Protein–protein interactions are important mechanisms for genes and gene networks to function. This study demonstrates that, although the PAIR database has limited coverage, representing ~24% of the entire interactome with ~40% precision, it is rich enough to capture many significant functional linkages within and between higher-order biological systems, such as pathways and biological processes.
Abstract
Predicted interactions are a valuable complement to experimentally reported interactions in molecular mechanism studies, particularly for higher organisms, for which reported experimental interactions represent only a small fraction of their total interactomes. With careful engineering consideration of the lessons from previous efforts, the Predicted Arabidopsis Interactome Resource (PAIR; ) presents 149,900 potential molecular interactions, which are expected to cover ~24% of the entire interactome with ~40% precision. This study demonstrates that, although PAIR still has limited coverage, it is rich enough to capture many significant functional linkages within and between higher-order biological systems, such as pathways and biological processes. These inferred interactions can nicely power several network topology-based systems biology analyses, such as gene set linkage analysis, protein function prediction, and identification of regulatory genes demonstrating insignificant expression changes. The drastically expanded molecular network in PAIR has considerably improved the capability of these analyses to integrate existing knowledge and suggest novel insights into the function and coordination of genes and gene networks.
INTRODUCTION
Protein–protein interactions are essential for almost all cellular processes. Deciphering the protein interaction network not only provides insights into protein functions but also advances our understanding of higher-level phenotypes and their regulation. In Saccharomyces cerevisiae (Ito et al., 2000; Uetz et al., 2000; Gavin et al., 2002; Ho et al., 2002; Yu et al., 2008), Homo sapiens (Rual et al., 2005; Stelzl et al., 2005), Drosophila melanogaster (Formstecher et al., 2005), and Caenorhabditis elegans (Li et al., 2004), genome-wide yeast two-hybrid screens and large-scale affinity purification/mass spectrometry studies have been reported. A number of databases, such as IntAct (Aranda et al., 2010), BioGRID (Stark et al., 2006), BIND (Alfarano et al., 2005), and TAIR (Swarbreck et al., 2008), have been established as repositories for interaction data. However, no experiment aiming to chart an entire plant interactome has been attempted. Even for the best-studied plant model, Arabidopsis thaliana, <6000 interactions currently can be found in the major interaction repositories.
Although current high-throughput interaction detection experiments are still prone to false-positives or false-negatives (Huang et al., 2007), the connection topology of many well-charted interactomes has been demonstrated to be robust enough to reflect significant functional linkages within and between higher-order biological systems, such as pathways and biological processes. These interactomes have been successfully explored to suggest potential coordination between pathways (Li et al., 2008; Dotan-Cohen et al., 2009), to predict novel gene functions (Vazquez et al., 2003; Sharan et al., 2007), and to identify robust expression signatures in response to perturbations at the interaction module level, including genes without detectable differential expression (Chuang et al., 2007). Unfortunately, the scarcity of the available Arabidopsis interactome limits the power of these approaches to produce novel hypotheses. Recognizing the need for a more comprehensive plant interactome, several efforts have been made to infer interactions using a variety of computational strategies. For instance, Geisler-Lee et al. (2007) predicted ~20,000 Arabidopsis interactions (interologs) based on homologous interactions in other species, De Bodt et al. (2009) filtered the interologs with functional association data to improve prediction reliability, Cui et al. (2008) predicted ~23,000 interactions from multiple types of indirect evidence using a relatively simple statistical learning tool, and Brandão et al. (2009) established a database integrating both experimentally reported and predicted interactions. However, none of these works rigorously assessed the coverage and reliability of the predicted interactions with externally reported experimental interactions. Likewise, their usefulness in supporting systems biology analyses was not evaluated.
The Predicted Arabidopsis Interactome Resource (PAIR) (Lin et al., 2009, 2011) is a dedicated effort to provide the most comprehensive and accurate Arabidopsis interactome inferred from multiple indirect lines of evidence, including coexpression, colocalization, coevolution, annotation similarity, domain interaction, and homologous interactions in other species. The last major release (V2) was assessed by an independent group and was shown to have the highest coverage of the known Arabidopsis interactome among all available predicted interactomes, more than doubling the second-best coverage (19% versus 9%) (Lee et al., 2010). The current version (V3.3) hosts a compilation of 5990 experimentally reported interactions collected from IntAct, BioGRID, TAIR, and BIND as of July 23, 2010 and 145,494 interactions that were predicted by an accurate evidence integration model. These predicted interactions were expected to cover ~24% of the entire Arabidopsis interactome, and their reliability was estimated to be ~40%. Two external benchmark data sets were used to verify the accuracy of PAIR, which contained only novel interactions reported after the time when all of the data used for PAIR predictions had been collected. Both external benchmark tests reported accuracies that were comparable to or higher than the expected measurements. Details on the PAIR prediction methods and the accuracy evaluation results are provided in the Supplemental Methods 1 online.
Because PAIR covers only 24% of the Arabidopsis interactome, it does not include many real interactions. However, below, we show evidence that it is rich enough to capture many significant functional linkages within and between higher-order biological systems, offering a practical basis for several systems biology approaches to generate new insights into the function and coordination of Arabidopsis genes and gene networks.
RESULTS AND DISCUSSION
Gene Set Linkage Analysis
It has been commonly assumed that proteins function together within a hierarchy of organized modules controlled by high-level biological processes. These modules, known as pathways or biological processes, are the building blocks for a complex network that is used to achieve diverse cellular objectives (Wang et al., 2008). Changes in the modular structure or intermodule connectivity are more frequently associated with altered or disrupted cell functions than with single gene mutations (Hartwell et al., 1999). A number of studies have been conducted to understand how different pathways work together to elicit a physiologic response at a systems level (Lu et al., 2007; Li et al., 2008; Dotan-Cohen et al., 2009). In this work, we demonstrate that in the predicted interactome, biologically meaningful gene sets were connected in a biologically meaningful manner, offering a framework to integrate the knowledge of biological processes and suggest novel insights into their coordination.
AraCyc Metabolic Pathway Network
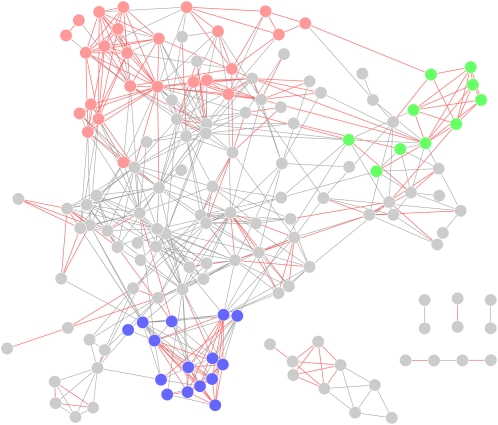
The AraCyc database (Mueller et al., 2003) contains the definitions of the Arabidopsis metabolic pathways. As detailed in Methods, two pathways were connected if the number of interpathway protein interactions was significantly larger (cutoff: P value<0.001) than was expected by chance. Figure 1 illustrates the resultant metabolic pathway linkage network (MPLN), containing 147 nodes (pathways; see Supplemental Data Set 1 online) and 544 edges (significant linkages; see Supplemental Data Set 2 online). This MPLN is well connected. Most of the pathway nodes (128 out of 147) are connected within the largest subnetwork. The remaining 19 pathway nodes form five subnetworks containing only 24 edges. The biological significance of the linkages in this MPLN is supported by two pieces of evidence: connected metabolic pathways often share common substrates, and functionally related pathways are frequently grouped together.
Figure 1.
The Metabolic Pathway Linkage Network Inferred from PAIR.
The three largest clusters identified by the Markov clustering algorithm are in red, blue, and green. The linkages between two pathways that shared one or more substrates are in red.
It has become increasingly clear that metabolism operates as a highly integrated network (Sweetlove et al., 2008). If the same substrate is shared between two pathways, then the scarcity or abundance of that substrate may affect the metabolic fluxes in both pathways, requiring a coordinated regulation mechanism to maintain flux balance. Enzymes often form large complexes in vivo to achieve this goal. The substrate information for each metabolic pathway was retrieved from AraCyc. Substrates involved in >20 pathways were ignored because they were considered too generic and did not reflect significant correlations between pathways. Results indicated that 216 of the 544 pathway connections shared one or more of these specific substrates, which was significantly larger than the expected number of pathway connections sharing a substrate in a randomized MPLN having the same topology (37.47 ± 7.79, P value = ~0).
The internal structure of our MPLN was analyzed by the Markov cluster algorithm (Enright et al., 2002). Twenty-four highly connected clusters were reported (see Supplemental Data Set 1 online). Pathways with related objectives were observed to share the same cluster. Using the largest three clusters as examples, the largest cluster was composed of 15 pathways involved in the metabolism of sugars and their derivatives, together with five nucleosides/nucleotides metabolism pathways, three pathways involved in the generation of precursor metabolites and energy, and two additional pathways. The second cluster contained pathways mainly related to the biosynthesis of plant hormones and defense compounds. The majority of these pathways required the involvement of cytochrome P450 family proteins (Hull et al., 2000; Bak and Feyereisen, 2001; Noordermeer et al., 2001; Takei et al., 2004; Nomura et al., 2005; Greer et al., 2007; Höfer et al., 2008). The third cluster was formed by four pathways for amino acid biosynthesis, four pathways for folate biosynthesis, and two additional pathways for photorespiration and de novo biosynthesis of pyrimidine deoxyribonucleotides. In plants, folates have been reported to play important roles in photorespiration and amino acid metabolism (Hanson and Roje, 2001).
AraCyc–Gene Ontology Biological Processes Network
Some linkages in the MPLN are intuitive, as discussed above, whereas others may not be straightforward. Therefore, it would be desirable to integrate knowledge of other biological processes to gain further insight from these pathway linkages, such as whether they represent coordinated regulation in response to certain stimuli. MPLN is conceptually a special case of a more general kind of network, a gene set linkage network (GSLN), in which all of the connected gene sets are metabolic pathways. A GSLN can be used to study the relationship between biologically significant gene sets, allowing the integration of any biological concept that can be represented as a gene set.
The Gene Ontology (GO) biological process annotations provide definitions of the gene sets important in most known biological processes, which may be useful to expand the MPLN for insights into metabolic pathway coordination. Significant linkages between AraCyc metabolic pathways and GO biological processes were computed using the same method. The resultant network, hereafter referred to as the AraCyc-BP GSLN, contains 4863 significant linkages between AraCyc metabolic pathways and GO biological processes (see Supplemental Data Set 3 online). Some linkages in this AraCyc-BP GSLN are between semantically similar gene sets, for example between the AraCyc abscisic acid biosynthesis pathway and the GO abscisic acid biosynthesis process (GO:0009688). Because the definitions of the same metabolic pathway in AraCyc and GO are expected to be highly similar if not identical, these linkages are trivial. However, there are also a significant number of nontrivial linkages between AraCyc pathways and GO biological processes involved in regulation, response to stimuli, and signaling. Some of these linkages represent novel knowledge of process coordination reported after the time when all of the data used to predict PAIR interactions had been collected.
Here, the AraCyc glucosinolate biosynthesis from Trp pathway was taken as an example to illustrate how the GO biological process concepts were integrated. Glucosinolates are secondary metabolites that are well known for their roles in plant resistance to insects and pathogens and their distinctive benefits to human nutrition (Grubb and Abel, 2006). As shown in Supplemental Data Set 4 online, this AraCyc pathway has linkages to 109 GO biological processes. GO biological processes have a hierarchical structure in which a conceptually more general parent biological process consists of several conceptually more specific child biological processes. Ninety-five biological processes were considered trivial because their more specific child biological processes were connected to this AraCyc pathway or because of semantic similarity. The majority of the remaining 14 nontrivial biological processes fell into three categories: defense responses, secondary metabolic pathways, and multicellular organismal development, as shown in Supplemental Data Set 5 online. Some of these relationships are well known, such as the relationship between glucosinolates and defense responses, whereas others are indirectly supported by the literature. The secondary metabolic processes connected to this AraCyc pathway are the biosynthesis processes for brassinosteroid, carotenoid, flavonoid, and jasmonic acid. It has been reported that cytochrome P450 monooxygenases (P450s) play important roles in the syntheses of diverse secondary metabolites, including Trp-derived glucosinolates (Hull et al., 2000; Bak and Feyereisen, 2001) and these four metabolites (Laudert et al., 1996; Winkel-Shirley, 2001; Fujioka and Yokota, 2003; Tian et al., 2004; Kim and DellaPenna, 2006). A recent study further indicated that the biosynthesis of these five secondary metabolites is coordinated by the circadian regulation of P450s (Pan et al., 2009). Other nontrivial linkages, such as those between this AraCyc pathway and GO multicellular organismal development processes, are less expected. However, it has been reported that the Arabidopsis TU8 mutant could developmentally alter meristem structure together with leaf glucosinolate profile (Kim et al., 2004). Further studies are therefore needed to clarify the relationship between glucosinolate synthesis and regulation of meristem structural organization (GO:0009934) or secondary shoot formation (GO:0010223). In addition, it is interesting to note that disruption of auxin transport results in a significant change in the abundance of several indolic compounds, including indole glucosinolates (Truman et al., 2010). It was reported previously that auxin synthesis is connected to glucosinolate synthesis, since both can be synthesized from Trp (Grubb and Abel, 2006), but a relationship between auxin transport and glucosinolate synthesis was reported much more recently (Truman et al., 2010). Using data prior to July 2009, PAIR was able to suggest this recently reported connection between glucosinolate biosynthesis and auxin transport (GO:0009926, auxin polar transport).
By integrating GO biological process concepts, the less intuitive connections between AraCyc pathways may be explained by shared GO process connections. For example, the connection between the AraCyc pathways glucosinolate biosynthesis from Trp and jasmonic acid biosynthesis may be explained by their shared connection to regulation of systemic acquired resistance (GO:0010112) (Truman et al., 2007; Clay et al., 2009). Similarly, the AraCyc metabolic pathways might also be useful as a mind map to assist in explaining the coordination between GO biological processes.
GO Biological Processes Network
The GO biological process ontology provides definitions of most known biological processes and their semantic relationships, such as whether one biological process is a kind of or is part of another biological process; however, the ontology does not provide information regarding the functional connections between processes, for example, whether two processes are coregulated to achieve a specific cellular objective. Using the same protocol that was used to develop the MPLN, we computed the significant linkages between the GO biological processes. The resultant biological process linkage network (BPLN) contains 26,836 linkages connecting 1237 biological process terms, as shown in Supplemental Data Set 6 online.
Many of the links in the BPLN were expected, reflecting semantic relationships between terms. Lin (1998) reported a method to measure semantic similarity between GO terms. The average semantic similarity between terms connected in the BPLN developed in this work is 0.28, which is significantly higher than that of a randomized BPLN (0.10, P value <1e-10, Wilcoxon test). This demonstrates the consistency between this BPLN and the GO semantic hierarchy. Our BPLN also contains a large number of unexpected links between terms that share small semantic similarities. Particularly, we found 8641 (31.95%) links between terms that only share the term biological process (the root term of the ontology) as their common ancestors (semantic similarity = 0). For example, the link between response to insect (GO:0009625) and jasmonic acid biosynthesis process (GO:0009695) reflected the well-known role of jasmonic acid in insect resistance (McConn et al., 1997); however, these two terms share no semantic similarity.
It is interesting to note that many linkages that were less intuitive at the time when the BPLN was computed were later supported by both processes sharing newly annotated genes. The PAIR interactions and the BPLN were both computed using the July 2009 version of GO annotation data. As shown in Supplemental Data Set 7 online, 193 process pairs that shared no common gene in this previous annotation have at least one shared gene in the recent update of the GO annotation for September 2010. Among these 193 links supported by both processes sharing newly annotated genes, 92 links were between terms with no semantic similarity. For example, the detoxifying efflux carrier 35 (DTX35; AT4G25640) was recently characterized. Mutant analysis demonstrated that the absence of the DTX35 transcript affects flavonoid levels, and the altered flavonoid metabolism affected anther dehiscence and pollen development (Thompson et al., 2010). Therefore, the DTX35 annotations were updated to include the terms flavonoid metabolic process (GO:0009812) and anther dehiscence (GO:0009901). DTX35 was the first gene that was annotated to both of these processes. However, the connection between these two processes could be successfully predicted by this BPLN prior to the availability of these annotations.
The above examples have shown how the strategy of gene set linkage analysis can be used to integrate different biological concepts and analyze potential functional linkages between them. A gene set linkage network is a higher-order abstraction of the underlining molecular network. By focusing on sets of related genes, instead of individual genes, this approach is expected to be more robust with the presence of missing and incorrect links in the underlining molecular network (Rives and Galitski, 2003; Li et al., 2008; Wang et al., 2008). Therefore, the predicted interactions in PAIR may offer a practically useful basis for this type of collective analyses. In gene set linkage analysis, the exact precision of each interaction becomes less critical; however, the overall coverage and precision of the underlining molecular network are still important for the discovery of novel linkages between concepts represented by gene sets. As shown in Supplemental Data Sets 5 and 7 online, the majority (80%) of the experimentally supported linkages discovered using PAIR interactions could not be predicted using any of the three other predicted interactomes (Geisler-Lee et al., 2007; Cui et al., 2008; De Bodt et al., 2009) and the experimentally reported Arabidopsis interactions up to July 2010.
In addition, it should be noted that the above examples all used gene sets derived from existing knowledge, for example, pathway definitions in AraCyc and biological process definitions in GO. This is not required because the strategy of gene set linkage analysis permits the use of any biologically meaningful gene set, whether it is previously well defined and associated with an existing biological concept or an observation in an experiment representing a group of genes forming a hypothetical process to respond to a specific perturbation. In fact, it is expected that not all de facto biological processes have been discovered nor can all be described by existing biological concepts. In this regard, gene set linkage analysis is potentially a powerful tool to link observation-driven hypothetical processes to the existing knowledge framework of biological processes, thereby facilitating further characterization of these hypothetical processes.
Protein Function Prediction
The functional characterization of proteins is one of the most fundamental issues in both experimental and computational biology. Despite the large-scale effort over previous decades, the functions for the majority of proteins in animal or plant genomes have remained either completely unknown or partially understood (Gollery et al., 2006, 2007). In 2008, it was estimated that >40% of the Arabidopsis genes were annotated as proteins of unknown function or lacked annotations in the GO system (Horan et al., 2008). It has been well recognized that functionally similar proteins tend to cluster together in protein interaction networks. With the advent of large-scale interaction data sets, many attempts have been made to predict gene functions by examining their interaction partners. This guilt-by-association strategy has been proven successful in many organisms with comprehensive interactome data (Brun et al., 2003; Vazquez et al., 2003; Deng et al., 2004; Nabieva et al., 2005; Chua et al., 2006).
To evaluate the accuracy of protein function predictions, a set of well-defined function terms are required. The GO terms are well-defined descriptions of gene functions. However, they include terms at different conceptual levels, making the interpretation of prediction results difficult. For example, a successful prediction of the term biological process (GO:0008150, the root term) is obviously less useful than a successful prediction of the term auxin polar transport (GO:0009926). A recent study on functional predictions in plants (Bradford et al., 2010) separated each ontology (i.e., molecular function, biological process, and cellular component) into six annotation levels implicit from the GO structure. Briefly, annotated genes were first clustered based on their functional similarity. By applying a threshold to the similarity score, a number of clusters were generated that corresponded to sets of functionally related genes at each annotation level. For each gene cluster, the most specific term(s) common to the genes’ annotations was used as a label for the function of the cluster. Six thresholds were chosen for the similarity score, resulting in six annotation levels (1 to 6), with level 1 containing general functional clusters and level 6 containing highly specific functional clusters. Therefore, the label terms of the gene clusters at each annotation level represent a set of GO terms that are of similar conceptual sizes and can cover all annotated genes. As expected, the authors demonstrated that the prediction of the general label terms at higher annotation levels was easier than the prediction of the specific label terms at lower annotation levels. In addition, the authors demonstrated that if the top five predicted terms were considered, the biological process terms were the most difficult to predict among the three ontologies (Bradford et al., 2010).
In this work, we examined the usefulness of PAIR in protein function prediction with its interactions to predict the most difficult functional terms: level 6 biological process terms (see Supplemental Data Set 8 online). As detailed in Methods, the biological processes of a gene were predicted as the significantly enriched biological processes among its interaction neighbors. By applying a threshold to the significance P value, a larger or smaller number of biological processes can be predicted for each gene. A lower cutoff is expected to increase the confidence of the predicted terms at the expense of missing known biological processes, whereas a higher cutoff may recover additional known biological processes but would include more false predictions. Therefore, a P value–independent approach, the precision recall curve, was used to measure the usefulness of PAIR interactions in biological process prediction. Precision measures the fraction of predicted biological processes that are known to be correct, while recall measures the fraction of known biological processes that are successfully predicted. Similar to the receiver operation characteristics curve analysis, this precision recall curve analysis is independent from the selection of cutoff P values. The higher a precision recall curve is, the more useful an interactome data set is in biological process prediction. In addition, whether or not a precision recall curve can reach the region of high recall indicates the power of an interactome to predict new biological processes. The ability to support biological process prediction was compared for four other interactome data sets and PAIR. The Experiment data set contains experimentally reported Arabidopsis interactions that were updated until July 2010; the Geisler-Lee data set is a collection of interologs (Geisler-Lee et al., 2007); the De Bodt data set contains high-confidence interologs filtered with functional association data (De Bodt et al., 2009); and the AtPID data set is an earlier interactome predicted by multiple lines of indirect evidence (Cui et al., 2008).
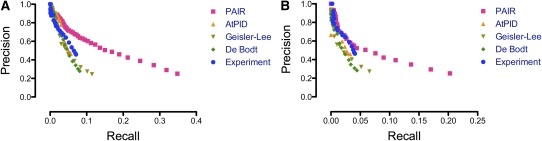
Based on the July 2009 version of GO annotation, Figure 2A compares the effectiveness of these interactomes to predict biological processes for every annotated gene, whether or not it was included in these interactomes or had any predicted processes based on these interactomes. The PAIR interactome dominated the results, and the other four interactomes, including the Experiment interactome, showed similar performances. These data demonstrated that the significantly expanded molecular network in PAIR (Lee et al., 2010) was able to considerably enhance the power of this guilt-by-association strategy to predict more known biological processes.
Figure 2.
The Precision Recall Curves Using Five Interactomes to Predict Biological Process Annotations.
(A) Precisions and recalls were calculated using the July 2009 version of GO annotation.
(B) Precisions were calculated using the September 2010 version of GO annotation. Recalls were calculated only for the 1575 newly added annotations in the September 2010 version of GO annotation.
However, it was noted that the GO biological process information was used in the prediction of PAIR interactions. This may lead to overestimation of recalls when the same set of biological process annotations was used to evaluate the ability of PAIR interactions to predict biological processes. For this reason, we downloaded the September 2010 version of GO annotation and evaluated the effectiveness of these interactomes to predict the 1575 newly added biological process annotations with experimental evidence. In this analysis, precision was calculated in the same way as the fraction of predicted biological processes that were included in the annotation, but recall only counted the fraction of the 1575 new biological process annotations that were successfully predicted.
As shown in Figure 2B, though the performance of PAIR interactions to predict new annotations was inferior to their performance predicting old annotations, PAIR continued to dominate the results. It is interesting to note that not only PAIR, but also all of the other four interactomes, including the Experiment interactome, showed reduced sensitivities (recalls) when they were used to predict recent annotations. One possible explanation might be that all the existing biological data are related. Therefore, even if biological processes are not explicitly used to infer an interactome, other data that are used in its inference may be related to biological process information and carry biological process information into the inference of the interactome, thereby introducing bias in predicting old biological process annotations. The case of the Experiment interactome could be similarly confounded. Several studies on the growth pattern of scientific knowledge have demonstrated that scientists prefer to study familiar subjects (Cokol et al., 2005; He and Zhang, 2009). Experimental testing of a potential interaction was often guided by hypotheses, which were formed based on existing biological data that carry information about known biological processes. Therefore, it might be expected that experiments would prefer to test interactions that would reconfirm the known biological process associations of a gene, rather than to test interactions that would explore potentially new biological process associations. This may lead to be observed sensitivity reduction when experimentally reported interactions were used to predict recent biological process annotations. In this analysis, PAIR interactions demonstrated again their strong capability to support biological process predictions.
Bradford et al. (2010) used a more intuitive way to measure the accuracy of biological process predictions: whether the top ranked prediction or top five predictions can recover the known biological processes of a gene. Using the July 2009 version of GO annotation, the top five predicted biological processes for Arabidopsis genes based on PAIR interactions are given in Supplemental Data Set 9 online. The September 2010 version of GO annotation contains 461 newly annotated genes with experimental evidence, which were not previously annotated to any biological process in the July 2009 version. Among these genes, 105 (23%) had predicted biological processes based on the PAIR interactome. If only the top predicted biological process was considered, we observed a success rate of 54% in recovering the newly annotated biological processes. If the top five predicted biological processes were considered, the success rate increased to 71%, which is noticeably higher than the observed success rate (56%) in Bradford et al. (2010) and is higher than the success rates using other two predicted interactomes (AtPID, 60%, and Geisler-Lee, 54%) (Table 1). Although this success rate was slightly lower than those of the experimentally determined Arabidopsis interactome and the De Bodt high-confidence interologs (71% versus 75% and 75%), the more comprehensive PAIR interactome predicted biological processes for a far greater number of genes (105 versus 12 and 12) (Table 1).
Table 1.
Prediction of the 461 New Biological Process Annotations Using Five Interactomes
| Interaction Data Set | No. of Genes That Have Predictions | Top Rank | Top Five | ||||
| No. of Successes | Success Rate | Expecteda | No. of Successes | Success Rate | Expecteda | ||
| PAIR | 105 | 57 | 0.54 | 0.03 | 75 | 0.71 | 0.06 |
| AtPID | 48 | 22 | 0.45 | 0.04 | 50 | 0.60 | 0.05 |
| Geisler-Lee | 85 | 30 | 0.35 | 0.04 | 46 | 0.54 | 0.04 |
| De Bodt | 12 | 9 | 0.75 | 0.02 | 9 | 0.75 | 0.02 |
| Experiment | 12 | 7 | 0.58 | 0.02 | 9 | 0.75 | 0.02 |
The expected success rate when random predictions are made.
In addition, it should be noted that predictions not overlapping with known biological processes were not necessarily wrong. Using PAIR interactions, there were 30 newly annotated genes for which the top five predicted biological processes did not recover the known ones. We manually examined these predicted processes and found that 11 predicted processes for nine genes were supported by literature (Table 2). For example, SPT16 (AT4G10710), a component of the facilitates chromatin transcription (FACT) complex (Duroux et al., 2004), was predicted to be involved in chromosome organization (GO:0051276). This prediction was intuitively correct considering its gene function. More interestingly, this gene was also predicted to be involved in regulation of flower development (GO:0009909), and it was recently reported that Arabidopsis FACT is critically involved in the transition to flowering (Lolas et al., 2010; Van Lijsebettens and Grasser, 2010).
Table 2.
Literature Supported Biological Process Predictions for the 30 Newly Annotated Genes, for Which the Top Five Predictions Did Not Recover the Annotated Biological Processes
| Gene | Predicted Annotation | Reference |
| AT4G10710 (SPT16) | Chromosome organization | Duroux et al. (2004) |
| AT4G10710 (SPT16) | Regulation of flower development | Lolas et al. (2010); Van Lijsebettens and Grasser (2010) |
| AT4G12560 (CPR30) | Ubiquitin-dependent protein catabolic process | Gou et al. (2009) |
| AT5G27740 (RFC3) | Response to DNA damage stimulus | Liu et al. (2010) |
| AT5G27740 (RFC3) | DNA repair | Liu et al. (2010) |
| AT1G59610 (DRP2B) | Response to hormone stimulus | Kline et al. (2010) |
| AT5G55920 (OLI2) | Translation | Fujikura et al. (2009) |
| AT5G19620 (OEP80) | Protein import into chloroplast stroma | Sun et al. (2009) |
| AT3G28730 (HMG) | Chromosome organization | Duroux et al. (2004) |
| AT3G13550 (FUS9) | Ubiquitin-dependent protein catabolic process | Lau and Deng (2009) |
| AT1G27840 (ATCSA-1) | Chromosome organization | Biedermann and Hellmann (2010) |
Identification of Regulatory Genes Demonstrating Insignificant Expression Changes
Microarrays are a useful and inexpensive research tool that are frequently used in studies of various regulatory mechanisms. However, regulation occurs at different levels, with some key regulators demonstrating no significant changes in expression. By exploring the interactions between genes demonstrating insignificant expression changes and genes demonstrating significant changes, it may be possible to identify these expressionally insignificant regulators in a biological process responsive to specific perturbation. This is another application of the guilt-by-association strategy in microarray analysis.
Goda et al. (2008) reported a hormone and chemical treatment microarray data set, which contains a group of experiments measuring the effect of methyl jasmonate (MJ), a stress hormone involved in development and defense responses. As detailed in Methods, the expression change for each gene was mapped to the PAIR interactome. Then, the Cytoscape plug-in, jActiveModules, was used to find active subnetworks (i.e., connected sets of genes with unexpectedly high levels of differential expression) (Ideker et al., 2002). It should be noted that an active subnetwork might include genes that demonstrate no significant expression changes but are tightly connected to other differentially expressed genes.
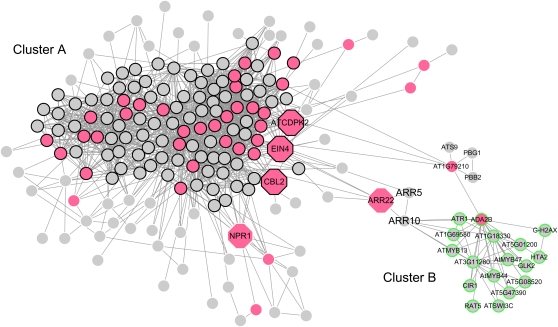
Figure 3 displays the active subnetwork found in response to MJ treatment. Using the MCODE clustering algorithm (Bader and Hogue, 2003), this active subnetwork can be divided into two clusters. The GO enrichment analysis (Boyle et al., 2004) revealed that the majority of the genes (80/115) in Cluster A were annotated to the biological process protein amino acid phosphorylation (GO:0006469), whereas Cluster B was primarily composed of the genes (11 of 17) involved in regulation of transcription (GO:0045449). This reflected that many hormone signals are transmitted and amplified through protein phosphorylation chains and eventually regulate transcription factors in the nucleus (McSteen and Zhao, 2008).
Figure 3.
The Identified Active Subnetwork in Response to MJ Treatment.
This subnetwork contained two densely connected clusters, Cluster A and Cluster B. Genes in Cluster A are marked with black borders. Genes in Cluster B are marked with green borders. Genes showing significant differential expressions in response to MJ treatment are in gray. Genes that show insignificant expression changes are in red. The expressionally insignificant genes that have experimental evidence to suggest their relations to MJ signaling process are shown as octagons.
In this active subnetwork, 44 genes without significant expression changes (P value > 0.05; see Supplemental Data Set 10 online) were included. A literature review revealed that five of these genes are related directly or indirectly to the MJ response. NPR1 (AT1G64280; P value = 0.21) is a well-known regulator in the salicylic acid–mediated systemic acquired resistance pathway. It was also reported that NPR1 modulates crosstalk between salicylate- and jasmonate-dependent defense pathways (Spoel et al., 2003). At-CDPK2 (AT1G35670; P value = 0.98) is a calcium-dependent protein kinase homologous to Nt-CDPK1 in Nicotiana tabacum, which is known to be regulated by jasmonic acid (Yoon et al., 1999), and it has been reported that the At-CDPK family of genes are generally subject to cross-regulation by the components of the jasmonic acid and brassinosteroids pathways (Harmon et al., 2000). CBL2 (AT5G55990; P value = 0.23) is a sensor relay protein. The CBL-CIPK network is the central system for decoding Ca2+ signals in response to a broad variety of stimuli (Dodd et al., 2010). A previous Arabidopsis molecular interaction network analysis indicated that Ca2+ signaling occupies separate and well-connected nodes within the subnetworks associated with jasmonic acid/ethylene signaling (Cui et al., 2008). EIN4 (AT3G04580; P value = 0.32) is an ethylene receptor. EIN4-ethylene binding could result in the rapid expression of ERF1, which is also induced by MJ and represents a point of intersection between the two hormone pathways (Alonso and Stepanova, 2004).
Furthermore, it is interesting to note that the Arabidopsis response regulator (ARR) family of genes were grouped together and formed a bridge linking Cluster A, which contained many phosphorylation related genes, to Cluster B, which contained many transcriptional regulators. ARRs are divided into three groups: the A-type ARRs, B-type ARRs, and a third group consisting of ARR22 and ARR24 (Imamura et al., 1999). In this active subnetwork, the expression of ARR5 and ARR10 was altered significantly (P value < 0.05), but ARR22 was not altered significantly (AT3G04280; P value = 0.90). However, Horák et al. (2008) reported that the ARR22 gene may be involved in several hormone response pathways, such as MJ, ethylene, and cytokinin.
It was noted that, in this type of analysis, the fraction of expressionally insignificant regulators with experimental support was significantly less than the fraction of literature-supported gene set linkages or predicted biological processes. In addition to methodological issues, this may also be due to the lack of experimental data on biological process regulators that do not demonstrate significant expression changes. Current mechanism studies are often guided by expression profile analyses. Regulation on other levels rarely is reported. This situation also creates the need for novel approaches that can capture the largely neglected expressionally insignificant regulators. Further dedicated research coupled with direct experimental verification is required to fully understand and evaluate the power of this guilt-by-association strategy for identification of expressionally insignificant regulators. In this application, comprehensive and accurate interactome data will be needed to perform the analysis. In our example, all of the five expressionally insignificant regulators supported by the literature could not be identified using other interactome data sets.
The Nature of the Predicted Interactions in PAIR
PAIR has been developed and presented as a network of predicted molecular interactions. Newly reported molecular interactions were used to rigorously assess its prediction accuracy (see Supplemental Methods 1 and Supplemental Data Sets 11 and 12 online). However, PAIR may also be perceived as a network of functional linkages. As detailed in the Supplemental Methods 1 online, the interactions were predicted by six types of indirect evidence, all of which were also indicators of functional linkages.
Unfortunately, currently there is no rigorous definition of functional linkage that has been universally agreed upon. Physical interaction is arguably the tightest functional linkage, although in general, all genes are linked functionally to maintain cell structure and function. The analysis of a functional linkage network requires rigorous criteria to filter out insignificant linkages in a consistent manner. Previous studies have successfully explored functional linkage networks based on coexpression data (Horan et al., 2008; Obayashi et al., 2009; Vandepoele et al., 2009). In these cases, the networks were generated using a single type of evidence data (i.e., expression profiles), which made it easy to implement a consistent measurement of the linkage significances (e.g., using a cutoff based on expression correlation). However, functional linkages can be suggested by multiple types of evidence data. Different types of data have different strengths to indicate functional linkages. On top of this complexity, some types of data are not independent (e.g., coexpression is likely to correlate to coevolution) (Fraser et al., 2004; Jordan et al., 2004). There is no intuitive method to integrate the native significance indicators for different data types, such as the correlation coefficient for expression similarity and that for evolution rate similarity, to produce a biologically meaningful and consistent overall measurement of the strengths of functional linkages suggested by multiple types of evidence data. The PAIR prediction model was trained with physical interactions; therefore, the attempt of PAIR can be viewed as constructing a functional linkage network suggested by multiple types of evidence, in which the strengths of linkages were similar to the functional linkages between physically interacting proteins. In this regard, PAIR implemented a consistent and biologically meaningful measurement for the strengths of functional linkages. A false-positive interaction predicted in PAIR could nonetheless represent a close functional relationship between two genes, such as a genetic interaction in a biological process. Consequently, these tight functional linkages might be expected to offer solid support for the systems-level network analyses discussed above.
However, this discussion on the nature of PAIR data is only qualitative and is based on relatively poorly defined concepts, such as the strength of functional linkages and the consistency between significance measurements. We are unaware of a method to examine these concepts directly with rigorous mathematical terms. Further research in this direction will undoubtedly advance our understanding of how different types of data can be integrated to validate each other and corroborate a holistic view of a biological system.
Systems-level network analysis approaches generally fall into two categories (Albert, 2007): the quantitative ones, which rely on the precise network structure and kinetics data to simulate the dynamics of a biological process; and the qualitative ones, which rely on collective topological features (often using the guilt-by-association strategy) to draw functional conclusions. PAIR significantly expanded the known Arabidopsis molecular interaction network with links that were tightly connected in function. Therefore, it was expected to offer a practically useful basis to support the systems biology approaches in the second category. The analyses discussed above, gene set linkage analysis, functional prediction, and identification of expressionally insignificant regulators, are examples of these network topology–based systems biology approaches.
Concluding Remarks
Although limited in many ways, it is our hope that this discussion on the abilities of PAIR will build confidence in predicted interactomes and attract more efforts to the study of how they can be used to help experimental research. Our data demonstrate that PAIR is not only a repository of potential interactions, but also a resource for plant systems biology to analyze relationships between higher-order biological systems. PAIR is committed to producing the most accurate and useful prediction of the Arabidopsis interactome. With this work, we promote the use of PAIR as a helpful and reliable resource and invite discussions and suggestions to improve its accuracy and capabilities.
METHODS
Protein–Protein Interaction Networks
PAIR version 3.3 contains 145,494 predicted interactions inferred from data before July 2009 and 5990 experimentally reported interactions integrated from BioGRID (Stark et al., 2006), TAIR (Swarbreck et al., 2008), IntAct (Aranda et al., 2010), and BIND (Alfarano et al., 2005) as of July 2010 (see Supplemental Methods 1 online). The PAIR data set in this work refers to the 145,494 predicted interactions. The Experiment data set refers to the 5990 experimentally reported interactions. The AtPID data set containing 23,396 predicted interactions was downloaded from its database website (Cui et al., 2008). The Geisler-Lee data set contains the 19,979 interologs generated by Geisler-Lee et al. (2007), and the De Bodt data set contains 18,674 high-confidence interologs (De Bodt et al., 2009).
Gene Set Data
Metabolic pathway definitions were collected from the AraCyc database (Mueller et al., 2003). Biological process definitions were collected from the July 2009 version of GO annotation. Gene sets that are less than five genes or more than 100 genes were excluded; this resulted in 206 AraCyc pathways and 1269 GO biological processes.
GO Annotations
The GO annotations were downloaded from the Gene Ontology Consortium (Kim and Subramaniam, 2006) website. The July 2009 version refers to version 1.1258 deposited on July 1, 2009. The September 2010 version refers to version 1.1330 deposited on September 1, 2010. To evaluate the capability of different interactomes for biological process prediction, only biological process annotations with experimental evidence codes, for example, EXP (inferred from experiment), IDA (inferred from direct assay), IPI (inferred from physical interaction), IMP (inferred from mutant phenotype), IGI (inferred from genetic interaction), IEP (inferred from expression pattern), and TAS (traceable author statement), were used. This resulted in 14,179 biological process annotations for 6707 genes in the July 2009 version and 16,105 biological process annotations for 7270 genes in the September 2010 version. Among the 1926 new experimental biological process annotations, 351 annotations appeared in the July 2009 version with computational evidence. Therefore, there were 1575 entirely new biological process annotations with experimental evidence. Of the 7270 annotated genes, 461 genes did not have any experimental or computational biological process annotation in the July 2009 version.
Measuring the Significance of Gene Set Linkages
Significant linkages between gene sets were detected using a method similar to the one described by Li et al. (2008). For each pair of gene sets, the number of interset protein interactions was first counted. A common gene shared by two gene sets was treated as two distinct delegate genes, with each delegate gene belonging to only one gene set. Any gene that interacted with the shared gene was considered to interact with both delegate genes in both gene sets. The fact that two gene sets sharing a gene was not considered an interset interaction. Then, the significance P value of the linkage between a pair of gene sets was calculated using the method outline here. The interaction network was randomized maintaining the same topology. Every gene in the network was swapped with a random gene with the same number of interactions. With 10,000 randomizations, the significance P value was the fraction of randomized networks in which the number of interset interactions was larger than that in the original interaction network.
Biological Process Prediction
All biological processes in the 2009 version of GO annotation were mapped to the level 6 terms as described by Bradford et al. (2010). These level 6 terms are listed in Supplemental Data Set 8 online. Annotation terms that were conceptually more general were excluded. To predict biological processes for a target gene, the annotations of the target gene were first removed. Then the interaction neighbors of the target gene were identified. The tool GO Term Finder (Boyle et al., 2004) was used to detect the significantly enriched terms associated with these neighboring genes and assign a significance P value to each term. This P value calculation assumes the distribution of GO terms in a random set of genes follows the hypergeometric distribution.
Expression Analysis
The MJ treatment microarray data set (Goda et al., 2008) was downloaded from the NASCArrays database (Craigon et al., 2004). The data set contained three time points of data for MJ treatments (30 min, 1 h, and 3 h). The Affymetrix CEL files were analyzed using GeneSpring version 7.2. The probe level intensities were processed with background adjustment, normalization, and log2 transformation of the perfect match values. Then, per-chip and per-gene normalization were performed. The statistical significance for the expression changes (P values) was evaluated using the two-way analysis of variance algorithm, which treated each experimental condition as a factor (three time points and MJ treatment/control, altogether six conditions). A total of 1564 genes showed significant expression changes (P value < 0.05). This level of expression change was similar to that reported by Nemhauser et al. (2006) (1518 differentially expressed genes). The P value of each gene was then mapped onto the interaction network. The Cytoscape plug-in, jActiveModules, was used to find active subnetworks (i.e., connected sets of genes with unexpectedly high levels of differential expression) (Ideker et al., 2002).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Methods 1. Inference and Evaluation of the Predicted Interactions in the Predicted Arabidopsis Interactome Resource.
Supplemental Data Set 1. The Pathways (Nodes) in the Metabolic Pathway Linkage Network.
Supplemental Data Set 2. The Linkages (Edges) in the Metabolic Pathway Linkage Network.
Supplemental Data Set 3. The Linkages in the AraCyc-BP Gene Set Linkage Network.
Supplemental Data Set 4. The Linkages between the AraCyc Glucosinolate Biosynthesis from Tryptophan Pathway and the GO Biological Processes.
Supplemental Data Set 5. The Nontrivial Linkages between the AraCyc Glucosinolate Biosynthesis from Tryptophan Pathway and the GO Biological Processes.
Supplemental Data Set 6. The Linkages in the GO Biological Process Linkage Network.
Supplemental Data Set 7. The Linkages in the GO Biological Process Linkage Network That Were Supported by Two Processes Sharing Newly Annotated Genes.
Supplemental Data Set 8. The Level 6 Biological Process Terms as in Bradford et al. (2010) That Were Used in the Evaluation of the Capabilities of Different Interactomes to Support Biological Process Prediction.
Supplemental Data Set 9. The Top Five Predicted Biological Processes for Arabidopsis Genes.
Supplemental Data Set 10. The Identified Active Subnetwork in Response to Methylene Jasmonate Treatment.
Supplemental Data Set 11. Rediscovery of Newly Curated Interactions in the BioGrid Database.
Supplemental Data Set 12. Rediscovery of Interactions between Cell Cycle Proteins.
Supplementary Material
Acknowledgments
This work is supported by National Natural Science Foundation of China Grants 31071161 and 30600039.
References
- Albert R. (2007). Network inference, analysis, and modeling in systems biology. Plant Cell 19: 3327–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfarano C., et al. (2005). The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 33(Database issue): D418–D424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., Stepanova A.N. (2004). The ethylene signaling pathway. Science 306: 1513–1515 [DOI] [PubMed] [Google Scholar]
- Aranda B., et al. (2010). The IntAct molecular interaction database in 2010. Nucleic Acids Res. 38(Database issue): D525–D531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader G.D., Hogue C.W. (2003). An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak S., Feyereisen R. (2001). The involvement of two p450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol. 127: 108–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedermann S., Hellmann H. (2010). The DDB1a interacting proteins ATCSA-1 and DDB2 are critical factors for UV-B tolerance and genomic integrity in Arabidopsis thaliana. Plant J. 62: 404–415 [DOI] [PubMed] [Google Scholar]
- Boyle E.I., Weng S., Gollub J., Jin H., Botstein D., Cherry J.M., Sherlock G. (2004). GO:TermFinder—Open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20: 3710–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford J.R., Needham C.J., Tedder P., Care M.A., Bulpitt A.J., Westhead D.R. (2010). GO-At: In silico prediction of gene function in Arabidopsis thaliana by combining heterogeneous data. Plant J. 61: 713–721 [DOI] [PubMed] [Google Scholar]
- Brandão M.M., Dantas L.L., Silva-Filho M.C. (2009). AtPIN: Arabidopsis thaliana protein interaction network. BMC Bioinformatics 10: 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun C., Chevenet F., Martin D., Wojcik J., Guénoche A., Jacq B. (2003). Functional classification of proteins for the prediction of cellular function from a protein-protein interaction network. Genome Biol. 5: R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua H.N., Sung W.K., Wong L. (2006). Exploiting indirect neighbours and topological weight to predict protein function from protein-protein interactions. Bioinformatics 22: 1623–1630 [DOI] [PubMed] [Google Scholar]
- Chuang H.Y., Lee E., Liu Y.T., Lee D., Ideker T. (2007). Network-based classification of breast cancer metastasis. Mol. Syst. Biol. 3: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay N.K., Adio A.M., Denoux C., Jander G., Ausubel F.M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokol M., Iossifov I., Weinreb C., Rzhetsky A. (2005). Emergent behavior of growing knowledge about molecular interactions. Nat. Biotechnol. 23: 1243–1247 [DOI] [PubMed] [Google Scholar]
- Craigon D.J., James N., Okyere J., Higgins J., Jotham J., May S. (2004). NASCArrays: A repository for microarray data generated by NASC’s transcriptomics service. Nucleic Acids Res. 32(Database issue): D575–D577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li P., Li G., Xu F., Zhao C., Li Y., Yang Z., Wang G., Yu Q., Li Y., Shi T. (2008). AtPID: Arabidopsis thaliana protein interactome database—an integrative platform for plant systems biology. Nucleic Acids Res. 36(Database issue): D999–D1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S., Proost S., Vandepoele K., Rouzé P., Van de Peer Y. (2009). Predicting protein-protein interactions in Arabidopsis thaliana through integration of orthology, gene ontology and co-expression. BMC Genomics 10: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M., Tu Z., Sun F., Chen T. (2004). Mapping Gene Ontology to proteins based on protein-protein interaction data. Bioinformatics 20: 895–902 [DOI] [PubMed] [Google Scholar]
- Dodd A.N., Kudla J., Sanders D. (2010). The language of calcium signaling. Annu. Rev. Plant Biol. 61: 593–620 [DOI] [PubMed] [Google Scholar]
- Dotan-Cohen D., Letovsky S., Melkman A.A., Kasif S. (2009). Biological process linkage networks. PLoS ONE 4: e5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duroux M., Houben A., Růzicka K., Friml J., Grasser K.D. (2004). The chromatin remodelling complex FACT associates with actively transcribed regions of the Arabidopsis genome. Plant J. 40: 660–671 [DOI] [PubMed] [Google Scholar]
- Enright A.J., Van Dongen S., Ouzounis C.A. (2002). An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30: 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formstecher E., et al. (2005). Protein interaction mapping: A Drosophila case study. Genome Res. 15: 376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser H.B., Hirsh A.E., Wall D.P., Eisen M.B. (2004). Coevolution of gene expression among interacting proteins. Proc. Natl. Acad. Sci. USA 101: 9033–9038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura U., Horiguchi G., Ponce M.R., Micol J.L., Tsukaya H. (2009). Coordination of cell proliferation and cell expansion mediated by ribosome-related processes in the leaves of Arabidopsis thaliana. Plant J. 59: 499–508 [DOI] [PubMed] [Google Scholar]
- Fujioka S., Yokota T. (2003). Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 54: 137–164 [DOI] [PubMed] [Google Scholar]
- Gavin A.C., et al. (2002). Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Geisler-Lee J., O’Toole N., Ammar R., Provart N.J., Millar A.H., Geisler M. (2007). A predicted interactome for Arabidopsis. Plant Physiol. 145: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H., et al. (2008). The AtGenExpress hormone and chemical treatment data set: Experimental design, data evaluation, model data analysis and data access. Plant J. 55: 526–542 [DOI] [PubMed] [Google Scholar]
- Gollery M., Harper J., Cushman J., Mittler T., Girke T., Zhu J.K., Bailey-Serres J., Mittler R. (2006). What makes species unique? The contribution of proteins with obscure features. Genome Biol. 7: R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollery M., Harper J., Cushman J., Mittler T., Mittler R. (2007). POFs: What we don’t know can hurt us. Trends Plant Sci. 12: 492–496 [DOI] [PubMed] [Google Scholar]
- Gou M., Su N., Zheng J., Huai J., Wu G., Zhao J., He J., Tang D., Yang S., Wang G. (2009). An F-box gene, CPR30, functions as a negative regulator of the defense response in Arabidopsis. Plant J. 60: 757–770 [DOI] [PubMed] [Google Scholar]
- Greer S., Wen M., Bird D., Wu X., Samuels L., Kunst L., Jetter R. (2007). The cytochrome P450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of Arabidopsis. Plant Physiol. 145: 653–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb C.D., Abel S. (2006). Glucosinolate metabolism and its control. Trends Plant Sci. 11: 89–100 [DOI] [PubMed] [Google Scholar]
- Hanson A.D., Roje S. (2001). One-carbon metabolism in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 119–137 [DOI] [PubMed] [Google Scholar]
- Harmon A.C., Gribskov M., Harper J.F. (2000). CDPKs - A kinase for every Ca2+ signal? Trends Plant Sci. 5: 154–159 [DOI] [PubMed] [Google Scholar]
- Hartwell L.H., Hopfield J.J., Leibler S., Murray A.W. (1999). From molecular to modular cell biology. Nature 402(6761, Suppl) C47–C52 [DOI] [PubMed] [Google Scholar]
- He X., Zhang J. (2009). On the growth of scientific knowledge: Yeast biology as a case study. PLoS Comput. Biol. 5: e1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y., et al. (2002). Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183 [DOI] [PubMed] [Google Scholar]
- Höfer R., Briesen I., Beck M., Pinot F., Schreiber L., Franke R. (2008). The Arabidopsis cytochrome P450 CYP86A1 encodes a fatty acid omega-hydroxylase involved in suberin monomer biosynthesis. J. Exp. Bot. 59: 2347–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horák J., Grefen C., Berendzen K.W., Hahn A., Stierhof Y.D., Stadelhofer B., Stahl M., Koncz C., Harter K. (2008). The Arabidopsis thaliana response regulator ARR22 is a putative AHP phospho-histidine phosphatase expressed in the chalaza of developing seeds. BMC Plant Biol. 8: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan K., Jang C., Bailey-Serres J., Mittler R., Shelton C., Harper J.F., Zhu J.K., Cushman J.C., Gollery M., Girke T. (2008). Annotating genes of known and unknown function by large-scale coexpression analysis. Plant Physiol. 147: 41–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Jedynak B.M., Bader J.S. (2007). Where have all the interactions gone? Estimating the coverage of two-hybrid protein interaction maps. PLoS Comput. Biol. 3: e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull A.K., Vij R., Celenza J.L. (2000). Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. USA 97: 2379–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideker T., Ozier O., Schwikowski B., Siegel A.F. (2002). Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics 18(Suppl. 1): S233–S240 [DOI] [PubMed] [Google Scholar]
- Imamura A., Hanaki N., Nakamura A., Suzuki T., Taniguchi M., Kiba T., Ueguchi C., Sugiyama T., Mizuno T. (1999). Compilation and characterization of Arabidopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol. 40: 733–742 [DOI] [PubMed] [Google Scholar]
- Ito T., Tashiro K., Muta S., Ozawa R., Chiba T., Nishizawa M., Yamamoto K., Kuhara S., Sakaki Y. (2000). Toward a protein-protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc. Natl. Acad. Sci. USA 97: 1143–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan I.K., Mariño-Ramírez L., Wolf Y.I., Koonin E.V. (2004). Conservation and coevolution in the scale-free human gene coexpression network. Mol. Biol. Evol. 21: 2058–2070 [DOI] [PubMed] [Google Scholar]
- Kim J., DellaPenna D. (2006). Defining the primary route for lutein synthesis in plants: The role of Arabidopsis carotenoid beta-ring hydroxylase CYP97A3. Proc. Natl. Acad. Sci. USA 103: 3474–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Durrett T.P., Last R.L., Jander G. (2004). Characterization of the Arabidopsis TU8 glucosinolate mutation, an allele of TERMINAL FLOWER2. Plant Mol. Biol. 54: 671–682 [DOI] [PubMed] [Google Scholar]
- Kim Y., Subramaniam S. (2006). Locally defined protein phylogenetic profiles reveal previously missed protein interactions and functional relationships. Proteins 62: 1115–1124 [DOI] [PubMed] [Google Scholar]
- Kline K.G., Barrett-Wilt G.A., Sussman M.R. (2010). In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc. Natl. Acad. Sci. USA 107: 15986–15991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W. (2009). Effect of Arabidopsis COP10 ubiquitin E2 enhancement activity across E2 families and functional conservation among its canonical homologues. Biochem. J. 418: 683–690 [DOI] [PubMed] [Google Scholar]
- Laudert D., Pfannschmidt U., Lottspeich F., Holländer-Czytko H., Weiler E.W. (1996). Cloning, molecular and functional characterization of Arabidopsis thaliana allene oxide synthase (CYP 74), the first enzyme of the octadecanoid pathway to jasmonates. Plant Mol. Biol. 31: 323–335 [DOI] [PubMed] [Google Scholar]
- Lee K., Thorneycroft D., Achuthan P., Hermjakob H., Ideker T. (2010). Mapping plant interactomes using literature curated and predicted protein-protein interaction data sets. Plant Cell 22: 997–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., et al. (2004). A map of the interactome network of the metazoan C. elegans. Science 303: 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Agarwal P., Rajagopalan D. (2008). A global pathway crosstalk network. Bioinformatics 24: 1442–1447 [DOI] [PubMed] [Google Scholar]
- Lin D. (1998). An information-theoretic definition of similarity. Proceedings of the 15th International Conference on Machine Learning, Shavlik J.W., (San Francisco, CA: Morgan Kaufmann Publishers; ), pp. 296–304 [Google Scholar]
- Lin M., Hu B., Chen L., Sun P., Fan Y., Wu P., Chen X. (2009). Computational identification of potential molecular interactions in Arabidopsis. Plant Physiol. 151: 34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M., Shen X., Chen X. (2011). PAIR: The predicted Arabidopsis interactome resource. Nucleic Acids Res. 39(Database issue): D1134–D1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Wang J., Miki D., Xia R., Yu W., He J., Zheng Z., Zhu J.K., Gong Z. (2010). DNA replication factor C1 mediates genomic stability and transcriptional gene silencing in Arabidopsis. Plant Cell 22: 2336–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolas I.B., Himanen K., Grønlund J.T., Lynggaard C., Houben A., Melzer M., Van Lijsebettens M., Grasser K.D. (2010). The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2. Plant J. 61: 686–697 [DOI] [PubMed] [Google Scholar]
- Lu L.J., Sboner A., Huang Y.J., Lu H.X., Gianoulis T.A., Yip K.Y., Kim P.M., Montelione G.T., Gerstein M.B. (2007). Comparing classical pathways and modern networks: Towards the development of an edge ontology. Trends Biochem. Sci. 32: 320–331 [DOI] [PubMed] [Google Scholar]
- McConn M., Creelman R.A., Bell E., Mullet J.E., Browse J. (1997). Jasmonate is essential for insect defense in Arabidopsis. Proc. Natl. Acad. Sci. USA 94: 5473–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P., Zhao Y. (2008). Plant hormones and signaling: Common themes and new developments. Dev. Cell 14: 467–473 [DOI] [PubMed] [Google Scholar]
- Mueller L.A., Zhang P., Rhee S.Y. (2003). AraCyc: A biochemical pathway database for Arabidopsis. Plant Physiol. 132: 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabieva E., Jim K., Agarwal A., Chazelle B., Singh M. (2005). Whole-proteome prediction of protein function via graph-theoretic analysis of interaction maps. Bioinformatics 21(Suppl. 1): i302–i310 [DOI] [PubMed] [Google Scholar]
- Nemhauser J.L., Hong F., Chory J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Nomura T., Kushiro T., Yokota T., Kamiya Y., Bishop G.J., Yamaguchi S. (2005). The last reaction producing brassinolide is catalyzed by cytochrome P-450s, CYP85A3 in tomato and CYP85A2 in Arabidopsis. J. Biol. Chem. 280: 17873–17879 [DOI] [PubMed] [Google Scholar]
- Noordermeer M.A., Veldink G.A., Vliegenthart J.F. (2001). Fatty acid hydroperoxide lyase: A plant cytochrome p450 enzyme involved in wound healing and pest resistance. ChemBioChem 2: 494–504 [DOI] [PubMed] [Google Scholar]
- Obayashi T., Hayashi S., Saeki M., Ohta H., Kinoshita K. (2009). ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37(Database issue): D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Michael T.P., Hudson M.E., Kay S.A., Chory J., Schuler M.A. (2009). Cytochrome P450 monooxygenases as reporters for circadian-regulated pathways. Plant Physiol. 150: 858–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rives A.W., Galitski T. (2003). Modular organization of cellular networks. Proc. Natl. Acad. Sci. USA 100: 1128–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual J.F., et al. (2005). Towards a proteome-scale map of the human protein-protein interaction network. Nature 437: 1173–1178 [DOI] [PubMed] [Google Scholar]
- Sharan R., Ulitsky I., Shamir R. (2007). Network-based prediction of protein function. Mol. Syst. Biol. 3: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S.H., et al. (2003). NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15: 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C., Breitkreutz B.J., Reguly T., Boucher L., Breitkreutz A., Tyers M. (2006). BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 34(Database issue): D535–D539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzl U., et al. (2005). A human protein-protein interaction network: A resource for annotating the proteome. Cell 122: 957–968 [DOI] [PubMed] [Google Scholar]
- Sun C.W., Huang Y.C., Chang H.Y. (2009). CIA2 coordinately up-regulates protein import and synthesis in leaf chloroplasts. Plant Physiol. 150: 879–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck D., et al. (2008). The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 36(Database issue): D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove L.J., Fell D., Fernie A.R. (2008). Getting to grips with the plant metabolic network. Biochem. J. 409: 27–41 [DOI] [PubMed] [Google Scholar]
- Takei K., Ueda N., Aoki K., Kuromori T., Hirayama T., Shinozaki K., Yamaya T., Sakakibara H. (2004). AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol. 45: 1053–1062 [DOI] [PubMed] [Google Scholar]
- Thompson E.P., Wilkins C., Demidchik V., Davies J.M., Glover B.J. (2010). An Arabidopsis flavonoid transporter is required for anther dehiscence and pollen development. J. Exp. Bot. 61: 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Musetti V., Kim J., Magallanes-Lundback M., DellaPenna D. (2004). The Arabidopsis LUT1 locus encodes a member of the cytochrome p450 family that is required for carotenoid epsilon-ring hydroxylation activity. Proc. Natl. Acad. Sci. USA 101: 402–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman W., Bennett M.H., Kubigsteltig I., Turnbull C., Grant M. (2007). Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc. Natl. Acad. Sci. USA 104: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman W.M., Bennett M.H., Turnbull C.G., Grant M.R. (2010). Arabidopsis auxin mutants are compromised in systemic acquired resistance and exhibit aberrant accumulation of various indolic compounds. Plant Physiol. 152: 1562–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P., et al. (2000). A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627 [DOI] [PubMed] [Google Scholar]
- Vandepoele K., Quimbaya M., Casneuf T., De Veylder L., Van de Peer Y. (2009). Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol. 150: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lijsebettens M., Grasser K.D. (2010). The role of the transcript elongation factors FACT and HUB1 in leaf growth and the induction of flowering. Plant Signal. Behav. 5: 715–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez A., Flammini A., Maritan A., Vespignani A. (2003). Global protein function prediction from protein-protein interaction networks. Nat. Biotechnol. 21: 697–700 [DOI] [PubMed] [Google Scholar]
- Wang X., Dalkic E., Wu M., Chan C. (2008). Gene module level analysis: Identification to networks and dynamics. Curr. Opin. Biotechnol. 19: 482–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon G.M., Cho H.S., Ha H.J., Liu J.R., Lee H.S. (1999). Characterization of NtCDPK1, a calcium-dependent protein kinase gene in Nicotiana tabacum, and the activity of its encoded protein. Plant Mol. Biol. 39: 991–1001 [DOI] [PubMed] [Google Scholar]
- Yu H., et al. (2008). High-quality binary protein interaction map of the yeast interactome network. Science 322: 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.