The transition from vegetative to reproductive development is regulated by the activity of graft-transmissible flowering hormone, florigen, which is encoded by the FLOWERING LOCUS T (FT) family of mobile proteins. This work identified, among many maize FT-like genes, a single gene, ZCN8, which has all required characteristics to function as florigen.
Abstract
The mobile floral-promoting signal, florigen, is thought to consist of, in part, the FT protein named after the Arabidopsis thaliana gene FLOWERING LOCUS T. FT is transcribed and translated in leaves and its protein moves via the phloem to the shoot apical meristem where it promotes the transition from vegetative to reproductive development. In our search for a maize FT-like floral activator(s), seven Zea mays CENTRORADIALIS (ZCN) genes encoding FT homologous proteins were studied. ZCN8 stood out as the only ZCN having the requisite characteristics for possessing florigenic activity. In photoperiod sensitive tropical lines, ZCN8 transcripts were strongly upregulated in a diurnal manner under floral-inductive short days. In day-neutral temperate lines, ZCN8 mRNA level was independent of daylength and displayed only a weak cycling pattern. ZCN8 is normally expressed in leaf phloem, but ectopic expression of ZCN8 in vegetative stage shoot apices induced early flowering in transgenic plants. Silencing of ZCN8 by artificial microRNA resulted in late flowering. ZCN8 was placed downstream of indeterminate1 and upstream of delayed flowering1, two other floral activator genes. We propose a flowering model linking photoperiod sensitivity of tropical maize to diurnal regulation of ZCN8.
INTRODUCTION
Early grafting experiments in plants suggested that the transition from vegetative to reproductive development is triggered by the movement of a hypothetical flowering hormone, named florigen (flower-former), from leaves to the shoot apical meristem (SAM; Chailakhyan, 1937). Until recently, the biochemical nature of florigen remained elusive until breakthrough experiments in tomato (Solanum lycopersicum; Lifschitz and Eshed, 2006; Lifschitz et al., 2006), Arabidopsis thaliana (Corbesier et al., 2007; Jaeger and Wigge, 2007; Notaguchi et al., 2008), and rice (Oryza sativa; Tamaki et al., 2007) revealed that the protein encoded by the FLOWERING LOCUS T (FT) gene had florigenic activity. The FT protein and other related proteins share sequence similarity with mammalian phosphatidylethanolamine binding protein (PEBP) (Kardailsky et al., 1999), which has been annotated as a kinase regulator (Banfield and Brady, 2000). The Arabidopsis FT and orthologous genes in other plant species were shown to behave as a flower-forming signal because they were transcribed and translated in leaves, but their proteins were shown to subsequently move through the phloem to the SAM where they induced the floral transition upon reaching a critical concentration (Tsuji et al., 2008; Turck et al., 2008; Zeevaart, 2008).
Several genetic pathways regulate the floral transition in Arabidopsis, including the autonomous, gibberellin, photoperiod, and vernalization pathways (Mouradov et al., 2002). FT is a key integrator because almost all pathways converge on it, and FT transmits the floral inductive signal to downstream floral identity genes (Mouradov et al., 2002). Therefore, the temporal and spatial expression of FT function is essential for proper flowering time regulation. Arabidopsis is sensitive to photoperiod with long days (LDs) promoting flowering. In the photoperiod pathway, FT transcription in the vasculature of the mature leaf is regulated by CONSTANS (CO) (Samach et al., 2000; An et al., 2004; Turck et al., 2008). CO encodes a transcription factor containing two B-box zinc finger domains and a CCT (CO, CO-Like, TOC1) domain near the C terminus (Putterill et al., 1995; Robson et al., 2001). CO transcripts predominantly accumulate in the shoot and leaves under LDs (Putterill et al., 1995) and are diurnally regulated with the peak at dusk under both LDs and short days (SDs) (Suárez-López et al., 2001). CO protein accumulation is stabilized toward the end of the day only under LDs, whereas SD conditions trigger CO degradation (Valverde et al., 2004). Subsequently, the accumulation of CO protein at dusk drives accumulation of FT mRNA at the end of day under LDs but not under SDs (Kardailsky et al., 1999; Suárez-López et al., 2001). FT mRNA levels stay low under SDs but can be induced to floral-promoting levels by a minimum 3-d exposure to LDs (Corbesier et al., 2007). Thus, transient FT mRNA accumulation caused by a short inductive photoperiod is sufficient to promote flowering (Corbesier et al., 2007). Using transgenic plants ectopically expressing fluorescent-tagged FT protein, it was shown that the FT protein moved from the vascular tissue to the SAM (Corbesier et al., 2007; Jaeger and Wigge, 2007; Lin et al., 2007; Mathieu et al., 2007; Tamaki et al., 2007; Giakountis and Coupland, 2008). There, the FT protein interacts with the bZIP transcription factor encoded by FLOWERING LOCUS D (FD) to induce expression of a number of floral identity genes (Abe et al., 2005; Wigge et al., 2005). Hence, the FT protein has all the required features expected for the hypothetical flowering hormone, florigen (Tsuji et al., 2008; Turck et al., 2008; Zeevaart, 2008).
An FT paralog, TWIN SISTER OF FT (TSF), is the closest homolog of FT in Arabidopsis and is thought to be an additional integrator of flowering time pathways (Yamaguchi et al., 2005). TSF appeared to be induced by cytokine and promotes flowering under SDs (Yamaguchi et al., 2005; D'Aloia et al., 2011).
The function of FT as a floral activator is widely conserved in plant species, although FT is regulated differently depending on the photoperiod sensitivity of the species (Turck et al., 2008). In contrast with Arabidopsis, which is a LD plant, most rice cultivars flower earlier under SDs due to their tropical origin. Heading Date 3a (Hd3a), a rice ortholog of FT, was identified as a floral activator under SDs (Kojima et al., 2002). Later, Rice flowering locus T1 (RFT1), the Hd3a paralog located within 6 kb of Hd3a on chromosome 6, was identified as a major floral activator under LDs conditions (Komiya et al., 2008, 2009). Both genes show a diurnal expression pattern with transcript abundance peaking at dawn (Kojima et al., 2002; Komiya et al., 2009). Both genes encode PEBP-like proteins that move from the leaf to shoot apex (Tamaki et al., 2007; Komiya et al., 2009). Rice is extremely sensitive to daylength variation, and flowering can be modulated by photoperiod differences as short as 30 min. Hd3a is highly expressed in photoperiods <13 h, but its expression decreases sharply in photoperiods >13.5 h and its expression is not detectable under longer photoperiods. This level of precision is achieved by the control of expression of two floral regulators by Hd3a: a floral activator, Early heading date1 (Ehd1) and a floral repressor, Grain number, plant height and heading date7 (Ghd7) (Itoh et al., 2010). Ehd1 encodes a protein with sequence similarity to B-type response regulators, and Ghd7 encodes a CCT domain protein (Itoh et al., 2010).
FT orthologs have been identified as floral activators in many species, including tomato single flower truss (Lifschitz and Eshed, 2006; Lifschitz et al., 2006), wheat (Triticum aestivum) Ta FT (Yan et al., 2006), barley (Hordeum vulgare) Hv FT (Faure et al., 2007), Pn FT1/2 (Pharbitis nil) (Hayama et al., 2007), sugar beet (Beta vulgaris) Bv FT2 (Pin et al., 2010), and sunflower (Helianthus annuus) Ha FT1 and Ha FT4 genes (Blackman et al., 2010).
Finding genes in maize (Zea mays) with florigenic function is critical for understanding the regulation of flowering time in this important model and agronomic grass species. However, this task is not trivial due to the expansion the PEBP gene family in maize (Danilevskaya et al., 2008a). There are 25 PEBP homologs in the maize genome named ZCN (Zea mays CENTRORADIALIS) after the first cloned PEBP plant gene from Antirrhinum (Danilevskaya et al., 2008a). Phylogenetic analyses divided the ZCN proteins into four clades, including TFL1-like, MFT-like, FT-like I, and FT-like II. Each of the 15 FT-like genes displays a unique spatial-temporal expression pattern, and of these 15, only six, ZCN7, ZCN8, ZCN12, ZCN14, ZCN18, and ZCN26, are expressed in leaves (Danilevskaya et al., 2008a). These six ZCN genes are potential floral activator candidates. It is important to point out that ZCN15, which maps to a region syntenic with the position on rice chromosome 6 region where Hd3a/b resides, is expressed predominantly in kernels and not in leaves, suggesting that it is not involved in control of flowering (Danilevskaya et al., 2008a). Therefore, functional analysis and experimental evidence are necessary to determine which of the ZCN genes possesses florigenic activity.
To identify maize FT orthologs, we took advantage of the tremendous natural diversity in photoperiod response and flowering time in maize. Maize was domesticated in Mexico from its wild ancestor, teosinte (Z. mays ssp parviglumis), a species that requires SDs to flower (Matsuoka et al., 2002). This ancestral photoperiod sensitivity is the most evident tropical maize that flowers normally under SDs but has a greatly delayed floral transition when grown under LDs (Colasanti and Muszynski, 2009). As a result of artificial selection by humans, maize is adapted to growth in an extraordinary large geographical area, from latitude 58° north to 35° south (Kuleshov, 1933). Most maize lines grown in temperate environments seem to have lost much of their photoperiod sensitivity as they flower, set seed, and mature under LDs.
We hypothesized that the transcription level of any maize FT-like gene that might possess florigenic activity would be elevated during the floral transition, similar to accumulation of FT transcripts in Arabidopsis and rice (Kardailsky et al., 1999; Kojima et al., 2002; Komiya et al., 2008). We examined expression of seven maize FT-like ZCN genes in the extremely early-flowering line Gaspé Flint (Brawn, 1968), the temperate inbred B73 (Troyer, 2000), and two photoperiod sensitive tropical lines, CML311 and CML436 (Russell and Stuber, 1983).
To place any putative FT orthologs into the maize flowering network, we used the loss-of-function indeterminate1 (id1) and delayed flowering1 (dlf1) late-flowering mutants (Colasanti et al., 1998; Muszynski et al., 2006). id1 is specific to monocots, and functional orthologs are not found in Arabidopsis. It encodes a zinc finger transcription factor and its mRNA is mainly expressed in the immature leaf before and after the floral transition (Colasanti et al., 2006). dlf1, the ortholog of Arabidopsis FD, encodes a basic Leu zipper (bZIP) transcription factor and its mRNA accumulates in the shoot apex (Muszynski et al., 2006). dlf1 acts downstream of id1 in the maize flowering time pathway (Muszynski et al., 2006).
Besides id1 and dlf1, a few other genes in maize have been identified to control the floral transition. Zea mays MADS 4 (ZMM4), a member of the maize MADS box gene family and an ortholog of Arabidopsis APETALA1/FRUITFUL is involved in the floral induction. ZMM4 mRNA is strongly expressed in the shoot apex at the onset of the floral transition and expression persists in reproductive organs. Transgenic plants overexpressing ZMM4 have an early-flowering phenotype. In addition, overexpression of ZMM4 in dlf1 or id1 mutants suppressed their late-flowering phenotypes, indicating ZMM4 likely functions downstream of id1 and dlf1 (Danilevskaya et al., 2008b). Rap2.7, an AP2-like transcription factor, is another flowering time gene and is thought to be a repressor of flowering time in maize (Salvi et al., 2007).
Our experimental approach for determining which ZCN gene might have florigenic function included analyses of the dynamics of transcript accumulation in four maize lines with different flowering times and photoperiod sensitivities and also in two maize flowering mutants, id1 and dlf1. A potential function as a floral activator was further tested by assessing their ectopic expression in transgenic plants. Our cumulative results are consistent with ZCN8 functioning as the primary floral activator. ZCN8 mRNA accumulation in leaves always preceded the floral transition of the SAM in all the four maize lines tested, regardless of photoperiod. ZCN8 transcription is diurnally regulated in photoperiod-sensitive tropical lines but showed an attenuated cycling pattern in day-neutral temperate lines. Moreover, only ZCN8 transcript is induced after a 7-d floral-inducing SD exposure in tropical lines, suggesting that ZCN8 is involved in photoperiod sensitivity. Genetically, ZCN8 was positioned downstream of id1 and upstream of dlf1. Based on our findings, we propose a genetic model for the flowering network in temperate and tropical maize.
RESULTS
Survey of FT-Like ZCN Gene Transcription in Leaves of Temperate and Tropical Lines under SD and LD Photoperiods
We assessed transcript accumulation of seven FT-like ZCN genes in leaves before and after the floral transition under SDs and LDs in four lines that vary significantly in their time to flower and sensitivity to photoperiod (see Supplemental Figure 1 online). To monitor the floral transition, shoot apices were dissected and images taken documenting the morphology of the SAM at each sampling time point. The transition from vegetative to flowering was judged by the extent of elongation of the apex and the appearance of branch meristems on the flanks of the SAM marking an early reproductive stage (Irish and Nelson, 1991).
Our survey of ZCN transcript accumulation is presented in Figure 1. Because the floral transition in the early-flowering temperate Gaspé Flint line takes place between the fifth and sixth days after sowing when plants are still at the VE stage (emergence), leaves wrapped in the coleoptile were collected (Figure 1A). Under SDs, only ZCN14 was expressed in leaves at the second and third days after sowing. When the leaves emerged from the soil on the fifth day after sowing and became photosynthetically competent, ZCN8 and ZCN18 mRNA was detected. Transcripts from both genes accumulated to moderately high levels after the floral transition at stages V1 and V2. ZCN26 transcript was only weakly detected in green leaves after the floral transition. Under LDs, the floral transition occurred at the same stage as in SDs, and the overall transcription patterns for the ZCN genes surveyed were similar to those under SDs. A low level of ZCN8 mRNA was detected earlier than in SDs in the coleoptile-wrapped leaf at the second day after sowing. After leaves fully emerged from the coleoptile and became green, ZCN8 expression was high at all stages. No transcripts of ZCN7, ZCN12, and ZCN15 were detected in leaves under either SDs or LDs (Figure 1A).
Figure 1.
Expression Patterns of Seven FT-Like ZCN Genes in Leaves of Four Maize Lines under SDs and LDs.
On the top of each panel the images represent the shoot apices at stages when leaf blades were collected. Black arrows and thin vertical lines demarcate the timing of the floral transition. Vegetative growth stages (V stages) were defined according to the full extension of the leaf collar of the uppermost leaf where the first V stage is designated as VE (emergence). The superscript numbers 1, 2, 3, and 4 mark early and later stages of the same V stage, as judged by different internode lengths between the uppermost leaf and the leaf below it. RT-PCR products were detected on ethidium bromide–stained agarose gels. The first and last lanes of each gel image are a DNA marker and genomic PCR product, respectively. Asterisks mark genes with unspliced transcripts. Bars = 100 μm.
(A) RT-PCR of FT-like ZCN genes in the early flowering Gaspé Flint line. Due to transition to reproductive development at the VE stage, leaves were sampled by days after seed sowing (DAS) starting with day 3. The tissues collected at the third and fourth DAS were leaves wrapped in the coleoptile. After that, the tissues collected were fully emerged leaf blades. Leaves at the fifth day after sowing were collected in the morning and the afternoon separately.
(B) RT-PCR of FT-like ZCN genes in the temperate line B73.
(C) RT-PCR of FT-like ZCN genes in the tropical line CML311.
(D) RT-PCR of FT-like ZCN genes in the tropical line CML436.
The floral transition in the temperate mid-maturity B73 line occurs at stages V4-V5 under both SDs and LDs (Figure 1B). ZCN8, ZCN18, and ZCN26 transcripts were detected before and after the floral transition in fully mature green leaves under both photoperiods. ZCN8 mRNA accumulation was low at early time points, increased modestly prior to the floral transition, and was abundant and stable thereafter. ZCN7 transcripts were not detected under SDs, but its unspliced form was found under LDs. ZCN12 was not expressed at stages V2-V5, but mRNA was detected later at stages V6-V10 that is consistent with our previous results (Danilevskaya et al., 2008a). No ZCN14 and ZCN15 mRNAs were detected at vegetative stages, but a low level of transcript could be seen after the floral transition. ZCN18 and ZCN26 were constitutively expressed in leaves across all stages under both SDs and LDs (Figure 1B).
Under SDs, the floral transition in the tropical lines CML311 and CML436 takes place around stages V4-V5, which is similar to the temperate B73 line (Figures 1C and 1D). Under SDs, the expression patterns of the ZCN genes were nearly identical in both lines and very similar to their patterns in B73. In the tropical line CML436, unspliced ZCN8 RNA was detected at early vegetative stages, but fully spliced mRNA was detected prior to and after the floral transition.
Under LDs, CML311 and CML436 transitioned at stages V7-V8 and V10-V11, respectively. The expression patterns of ZCN8, ZCN12, ZCN15, ZCN18, and ZCN26 were similar to their patterns under SD conditions. Unspliced ZCN7 mRNA was present only in CML436 grown under LDs. However, under LDs, a novel ZCN14 mRNA pattern was observed in both tropical lines. The abundance of ZCN14 mRNA gradually increased before the floral transition preceding the onset of the ZCN8 expression, suggesting a possible role of ZCN14 in promoting flowering under LDs.
Overall, our transcriptional analyses revealed that three genes, ZCN8, ZCN18, and ZCN26, displayed expression patterns in the mature leaf before and after the floral transition that are consistent with a hypothetical function as a floral activator. However, the expression pattern of ZCN14 suggests it may also play a role in the floral transition under LDs in some tropical lines.
Responses of FT-Like ZCN Genes to a Floral Inductive SD Treatment
Arabidopsis plants grown under nonpermissive SDs but exposed to three inductive LDs will express FT and flower (Corbesier et al., 2007). Conversely, exposure of the obligate SD plant goosefoot (Chenopodium rubrum) to LDs will induce FT-like gene expression resulted in early flowering (Cháb et al., 2008). Tropical maize varieties are either moderately or highly sensitive to photoperiod and will flower earlier under SDs than under LDs. We conducted an SD inductive experiment to assess the response of two tropical lines to a limited exposure of SD photoperiod.
Pilot experiments indicated that an SD treatment of either 3 or 5 d was not long enough to induce an earlier floral transition in the tropical lines CML311 and CML436 grown under continuous LDs (see Supplemental Figure 2 online). However, after a treatment of seven SDs, the SAM started transitioning in both CML311 and CML436 at growth stage V5, which is earlier than controls grown under continuous LDs (Figure 2). The inflorescence meristem continued to develop into tassel primordial, indicating that the transition to reproductive growth was complete and irreversible. The expression patterns of the FT-like ZCN genes were determined in leaf blade tissue and shoot apices dissected from plants grown under continuous LDs but treated with seven SDs (Figure 2). Out of seven ZCN genes assayed, only ZCN8 and ZCN12 mRNA were induced in leaves of plants induced by a transient SD treatment (Figures 2A and 2B). ZCN8 mRNA was detectable after one (CML436) or three (CML311) SD treatments, and its abundance sharply increased during the rest of the treatment time period. The induction of ZCN12 mRNA was observed 2 to 3 d later than ZCN8. After return to LDs, ZCN8 mRNA level dropped significantly but was still detectable, while ZCN12 expression was not detectable.
Figure 2.
Effect of an Inductive SD Treatment on Gene Expression in Tropical Lines.
(A) and (B) RT-PCR of FT-like ZCN genes in leaf blade of CML311and CML436 plants grown for 4 weeks under LDs, followed by seven SDs and then transfer back to LDs for 1 month. The images on the top of each panel represent development of shoot apices at specific growth stages when leaf blades were collected. The images marked by the superscript letter “a” indicate leaves collected every other day. The superscript numbers 1, 2, and 3 mark early and later growth stages of the same V stage, as judged by internode lengths between the uppermost leaf and the leaf below it. The bracket and the solid lines indicate the growth stages that underwent the SD treatment. RT-PCR products were detected on ethidium bromide–stained agarose gels. The first and last lanes of each gel image are marker DNA and genomic PCR, respectively.
(C) and (D) RT-PCR of FT-like ZCN genes and the flowering MADS box gene ZMM4 in shoot apices of the same plants as described above.
Bars = 100 μm.
Transcript accumulation of ZMM4, a floral transition MADS box gene (Danilevskaya et al., 2008b), was assayed in shoot apices sampled from the same plants given the SD inductive photoperiod treatment (Figures 2C and 2D). ZMM4 transcript was detected in shoot apices during and after the SD treatment. Unspliced ZCN7 and ZCN8 transcripts were always present in shoot apices, but mature ZCN8 mRNA was only detected in leaves. This observation hints that ZCN8 expression might be also regulated via pre-mRNA splicing. ZCN15, ZCN18, and ZCN26 transcripts were not detected in shoot apices. Notably, ZCN14 was the only FT-like ZCN gene whose fully spliced transcript was detected in shoot apices. Mature ZCN14 mRNA was detected in shoot apices after the floral transition, and this expression pattern placed this gene apart from the other FT-like ZCNs, as none produced fully spliced mRNA in the shoot apex.
To quantify the effect of the inductive SD treatment on flowering time, leaf number was counted for plants grown under LDs and treated for seven SDs (Table 1). CML311 and CML436 plants produced an average of 21 and 23 leaves, respectively, when grown under continuous LDs. Plants of both lines given the 7-d SD treatment produced 19 leaves on average, which was significantly fewer than those of controls. Thus, the SD treatment induced an earlier floral transition. The inductive SD experiment narrowed down the list of candidate floral activators to ZCN8 as the only FT-like gene whose expression pattern was the most consistent as functioning as a floral activator under SDs.
Table 1.
Leaf Number of CML311 and CML436 Grown under an Inductive Seven SD Treatment Compared with Continuous LDs
| Tropical Lines | Leaf No. | No. of Plants |
| CML311-SDs inductive | 19 ± 0.48 | 10 |
| CML311-LDs continuous | 21 ± 0.87a | 10 |
| CML436-SDs inductive | 19 ± 0.67 | 10 |
| CML436-LDs continuous | 23 ± 2.21a | 10 |
Leaf numbers were collected from at least 10 individual plants. Mean values and standard deviations were calculated by analyses of variance from the Minitab statistical program.
Means are statistically significant with P < 0.05.
Diurnal Expression of Three FT-Like ZCN Genes under SD and LD Photoperiods
A hallmark of Arabidopsis FT gene expression, which directly contributes to its function as a floral activator, is its diurnal, circadian pattern with transcript abundance peaking at dusk under permissive LDs (Turck et al., 2008). Rice Hd3a mRNA accumulation is also diurnally regulated under SDs with a peak of expression just before dawn (Kojima et al., 2002). ZCN8, ZCN18, and ZCN26 transcripts accumulate in leaves, before and after the floral transition, from plants grown under either SD or LD photoperiods. We investigated whether their mRNA accumulation showed a diurnal expression pattern in leaf blades of Gaspé Flint, B73, and CML436 by sampling plants every 4 h during 3 d of vegetative growth for a set of plants grown under LDs and a set of plants grown under SDs (Figure 3). We also tested expression of the maize homologs of GIGANTEA (Gigz1) and CO (Conz1) in the same tissues (Miller et al., 2008). In our experiments, these genes showed a diurnal pattern of expression as was described previously (Miller et al., 2008) (see Supplemental Figure 3 online).
Figure 3.
Diurnal Expression Patterns of Three FT-Like ZCN Genes in Leaf Blades of Three Inbred Lines under SDs or LDs during Vegetative Growth.
Relative expression levels were determined by quantitative RT-PCR normalized to ubiquitin5 (the y axis). The x axis represents time point (hours). Data points represent an average of three biological replicates with three technical replicates. Error bars represent sd. The shaded bars over each chart represent dark periods.
In the extremely early flowering day-neutral genotype Gaspé Flint, ZCN8 mRNA accumulation did not display a consistent diurnal oscillation under SDs or LDs, although accumulation tended to be higher at night (Figure 3). In the day-neutral line B73, ZCN8 mRNA expression had a weak diurnal expression pattern with low amplitude under both photoperiods but with expression peaking just before dawn. In B73, the oscillation of ZCN8 under SDs was more evident than that under LDs; however, the lowest level of ZCN8 transcript accumulation never reached zero.
In the tropically adapted, photoperiod-sensitive CML436 line, expression of ZCN8 under SDs was strikingly distinct (Figure 3). ZCN8 transcript abundance under SDs showed a clear diurnal pattern peaking at dawn. During the day, ZCN8 transcript accumulation sharply declined and 8 h after dawn ZCN8 transcript was too low to be detected. Under LDs at stage V5, when the line is in a vegetative growth stage, ZCN8 transcript accumulation was very low but was still diurnally expressed if graphed at an expanded scale (see Supplemental Figure 4A online). However, when leaves were sampled during the floral transition under the LDs, the amplitude of the diurnal level of ZCN8 mRNA was comparable to amplitude under SDs. In addition, the peak of transcript accumulation shifted to 4 h after dawn even though transcript accumulation at dusk remained undetectable (see Supplemental Figure 4B online).
The expression of ZCN18 did not exhibit any consistent diurnal patterns in any inbred lines under either photoperiod (Figure 3). Notably, its expression level was very high in all lines except B73. Under SDs, ZCN26 displayed a diurnal expression pattern in B73 and CML436 with expression peaking at dawn. This was in contract with expression in Gaspé Flint where ZCN26 mRNA was barely detectable. ZCN26 mRNA did not show a diurnal expression pattern under LDs in any of the three lines tested. Thus, only ZCN8 mRNA displayed an obvious and consistent diurnal expression pattern in the tropical line CML436 under both SD and LD photoperiods.
Expression of FT-Like ZCN Genes in B73 Wild Type and dlf1 and id1 Late-Flowering Mutants
We examined expression of seven FT-like ZCN genes in both id1-m1 and dlf1-N2461A late-flowering mutants to establish their genetic interactions. Homozygous mutants introgressed into the B73 inbred background were grown and tissues sampled as described previously (Danilevskaya et al., 2008b). The morphological development of the SAM of wild-type B73, dlf1, and id1 at different stages are shown in Figure 4A. The floral transition occurs 2 weeks later in dlf1 and 1 month later in id1 compared with the wild type. RT-PCR was performed using leaf blade tissue (Figure 4B). ZCN7, ZCN14, and ZCN15 mRNA were not detectable in any genotype. ZCN26 transcript was detected in all genotypes with a similar accumulation pattern, indicating its expression is independent of the id1-dlf1 pathway. ZCN18 showed an inconsistent pattern of expression that was difficult to interpret. Interestingly, ZCN12 was ectopically expressed in the dlf1 mutant, pointing to the possibility of regulation by dlf1.
Figure 4.
FT-Like ZCN Gene Expression in Leaves of the Wild Type and Late-Flowering Mutants.
(A) Images of shoot apices at growth stages (days after sowing) when leaves were collected. Red arrows mark the floral transition. WT, wild type.
(B) Expression of FT-like ZCN genes in leaf blades from wild-type B73, dlf1-N2461A, and id1-m1. The first and last lanes of each gel image are marker DNA and genomic template DNA, respectively.
(C) Quantitative RT-PCR of ZCN8 and id1 in dissected sections of B73 plants at V5. The images on the top show sections numbered from 1 to 17. Section 1 represents the shoot apex with the youngest enclosing leaf primordia. Section 2 is 1 cm of immature leaves around the shoot apex. Sections 3 to 6 are immature leaves, each of 4 cm length where 3 is the closest one to shoot apex. Section 7 is the transitional part of the sixth and seventh leaves. Section 8 is mature, photosynthetically competent blade of 6th leaf. Section 9 is the sheath of the 5th leaf. Sections 10 to 12 are parts of the mature leaf blade of the fifth leaf, each 12 cm in length. Section 13 is the tip of the mature blade of the fifth leaf. Section 14 is the sheath of the 4th leaf. Sections 15 to 16 are sections of the mature leaf blade of the 4th leaf, each 12 cm in length. Section 17 is the tip of the 4th leaf blade. On the x axis the numbers are marking the dissected tissue sections. The y axis represents the relative expression level normalized to ubiquitin5. Error bars represent sd. Bars = 18 cm.
Of the ZCN genes assessed, only ZCN8 displayed a clear and distinct pattern of expression in the two late-flowering mutants. In B73 plants, transcript accumulation in leaves was first detected on the 17th day after sowing, and accumulation increased during development, which is consistent with results shown in Figure 1B. In the dlf1 mutant, ZCN8 was expressed in leaves at all time points tested. In the id1 mutant, ZCN8 mRNA was not detected until the last stage that was well after the floral transition. This result suggested that ZCN8 is downstream of id1 and upstream or in parallel to dlf1.
Given the placement of ZCN8 downstream of id1, we asked if ZCN8 or any of the other FT-like ZCNs surveyed were coexpressed with id1 in immature leaves, the tissue where id1 mRNA is predominantly produced (Colasanti et al., 1998). For this reason, expression of the seven FT-like ZCN genes was tested in immature leaves. None of the seven ZCN genes tested was expressed in immature leaves (see Supplemental Figure 5 online). Because ZCN14 mRNA was expressed in the shoot apex, its expression was examined in wild-type and id1 mutant apices. ZCN14 transcript was abundant in both wild-type and mutant shoot apices after the floral transition, indicating its expression is independent of the id1-dlf1 pathway (see Supplemental Figure 6 online).
id1 is expressed in the immature leaf, but ZCN8 transcripts are mainly detected in the mature leaf. To determine if expression of both genes overlaps at some position along the gradient of the developing leaf, leaves from B73 plants at stage V5 were dissected into precise sections. From stage V5 plants, leaf 4 and leaf 5, the youngest, uppermost leaf with an exposed auricle/ligule (collar) region, were dissected into basal leaf sheath tissue and 12-cm sections of mature, distal leaf blade tissue (Figure 4C, sections 9 to 17). The next youngest leaf, leaf 6, with a photosynthetically competent distal blade tip but still immature basal region, was dissected into 4-cm sections of basal immature leaf tissue, a 20-cm section of transitional leaf tissue, and a 20-cm section of mature leaf blade tissue (sections 3 to 8). The youngest leaf primordia enclosing the SAM and a section with the adjacent one to two more developed leaves above the apex comprise the last two samples (sections 1 to 2). The abundance of ZCN8 and id1 transcript in each section was analyzed by quantitative RT-PCR (Figure 4C). Transcripts for both genes were not detected in the shoot apex and next adjacent section of immature leaf (sections 1 and 2). In the sixth leaf, id1 was expressed in the immature sections (sections 3 to 6) and the transitioning section (section 7) with expression barely detectable in the green, mature leaf blade (section 8). In the more mature fourth and fifth leaves, no id1 mRNA was detected in either the leaf sheath or leaf blade (sections 9 to 17). Overall, id1expresion was highest in immature leaf sections 4 to 8 cm above the SAM with expression decreasing in more mature sections of the leaf.
ZCN8 expression, on the other hand, displayed an expression pattern complimentary to that of id1. No expression was detected in the immature leaf sections up to 4 cm above the shoot apex (sections 1 to 3). Rather, transcripts were first detected in immature leaf sections 4, 5, and 6. Transcripts were also detected in transitioning leaf tissue (section 7) with the highest accumulation in the mature leaf tissue of section 8. ZCN8 mRNA was consistently detected in the mature leaf blade sections (10 to 13 and 15 to 17) of leaf 4 and 5 but not the leaf sheath. Overall, throughout the basal to distal developmental gradient of the leaf, id1 expression is highest in basal immature leaf sections and diminishes in more distal mature blade, while ZCN8 expression is low in basal sections and increases in more mature distal tissue. Hence, during leaf development, expression of id1 precedes expression of ZCN8.
Interaction of FT-Like ZCN Proteins with the DLF1 Protein
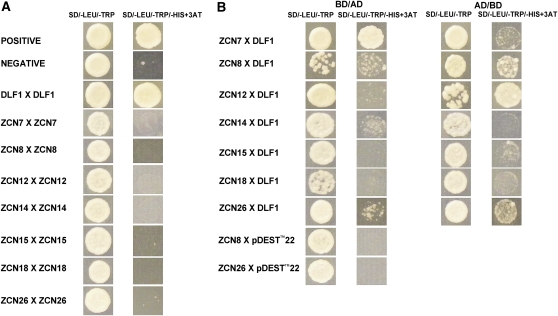
In Arabidopsis, the FT protein interacts in the SAM with the bZIP transcription factor FD to activate flower morphogenesis (Abe et al., 2005; Wigge et al., 2005; Corbesier et al., 2007; Jaeger and Wigge, 2007). Previously, we have shown that the ZCN8 protein interacts with the DLF1 protein, which is the presumed maize FD ortholog (Danilevskaya et al., 2008a). Yeast two-hybrid analysis was used to test the seven candidate FT-like ZCN proteins for interactions with DLF1. DLF1 showed self-interaction, which is typical for bZIP transcription factors that normally function as dimers. None of the ZCN proteins showed self-interactions (Figure 5A). ZCN7, ZCN8, and ZCN26 showed interactions with DLF1 in reciprocal yeast two-hybrid reactions, whereas, ZCN12, ZCN15, and ZCN18 only interacted with DLF1 in one bait-prey combination, indicating that the later three ZCNs might interact weakly with DLF1 (Figure 5B).
Figure 5.
Yeast Two-Hybrid Interactions between FT-Like ZCN Proteins and the DLF1 Protein.
(A) FT-like ZCN and DLF1 self-interaction in yeast.
(B) FT-like ZCN and DLF1 reciprocal interactions in yeast.
BD, binding domain; AD, activation domain; SD, synthetic dextrose minimal medium. (More details are in Methods.)
[See online article for color version of this figure.]
Spatial Expression Patterns of Select FT-Like ZCN Genes in Temperate and Tropical Maize during the Floral Transition
The transcripts of Arabidopsis FT and rice Hd3a and RFT1 are mainly detected in the phloem of the leaf (An et al., 2004; Tamaki et al., 2007; Komiya et al., 2009). Based on our analyses, ZCN8 was the most favorable candidate for a floral activator in maize. Thus, we determined the spatial expression pattern of ZCN8 using in situ hybridization analysis. Because ZCN7 and ZCN26 interact with the DLF1 protein in the yeast two-hybrid analyses, ZCN7 and ZCN26 were also included in the in situ hybridization experiment. ZCN7 and ZCN8 are duplicated genes sharing 94% identity at the nucleotide level (Danilevskaya et al., 2008a). Because mature, spliced ZCN7 mRNA was never detected in leaf tissues, sequences from the third intron of ZCN7 were used as an RNA probe for in situ hybridization to distinguish between these two paralogs. No signal was detected in any leaf tissues using the ZCN7 intron probe (see Supplemental Figure 7A online). Because ZCN8 transcript is abundant in the distal region of mature leaf blades, this tissue was used for in situ hybridization. The distal leaf blade region from the temperate lines Gaspé Flint and B73 were harvested before and after the floral transition from plants grown under LDs. Similar tissues were harvested from the tropical line CML436 before the floral transition from plants grown under SDs. Hybridization with the ZCN8 RNA antisense probe revealed signal over vascular bundles within transverse leaf sections in all genotypes tested (Figures 6A, 6C, and 6E). At higher magnification, signal was detected primarily in phloem, xylem parenchyma, and epidermal cells (Figures 6B, 6D, and 6F). Stages before (V4) and after (V6) the floral transition in B73 were also examined (see Supplemental Figure 8 online). Before the floral transition, the ZCN8 signal was detected in the phloem (see Supplemental Figures 8B and 8C online). At later stages, after the floral transition, signal was detected in additional cells, including the xylem parenchyma and sclerenchyma cells on the adaxial and abaxial sides of the vascular bundles (see Supplemental Figures 8E and 8F online). In Gaspé Flint, when the SAM had already developed into tassel primordia, long after the floral transition, ZCN8 signal was detected only in sclerenchyma cells on the both sides of the vascular bundles (see Supplemental Figures 9A and 9B online).
Figure 6.
RNA in Situ Hybridization of ZCN8 and ZCN26 in Leaf Tissue from Gaspé Flint, B73, and CML436 during the Floral Transition.
(A), (C), and (E) Hybridization signal of ZCN8 DIG-labeled antisense probe in transverse sections of the leaf blade tip from Gaspé Flint at 5 d after sowing (A), B73 at stage V4 (C), and CML436 at stage V5 (E) under SDs. The negative controls for hybridization with sense strand are shown in Supplemental Figure 7B online.
(B), (D), and (F) The ×40 magnification of the individual vascular bundle from near the midrib (marked by circles in [A], [C], and [E]) showing signal in the phloem, xylem parenchyma cells, and in the sclerenchyma fibers ([B] and [D]).
(G), (I), and (K) Hybridization signal of the ZCN26 DIG-labeled RNA antisense probes in transverse sections of the leaf blade tip described above.
(H), (J), (L), and (M) The ×40 magnification of an individual vascular bundle near the midrib or major vein (marked by circles in [G], [I], and [K]) showing signal in the phloem, the xylem parenchyma cells, and the sclerenchyma fibers ([H] and [J]).
bs, bundle shealth; m, metaxylem vessels; pxv, protoxylem vessels; p, phloem; sf, sclerenchyma fibers; xp, xylem parenchyma. Bars = 200 μm in (E) and (K), 100 μm in (A), (C), (G), and (I), and 50 μm in (B), (D), (F), (H), (J), and (M).
ZCN26 displayed spatial expression patterns similar to ZCN8. Hybridization signal was detected over the vascular bundles (Figures 6G, 6I, and 6K). Detailed images at a higher magnification showed signal over phloem, xylem parenchyma, and sclerenchyma cells (Figures 6H, 6J, 6L, and 6M).
Flowering Phenotype of Transgenic Plants Ectopically Expressing ZCN Genes
As a functional test of the candidate ZCN genes, their ectopic expression in transgenic plants was exploited. If the candidate gene functions as a floral activator and is ectopically expressed earlier in development in the shoot apex, an early flowering phenotype should be observed. We previously attempted to overexpress most of the FT-like ZCN genes using the ubiquitin (Ubi) promoter, which is a strong constitutive promoter routinely used for transgenic studies in maize (McElroy and Brettell, 1994). To our surprise, only ProUBI:ZCN18 and ProUBI:ZCN26 transgenic plants were obtained as plants presumably expressing the other ZCN constructs failed to regenerate plants from callus (Danilevskaya et al., 2010). We also tried using a weaker, constitutive promoter, ZM-GOS2, to ectopically express the ZCN genes that failed to regenerate plants using the Ubi promoter (Barbour et al., 2003). However, transgenic plants were only generated with this weaker promoter driving expression of the genomic sequence of ZCN7 (ZCN7g). Even though endogenous ZCN7 does not produce fully spliced mRNA in any of the tissues we tested, ProZM-GOS :ZCN7g transgenic plants did unexpectedly produce three independent events with spliced mature mRNAs (see Supplemental Figure 11A online). Transgenic plants bearing ProUBI:ZCN18, ProUBI:ZCN26, or ProZM-GOS :ZCN7g constructs produced the same number of leaves as their nontransgenic siblings. Hence, ectopic expression of these three ZCN genes did not alter the floral transition and appear to play no apparent role in flowering time regulation.
To recover transgenic plants ectopically expressing a few of the ZCN genes, we next tried two tissue-specific promoters, ProZMM4 and ProZM-ADF4 (Zea mays Actin Depolymerizing Factor 4) (Bate and Reimann, 2009). ProZMM4 will condition expression in the shoot apex, including the SAM and nascent leaf primordia near the time of the floral transition (Danilevskaya et al., 2008b). Using a ProZM-ADF4:GUS construct, we found this promoter drives strong expression of the reporter β-glucuronidase (GUS) in the shoot apex as early as stage V3 and expression continued in these tissues at later stages (see Supplemental Figure 10 online). We elected to try both promoters to drive expression of the most favorable candidate ZCN8. ProZMM4:ZCN8 and ProZM-ADF4:ZCN8 constructs were made, transgenic T0 plants regenerated, and single transgene copy events were outcrossed to produce segregating T1 progeny, which were evaluated for flowering time phenotypes. Expression of the transgene was confirmed by RT-PCR in all events selected for phenotyping (see Supplemental Figure 11A online). Leaf numbers were counted as a measure of the timing of the floral transition. Transgenic plants with either construct transitioned earlier, resulting in plants with fewer leaves (Table 2). The average leaf number of ProZMM4:ZCN8 plants was 18, one leaf fewer than their nontransgenic siblings, while the average leaf number of ProZM-ADF4:ZCN8 plants was 17, two leaves fewer than their nontransgenic siblings. These differences were statistically significant. The earlier flowering phenotype of the ProZM-ADF4:ZCN8 plants are consistent with a relatively higher and earlier ectopic ZCN8 expression in the shoot apex compared with the ProZMM4:ZCN8 plants (see Supplemental Figures 10 and 11B online). A representative image of ProZM-ADF4:ZCN8 transgenic plant is shown in Supplemental Figure 12 online. These results demonstrate that ectopic expression of ZCN8 in shoot apices prior the floral transition promotes early flowering. To downregulate ZCN8, we designed an artificial microRNA (Schwab et al., 2006). ZCN8 mRNA level is reduced in transgenic amiR plants (see Supplemental Figure 11 online). amiRNA transgenic plants showed the late-flowering phenotype producing three to four more leaves relative to a nontransgenic control (Table 2). Thus, transgenic results strongly support ZCN8 function as a floral activator.
Table 2.
Leaf Number of Transgenic Maize Plants
| Constructs | Background | No. of Plants | Leaf No. |
| NTG siblings | Temperate | 32 | 19 ± 0.6 |
| TG ProZMM4:ZCN8 | Temperate | 34 | 18 ± 0.6a |
| NTG siblings | Temperate | 12 | 19 ± 0.6 |
| TG ProZM-ADF4:ZCN8 | Temperate | 13 | 17 ± 0.6b |
| NTG siblings | Temperate | 30 | 19 ± 0.4 |
| TG ProGOS2:ZCN7g | Temperate | 30 | 19 ± 0.6 |
| NTG siblings | Early temperate | 28 | 10 ± 0.8 |
| TG ProUBI:ZCN18 | Early temperate | 20 | 10 ± 0.8 |
| NTG siblings | Temperate | 32 | 19 ± 0.5 |
| TG ProUBI:ZCN26 | Temperate | 36 | 19 ± 0.6 |
| NTG siblings | Early temperate | 30 | 9 ± 0.6 |
| TG ProUBI:amiR-ZCN8 | Early temperate | 27 | 13 ± 0.6 b |
All constructs were expressing cDNA with exception of ZCN7g, which was a genomic fragment including exons and introns. Leaf numbers were collected from at least 10 individual plants from two independent events. Mean values and standard deviations were calculated by the analyses of variance from the Minitab statistical program. NTG, nontransgenic sibling plants; TG, transgenic plants.
Means are statistically significant with P < 0.1.
Means are statistically significant with P < 0.05.
DISCUSSION
FT-Like ZCN Genes Display Functional Diversification
The flowering hormone florigen appears to be universal in flowering plants (Shalit et al., 2009), and its genetic regulation and a mode of action have been deciphered in a number of species including Arabidopsis, rice, and tomato. According to the current model established in these species, florigen is a mobile PEBP protein encoded by one (or more) FT-like gene that is transcribed and translated in the leaf vasculature and then moves via the phloem to the SAM where it interacts with an FD-like bZIP transcription factor to induce flower development (Abe et al., 2005; Wigge et al., 2005; Turck et al., 2008). However, it is still unclear which FT orthologous gene(s) in maize function as the mobile flowering signal. This knowledge is essential for understating the genetic regulation of flowering time in this important agronomic and model crop species. Here, we provided evidence that at least one maize gene, ZCN8, possesses nearly all the characteristics consistent with florigenic function.
Assuming the mechanism of florigen activity and movement is evolutionarily conserved, we defined a number of criteria for the functional analysis of a number of FT-like ZCN genes in maize. A ZCN gene with florigenic activity should (1) be expressed in leaves, (2) be transcriptionally responsive to inductive photoperiods in sensitive genotypes, (3) induce early flowering when ectopically expressed in the shoot apex of transgenic plants, (4) encode a protein that interacts with the DLF1 protein (the maize FD ortholog), (5) function genetically within a defined flowering network, and (6) encode a protein that moves through the phloem to the shoot apex. We were able to apply five of these six criteria to the analyses of seven maize FT-like ZCN genes. Six of them, ZCN7, ZCN8, ZCN12, ZCN14, ZCN18, and ZCN26, were chosen due to their expression in leaves in at least one developmental stage near the time of the floral transition (Danilevskaya et al., 2008a). In addition, the ZCN15 gene was included in this study as it is syntenic to the rice flowering QTL Hd3, which harbors the rice FT-like genes Hd3a and Hd3b (also known as RFT1) (Tsuji et al., 2008; Komiya et al., 2009). A summary of our functional analyses of the seven FT-like ZCN genes is shown in Table 3.
Table 3.
Summary of the Functional Analysis of Seven FT-Like ZCN Genes
| Gene | Tissue mRNA Detected | Timing mRNA Detected | Diurnal Pattern | SD Induction | mRNA in id1 Mutant | mRNA in dlf1 Mutant | DLF1 Interaction | TG* |
| ZCN7 | Unspliced | n/a | n/a | n/a | n/a | n/a | Weak | Neutral |
| ZCN8 | Leaf | Prior FT | SD; LD | Induced | NO | Detected | Strong | Early |
| ZCN14 | SAM | After FT | n/a | Induced | Detected | Detected | Weak | n/a |
| ZCN15 | Kernel | n/a | n/a | n/a | n/a | n/a | NO | n/a |
| ZCN12 | Leaf | After FT | n/a | Induced | n/a | Induced | Weak | n/a |
| ZCN18 | Leaf | All times | LD | NO | Detected | Detected | NO | Neutral |
| ZCN26 | Leaf | All times | SD | NO | Detected | Detected | Strong | Neutral |
FT, floral transition; n/a, not applicable; TG*, flowering phenotype of transgenic plants that ectopically expressed ZCN genes.
Out of seven ZCN genes selected for functional analyses, only ZCN8 satisfied all five of our criteria for possessing florigenic activity. ZCN8 transcript gradually accumulates in leaves at vegetative stages and reaches its highest expression level at or near the floral transition in all genotypes tested. ZCN8 mRNA accumulates rapidly in response to a floral-inductive SD treatment in photoperiod-sensitive tropical lines, correlating with an earlier floral transition. The ZCN8 protein interacts with the DLF1 protein in the yeast two-hybrid assay and functions genetically downstream of the id1 floral activator in the id1-dlf1 flowering network. Furthermore, ectopic expression of ZCN8 in the shoot apex during vegetative growth stages induced early flowering in transgenic plants. Collectively, these data are consistent with ZCN8 functioning as a floral activator. Notably, its duplicate, the ZCN7 gene, appears to be nonfunctional, as fully spliced ZCN7 mRNA was not detected by RT-PCR from any of the tissues we tested under normal growth conditions. Nevertheless, in transgenic plants overexpressing a genomic ZCN7 sequence, properly spliced mRNA was produced in leaves. This finding points toward the possibility that under some specific conditions, ZCN7 might play a functional role and its function might be regulated by RNA splicing. However, we observed that properly spliced ZCN7 mRNA in transgenic plants did not have an impact on flowering time, suggesting this gene appears to not play a role in the floral transition.
Surprisingly, the ZCN15 gene, which is syntenic to the major rice flowering quantitative trait loci on chromosome 6, Hd3a and Hd3b, also appears not to be involved in regulating flowering time. Consistent with our previous data, no ZCN15 mRNA was detected in leaves from four diverse maize genotypes, as its expression is rather restricted to early stages of kernel development (Danilevskaya et al., 2008a). When during the evolution of the Poaceae ZCN15 might have acquired a function in developing kernels is an intriguing question to study. Identification of functional FT equivalents in teosinte and related species like sorghum (Sorghum bicolor) might help to answer this question. The second closest rice FT homolog, ZCN14, is expressed predominantly in the shoot apex after the floral transition in temperate maize lines and in tropical lines grown under SDs. However, in tropical lines grown under LDs, its mRNA was detected in leaves prior to the floral transition and in the apex after the transition. This observation suggests a putative function for ZCN14 in LD photoperiod response in tropical lines, but this hypothesis requires more rigorous study.
ZCN12 mRNA is detected in leaves at reproductive stages in temperate and tropical lines under SDs. Its expression is induced by the SD treatment in tropical lines, indicating that ZCN12 expression is regulated by SD photoperiods in these genotypes. Interestingly, its mRNA is ectopically expressed in leaves in the dlf1 mutant, suggesting a genetic interaction between ZCN12 and dlf1 where dlf1 may be a repressor of ZCN12 expression. Moreover, a weak interaction was observed between the ZCN12 and DLF1 proteins using yeast two-hybrid assay. This finding suggests ZCN12 might have a role in reproductive development and is therefore an unlikely candidate for functioning as a floral activator.
Both ZCN18 and ZCN26 appear to be constitutively expressed in leaves in all growth stages and genotypes studied. Although ZCN18 transcript accumulated to a higher level than the other leaf-expressed ZCNs, its diurnal regulation was inconsistent, its ectopic expression did not affect flowering time, it did not interact genetically with the id1-dlf1 network, and its protein did not interact with the DLF1 protein. Thus, ZCN18 function appears to be independent of the flowering time network and remains to be understood. Although ectopic expression of ZCN26 did not affect flowering time, endogenous expression showed a distinct diurnal pattern under SDs, and the ZCN26 protein interacts strongly with DLF1. ZCN26 transcripts accumulate in leaf phloem cells but are not regulated by either id1 or dlf1. Thus, ZCN26 is an intriguing candidate for further functional study.
Each of the seven ZCN genes displays a unique combination of spatial, temporal, and photoperiod-specific expression that implies functional diversification. However, only ZCN8 has the requisite characteristics that are consistent with it possessing florigenic activity. A recent report from beet showed divergent roles of FT orthologs (Pin et al., 2010). However, ZCN8 has different amino acids in the critical position that defines the antagonistic function of Bv FT1 and Bv FT2 proteins (see Supplemental Figure 13 online). Those residues are neither common to the Bv FT2 activator nor to the Bv FT1 repressor. This could be interpreted that this position has evolved enough flexibility for FT proteins function as a repressor or an activator of flowering.
ZCN8 Is Expressed in Leaf Phloem and Induces Early Flowering When Ectopically Expressed in the Shoot Apex
One essential functional feature of FT genes is their expression in the phloem of the leaf vasculature and transport of the translated protein to the shoot apex. Due to low FT expression in Arabidopsis and rice, indirect techniques, such as promoter-GUS/green fluorescent protein fusions, were employed to visualize cell-specific FT expression (Corbesier et al., 2007; Tamaki et al., 2007; Komiya et al., 2009). By contrast, we were able to detect ZCN8 expression in the leaf directly by RNA in situ hybridization. An obvious ZCN8 signal was reproducibly observed in the major and minor veins of leaves sampled from different genotypes at or near the floral transition. At a higher magnification, signal was evident in the phloem, xylem parenchyma, and sclerenchyma cells of the vascular bundles. We investigated the dynamics of ZCN8 spatial expression in the vasculature at several developmental stages in B73. ZCN8 signal appeared first in the phloem at vegetative stage V4. Later, during the time of the floral transition, ZCN8 signal was not only seen in the phloem but was also detected in the xylem parenchyma and the sclerenchyma cells. At an even later reproductive stage (in the early temperate line Gaspé Flint), ZCN8 signals persisted only in the sclerenchyma fiber cells. We speculate that ZCN8 expression in the phloem is required prior to and during the floral transition but at later reproductive stages, ZCN8 is expressed in the sclerenchyma cells where its function, if any, is not clear.
To induce the floral transition, the FT protein must move to the shoot apex. Ectopic expression of both FT and Hd3a driven by phloem-specific promoters was used to induce early flowering in transgenic Arabidopsis and rice, respectively (An et al., 2004; Tamaki et al., 2007; Turck et al., 2008). Unfortunately, phloem-specific promoters are not available in maize. For this reason, two well-characterized promoters, ProZMM4 and ProZM-ADF4, were used to drive expression in the shoot apex. Ectopic expression of ZCN8 in the shoot apex at vegetative stages before the floral transition using the ProZM-ADF4:ZCN8 construct promotes early flowering. Conversely, the amiR downregulation of ZCN8 mRNA resulted in the late-flowering phenotype. These findings are direct evidence that ZCN8 has floral-inductive activity in maize. Although ZCN8 expression was very high in the shoot apex with this construct (see Supplemental Figure 11B online), it did not produce an extremely early flowering phenotype similar to Gaspé Flint. To induce the floral transition, ZCN8 must interact with its partner, the DLF1 protein, in the shoot apex. The amount of DLF1 in the apex may be limiting in these early vegetative stages, as we know dlf1 transcripts accumulate to high levels in stages just preceding the floral transition (Muszynski et al., 2006). The early-flowering phenotype of the ProZMM4:ZCN8 transgenic plants were more moderate, likely due to a weaker and later activity of this promoter.
Consistent with the florigen model, ZCN8 transcript is not detected in the shoot apex at any stage of development, but ZCN8 mRNA accumulates in the leaf phloem at or near the floral transition. However, a critical but more difficult experiment is required to prove the ZCN8 protein does indeed move from the leaf to the shoot apex and thus functions as florigen in maize.
ZCN8 Transcription Is Diurnally Regulated in Photoperiod-Sensitive Tropical Lines but Not in Day-Neutral Lines
The diurnal expression of FT genes plays a key role in how flowering is regulated in photoperiod-sensitive species. In Arabidopsis, an LD plant, FT is regulated by the circadian clock through the GI-CO photoperiod pathway (Turck et al., 2008). In rice, an SD plant, there are two FT-like genes that produce a mobile florigen signal, Hd3a and RFT1 (also known as Hd3b). Under SDs, rice flowering is regulated by an SD activation pathway, predominantly via Hd3a. Under LDs, flowering is regulated by an LD suppression and activation pathway, via RFT1 (Komiya et al., 2008, 2009). Accurate daylength regulation of both FT-like genes is critical for the fine-tuning of flowering in rice to achieve the most optimal reproductive outcome (Itoh et al., 2010).
Widely grown throughout the temperate regions of the globe, modern maize varieties have been adapted to be day-neutral, although maize was originally domesticated from the obligate SD plant teosinte (Colasanti and Muszynski, 2009). Temperate maize is considered as daylength insensitive even though it produced one to two fewer leaves when grown under SD compared with LD photoperiods (Russell and Stuber, 1983). Maize genotypes adapted to growth in tropical environments retain more photoperiod sensitivity, and the most sensitive lines produce up to 30 leaves under LDs compared with 19 to 20 leaves under SDs (Russell and Stuber, 1983).
We compared ZCN8 diurnal expression patterns during vegetative growth in the temperate early flowering cultivar Gaspé Flint, the mid-maturity temperate inbred B73, and the photoperiod-sensitive tropical inbred CML436 under both SD and LD photoperiods. In Gaspé Flint, ZCN8 shows no diurnal oscillation under either daylength. In B73, ZCN8 mRNA has a weak diurnal expression pattern under SDs but not under LDs. Thus, we hypothesize diurnal oscillations of ZCN8 transcript levels may not be required in these genotypes to induce the floral transition. By contrast, in CML436, under inductive SDs, ZCN8 transcript exhibits a strong diurnal expression pattern with the maximum amplitude of expression occurring at the end of the dark period, just before dawn. After its peak in expression, ZCN8 transcript accumulation plummets to zero and stays undetectable during the rest of the light period (Figure 3A). In CML436 grown in LDs, the ZCN8 mRNA level is very low, but it still retains a diurnal rhythm with a peak of expression around dawn. During the floral transition under LDs, the amplitude of ZCN8 expression increased 50- to 100-fold compared with expression at vegetative stages under LDs. These results indicate that ZCN8 expression in photoperiod-sensitive tropical maize is tightly regulated by photoperiod. Under less inductive LDs, ZCN8 transcription is suppressed, suggesting the existence of a LD suppression pathway.
We also investigated the diurnal expression of the maize homologs of CO (conz1) and GI (Gigz1A and 1B), which are upstream of the FT-like genes in the photoperiod pathway (Miller et al., 2008). All three genes are diurnally regulated in temperate and tropical lines, suggesting that the photoperiod pathway upstream of ZCN8 is intact in temperate and tropical genotypes. The circadian clock is also intact and functional in temperate maize as was recently published for the inbred B73 (Hayes et al., 2010). This finding suggests that temperate maize may have lost photoperiod sensitivity downstream of the circadian clock but upstream of the regulation of ZCN8, which might explain why the floral transition in temperate lines is less sensitive to differences in daylength.
Genetic Position of ZCN8 in the Maize Flowering Pathway
In maize, only two late-flowering mutants, id1 and dlf1, have been cloned, and id1 was placed upstream of dlf1 in the genetic pathway for flowering (Muszynski et al., 2006). We characterized ZCN8 transcript accumulation in the leaf blade of both mutants, the tissue with the highest level of ZCN8 expression. No ZCN8 transcript was detected in id1 mutant leaves, but in the dlf1 mutant, ZCN8 expression was similar to the wild type. Therefore, we placed ZCN8 downstream of id1 and upstream or in parallel to dlf1 in the maize flowering pathway. However, id1 is exclusively transcribed in immature leaves and was shown to function non-cell-autonomously to regulate the floral transition (Colasanti et al., 1998). The ID1 protein appears to not be mobile; it stays in immature leaves (Wong and Colasanti, 2007). id1 was proposed to control the production of a mobile florigenic (F) signal that moves to the shoot apex (Wong and Colasanti, 2007; Colasanti and Muszynski, 2009). Our results show that ZCN8 is likely the mobile signal (F) that functions downstream of id1. The genetic interaction of id1 and ZCN8 suggests that id1 activity is required for ZCN8 transcription.
How id1 regulates transcription of ZCN8 is not clear. It is unlikely that ZCN8 is a direct target of id1 as there are no obvious ID1 binding sites near the ZCN8 gene sequence (Kozaki et al., 2004). In addition, the two genes are most highly expressed in nonoverlapping domains of developing leaves, although both genes are moderately expressed in the transition zone of the leaf where id1 expression diminishes and ZCN8 expression initiates (Figure 4C). We think it is more probable that id1 regulates expression of at least one other gene that is required for transcription of ZCN8. ID1 may turn on expression of another transcription factor that, in turn, activates transcription of ZCN8 in the transition zone and more distal parts of the emerging leaf. Alternatively, ID1 or another input pathway may control expression of an epigenetic regulatory factor that affects the chromatin state of cis-regulatory sequences near ZCN8 and therefore affects its transcriptional competence. This may explain how ZCN8 expression is maintained in the vascular cells of the mature leaf blade, which are many mitotic generations removed from when id1 was active. Recently, several studies have shown that FT is subject to epigenetic regulation as one of several inputs required for its expression (Jiang et al., 2008; Jeong et al., 2009; Adrian et al., 2010; Yang et al., 2010), but this idea remains to be studied in maize.
A Model Integrating the Autonomous and Photoperiod Pathways for Flowering in Maize
The maize flowering network is relatively rudimentary due to a limited number of flowering mutants (Colasanti and Muszynski, 2009). Our findings allow for the further elaboration of the maize flowering network (Figure 7). In this model, id1 represents the autonomous inductive pathway in maize as neither id1 transcript nor ID1 protein are diurnally regulated (Wong and Colasanti, 2007). id1 indirectly activates expression of ZCN8 in the transition zone of the immature leaf. Hence, ZCN8 is regulated by the autonomous pathway in developing leaves.
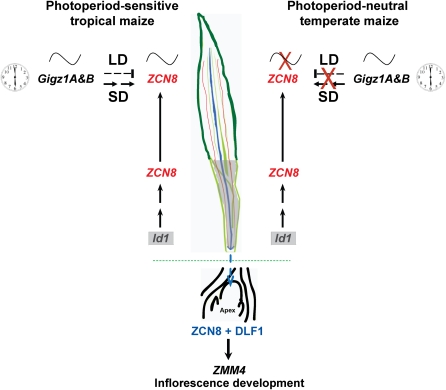
Figure 7.
A Model Integrating the Autonomous and Photoperiod Pathways in Tropical and Temperate Maize.
In the autonomous pathway, id1 indirectly controls ZCN8 transcription in the immature region of leaves that are hidden in the whorl (marked as a gray area on the leaf image). ZCN8 but not id1 is transcribed in the green, photosynthetically competent parts of the leaf blade. In SD tropical maize, leaf-expressed ZCN8 is diurnally regulated under SDs and LDs, but under nonpermissive LDs, the amplitude of ZCN8 expression is severely attenuated, resulting in later flowering. This suggests the possible existence of an LD suppression pathway in maize similar to that in rice (Komiya et al., 2009). In day-neutral temperate maize, regulation of ZCN8 transcription is disconnected from the circadian clock and from the hypothetical LD suppression pathway (marked by X). Disconnection from the circadian clock results in ZCN8 transcription displaying no diurnal regulation, even though the upstream genes, gigz1A, 1B (maize homolog of GIGANTEA) show diurnal transcript regulation. In both tropical and temperate maize, ZCN8 transcripts are localized in the phloem of the vascular bundles (fine red lines over the leaf image), which allows the ZCN8 protein to move to the shoot apex (blue line and blue arrow), although this movement has not yet been directly demonstrated. In the shoot apex, the ZCN8 protein interacts with the DLF1 bZIP transcription factor to activate expression of floral identity genes like ZMM4. The onset of ZMM4 expression marks the floral transition and initiation of reproductive development (inflorescence development) in maize.
In temperate, day-neutral maize lines, ZCN8 transcript accumulates at early stages of development and does not exhibit a diurnal expression pattern, which is especially evident in the extremely early flowering cultivar Gaspé Flint. This finding suggests that in temperate maize genotypes, ZCN8 transcription is disconnected from the circadian clock and, in particular, is not responsive to negative LD regulation, even though the circadian machinery appears to be intact (Hayes et al., 2010). On the other hand, in tropical genotypes, ZCN8 is regulated by signals from the photoperiod pathway. Under floral-inductive SDs, ZCN8 transcript accumulation is upregulated at early vegetative stages to ensure an earlier floral transition and earlier flowering under the relatively unfluctuating tropical daylengths. However, when these genotypes are grown in LDs, the amplitude of ZCN8 expression is suppressed, possibly by a putative LD suppression pathway. After an extended period of vegetative growth, the amplitude of ZCN8 expression gradually increases, allowing plants transitioning to reproductive development and flower later.
According to the proposed model, during selection for adaptation to growth at higher latitudes, several genetic events could have occurred, including a relaxation of the diurnal regulation of ZCN8 expression under LDs and its disconnection from the circadian machinery. This might involve mutations in the ZCN8 gene per se or in the upstream regulators connecting ZCN8 expression to the circadian clock. How the regulation and/or activity of ZCN8 have been altered between photoperiod sensitive and insensitive lines is of interest to both evolutionary biology and plant breeding. Our finding reveals a putative target gene for maize adaption outside of the tropical regions of the globe and provides guidance in the study of photoperiod sensitivity in maize.
METHODS
Plant Materials and Growth Conditions
Gaspé Flint, B73, CML311, and CML436 maize (Zea mays) lines were chosen for this research due to their distinct photoperiod sensitivities and differences in flowering times. Gaspé Flint is an extremely early temperate variety producing eight to nine leaves. B73 is a temperate inbred line producing 19 to 20 leaves. Gaspé Flint and B73 are considered daylength insensitive. CML311 and CML436 are two tropical inbred lines. CML311 is a moderately photoperiod-sensitive line producing 19 to 20 leaves under SDs and 23 to 24 leaves under LDs. CML436 is a late highly photoperiod-sensitive line producing 19 to 20 leaves under SDs and 27 to 30 leaves under LDs. Seeds were germinated directly in soil and seedlings grown in growth chambers. LDs were set up for 16 h light and 8 h dark, and SDs were 8 h light and 16 h dark. The temperature varied from 25 to 27°C during the day and 23 to 24°C during the night.
Tissue Collection
Tissues were collected at different vegetative growth stages, mainly before and after the floral transition. Vegetative growth stages (V stages) were defined according to the full extension of the leaf collar of the uppermost leaf (Ritchie et al., 1997). Stage VE is defined as the time when the first true leaf emerges from the coleoptile. The floral transition was detected when the SAM was elongated compared with its width. Collection of Gaspé Flint tissue began after 5 d of growth under SDs or LDs. Collection of B73, CML311, and CML436 tissue began after 2 weeks of growth under SDs or LDs. B73 tissue collection stopped after elongation of the SAM was observed under SDs and after the apex of the inflorescence finished spikelet initiation under LDs. Collection of CML311 and CML436 tissue was stopped after spikelet initiation was observed on the apical inflorescence under both SDs and LDs. All tissue sampling was performed in the morning and tissue from at least three plants was pooled together.
Diurnal Experiment
For the diurnal experiment under SDs, tissues from Gaspé Flint, B73, and CML436 were collected near the time of the floral transition as determined by examination of the SAM of representative plants. CML436 was planted first, B73 2 weeks later, and Gaspé Flint last, such that all three lines would transition at the same time under SDs. After one more week of growth, tissue from all genotypes were harvested every 4 h for over a 68-h period. Tissues sampled at each time point included leaf blades from the uppermost leaf, immature leaves, and shoot apices of two randomly selected plants. Three biological replicates were collected at the same time.
For the diurnal experiment under LDs, tissue of Gaspé Flint and B73 was collected around the floral transition. CML436 was harvested at V5 when CML436 was still at a vegetative growth stage and at V9 when CML436 started the floral transition. All other procedures were the same as those under SDs.
SD Inductive Experiment
The two tropical varieties were grown under LDs until stage V4-V5, were exposed to seven SDs, and then returned to LDs. Control plants were kept continuously in LDs. Before the SD treatment, blades of the uppermost leaf of three plants were harvested. During the SD treatment and next 7 d after returning to LD conditions, blades of the uppermost leaf of five plants were harvested every other day. After that, leaf blades of five plants were harvested according to their V stages. At the time leaves were harvested, plants were dissected to check development of their SAM. All tissue collections were performed in the morning.
RNA Isolation, RT-PCR, and Quantitative RT-PCR
Total RNA was isolated with TRIzol Reagent (Roche Applied Science). The Turbo DNA-free kit (Applied Biosystems) was used to remove DNA from the RNA samples. cDNA synthesis was performed with Superscript III First-Strand Synthesis system (Invitrogen). All processes were done following the manufacturers’ instructions. PCR amplifications were performed using Expand Long Template DNA polymerase with 35 cycles for all transcripts (Roche Applied Science). PCR products were detected by ethidium bromide on agarose gels. Quantitative RT-PCR amplifications were performed using the TaqMan probe–based detection system (Applied Biosystems). Primers and probes were designed according to nucleotide sequences of FT-like genes published in GenBank. All primers and probes are shown in Supplemental Table 1 online. In quantitative RT-PCR reactions, three biological replicates were used and each PCR was performed in triplicate. The relative expression level of genes was calculated by their quantification normalized to ubiquitin5. The processes were performed following the User Bulletin #2 from Applied Biosystems at http://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_040980.pdf.
In Situ Hybridization
The procedures of tissue fixation, probe preparation, and in situ hybridization were performed following the protocols described previously with minor modifications (Kidner and Timmermans, 2006). The templates for in vitro RNA syntheses were generated by PCR using primers listed in Supplemental Table 1 online. Antisense and sense probes were labeled with digoxigenin. Sections were viewed under a Leica DMRXA research microscope, and images were taken with a Leica digital camera DC500.
Tissue Imaging
Shoot apices and inflorescence meristems were dissected and fixed in 50% acetic acid. Images were taken with a Nikon SMZ1500 microscope and attached Nikon digital camera DXM1200.
Yeast Two-Hybrid Assay
Yeast two-hybrid analysis was performed using the ProQuest Two-Hybrid system (Invitrogen) following the manufacturer’s instructions. cDNAs of genes were cloned into both pDEST 32 (the bait vector) and pDEST 22 (the prey actor) to test for reciprocal interactions. All plasmids were transformed into the MaV203 yeast strain. The yeast cells were plated on dropout media SD/-Leu/-Trp. Individual clones were suspended in sterile saline and diluted 10 fold. Two microliters of diluted clones were spotted onto plates of SD/-Leu/-Trp and SD/-Leu/-Trp/-His+3AT.
T-DNA Constructs and Plant Transformation
Gateway technology (Invitrogen) was used for vector construction. The expression cassette comprising the promoters, the full-length cDNA sequences, and the PINII terminator were assembled in pENTRTM/D-TOPO vector (Invitrogen). Then LR recombination reaction was performed between the pENTR ATT-L sites and the destination Agrobacterium tumefaciens JT vectors containing ATT-R recombination sites. The cointegrated JT vectors were introduced into Agrobacterium strain LBA4404 and used to transform Hi-Type II maize embryos (Unger et al., 2001; Cigan et al., 2005). Typically, 20 independent events were generated for each construct. Single-copy T-DNA integration events were used for further characterization. Transgene-expressing plants were identified by RT-PCR at the T0 or T1 generations. In the T1 generation, 15 plants from two independent events per each construct were chosen to count a leaf number. The fifth and tenth leaves were marked during the plant growth, and the total leaf number was counted at maturity. The 21-mer (amiRNA) 5′-TCTCATAAAATATTAGCTCTT-3′ was designed for ZCN8 according the rules for artificial microRNA published by Schwab et al. (2006). The 21 amiRNA was incorporated into the zma-miR396h backbone (Zhang et al., 2009) and then was cloned into the pENTR vector for generated the cointegrated JT vectors.
Statistical Analysis
Mean values and standard deviations were calculated by linear regression using Minitab 15 statistical software. The difference in leaf number was tested by a one-way analysis of variance taking the phenotype and the presence or absence of the transgene as the sources of variation. Tukey’s family error rate was chosen for one-way multiple comparisons with a P value level of significance (α-level) equal to 0.05 and 0.1.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: EU241923 (ZCN7), EU241924 (ZCN8), EU241928 (ZCN12), EU241929 (ZCN14), EU241930 (ZCN15), EU241933 (ZCN18), and EU241937 (ZCN26).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Whole Plant Images of the Four Varieties Used in This Study.
Supplemental Figure 2. Shoot Apices of Tropical Varieties after Three and Five SD Treatments.
Supplemental Figure 3. Diurnal Expression of Gigz1 and Conz1 in Leaf Blades of Plants Grown under Either SDs or LDs.
Supplemental Figure 4. ZCN8 Diurnal Expression in the Tropical Line CML436.
Supplemental Figure 5. FT-Like ZCN Genes Are Not Expressed in the Immature Leaf of the Wild Type and Late-Flowering Mutants.
Supplemental Figure 6. ZCN14 Expression Patterns in the Shoot Apices of the Wild Type and Late-Flowering Mutants.
Supplemental Figure 7. Negative Controls for RNA in Situ Hybridization.
Supplemental Figure 8. ZCN8 RNA in Situ Hybridization to the Leaf Tip of B73 at Stages V4 and V6.
Supplemental Figure 9. ZCN8 RNA in Situ Hybridization to Leaf Blade Tissue from Gaspé Flint at Stage V2 after the Floral Transition.
Supplemental Figure 10. GUS Staining of the Shoot Apices of ProZM-ADF4:GUS Transgenic Plants at Stages V3 to V6.
Supplemental Figure 11. Transgene and Native Gene Expression in Transgenic Plants.
Supplemental Figure 12. Images of Nontransgenic and Transgenic ProZM-ADF4ZCN8 Plants Grown in the Field.
Supplemental Figure 13. Partial Amino Sequence Alignment of Arabidopsis, Sugar Beet, and Maize FT-Like Proteins.
Supplemental Table 1. The Sequences of Primers and Probes.
Supplementary Material
Acknowledgments
We thank Brian McGonigle for designing the amiR construct, Pedro Hermon for images of id1 and dlf1 shoot apices, Will Stork for technical assistance, and the transformation group for generating transgenic plants.
References
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Adrian J., Farrona S., Reimer J.J., Albani M.C., Coupland G., Turck F. (2010). cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22: 1425–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H., Roussot C., Suárez-López P., Corbesier L., Vincent C., Piñeiro M., Hepworth S., Mouradov A., Justin S., Turnbull C., Coupland G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Banfield M.J., Brady R.L. (2000). The structure of Antirrhinum centroradialis protein (CEN) suggests a role as a kinase regulator. J. Mol. Biol. 297: 1159–1170 [DOI] [PubMed] [Google Scholar]
- Barbour E., Meyer T.E., Saad M.E. (2003). Maize GOS-2 Promoters. U.S. Patent No. 6504083 B1, Pioneer Hi-Bred International [Google Scholar]
- Bate N.J., Reimann K. (2009). A Plant Regulatory Region That Directs Transgene Expression in the Maternal and Supporting Tissue of Maize Ovules and Pollinated Kernels. International Patent No. WO2009/021004 A1, Pioneer Hi-Bred International [Google Scholar]
- Blackman B.K., Strasburg J.L., Raduski A.R., Michaels S.D., Rieseberg L.H. (2010). The role of recently derived FT paralogs in sunflower domestication. Curr. Biol. 20: 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawn R.I. (1968). Breeding corn for earliness. Proc. Annu. Corn Sorghum Res. Conf. 23: 59–66 [Google Scholar]
- Cháb D., Kolár J., Olson M.S., Storchová H. (2008). Two flowering locus T (FT) homologs in Chenopodium rubrum differ in expression patterns. Planta 228: 929–940 [DOI] [PubMed] [Google Scholar]
- Chailakhyan M.K. (1937). Concerning the hormonal nature of plant development processes. Dokl. Akad. Nauk SSSR 16: 227–230 [Google Scholar]
- Cigan A.M., Unger-Wallace E., Haug-Collet K. (2005). Transcriptional gene silencing as a tool for uncovering gene function in maize. Plant J. 43: 929–940 [DOI] [PubMed] [Google Scholar]
- Colasanti J., Muszynski M. (2009). The Maize Handbook. (New York: Springer-Verlag). [Google Scholar]
- Colasanti J., Tremblay R., Wong A.Y., Coneva V., Kozaki A., Mable B.K. (2006). The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics 7: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J., Yuan Z., Sundaresan V. (1998). The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93: 593–603 [DOI] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- D'Aloia M., Bonhomme D., Bouche F., Tamseddak K., Ormenese S., Torti S., Coupland G., Perilleux C. (2011). Cytokinin promotes flowering of Arabidopsis via transcriptional activation of the FT paralogue TSF. Plant J. 65: 972–979 [DOI] [PubMed] [Google Scholar]
- Danilevskaya O.N., Meng X., Ananiev E.V. (2010). Concerted modification of flowering time and inflorescence architecture by ectopic expression of TFL1-like genes in maize. Plant Physiol. 153: 238–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya O.N., Meng X., Hou Z., Ananiev E.V., Simmons C.R. (2008a). A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol. 146: 250–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya O.N., Meng X., Selinger D.A., Deschamps S., Hermon P., Vansant G., Gupta R., Ananiev E.V., Muszynski M.G. (2008b). Involvement of the MADS-box gene ZMM4 in floral induction and inflorescence development in maize. Plant Physiol. 147: 2054–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure S., Higgins J., Turner A., Laurie D.A. (2007). The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare). Genetics 176: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giakountis A., Coupland G. (2008). Phloem transport of flowering signals. Curr. Opin. Plant Biol. 11: 687–694 [DOI] [PubMed] [Google Scholar]
- Hayama R., Agashe B., Luley E., King R., Coupland G. (2007). A circadian rhythm set by dusk determines the expression of FT homologs and the short-day photoperiodic flowering response in Pharbitis. Plant Cell 19: 2988–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes K.R., Beatty M., Meng X., Simmons C.R., Habben J.E., Danilevskaya O.N. (2010). Maize global transcriptomics reveals pervasive leaf diurnal rhythms but rhythms in developing ears are largely limited to the core oscillator. PLoS ONE 5: e12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish E., Nelson T. (1991). Identification of multiple stages in the conversion of vegetative to floral development. Development 112: 891–898 [Google Scholar]
- Itoh H., Nonoue Y., Yano M., Izawa T. (2010). A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat. Genet. 42: 635–638 [DOI] [PubMed] [Google Scholar]
- Jaeger K.E., Wigge P.A. (2007). FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 17: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Jeong J.H., Song H.R., Ko J.H., Jeong Y.M., Kwon Y.E., Seol J.H., Amasino R.M., Noh B., Noh Y.S. (2009). Repression of FLOWERING LOCUS T chromatin by functionally redundant histone H3 lysine 4 demethylases in Arabidopsis. PLoS ONE 4: e8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Wang Y., Wang Y., He Y. (2008). Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS ONE 3: e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I., Shukla V.K., Ahn J.H., Dagenais N., Christensen S.K., Nguyen J.T., Chory J., Harrison M.J., Weigel D. (1999). Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kidner C., Timmermans M. (2006). In situ hybridization as a tool to study the role of microRNAs in plant development. In MicroRNA Protocols, Ying S.-Y., (Totowa, NJ: Humana Press; ), pp. 159–179 [DOI] [PubMed] [Google Scholar]
- Kojima S., Takahashi Y., Kobayashi Y., Monna L., Sasaki T., Araki T., Yano M. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43: 1096–1105 [DOI] [PubMed] [Google Scholar]
- Komiya R., Ikegami A., Tamaki S., Yokoi S., Shimamoto K. (2008). Hd3a and RFT1 are essential for flowering in rice. Development 135: 767–774 [DOI] [PubMed] [Google Scholar]
- Komiya R., Yokoi S., Shimamoto K. (2009). A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136: 3443–3450 [DOI] [PubMed] [Google Scholar]
- Kozaki A., Hake S., Colasanti J. (2004). The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res. 32: 1710–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov N.N. (1933). World's diversity of phenotypes of maize. J. Am. Soc. Agron. 25: 688–700 [Google Scholar]
- Lifschitz E., Eshed Y. (2006). Universal florigenic signals triggered by FT homologues regulate growth and flowering cycles in perennial day-neutral tomato. J. Exp. Bot. 57: 3405–3414 [DOI] [PubMed] [Google Scholar]
- Lifschitz E., Eviatar T., Rozman A., Shalit A., Goldshmidt A., Amsellem Z., Alvarez J.P., Eshed Y. (2006). The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 103: 6398–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M.K., Belanger H., Lee Y.J., Varkonyi-Gasic E., Taoka K., Miura E., Xoconostle-Cázares B., Gendler K., Jorgensen R.A., Phinney B., Lough T.J., Lucas W.J. (2007). FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell 19: 1488–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J., Warthmann N., Küttner F., Schmid M. (2007). Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17: 1055–1060 [DOI] [PubMed] [Google Scholar]
- Matsuoka Y., Vigouroux Y., Goodman M.M., Sanchez G.J., Buckler E., Doebley J. (2002). A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. USA 99: 6080–6084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy D., Brettell R.I.S. (1994). Foreign gene expression in transgenic cereals. Trends Biotechnol. 12: 62–68 [Google Scholar]
- Miller T.A., Muslin E.H., Dorweiler J.E. (2008). A maize CONSTANS-like gene, conz1, exhibits distinct diurnal expression patterns in varied photoperiods. Planta 227: 1377–1388 [DOI] [PubMed] [Google Scholar]
- Mouradov A., Cremer F., Coupland G. (2002). Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14(suppl.): S111–S130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muszynski M.G., Dam T., Li B., Shirbroun D.M., Hou Z., Bruggemann E., Archibald R., Ananiev E.V., Danilevskaya O.N. (2006). delayed flowering1 Encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol. 142: 1523–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notaguchi M., Abe M., Kimura T., Daimon Y., Kobayashi T., Yamaguchi A., Tomita Y., Dohi K., Mori M., Araki T. (2008). Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol. 49: 1645–1658 [DOI] [PubMed] [Google Scholar]
- Pin P.A., Benlloch R., Bonnet D., Wremerth-Weich E., Kraft T., Gielen J.J., Nilsson O. (2010). An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330: 1397–1400 [DOI] [PubMed] [Google Scholar]
- Putterill J., Robson F., Lee K., Simon R., Coupland G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- Ritchie S.W., Hanway J.J., Benson G.O. (1997). How a Corn Plant Develops. Special Report 48. (Ames, IA: Iowa State University Cooperative Extension Service) [Google Scholar]
- Robson F., Costa M.M., Hepworth S.R., Vizir I., Piñeiro M., Reeves P.H., Putterill J., Coupland G. (2001). Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 28: 619–631 [DOI] [PubMed] [Google Scholar]
- Russell W.K., Stuber C.W. (1983). Inheritance of photosensitivity in maize. Crop Sci. 23: 935–939 [Google Scholar]
- Salvi S., et al. (2007). Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc. Natl. Acad. Sci. USA 104: 11376–11381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalit A., Rozman A., Goldshmidt A., Alvarez J.P., Bowman J.L., Eshed Y., Lifschitz E. (2009). The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc. Natl. Acad. Sci. USA 106: 8392–8397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P., Wheatley K., Robson F., Onouchi H., Valverde F., Coupland G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H.L., Yokoi S., Shimamoto K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Troyer A.F. (2000). Temperate corn: Background, behavior, and breeding. Specialty Corns, Hallauer A.R., (Boca Raton, FL: CRC Press; ), pp. 395–455 [Google Scholar]
- Tsuji H., Tamki S., Komiya R., Shimamoto K. (2008). Florigen and the photoperiodic control of flowering in rice. Rice 1: 25–35 [Google Scholar]
- Turck F., Fornara F., Coupland G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59: 573–594 [DOI] [PubMed] [Google Scholar]
- Unger E., Betz S., Xu R., Cigan A.M. (2001). Selection and orientation of adjacent genes influences DAM-mediated male sterility in transformed maize. Transgenic Res. 10: 409–422 [DOI] [PubMed] [Google Scholar]
- Valverde F., Mouradov A., Soppe W., Ravenscroft D., Samach A., Coupland G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Wigge P.A., Kim M.C., Jaeger K.E., Busch W., Schmid M., Lohmann J.U., Weigel D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Wong A.Y., Colasanti J. (2007). Maize floral regulator protein INDETERMINATE1 is localized to developing leaves and is not altered by light or the sink/source transition. J. Exp. Bot. 58: 403–414 [DOI] [PubMed] [Google Scholar]
- Yamaguchi A., Kobayashi Y., Goto K., Abe M., Araki T. (2005). TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 46: 1175–1189 [DOI] [PubMed] [Google Scholar]
- Yan L., Fu D., Li C., Blechl A., Tranquilli G., Bonafede M., Sanchez A., Valarik M., Yasuda S., Dubcovsky J. (2006). The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. USA 103: 19581–19586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Jiang D., Jiang J., He Y. (2010). A plant-specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis. Plant J. 62: 663–673 [DOI] [PubMed] [Google Scholar]
- Zeevaart J.A. (2008). Leaf-produced floral signals. Curr. Opin. Plant Biol. 11: 541–547 [DOI] [PubMed] [Google Scholar]
- Zhang L., Chia J.M., Kumari S., Stein J.C., Liu Z., Narechania A., Maher C.A., Guill K., McMullen M.D., Ware D. (2009). A genome-wide characterization of microRNA genes in maize. PLoS Genet. 5: e1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.