinity (neighborhood) of a given gene (network node), we employ the optimality principle in selecting the set of best-performing parameter values with respect to the guilty-by-association criterion. This is in line with the principle objective of this framework to provide knowledge transfer from well-studied model species to other species of scientific interest. (4) Lastly, to minimize the number of disconnected nodes in the coexpression networks, we analyzed the influence of highest reciprocal rank (HRR) cutoff on network density.
Figure 1.
PlaNet Framework for Statistically Significant and Robust Knowledge Transfer between Species.
The different steps in the analysis are discussed in Methods.
Affymetrix GeneChip microarray data sets (summarized in Table 1) for seven plant species (Arabidopsis, barley [Hordeum vulgare], rice, Medicago, poplar [Populus spp], wheat [Triticum aestivum], and soybean [Glycine max]) were obtained from the Gene Expression Omnibus (Edgar et al., 2002) and ArrayExpress (Parkinson et al., 2009), quality controlled to remove possible array errors (Persson et al., 2005), and preselected for most mutually independent experiments (see Column Selection in Methods). Phenotypic data for Arabidopsis and rice have been obtained from www.Arabidopsis.org and Ficklin et al. (2010). All data associated with PlaNet database can be downloaded from http://aranet.mpimp-golm.mpg.de/download.
For visualization of the expression relationships, we used the HRR between any two genes as a measure (Mutwil et al., 2010), as rank-based associations produce robust coexpression analyses (Obayashi and Kinoshita, 2009). To find statistically significant HRR values, we investigated the distribution of HRR values over 100 permutations of the microarray data set (see Statistical Significance of Reciprocal Ranks in Methods). While the analysis revealed that values of ≤228 are significant (P < 0.01), it is important to keep in mind that statistical significance of coexpression relationships often does not reflect biological relevance (Usadel et al., 2009). We therefore determined the HRR value that optimized the biological relevance (described in Optimality Principal in Methods) and found that 10≤HRR≤30 produced biologically relevant networks. Lastly, while >80% of the nodes were disconnected for HRR=10, and consequently excluded from any further coexpression analysis, the number of disconnected nodes decreased to 25% for HRR=30 (see Optimality Principal in Method). Thus, combining the statistical significance, biological relevance, and inclusion of maximum number of the nodes connected in the network, we found that HRR=30 resulted in the good compromise between the three parameters for the seven species. However, since the preselected parameters do not necessarily correspond to the type of analysis of interest for some users, we also provide a downloadable, stand-alone version of PlaNet (available at http://aranet.mpimp-golm.mpg.de/download). In this version, users can construct HRR-based coexpression networks using any microarray data and apply self-selected parameters for the analyses.
As whole-genome-scale networks are too large and complex for comprehensive visualization, we first partitioned the networks into manageable clusters using Heuristic Cluster Chiseling Algorithm (HCCA) with three-step NVN (Mutwil et al., 2010). HCCA finds clusters by generating putative clusters for every node in the graph and then iteratively removes nodes that show higher connectivity to nodes outside of a cluster compared with nodes within the cluster. During each iteration, new clusters are generated until all nodes are assigned to clusters (see Supplemental Figure 1 online; Mutwil et al., 2010). HCCA was chosen since this algorithm supports weighted edge graphs and permits the user to specify their own desired cluster size. The latter is crucial for visualization of large networks since large clusters (>400 nodes) often are too dense for visual inspection, and conversely small networks (<10 nodes) are often biologically meaningless. When the desired cluster size interval is set to 40 to 200 nodes, the algorithm yields between 215 and 565 clusters for the seven plant species (Table 1). Following this example further, Graphviz (www.graphviz.org) was used to calculate the spring model layout of the networks. The resulting interactive clusters (available at http://aranet.mpimp-golm.mpg.de) represent coexpressed genes, presumably involved in related biological processes.
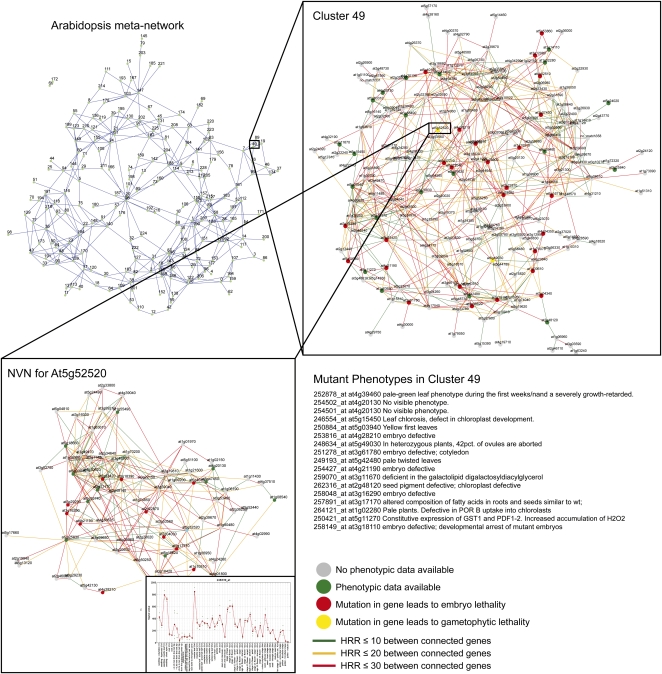
To interrogate the resulting networks, the user can specify their gene of interest by probe set ID, gene ID, nucleotide/protein sequence, or keyword, which redirects the user to the corresponding gene cluster or to an individual gene specific page (Figure 2). The latter page contains the expression profile of the gene across different tissues, a two-step NVN surrounding the gene (step selection is discussed in Optimality Principal in Methods), phenotypes found in the NVN, and MapMan Ontology analysis (Figure 2). The phenotype associations are displayed as color-coded nodes, where red, yellow, and green represent embryo lethal, gametophytic lethal, and nonlethal phenotypes, respectively, and are available for Arabidopsis and rice. Moving the mouse pointer over a node opens a pop-up window displaying annotation and phenotypic information of a gene, while clicking a node redirects the user to a page dedicated to the corresponding gene.
Figure 2.
Outline of the PlaNet Platform.
Coexpression relationships were calculated using quality controlled microarray data for Arabidopsis, rice, barley, poplar, Medicago, wheat, and soybean. The relationships were clustered using HCCA, and the resulting clusters were connected based on the coexpression between individual genes retained in the clusters, resulting in a MetaNetwork (top left). Each node in this network represents a cluster of coexpressed genes (top right). Available phenotypic data were mapped onto the genes (inset top right; red indicates embryo lethality, yellow indicates gametophytic lethality, and green indicates other reported phenotypes that result when the gene is mutated). The colored edges indicate strength of the coexpression based on HRRs between the individual gene pairs (green indicates a HRR ≤ 10, orange indicates a mutual rank between 11 and 20, and red indicates a mutual rank between 21 and 30). Each of the genes in the cluster can also be displayed with its NVN in which the gene of interest is centered, and the surrounding coexpressed genes are displayed (bottom left). This layout also includes the expression profile of the gene across different tissues (bottom left). All pages also give information about phenotypic data for the genes in the cluster (bottom right) and enriched ontology terms (see Supplemental Data Set 1 online).
Partitioning of any object into smaller units indisputably removes information about how the units are arranged to make up the object. To avoid loss of such valuable information, we connected the clusters based on mutual coexpression relationships to form a network of clusters, which should reflect the organization of the genome-wide coexpression network. The resulting meta-networks rendered from this analysis thus depict relations between coexpressed gene clusters (Figure 2) (i.e., a node in this type of network is a HCCA obtained cluster of coexpressed genes) (Mutwil et al., 2010). Any two clusters in the meta-network are connected if the sum of shared edge weights between them are significant (P < 0.0025; see Methods).
Given that any cluster in the meta-network contains genes that are coexpressed with one another, it might be anticipated that the majority of these genes should be involved in related biological processes. However, inferring such relations is not trivial as many genes are not associated with useful annotations. We attempted to get around this problem by combining MapMan ontology analysis, available phenotypic data, and tissue-dependent expression profiling. For example, the majority of genes in the Arabidopsis cluster 49 show ubiquitous expression profiles, and mutations in the genes often show pale-green phenotypes or are embryo lethal (Figure 2). MapMan analysis revealed that this cluster is strongly enriched for genes associated with chloroplast protein targeting and to a lesser degree with amino acid synthesis (see Supplemental Data Set 1 online; http://aranet.mpimp-golm.mpg.de/aranet/ac49). Based on the combined information from these analyses, we predict that cluster 49 holds genes involved in amino acid synthesis and chloroplast development.
Analogous to the enrichment of certain biological processes within a cluster, connected clusters also share coexpressed gene pairs and may therefore also be involved in related processes. One such example is evident for genes grouped in the connected clusters 7, 19, 37, 46, 49, 76, 89, 99, and 134 in Arabidopsis. Mutations in many of these genes display pale-green phenotypes or result in embryo lethality. Most of the genes associated with these clusters are also ubiquitously expressed with the exception of roots, and the clusters are enriched for MapMan ontology terms such as protein targeting to chloroplast, plastid protein synthesis, and photosystem light reaction. Therefore, we find it likely that many of these clusters are associated with chloroplast development and photosynthesis. Many other groups of clusters are also enriched for certain biological functions, such as cell division, protein synthesis, defense, and tissue-specific development. Therefore, it may be useful not only to explore the direct NVN of the gene of interest for functional context but also to evaluate neighboring clusters for a higher contextual order.
Thus, the interactive gene-related networks in PlaNet may be browsed on three different levels for each of the individual species: as meta-networks displaying interconnectivity between gene clusters, as individual gene clusters, and as single gene pages with surrounding gene NVNs (Figure 2).
Comparative Coexpression Relationships across Seven Plant Species
The coexpressed network arrangements of the individual species may reveal genes and processes that are associated with the function of a gene of interest. However, due to incomplete coverage of the arrays, and also of the analyzed data sets, the individual networks most likely lack some candidate genes as well as containing false positive candidates. Here, we argue that by comparing network structures across species we may enrich for genes related to the particular biological function of interest. In addition, considering the comparably high knowledge about gene functions in Arabidopsis, we anticipated that by comparing NVNs to other species we would be able to readily infer functional homologs in other species that are likely to be of more economic importance.
To compare different network vicinities, we included a NetworkComparer pipeline, which can score and display conserved coexpression network structures across species by combining gene sequences with coexpression network structure (see Supplemental Figure 2 online). To obtain a sequence-based classification of genes into families, we proceeded as follows.
For every probe set present on the seven microarray platforms, we obtained the identity of the representative target sequence from MapManStore (http://mapman.gabipd.org/web/guest/mapmanstore) and downloaded corresponding coding sequences from sources shown in Table 1. Every transcript sequence found was then associated to the family sequences represented in the manually curated database Pfam (Finn et al., 2008; v24.0) using RPS-BLAST (Marchler-Bauer et al., 2007). Sequences with no hit below the E-value threshold of 10−5 were excluded from further analysis. Since every Pfam sequence is associated with a specific PFAM family, the whole procedure effectively bins genes and their expression measurements into gene families.
The NetworkComparer pipeline can compare either user-defined genes to each other or a specific gene to all members of the associated protein family (see Supplemental Figure 2 online). Here, we illustrate the principle and application of the pipeline by two examples, involving photosynthesis and polyketide synthase family-related pathways. Notably, this approach can be adopted for any gene/pathway/process of interest.
Photosynthesis: At-PSA-D1 and At-PSA-D2
The two Arabidopsis gene products At-PSA-D1 (At4g02770) and At-PSA-D2 (At1g03130) belong to the photosystem I reaction center (PSA-D) family (see Supplemental Figure 3 online; see Table 2 for acronym explanations) necessary for assembly of the photosystem I complex (Ihnatowicz et al., 2004). BLAST and phylogenetic analysis of the PSA-D family revealed two, two, two, and one members from Arabidopsis, Medicago, poplar, and rice, respectively (Figure 3A). As only a single PSA-D–related copy is present in rice, it appears easy to infer that this gene should represent the functional homolog to the Arabidopsis PSA-D1. Consistent with this idea, the NVN for rice PSA-D contains many genes for which homologs are also found in the At-PSA-D1 NVN (see Supplemental Figure 3 online). However, the sequence divergence between the At-PSA-D proteins and the two Medicago PSA-D proteins is minute (Medtr5g006220, BLAST score: 295; Medtr1g132590, BLAST score: 246). Similarly, the two poplar PSA-Ds (564827, BLAST score: 308; 566261, BLAST score: 301) are also at approximately equal sequence distance to the At-PSA-D proteins (Figure 3A). Therefore, it is difficult to predict which of the PSA-D proteins in poplar and Medicago have the most closely related biological function to the At-PSA-Ds.
Table 2.
Mutant and Gene Names
| Acronym | Full Name/Explanation |
| A3G2′′XT | Anthocyanin 3-O-glucoside 2′′-O-xylosyltransferase |
| A3GCoT | Anthocyanin 3-O-glucoside 6′′-O-p-coumaroyltransferase |
| A5GMaT | Anthocyanin 5-O-glucoside 6′′′-O-malonyltransferase |
| A5GT | Anthocyanin 5-O-glucosyltransferase |
| AAT | Anthocyanin-acyltransferase |
| ACOS | Acyl-CoA synthetase |
| ANS | Anthocyanidin synthese |
| AtSHT | Spermidine hydroxycinnamoyl transferase |
| AtTSM1 | Tri-hydroxy feruloyl-spermidin methyltransferase |
| CCoAOMT1 | Caffeoyl-CoA 3-O-methyltransferase |
| CHI | Chalcone isomerase |
| CHR | Chalcone reductase |
| CHS | Chalcone synthese |
| 4CL | 4-Coumarate CoA ligase |
| CRY | Cryptochrome |
| CYP | Cytochrome P450 |
| DFR | Dihydroflavonol reductase |
| DMID | 7,2′-Dihydroxy-4′-methoxy-isoflavanol dehydratase |
| ELIP | Early light-inducible protein |
| F3H | Flavanone 3-hydroxylase |
| F3′H | Flavonoid 3′-hydroxylase |
| F3RT | Flavonol 3-O-rhamnosyltransferase |
| F5′H | Flavonoid 5′-hydroxylase |
| F7RT | Flavonol 7-O-rhamnosyltransferase |
| Fd3GT | Flavonoid 3-O-glucosyltransferase |
| FLS | Flavonol synthese |
| FOMT | Flavonoid-O-methyltransferase |
| FS | Flavone synthase |
| GST | Glutathione S-transferase |
| HIDH | 2-Hydroxyisoflavanone dehydratase |
| HY | Elongated hypocotyl |
| IFOH | Isoflavone hydroxylase |
| IFR | Isoflavone reductase |
| IFS | Isoflavone synthase |
| LAP | Less adhesive pollen |
| MS2 | Male sterility 2 |
| O-UGT | O-Glycosyltransferase |
| O-UGT | C-Glycosyltransferase |
| pCHI | Putative chalcone isomerase |
| PSAD | Photosystem I Subunit D |
| PSAK | Photosystem I Subunit K |
| TTG2 | Transparent testa glabra 2 |
| VR | Vestitone reductase |
| TT4/tt4 | Chalcone synthase, At5g13930/transparent testa 4 |
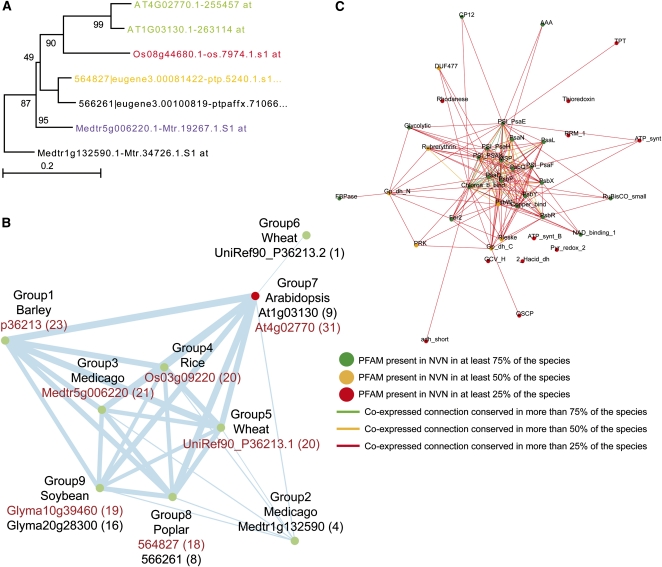
Figure 3.
Comparative Analysis of PSA-D–Related NVNs across Seven Plant Species.
(A) Phylogenetic tree of PSA-D–related proteins in Arabidopsis, poplar, rice, and Medicago. Color-coded proteins/probe sets corresponds to the following: green, Arabidopsis PSA-D1 and D2; red, rice PSA-D with a similar NVN to AtPSA-D1; yellow, poplar PSA-D with a similar NVN to AtPSA-D1; purple, Medicago PSA-D with a similar NVN to AtPSA-D1; and black, proteins for which the gene NVNs have low similarity to the AtPSA-D1 NVN. Protein sequences were aligned in MEGA4 (Tamura et al., 2007) using ClustalW, and phylogenetic trees were constructed using the bootstrapped neighbor-joining method (1000 runs). Values on the branches indicate bootstrap support in percentages.
(B) NVN similarity network of PSA-D–related genes across the seven species. A node represents an NVN of a PSA-D gene found in the seven species. If any NVNs are overlapping (i.e., are strongly coexpressed and share same genes in their NVNs), they are collapsed into one group, as a comparison of overlapping networks would result in artificially high similarity values. For PSA-D NVN analysis, group 7 (Arabidopsis At1g03130 and At4g02770), group 8 (poplar 564827 and 566261), and group 9 (soybean Glyma10g39460 and Glyma20g28300) represent coexpressed NVNs and are therefore grouped. Genes marked with red scored highest to AtPSA-D1 (i.e., their NVNs were most similar to the NVN of AtPSA-D1) and were selected for generation of the ancestral network. Thickness of edges between the nodes is proportional to the similarity score between the NVNs. The numbers in parentheses indicate the amount of PFAMs each PSAD-1–related gene has in common with the NVN of AtPSAD-1.
(C) Ancestral network depicts similarities between NVNs selected in the previous step. Color-coded nodes and edges represent presence of certain gene families and connections across species. Green, orange, and red nodes indicate that a given PFAM is found in the NVN of >75, 50, and 25% of the selected NVNs, respectively. Similarly, green, orange, and red edges indicate that an edge is found between the any two PFAMs in >75, 50, and 25% of the selected NVNs, respectively.
In an attempt to predict which of the PSA-D genes most closely resemble At-PSA-D genes in terms of function, we used At-PSA-D1 (At4g02770) as query for the NetworkComparer (see Supplemental Figure 2.1 online). The pipeline found 12 PSA-D associated probe sets in the seven plant species, which were passed on to the comparative analysis. The algorithm implemented in the NetworkComparer first generated two-step NVNs for each of the 12 probe sets (see Supplemental Figure 2.2 online) and then compared them in a pairwise fashion, where the score value between any two NVNs equals the number of PFAM families they have in common (see Supplemental Figure 2.3 online). Thus, NVNs with highly similar PFAM content should show high mutual scores. Results of this comparison are shown as a network and a table. The network graphically reveals the similarity scores of NVNs of all probe sets in the protein family of the query gene. The table shows the similarity scores and associated P value of the query gene to all genes from the analyzed PFAM (see Supplemental Figure 2.4 online).
As some members from the same gene family may be coexpressed (i.e., present in one another’s NVNs), comparison of such genes will return an artificially large comparison score. To avoid such artificial enrichments, the pipeline bins the overlapping NVNs into coexpression groups. For the PSA-D family, nine such coexpression groups were found across the seven plant species (Figure 3B). Of the two corresponding PSAD-1 genes from Medicago, Medtr5g006220.1 (represented by probe set Mtr19267.1.S1_at) showed the highest score to coexpression network of At-PSA-D1. In addition, of the two PSA-D genes from poplar, gene model 564827|eugene3.00081422 (represented by probe set ptp.5240.1.s1_s_at) showed the highest score to At-PSA-D1 (Figure 3B). Taken together, these results suggest that the second step of the NetworkComparer pipeline can be used to identify potential functional homologs across the different species.
To analyze the commonalities between the PSA-D–associated networks, we chose the highest scoring PSA-D genes (i.e., those with the most similar NVNs to At-PSA-D1) from each species (see Supplemental Figure 2.4 online) and sent these to the final step of the analysis (see Supplemental Figure 2.5 online). In this step, the pipeline extracts and displays the common features of the selected gene NVNs in form of a combined network, termed ancestral network and a table (Figure 3C; see Supplemental Data Set 2 online). The ancestral network (as the network represents conserved transcriptional program across analyzed species) depicts the frequency for which a given protein family is found in the selected NVNs, where green, orange, and red nodes and connections correspond to PFAMs and associations between PFAMs found in ≥75, ≥50, and ≥25% of the networks, respectively.
The coexpression networks of the selected PSA-D genes showed strong enrichment in PSA and PSB gene families, which encode components of photosystem I and II complexes (Figure 3C; Nelson and Yocum, 2006). In addition, several other genes encoding proteins not directly associated with the photosystem complexes, but with ATP generation, such as ATP synthase, photosynthetic glyceraldehyde 3-phosphate dehydrogenase, and triose phosphate transporter family were also present in the network (see Supplemental Data Set 2 online). Interestingly, a Domain of Unknown Function family, DUF477, was present in three of the coexpression networks analyzed, suggesting association of this family with the biological function of the PSA-D gene products.
While the ancestral network depicts enrichment and associations between PFAMs in the analyzed coexpression networks for the individual species, the associated table provides detailed information regarding the identity of probe sets associated with the families (see Supplemental Data Set 2 online). For example, Arabidopsis GAPDH (At1g42970) encodes a putative glyceraldehyde 3-phosphate dehydrogenase and is present in the Arabidopsis PSA-D NVN. Not surprisingly, close homologs to At-GAPDH are also present in the barley, rice, and wheat PSA-D NVNs (see Supplemental Data Set 2 online). Since the expression of these GAPDH-related genes is associated with the PSA-D NVNs in all of these species, we propose that the function of the gene product in each case is relevant for photosystem I function. We might also anticipate that a gene homolog would be coexpressed with, for example, the poplar PSA-D, but this was not found to be the case (see Supplemental Data Set 2 online). One explanation for this could be that no corresponding probe set for the gene is present on the poplar microarray. To assess this, we identified the closest GAPDH-related sequence in poplar via BLASTN using At1g42970 as query. The best hit was obtained for the poplar gene jgi|Poptr1_1|422935|gw1.II.3569.1 (e-value = 10e-165), which was not represented on the poplar microarray, suggesting that, indeed, the most likely functional homolog of At-GAPDH is not represented on the microarray. We therefore propose that genes that are not represented on the Genechips can be inferred to be associated with certain NVNs in a species based on occurrence in the related NVNs in other species.
Apart from predicting functionally related genes and putative functional homologs across species, the table can also reveal functional redundancies. For example, using PSA-D1 from Arabidopsis as query for the analysis, the pipeline placed At-PSA-D1 and At-PSA-D2 into one group, indicating that the two genes are coexpressed (Figure 3B). Importantly, while mutations in At-PSA-D1 affect the photosynthetic electron flow, disruption of At-PSA-D2 results in no observable phenotype (Ihnatowicz et al., 2004). This could perhaps be due to functional redundancy between the two gene products. Intuitively, redundant genes should have similar function but also be expressed with same spatio-temporal pattern. Indeed, atpsa-d1 atpsa-d2 double mutants result in an additive phenotype (i.e., seedling lethality) (Ihnatowicz et al., 2004), supporting this hypothesis.
Flavonol and Flavonoid Synthesis: CHSs
The PSA-D gene family is relatively small; therefore, we also chose to approach a considerably larger gene family: the polyketide synthase family (PKSs; Austin and Noel, 2003; Abe and Morita, 2010). From within this family, we chose the CHS subfamily, which is one of the larger subfamilies of the PKSs. While Arabidopsis only contains four CHS-related genes, rice and Medicago have at least 20 CHS homologs each (see Supplemental Figure 4 online). The CHS gene products are associated with flavonoid-related biosynthetic pathways in which they catalyze the conversion of coumaroyl-CoA into naringenin chalcone (Austin and Noel, 2003; Abe and Morita, 2010; see Supplemental Figure 5 online). One of the more prominent CHS members in Arabidopsis (At5g13930; TT4) has been experimentally associated with the main flavonol/flavonoid biosynthetic route (Feinbaum and Ausubel, 1988) and is coexpressed with many of the genes for which the gene products work either up- or downstream of the CHS (Tohge et al., 2007; Yonekura-Sakakibara et al., 2008; Tohge and Fernie, 2010). These relationships may readily be seen in Supplemental Figure 5 online, in which the general flavonoid biosynthetic genes, such as CHS (TT4, At5g13930; see Table 2 for acronyms), CHI (TT5, At3g55120), F3H (TT6, At3g51240), F3′H (TT7, At5g07990), Fd3GT (UGT78D2, At5g17050), and 4CL3 (At4CL3, At1g65060), are found in a central coexpressed cluster. In addition, this central network is connected to genes involved in phenylpropanoid pathway, such as 4CL (At1g65060), CAD (At3g19450), CCoAOMT (At4g34050), PAL (At3g53260, At2g37040), SGT (At3g21560), OMT (At5g54160), and CCR (At1g15950), and also genes associated with flavonol production, such as FLS (At5g08640), F7RT (At1g06000), and MYB12 (At2g47460). Thus, the CHS NVN reveals links between various natural products and between transcriptional activators and biosynthetic genes.
To assess similarities across the different species for CHS-related processes, we used TT4 as query gene for the NetworkComparer platform. The output from the tool resulted in 57 coexpression groups, with varying degrees of NVN similarities to the NVN of the query gene CHS (Figure 4A). The most similar NVN from each species (depicted by color-coded boxes) was selected and compared with the query gene NVN (light-green box, Figure 4A), which yielded a ancestral network for the CHSs across the seven species (Figure 4B). Many of the NVNs contain genes associated with the general flavonoid biosynthesis, including CHSs, CHIs, F3Hs, FLSs, and DFRs (Figure 4C; see Supplemental Data Set 3 online). Also, several genes that encode proteins that transport and modify flavonoids, such as O-methyltransferases, glycosyltransferases (UDPGT), ABC transporters, glutathione S-transferases (GSTs), and sugar transporters, are present in the NVNs in multiple species (Figures 4B and 4C). Flavonoids are generally accumulated as glycosylated forms in the vacuole. The conservation of genes encoding both glycosyltransferses and transporters in the NVNs across species suggests that both glycosylation events and the vacuolar transporting systems occur in all the species we studied here.
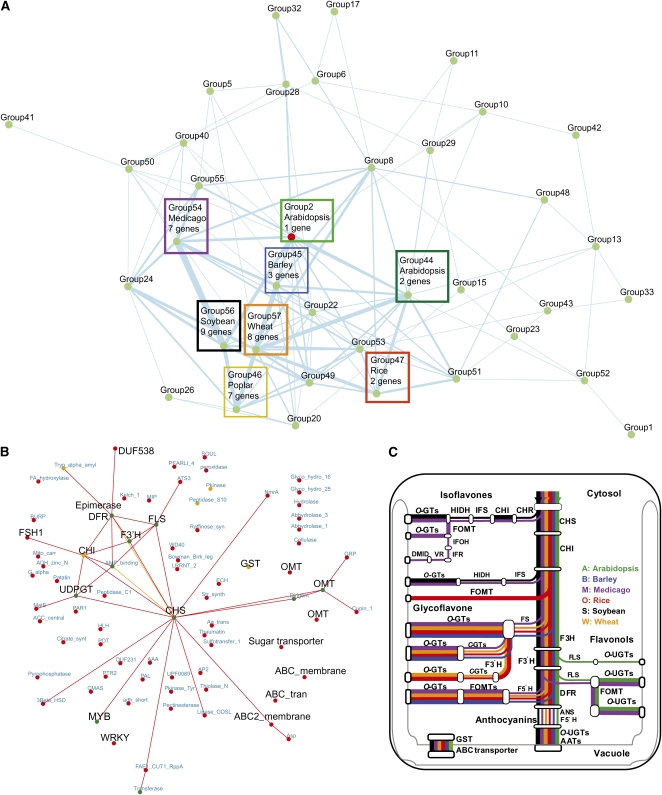
Figure 4.
Combined NVNs for CHS-Related Genes across Seven Species.
(A) NVN similarity network of CHS-related genes across the seven species. A node represents an NVN of a CHS gene found in the seven species. Red node indicates the group containing AtCHS, and boxed nodes represent coexpression groups most similar to CHS NVN from Arabidopsis (group 2). Green, blue, purple, yellow, red, black, and orange boxes represent coexpression groups from Arabidopsis, barley, Medicago, poplar, soybean, and wheat. Number of CHS-related NVNs constituting each coexpression group is indicated in each selected box.
(B) Combined NVNs (ancestral network) for CHS-related genes across the seven species using CHS (At5g13930) as bait (see Supplemental Data Set 3 online). Different colored nodes correspond to number of species in which a homolog is found in the gene NVN (see explanation in Figure 2). Similarly, the different colored edges correspond to number of species in which the two connected nodes are connected (see Figure 2). PFAMs that directly correspond to pathway members are highlighted in black.
(C) Schematic pathway structure of the anthocyanin/flavone biosynthesis in the different species. Different colors correspond to the different species as indicated. See Table 2 for acronym annotations.
Genes encoding sugar converting enzymes and certain transcription factors are also included in the combined network. NDP-sugar converting enzymes, such as UDP-rhamnose synthases, can provide substrates for the glycosylation of flavonoids (Yonekura-Sakakibara et al., 2008). Transcription factors, on the other hand, transcriptionally activate the biosynthetic pathway genes. It is important to note that several of these annotations have previously been directly associated with flavonoid-related biosynthesis in Arabidopsis. For example, overexpression of the MYB transcription factor PAP1 resulted in accumulation of cyanidin and quercetin derivates and led to the activation of genes associated with the anthocyanin production (Tohge et al., 2005). Figure 4C shows a schematic pathway outline of the conserved flavonoid biosynthesis pathway, including anthocyanin, flavonol, glycoflavone, and isoflavone synthesis, based on literature survey and KEGG pathways, for Arabidopsis (Tohge et al., 2005, 2007; Yonekura-Sakakibara et al., 2008), barley (Nørbaek et al., 2003; Brazier-Hicks et al., 2009; Klausen et al., 2010), Medicago (Kowalska et al., 2007; Farag et al., 2008), rice (Han et al., 2009; Kim et al., 2009), soybean (Steele et al., 1999; Choung et al., 2001; Latunde-Dada et al., 2001), and wheat (Ioset et al., 2007). Subclasses of flavonoids and anthocyanins have been detected and reported in all six plant species, but none of the flavonoid subfamilies, flavonol, glycoflavone, and isoflavone, has been reported. By comparing Figures 4B and 4C it is clear that many of the enriched protein family annotations in Figure 4B are prominent in the flavonoid pathway structure and are conserved across the species.
To obtain further information about the specific genes in the different NVNs for the CHS-related genes, we looked at the respective gene pages (Figure 5; see Supplemental Figures 6 and 7 online). One prominent example is the CHS-related gene (At-PKS-B, LAP5, At4g34850; Mizuuchi et al., 2008; Dobritsa et al., 2010) in Arabidopsis. The NVN for this gene contains some genes that could be associated with flavonoid-related processes, such as a dihydroflavonol reductase family (DRL1, At4g35420; Tang et al., 2009), a 4-coumarate CoA ligase family (At1g62940), and a glycosyltransferase (At1g33430; see Supplemental Figure 6 online). However, the NVN does not contain the characterized flavonoid biosynthesis genes nor the flavonol arabinosyltransferase F3AbT (UGT78D3, At5g17030), which convert the flower specific flavonol. In addition, flavonoid profiling of tt4 mutant flowers showed that no flavonoids were detected (Yonekura-Sakakibara et al., 2008). These results indicate that the majority of flavonoids is produced via TT4 in Arabidopsis flowers. This in turn suggests that At-PKS-B is part of a different biosynthetic pathway. Consistent with this observation, Mizuuchi et al. (2008) concluded that At-PKS-A (At1g02050) and At-PKS-B could accept fatty acyl-CoAs as a starting substrate. In agreement with this, mutant analyses and enzymatic assays by Dobritsa et al. (2010) showed that At-PKS-B plays a role in both synthesis of fatty acids and phenolics for pollen exine development in Arabidopsis. Furthermore, recent studies have shown that several of the genes in the NVN of At-PKS-B encode proteins that participate in the synthesis of polyamines, such as N1,N5-di(hydroxyferuloyl)-N10-sinapoylspermidine, being a part of the sporopollenin surrounding the pollen grains (Ehlting et al., 2008; Matsuno et al., 2009; Dobritsa et al., 2010). Several MYB and bHLH transcription factor encoding genes are also present in the At-PKS-B NVN and may be good candidates for transcriptional regulators of the pathway (see Supplemental Figure 6 online). In addition, At-PKS-A and At-PKS-B are present in the same NVN, suggesting possible functional redundancy (see Supplemental Figure 6 online). Interestingly, Os07g22850 is a close rice homolog to AtPKS-B (see Supplemental Figure 4 online), making it a good candidate for related functions in rice. Indeed, the Os07g22850 gene appears to be exclusively expressed in floral tissues, and the NVN contains genes that are associated with polyamine-related processes (http://aranet.mpimp-golm.mpg.de/ricenet/r31036). These gene annotations include UDP-glucosyl transferase (Os03g11350), dihydroflavonol reductase (Os08g40440), 4-coumarate CoA ligase (ACOS12, Os04g24530; de Azevedo Souza et al., 2009), cytochrome P450 (Os08g03676), and stilbene synthase (Os10g34360 and Os07g22850) among other genes (http://aranet.mpimp-golm.mpg.de/ricenet/r31036). We propose that these and other genes in the Os07g22850 NVN represent a flower-specific gene module in rice involved in sporopollenin formation.
Figure 5.
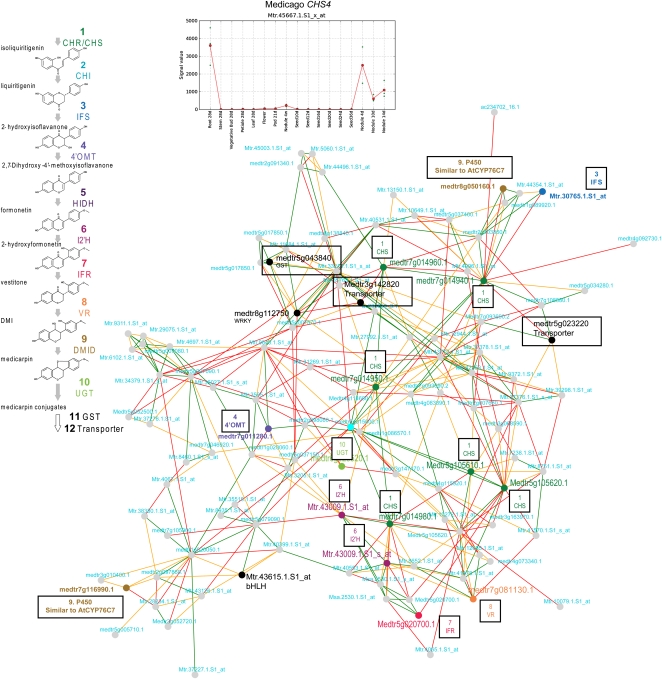
NVN for Putative CHS-Mediated (CHS4) Medicarpin Biosynthesis.
Nodes in the network resemble individual genes, and the connecting edges represent coexpressed links. The coloration of nodes and edges is explained in Figure 2. The different steps in the medicarpin biosynthetic pathway are indicated in different colors according to the pathway structure on the left. The P450-related genes indicated in gold could encode the missing DMID step in the pathway. The interactive networks can be found at http://aranet.mpimp-golm.mpg.de/medinet/m15662. The expression profile for CHS4 is indicated above the network. See Table 2 for acronym annotations.
Since many of the flavonoid-related processes have been relatively well characterized in Arabidopsis, we inspected NVNs from Medicago with high similarity to the Arabidopsis TT4 NVN. The highest scoring Medicago NVN is associated with the probe ID Mtr.40122.1.s1_s_at (see Supplemental Figure 7 online). Not surprisingly, this NVN also contains many genes with annotations related to isoflavone/flavonoid synthesis, including cytochrome P450s, IFRs, and O-methyltransferases (see Supplemental Figure 7 online). Most of the genes in this NVN are relatively low but ubiquitously expressed (http://aranet.mpimp-golm.mpg.de/medinet/m19811). This NVN also holds MYB and WRKY transcription factors, which, given the importance for MYB-related transcription factors in the activation of flavonoid-related genes in Arabidopsis (Borevitz et al., 2000; Stracke et al., 2001; Mehrtens et al., 2005; Tohge et al., 2005), may represent good candidates for transcriptional activators of the pathway in Medicago.
Interestingly, another high-scoring NVN surrounds the probe ID (Mtr.45667.1.s1_x_at), which is exclusively expressed in roots and root nodules (Figure 5). The flavonoid-derived metabolite medicarpin is a phytoalexin that is used by plant roots as protection from fungus and insects (Dakora et al., 1993; Dixon and Sumner, 2003; Naoumkina et al., 2007). Closer inspection of the NVN revealed that many genes that encode proteins tentatively involved in the synthesis of medicarpin-conjugates were found coexpressed with the TT4-related Medicago gene. These gene products represent almost all the pathway steps from isoliquiritgenin to medicarpin and its downstream conjugates (Figure 5) and include IFSs, 4O’MTs, HIDHs, I2’Hs, IFRs, VRs, DMIDs, UGTs, GSTs, transporters, WRKY, and bHLH (Figure 5). While several of the proteins responsible for the catalysis of the constituent pathway steps have been identified or anticipated (Naoumkina et al., 2007), we propose that many of the genes associated with the NVN may qualify as good candidates for biosynthetic and regulatory gene products for this pathway.
MapMan Ontology Networks
Although coexpression analysis can suggest gene function and help unravel novel components of biological machineries, another important quest in biology is to understand how different biological functions are orchestrated to fulfill cellular processes. We observed that genes involved in distinct yet related biological functions are often associated in coexpression networks. For example, ontology analysis of PSA-D genes used in one of the above case studies revealed that genes associated with photosystem I/II complexes, ATP synthesis, and the Calvin cycle are coexpressed in Arabidopsis (http://aranet.mpimp-golm.mpg.de/aranet/a11983). To similarly evaluate co-occurrence of ontology terms, we examined the HCCA clusters generated for the different organisms for MapMan ontology terms annotated to genes in the NVN. Ontology terms showing a significant enrichment or depletion (Fisher test P < 0.05) were then extracted. Subsequently, the co-occurrence of pairs of terms was determined for all clusters and was tested for overrepresentation using a Fisher’s exact test. Pairs of terms that were overrepresented were then connected, and the resulting networks were visualized as interactive networks for the seven species. Many ontology terms that were anticipated to be functionally connected also occurred in close vicinity in the networks. For example, mitochondrial ATP synthesis/electron transport and tricarboxylic acid cycle–related ontology terms and terms such as photosynthesis, Calvin cycle, and tetrapyrrole synthesis are connected in the network for Medicago (see Supplemental Figure 8 online).
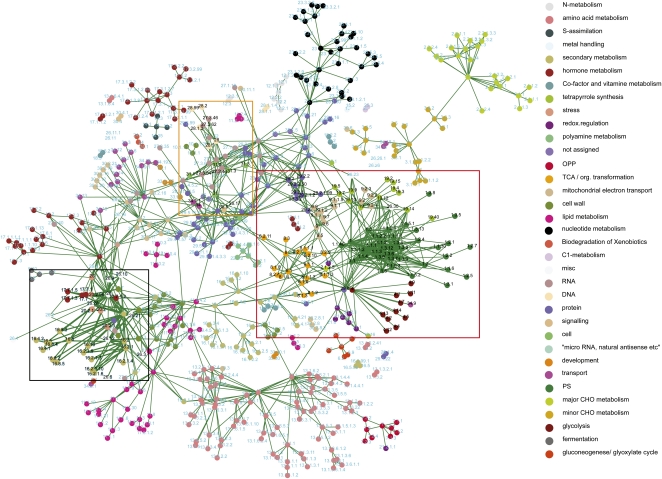
Similar to the comparative network approach for individual gene networks described above, we argued that ontology terms that are connected in two or multiple species may more reliably reflect noteworthy links between the terms. To produce such a network, we identified terms that were associated in at least two monocots and two dicots. Therefore, the resulting network represents conserved ontology term associations across at least four of the plant species (Figure 6). Visual inspection of the network reveals that related processes are readily connected and often form clusters. For example, photosynthesis-related terms are associated with terms such as Calvin cycle and tetrapyrrole biosynthesis, but these are also associated with glycolysis, tricarboxylic acid cycle, and various mitochondrial processes (Figure 6, red box). The latter associations could be viewed as reflecting the crosstalk between the chloroplast and mitochondria, for example, in the form of different redox-related metabolites, such as malate and oxaloacetate, and in the exchange of ATP (Raghavendra and Padmasree, 2003; Sweetlove et al., 2006; Nunes-Nesi et al., 2007). In addition, terms associated with cell division (e.g., various cell cycle–related terms, histone and DNA biosynthesis, and chromatin structure) are closely linked to various vesicle trafficking terms (e.g., p- and v-ATPases, G-protein signaling, and dynamins) (Figure 6, orange box). Finally, flavonoid synthesis-related terms are directly connected to several stress related terms, which in turn connects to cytochrome P450s, GSTs, peroxidizes, and jasmonate biosynthesis (Figure 6, black box). The latter relationship is substantiated by the induction of anthocyanin production and of flavonoid-related genes by methyl jasmonates (Franceschi and Grimes, 1991). Hence, the ontology associations captured in the networks recapitulate known biological connections and may therefore also be used as a guide to discover and establish new relationships between different biological pathways and functions.
Figure 6.
Combined MapMan Ontology Network for the Seven Species.
Coexpressed gene MapMan ontology terms in at least two monocots and two dicots. Exemplified associations between the ontology terms are represented by black fonts in the network and indicated by orange, red, and black boxes for cell cycle, photosynthesis, and flavonoid-related processes, respectively. Key to the MapMan ontology is displayed on the right.
Summary and Future Prospects
The PlaNet platform integrates transcriptomic, genomic, phenomic, and ontology terms for seven plant species with the aim to rapidly transfer knowledge across the species. While other comparative cross-species tools do exist, they only allow for pairwise comparisons. By contrast, the pipeline implemented in PlaNet permits simultaneous comparison of the seven species, thus enabling extraction of conserved coexpression relationships. Current functional homology predictions rely heavily on sequence comparisons (i.e., phylogenetic relationships). While such inference may yield useful information, we argue that the combination of sequence information with transcriptional relationship is likely to improve such inferences. This has been substantiated in examples using genes involved in the assembly of the photosystems and in PKS gene family–related processes. These analyses revealed many gene products that we predict would be closely linked to the respective pathways but currently are uncharacterized. Zooming out from the individual gene level, we took advantage of coexpression between ontology terms and provided networks displaying how different terms are transcriptionally linked. The combination of the tools presented here will allow researchers to predict genes involved in highly diverse pathways and processes across diverse species and to contextualize biological processes by ontology term associations. This tool may be extended readily to include other species, provided that sufficient expression data sets exist. Inclusion of additional plant species may also allow for more detailed analyses, for example, more detailed assessments of transcriptional differences between monocots and dicots. Even in its current format, PlaNet should represent a highly useful resource for the many groups currently attempting to transfer knowledge gleaned from Arabidopsis to species with greater agricultural, ecological, and pharmaceutical value.
METHODS
Column Selection
The Column Subset Selection is a fundamental problem in numerical linear algebra, whereby, for a given number k and a matrix A, one is to find the subset of k columns from A that are most mutually independent; for example, the submatrix C given by the k columns (selected from A) is as nonsingular as possible. We employed a two-step hybrid algorithm (see Supplemental Figure 9 online) that combines random with deterministic column selection to determine a given number, k, of columns satisfying the requirements in the problem definition. Randomized sampling of the matrix for column selection was based on considerations of Anderson et al. (1999), Boutsidis et al. (2009), and Golub (1965), as described in further detail in Supplemental Figures 9 and 10 online.
Statistical Significance of Reciprocal Ranks
To determine the statistical significance of establishing an edge in the network, we considered two aspects: (1) the significance of the Pearson correlation coefficients used in determining the rank, and (2) the significance of a given reciprocal rank. To address these issues, we provide values for the employed thresholds (for correlation coefficients and reciprocal ranks) that ensure a given level of statistical significance. First, we investigate the distribution of values for the Pearson correlation coefficient corresponding to values for the reciprocal rank in the range from [1, r], where r ∈ {100, 500, 1000, 2000, 3000, 4000} in the data set (see Supplemental Figure 11 online). The order statistics on the Pearson correlation coefficients used in establishing the reciprocal ranks obscure the ability to determine a reliable cutoff for the Pearson correlation. For instance, a rank cutoff of 1000 already includes the values of the Pearson correlation in the ~75% from the observed correlation distribution. Therefore, the statistical significance is inferred directly for the reciprocal ranks without propagating it from the significance of the underlying correlations.
As a result, establishing the statistical significance of reciprocal ranks directly is sufficient for robust network reconstruction. Therefore, we investigated the distribution of reciprocal ranks over 100 permutations of the data set. This distribution approaches the uniform across all ranks with an average occurrence per rank of m/2, where m = 22,810 number of genes for Arabidopsis thaliana, resulting in a statistically significant cutoff value for the reciprocal ranks of 228 at level 0.01. This value is derived by inspecting the area under the curve (shaded in Supplemental Figure 12 online).
Optimality Principle
Here, we provide a simple measure of the biological relevance of a constructed network that can be used to investigate the effect of varying the two parameters: reciprocal rank cutoff r and neighborhood n employed for knowledge transfer. Given functional annotations (i.e., MapMan terms) for the genes in the network and a seed node u, we determine the proportion of connected node pairs (edges) in the neighborhood of u which share its function (i.e., are annotated with the same term) compared with the total number of all edges in the given neighborhood of u. Note that edges incident to at least one unannotated gene are ignored in the computation. Summation over all scores for each gene results in a final assessment of the biological relevance for the investigated network. Finally, analysis for all considered rank cutoffs r and neighborhoods n leads to the selection of an optimal parameter set. This optimality principle indicates that a reciprocal rank cutoff in the range of r = [10,30] is on average optimal for the seven species (see Supplemental Figure 13 online). The analysis revealed that one-step neighborhood is optimal in optimizing the NVN for genes belonging to same MapMan terms. However, note that while a one-step NVN captures more biological information, the drawback of obscuring the modular structure of highly coexpressed gene clusters (e.g., a cluster structure is clearly visible when two-step NVN is used, http://aranet.mpimp-golm.mpg.de/aranet/a21001, which would not be the case if one-step NVN was used) coupled with possible omission of genes belonging to distinct yet relevant biological process leads to the selection of a two-step neighborhood for the performed analysis. The robustness of the HRR cutoff values under both values indicates the strength of applying measures of biological relevance for determining the optimal parameter values.
While this analysis performed indicated that in most cases HRR cutoff of 10 produced most biologically relevant networks, it is important to note that too stringent a cutoff produces a network where a majority of the nodes are disconnected and thus cannot be used for further analysis. The number of disconnected nodes as a function of the HRR cutoff can be seen in Supplemental Figure 14 online. While an HRR cutoff of 10 on average resulted in 80% of the nodes being disconnected, an HRR cutoff of 30 is placed close to a plateau, where on average only 25% of the nodes are disconnected. Taken together, based on the results obtained as described above, an HRR network with cutoff of 30, with two-step NVN, was used in this study.
Determination of Significantly Connected Clusters in the Meta-Network
Many clusters contain nodes that form intercluster edges, connecting the clusters. We assume that cluster C1 has n outgoing edges and cluster C2 has m outgoing edges, with a population of o edges, each with weight wi, between them. To determine whether clusters C1 and C2 are significantly connected, we again derive an empirical P value by a permutation test: first, we calculated the sum of weights of edges in the entire population. To derive a P value of the score between C1 and C2, we sampled m edges randomly from the network with uniform probability, assigned them to cluster C2, and calculated the sum of weights of edges shared between the two clusters. We repeat this sampling procedure 10,000 times. Two clusters were judged as being not significantly connected at a significance level of 1%, if the sum of edge weights resulting from random sampling exceeded the observed value more than 50 times in 10,000 iterations (two-tailed test).
Phylogenetic Analysis
Protein sequences were aligned in MEGA4 (Tamura et al., 2007) using ClustalW with gap opening penalty of 10 and gap extension penalty of 0.1 for pairwise alignment. For multiple alignment, gap opening penalty of 10 and gap extension penalty of 0.2 was used. BLOSUM protein weight matrix was selected with gap separation distance of 4 and residue specific and hydrophobic penalties enabled, while end gap separation was disabled. Delay divergent cutoff was set to 30%, and negative matrix was disabled. The alignment is available as Supplemental Data Set 4 online. Phylogenetic trees were constructed using the bootstrapped neighbor-joining method (1000 runs) with parameter (gaps/missing data: complete deletion). Substitution model Poisson correction was selected with parameters (substitution to include: all), (pattern among lineages: same), and (rates among sites: uniform rates). Values on the branches indicate bootstrap support in percentages.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Principles of Heuristic Cluster Chiseling Algorithm.
Supplemental Figure 2. Outline for the NetworkComparer Pipeline.
Supplemental Figure 3. Coexpressed Gene Vicinity Networks for PSA-D Genes in Arabidopsis and Rice.
Supplemental Figure 4. Phylogenetic Tree of CHS-Related Proteins in Arabidopsis, Poplar, Rice, and Medicago.
Supplemental Figure 5. Coexpressed Gene Vicinity Network for Chalcone Synthase (At5g13930) in Arabidopsis.
Supplemental Figure 6. Coexpressed Gene Vicinity Network for CHS-Related Gene (At4g34850) in Arabidopsis.
Supplemental Figure 7. Coexpressed Gene Vicinity Network for Chalcone Synthase 4 in Medicago.
Supplemental Figure 8. Coexpressed MapMan Ontology Network for Medicago.
Supplemental Figure 9. Pseudocode for Column Selection.
Supplemental Figure 10. Spectral Norm of Optimal k Column Submatrices for Arabidopsis in the Range of 20 to 1400.
Supplemental Figure 11. Distribution of Correlation Coefficients Observed for Specific Ranks on the Arabidopsis Data Collection.
Supplemental Figure 12. Distribution of Reciprocal Ranks on the Arabidopsis Data Collection.
Supplemental Figure 13. Influence of HRR Cutoff and NVN Step Size on the Biological Relevance for the Seven Analyzed Species.
Supplemental Figure 14. Influence of HRR Cutoff on the Percentage of Connected Nodes in the Networks of the Seven Analyzed Species.
Supplemental Data Set 1. MapMan Terms Associated with Cluster 49 in Arabidopsis.
Supplemental Data Set 2. Identity Information from the Network Comparer Analysis for PSA-D Genes.
Supplemental Data Set 3. Identity Information from the Network Comparer Analysis for CHS Genes.
Supplemental Data Set 4. Amino Acid Alignment Used to Generate the Phylogenetic Tree in Supplemental Figure 4.
Supplementary Material
Acknowledgments
We thank Nicholas Provart for aid in analyzing the poplar microarray data sets and Marc Lohse for providing MapMan bins. O.W. was generously supported by a Natural Science and Engineering Research Council of Canada (NSERC) Canadian Graduate Scholarship. Z.N. was supported by the GoFORSYS project funded by the Federal Ministry of Education and Research (Grant 0313924). The poplar microarray work was generously supported by funding from NSERC, the Canada Foundation for Innovation, and the University of Toronto to M.M.C. We thank the Max-Planck-Gesellschaft for financial support. Results have been achieved within the framework of the Transnational (Germany, France, and Spain) Cooperation within the PLANT-KBBE Initiative, with funding from Agence Nationale pour la Recherche, Bundesministerium für Bildung und Forschung, and Ministerio de Ciencia e Innovación.
References
- Abe I., Morita H. (2010). Structure and function of the chalcone synthase superfamily of plant type III polyketide synthases. Nat. Prod. Rep. 27: 809–838 [DOI] [PubMed] [Google Scholar]
- Anderson E., Bai Z., Sischof C. (1999). LAPACK Users’ Guide, 3rd ed (Philadelphia: Society for Industrial and Applied Mathematics; ). http://www.netlib.org/lapack/lug/ [Google Scholar]
- Austin M.B., Noel J.P. (2003). The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 20: 79–110 [DOI] [PubMed] [Google Scholar]
- Bergmann S., Ihmels J., Barkai N. (2004). Similarities and differences in genome-wide expression data of six organisms. PLoS Biol. 2: E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz J.O., Xia Y., Blount J., Dixon R.A., Lamb C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutsidis C., Mahoney M.W., Drineas P. (2009). An improved approximation algorithm for the column subset selection problem. SODA ’09 Proceedings of the Twentieth Annual ACM-SIAM Symposium on Discrete Algorithm, Mathieu C., (Philadelphia: Society for Industrial and Applied Mathematics; ), pp. 968–977 [Google Scholar]
- Brazier-Hicks M., Evans K.M., Gershater M.C., Puschmann H., Steel P.G., Edwards R. (2009). The C-glycosylation of flavonoids in cereals. J. Biol. Chem. 284: 17926–17934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choung M.G., Baek I.Y., Kang S.T., Han W.Y., Shin D.C., Moon H.P., Kang K.H. (2001). Isolation and determination of anthocyanins in seed coats of black soybean (Glycine max (L.) Merr.). J. Agric. Food Chem. 49: 5848–5851 [DOI] [PubMed] [Google Scholar]
- Clemente H.S., Pont-Lezica R., Jamet E. (2009). Bioinformatics as a tool for assessing the quality of sub-cellular proteomic strategies and inferring functions of proteins: Plant cell wall proteomics as a test case. Bioinform. Biol. Insights 3: 15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakora F.D., Joseph C.M., Phillips D.A. (1993). Alfalfa (Medicago sativa L.) root exudates contain isoflavonoids in the presence of Rhizobium meliloti. Plant Physiol. 101: 819–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Azevedo Souza C., Kim S.S., Koch S., Kienow L., Schneider K., McKim S.M., Haughn G.W., Kombrink E., Douglas C.J. (2009). A novel fatty Acyl-CoA Synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21: 507–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R.A., Sumner L.W. (2003). Legume natural products: Understanding and manipulating complex pathways for human and animal health. Plant Physiol. 131: 878–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A.A., Lei Z., Nishikawa S., Urbanczyk-Wochniak E., Huhman D.V., Preuss D., Sumner L.W. (2010). LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant Physiol. 153: 937–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J., Sauveplane V., Olry A., Ginglinger J.F., Provart N.J., Werck-Reichhart D. (2008). An extensive (co-)expression analysis tool for the cytochrome P450 superfamily in Arabidopsis thaliana. BMC Plant Biol. 8: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag M.A., Huhman D.V., Dixon R.A., Sumner L.W. (2008). Metabolomics reveals novel pathways and differential mechanistic and elicitor-specific responses in phenylpropanoid and isoflavonoid biosynthesis in Medicago truncatula cell cultures. Plant Physiol. 146: 387–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinbaum R.L., Ausubel F.M. (1988). Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Mol. Cell. Biol. 8: 1985–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficklin S.P., Luo F., Feltus F.A. (2010). The association of multiple interacting genes with specific phenotypes in rice using gene coexpression networks. Plant Physiol. 154: 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D., Tate J., Mistry J., Coggill P.C., Sammut S.J., Hotz H.R., Ceric G., Forslund K., Eddy S.R., Sonnhammer E.L., Bateman A. (2008). The Pfam protein families database. Nucleic Acids Res. 36(Database issue): D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi V.R., Grimes H.D. (1991). Induction of soybean vegetative storage proteins and anthocyanins by low-level atmospheric methyl jasmonate. Proc. Natl. Acad. Sci. USA 88: 6745–6749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub G. (1965). Numerical methods for solving linear least squares problems. Numerische Mathematik 7: 206–216 [Google Scholar]
- Han R.M., Tian Y.X., Liu Y., Chen C.H., Ai X.C., Zhang J.P., Skibsted L.H. (2009). Comparison of flavonoids and isoflavonoids as antioxidants. J. Agric. Food Chem. 57: 3780–3785 [DOI] [PubMed] [Google Scholar]
- Ihnatowicz A., Pesaresi P., Varotto C., Richly E., Schneider A., Jahns P., Salamini F., Leister D. (2004). Mutants for photosystem I subunit D of Arabidopsis thaliana: Effects on photosynthesis, photosystem I stability and expression of nuclear genes for chloroplast functions. Plant J. 37: 839–852 [DOI] [PubMed] [Google Scholar]
- Ioset J.R., Urbaniak B., Ndjoko-Ioset K., Wirth J., Martin F., Gruissem W., Hostettmann K., Sautter C. (2007). Flavonoid profiling among wild type and related GM wheat varieties. Plant Mol. Biol. 65: 645–654 [DOI] [PubMed] [Google Scholar]
- Jupiter D.C., Chen H., VanBuren V. (2009). STARNET 2: A web-based tool for accelerating discovery of gene regulatory networks using microarray co-expression data. BMC Bioinformatics 10: 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Kim S.K., Kim J.H., Kim B.G., Ahn J.H. (2009). Molecular characterization of flavonoid malonyltransferase from Oryza sativa. Plant Physiol. Biochem. 47: 991–997 [DOI] [PubMed] [Google Scholar]
- Klausen K., Mortensen A.G., Laursen B., Haselmann K.F., Jespersen B.M., Fomsgaard I.S. (2010). Phenolic compounds in different barley varieties: identification by tandem mass spectrometry (QStar) and NMR; quantification by liquid chromatography triple quadrupole-linear ion trap mass spectrometry (Q-Trap). Nat. Prod. Commun. 5: 407–414 [PubMed] [Google Scholar]
- Klie S., Nikoloski Z., Selbig J. (2010). Biological cluster evaluation for gene function prediction. J. Comput. Biol. 17: 1–18 [DOI] [PubMed] [Google Scholar]
- Kowalska I., Stochmal A., Kapusta I., Janda B., Pizza C., Piacente S., Oleszek W. (2007). Flavonoids from barrel medic (Medicago truncatula) aerial parts. J. Agric. Food Chem. 55: 2645–2652 [DOI] [PubMed] [Google Scholar]
- Latunde-Dada A.O., Cabello-Hurtado F., Czittrich N., Didierjean L., Schopfer C., Hertkorn N., Werck-Reichhart D., Ebel J. (2001). Flavonoid 6-hydroxylase from soybean (Glycine max L.), a novel plant P-450 monooxygenase. J. Biol. Chem. 276: 1688–1695 [DOI] [PubMed] [Google Scholar]
- Liolios K., Chen I.M., Mavromatis K., Tavernarakis N., Hugenholtz P., Markowitz V.M., Kyrpides N.C. (2010). The Genomes On Line Database (GOLD) in 2009: Status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 38(Database issue): D346–D354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfield I.W., Jen C.H., Pinney J.W., Michalopoulos I., Bradford J.R., Gilmartin P.M., Westhead D.R. (2006). Arabidopsis Co-expression Tool (ACT): Web server tools for microarray-based gene expression analysis. Nucleic Acids Res. 34(Web Server issue): W504–W509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L., Van Hemert J.L., Dash S., Dickerson J.A. (2009). Arabidopsis gene co-expression network and its functional modules. BMC Bioinformatics 10: 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., et al. (2007). CDD: A conserved domain database for interactive domain family analysis. Nucleic Acids Res. 35(Database issue): D237–D240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno M., et al. (2009). Evolution of a novel phenolic pathway for pollen development. Science 325: 1688–1692 [DOI] [PubMed] [Google Scholar]
- Mehrtens F., Kranz H., Bednarek P., Weisshaar B. (2005). The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 138: 1083–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi Y., Shimokawa Y., Wanibuchi K., Noguchi H., Abe I. (2008). Structure function analysis of novel type III polyketide synthases from Arabidopsis thaliana. Biol. Pharm. Bull. 31: 2205–2210 [DOI] [PubMed] [Google Scholar]
- Mutwil M., Obro J., Willats W.G.T., Persson S. (2008). GeneCAT—Novel webtools that combine BLAST and co-expression analyses. Nucleic Acids Res. 36(Web Server issue): W320–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M., Ruprecht C., Giorgi F.M., Bringmann M., Usadel B., Persson S. (2009). Transcriptional wiring of cell wall-related genes in Arabidopsis. Mol. Plant 2: 1015–1024 [DOI] [PubMed] [Google Scholar]
- Mutwil M., Usadel B., Schütte M., Loraine A., Ebenhöh O., Persson S. (2010). Assembly of an interactive correlation network for the Arabidopsis genome using a novel heuristic clustering algorithm. Plant Physiol. 152: 29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoumkina M., Farag M.A., Sumner L.W., Tang Y., Liu C.J., Dixon R.A. (2007). Different mechanisms for phytoalexin induction by pathogen and wound signals in Medicago truncatula. Proc. Natl. Acad. Sci. USA 104: 17909–17915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson N., Yocum C.F. (2006). Structure and function of photosystems I and II. Annu. Rev. Plant Biol. 57: 521–565 [DOI] [PubMed] [Google Scholar]
- Nørbaek R., Aaboer D.B., Bleeg I.S., Christensen B.T., Kondo T., Brandt K. (2003). Flavone C-glycoside, phenolic acid, and nitrogen contents in leaves of barley subject to organic fertilization treatments. J. Agric. Food Chem. 51: 809–813 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A., Sweetlove L.J., Fernie A.R. (2007). Operation and function of the tricarboxylic acid cycle in the illuminated leaf. Physiol. Plant. 129: 45–56 [Google Scholar]
- Obayashi T., Hayashi S., Saeki M., Ohta H., Kinoshita K. (2009). ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37(Database issue): D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T., Kinoshita K. (2009). Rank of correlation coefficient as a comparable measure for biological significance of gene coexpression. DNA Res. 16: 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata Y., Suzuki H., Sakurai N., Shibata D. (2010). CoP: A database for characterizing co-expressed gene modules with biological information in plants. Bioinformatics 26: 1267–1268 [DOI] [PubMed] [Google Scholar]
- Parkinson H., et al. (2009). ArrayExpress update—From an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 37(Database issue): D868–D872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S., Wei H., Milne J., Page G.P., Somerville C.R. (2005). Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl. Acad. Sci. USA 102: 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra A.S., Padmasree K. (2003). Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci. 8: 546–553 [DOI] [PubMed] [Google Scholar]
- Saito K., Hirai M.Y., Yonekura-Sakakibara K. (2008). Decoding genes with coexpression networks and metabolomics - ‘Majority report by precogs’. Trends Plant Sci. 13: 36–43 [DOI] [PubMed] [Google Scholar]
- Srinivasasainagendra V., Page G.P., Mehta T., Coulibaly I., Loraine A.E. (2008). CressExpress: A tool for large-scale mining of expression data from Arabidopsis. Plant Physiol. 147: 1004–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele C.L., Gijzen M., Qutob D., Dixon R.A. (1999). Molecular characterization of the enzyme catalyzing the aryl migration reaction of isoflavonoid biosynthesis in soybean. Arch. Biochem. Biophys. 367: 146–150 [DOI] [PubMed] [Google Scholar]
- Steinhauser D., Usadel B., Luedemann A., Thimm O., Kopka J. (2004). CSB.DB: A comprehensive systems-biology database. Bioinformatics 20: 3647–3651 [DOI] [PubMed] [Google Scholar]
- Stracke R., Werber M., Weisshaar B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Stuart J.M., Segal E., Koller D., Kim S.K. (2003). A gene-coexpression network for global discovery of conserved genetic modules. Science 302: 249–255 [DOI] [PubMed] [Google Scholar]
- Sweetlove L.J., Lytovchenko A., Morgan M., Nunes-Nesi A., Taylor N.L., Baxter C.J., Eickmeier I., Fernie A.R. (2006). Mitochondrial uncoupling protein is required for efficient photosynthesis. Proc. Natl. Acad. Sci. USA 103: 19587–19592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tang L.K., Chu H., Yip W.K., Yeung E.C., Lo C. (2009). An anther-specific dihydroflavonol 4-reductase-like gene (DRL1) is essential for male fertility in Arabidopsis. New Phytol. 181: 576–587 [DOI] [PubMed] [Google Scholar]
- Tohge T., Fernie A.R. (2010). Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nat. Protoc. 5: 1210–1227 [DOI] [PubMed] [Google Scholar]
- Tohge T., et al. (2005). Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42: 218–235 [DOI] [PubMed] [Google Scholar]
- Tohge T., Yonekura-Sakakibara K., Niida R., Watanabe-Takahashi A., Saito K. (2007). Phytochemical genomics in Arabidopsis thaliana: A case study for functional identification of flavonoid biosynthsis genes. Pure Appl. Chem. 79: 811–823 [Google Scholar]
- Usadel B., Obayashi T., Mutwil M., Giorgi F.M., Bassel G.W., Tanimoto M., Chow A., Steinhauser D., Persson S., Provart N.J. (2009). Co-expression tools for plant biology: Opportunities for hypothesis generation and caveats. Plant Cell Environ. 32: 1633–1651 [DOI] [PubMed] [Google Scholar]
- Wei H., Persson S., Mehta T., Srinivasasainagendra V., Chen L., Page G.P., Somerville C., Loraine A. (2006). Transcriptional coordination of the metabolic network in Arabidopsis. Plant Physiol. 142: 762–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K., Tohge T., Matsuda F., Nakabayashi R., Takayama H., Niida R., Watanabe-Takahashi A., Inoue E., Saito K. (2008). Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20: 2160–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.