Plants synthesize the growth hormone indole-3-acetic acid using a variety of incompletely understood pathways. This work demonstrates that the conversion of indole-3-butyric acid to indole-3-acetic acid contributes to wide-ranging auxin-regulated processes in Arabidopsis seedlings and that shutting off this auxin supply renders seedlings less able to respond to exogenous auxin.
Abstract
Levels of auxin, which regulates both cell division and cell elongation in plant development, are controlled by synthesis, inactivation, transport, and the use of storage forms. However, the specific contributions of various inputs to the active auxin pool are not well understood. One auxin precursor is indole-3-butyric acid (IBA), which undergoes peroxisomal β-oxidation to release free indole-3-acetic acid (IAA). We identified ENOYL-COA HYDRATASE2 (ECH2) as an enzyme required for IBA response. Combining the ech2 mutant with previously identified iba response mutants resulted in enhanced IBA resistance, diverse auxin-related developmental defects, decreased auxin-responsive reporter activity in both untreated and auxin-treated seedlings, and decreased free IAA levels. The decreased auxin levels and responsiveness, along with the associated developmental defects, uncover previously unappreciated roles for IBA-derived IAA during seedling development, establish IBA as an important auxin precursor, and suggest that IBA-to-IAA conversion contributes to the positive feedback that maintains root auxin levels.
INTRODUCTION
Auxin is a key phytohormone that directs both cell division and cell elongation, thus regulating critical aspects of plant growth and development (reviewed in Perrot-Rechenmann, 2010). The active auxin indole-3-acetic acid (IAA) is a potent growth regulator, and its levels are modulated through synthesis, regulated transport, and storage forms (reviewed in Woodward and Bartel, 2005b). However, the relative importance of various pathways that contribute to the active auxin pool is not well understood.
The side chain of the auxin precursor indole-3-butyric acid (IBA) is two carbons longer than the IAA side chain and is shortened to IAA in numerous plants (Fawcett et al., 1960; reviewed in Epstein and Ludwig-Müller, 1993) in a peroxisome-dependent manner (Strader et al., 2010). Genetic screens in Arabidopsis thaliana have revealed that the auxin activity of IBA requires conversion to IAA through a multistep process similar to fatty acid β-oxidation, which removes two-carbon units from fatty acyl-CoA molecules. Proteins required for full IBA responsiveness include the ATP binding cassette (ABC) transporter PEROXISOMAL ABC TRANSPORTER1 (PXA1), which may transport IBA into the peroxisome for β-oxidation (Zolman et al., 2001), and proteins required for peroxisome biogenesis, such as PEROXIN4 (PEX4) (Zolman et al., 2005), PEX5 (Zolman et al., 2000), PEX6 (Zolman and Bartel, 2004), and PEX7 (Woodward and Bartel, 2005a; Ramón and Bartel, 2010). In addition, several peroxisomal enzymes are required for IBA responsiveness, including a 3-ketoacyl-CoA thiolase (Zolman et al., 2000) and several acyl-CoA oxidase (ACX) (Adham et al., 2005) enzymes, which also act in fatty acid β-oxidation, and the apparent IBA β-oxidation enzymes INDOLE-3-BUTYRIC ACID RESPONSE1 (IBR1) (Zolman et al., 2008), IBR3 (Zolman et al., 2007), and IBR10 (Zolman et al., 2008).
Mutations in IBR1, IBR3, or IBR10 confer IBA resistance without altering IAA response or causing dependence on exogenous carbon sources for postgerminative growth (Zolman et al., 2000, 2007, 2008), consistent with the possibility that the encoded peroxisomal enzymes act directly in IBA-to-IAA conversion. IBR1 is predicted to be a short-chain dehydrogenase/reductase (Zolman et al., 2008), IBR3 resembles acyl-CoA dehydrogenases/oxidases (Zolman et al., 2007), and IBR10 appears to be an enoyl-CoA hydratase (Zolman et al., 2008). A triple mutant defective in these three IBR enzymes displays enhanced IBA response defects (Zolman et al., 2008), converts IBA to IAA inefficiently, and exhibits expansion defects in root hairs and cotyledons suggestive of lowered IAA levels (Strader et al., 2010).
Here, we describe the identification of an IBA response mutant, ech2-1, from a new screen using dark-grown seedlings. The peroxisomally localized ENOYL-COA HYDRATASE2 (ECH2) enzyme is required for full response to applied IBA, and ech2 phenotypes are synergistic with previously identified ibr mutants. Moreover, combining ech2 with other ibr mutations results in multiple striking developmental defects, reduced responsiveness to both IBA and IAA, and decreased IAA levels in root tips, revealing important contributions of IBA to auxin levels in developing seedlings.
RESULTS
ECH2 Is Required for IBA Responsiveness
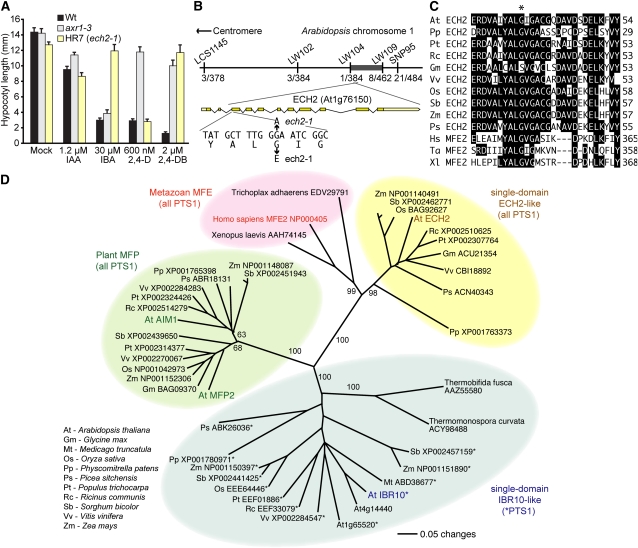
Endogenous auxin generally promotes cell expansion, but supraoptimal auxin levels can inhibit cell and consequently organ expansion in both roots and hypocotyls. Classical auxin response mutant screens are based on resistance to natural or synthetic auxins in light-grown root elongation assays (reviewed in Woodward and Bartel, 2005b). Because several mutants exhibiting strongly auxin-resistant root elongation are not markedly IBA resistant in dark-grown hypocotyl assays (Strader et al., 2008a), we employed a hypocotyl resistance (HR) screen for genes necessary for IBA response in dark-grown seedlings and isolated mutants with long hypocotyls on an inhibitory IBA concentration. In dark-grown wild-type seedlings, hypocotyl elongation is inhibited in the presence of the naturally occurring active auxin IAA, the auxin precursor IBA, the synthetic auxin 2,4-D, and the synthetic auxin precursor 2,4-dichlorophenoxybutyric acid (2,4-DB; Figure 1A). Hypocotyl elongation of the auxin-resistant mutant axr1-3 (Estelle and Somerville, 1987; Leyser et al., 1993) was strongly resistant to IAA, 2,4-D, and 2,4-DB, but only slightly resistant to IBA, whereas dark-grown hypocotyl elongation of the HR7 isolate was strongly resistant to the auxin precursors IBA and 2,4-DB but sensitive to the active auxins IAA and 2,4-D (Figure 1A). Because both IBA (Zolman et al., 2000, 2007; Strader et al., 2010) and 2,4-DB (Hayashi et al., 1998) require peroxisomal carboxyl side chain shortening to release active auxins, the specific resistance of HR7 to IBA and 2,4-DB suggested a peroxisomal defect.
Figure 1.
ECH2 Is Required for Hypocotyl IBA Response.
(A) Mean hypocotyl lengths (+se; n ≥ 14) of dark-grown wild type (Wt), axr1-3, and isolate HR7 (ech2-1) on various natural and synthetic auxins.
(B) Recombination mapping with the indicated markers (see Supplemental Table 2 online) localized HR7 to a 2-Mb region on chromosome 1 (dark bar) containing 592 predicted genes between LW104 and LW109 with 1/384 and 8/462 flanking recombinants. Examination of the ECH2 (At1g76150) gene in this region revealed a G-to-A mutation at position 371 in HR7 DNA that results in a Gly36-to-Glu substitution.
(C) The ech2-1 mutation disrupts a conserved Gly (asterisk). Sequences from predicted ECH2 homologs (accession numbers in [D]) and MFE2 homologs were aligned using the MegAlign program (DNAStar; full alignment and accession numbers are in Supplemental Figure 5 online).
(D) Phylogenetic tree of ECH2, IBR10, MFE2, MFP2, AIM1, and relatives. Protein portions corresponding to the hydratase domains were aligned using ClustalW (alignment in Supplemental Figure 6 online and Supplemental Data Set 1 online), and the unrooted phylogram was generated using PAUP 4.05b (Swofford, 2001) by performing the bootstrap method with 500 replicates. Bootstrap values are shown at the nodes.
We mapped the causative mutation in HR7 to a 592-gene region on the lower arm of chromosome 1 (Figure 1B) and looked for peroxisomally targeted enzymes in the mapping interval. One such candidate was ECH2 (At1g76150), which encodes a peroxisomal enoyl-CoA hydratase implicated in the β-oxidation of unsaturated fatty acids (Goepfert et al., 2006). We sequenced ECH2 from HR7 genomic DNA and identified a G-to-A base change at position 371 (using the A of the ATG as position 1) that causes a Gly36-to-Glu missense mutation (Figure 1B) in a residue conserved (Figure 1C) in many ECH2 homologs (Figure 1D). We named the mutation in HR7 ech2-1.
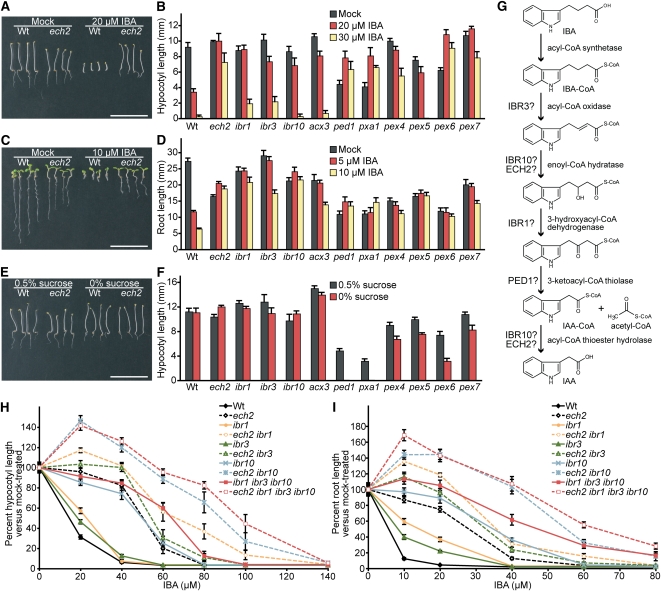
Because ech2-1 carried a mutation in a putative peroxisomal enzyme, we compared the IBA responsiveness of ech2-1 in hypocotyl and root elongation assays to mutants identified in screens for IBA-resistant root elongation (see Supplemental Table 1 online) (Zolman et al., 2000), including mutants defective in peroxisomal enzymes IBR1 (Zolman et al., 2008), IBR3 (Zolman et al., 2007), IBR10 (Zolman et al., 2008), ACX3 (Adham et al., 2005), and PEROXISOME DEFECTIVE1 (PED1) (Lingard and Bartel, 2009); a mutant defective in the peroxisomal transporter PXA1 (Zolman et al., 2001); and mutants defective in peroxisome biogenesis factors PEX4 (Zolman et al., 2005), PEX5 (Zolman et al., 2000), PEX6 (Zolman and Bartel, 2004), and PEX7 (Ramón and Bartel, 2010). Like ech2-1, these mutants displayed IBA-resistant hypocotyls: ped1-96, pxa1-1, pex4-1, pex6-1, and pex7-2 displayed IBA resistance similar to ech2-1, whereas ibr1-2, ibr3-1, ibr10-1, acx3-6, and pex5-1 displayed more moderate resistance (Figure 2B). In addition, ech2-1 displayed strongly IBA-resistant roots, similar to the ibr10-1, ped1-96, pxa1-1, pex5-1, and pex6-1 mutants (Figures 2C and 2D). Although ech2 RNA interference (RNAi) lines have normal 2,4-DB responsiveness in spite of 5- to 10-fold reductions in ECH2 mRNA levels (Goepfert et al., 2006), we found that ech2-1 hypocotyl (Figure 1A) and root (see Supplemental Figure 1A online) elongation were 2,4-DB resistant. This 2,4-DB resistance indicated that the ech2-1 defect was general to chain-elongated auxins and not IBA specific and suggested that the ech2-1 lesion more severely impaired ECH2 function than the previously characterized ech2 RNAi lines (Goepfert et al., 2006).
Figure 2.
Hormone Responses of ech2 and Other Peroxisomal Mutants.
(A) Five-day-old wild-type (Wt) and ech2-1 seedlings following growth in the dark on medium supplemented with ethanol (Mock) or 20 μM IBA. Bar = 1 cm.
(B) Mean hypocotyl lengths (+se; n ≥ 12) of 5-d-old dark-grown wild-type, ech2-1, ibr1-2, ibr3-1, ibr10-1, acx3-6, ped1-96, pxa1-1, pex4-1, pex5-1, pex6-1, and pex7-2 grown on medium supplemented with ethanol (Mock) or IBA at concentrations indicated.
(C) Eight-day-old wild-type and ech2-1 seedlings following growth in the light on medium supplemented with ethanol (mock) or 10 μM IBA. Bar = 1 cm.
(D) Mean root lengths (+se; n ≥ 10) of 8-d-old seedlings listed in (B) grown in the light on medium supplemented with ethanol (Mock) or IBA at concentrations indicated.
(E) Five-day-old wild-type and ech2-1 seedlings following growth in the dark in the presence (0.5%) and absence of an exogenous carbon source (sucrose). Bar = 1 cm.
(F) Mean hypocotyl lengths (+se; n ≥ 12) of 5-d-old seedlings listed in (B) grown as in (E).
(G) Schematic of a proposed IBA-to-IAA conversion pathway (Zolman et al., 2008) showing possible enzymatic activities for ECH2, IBR1, IBR3, and IBR10, along with the PED1 thiolase, which may act in both IBA and fatty acid β-oxidation.
(H) ech2 enhances IBA resistance of ibr mutant hypocotyls. Mean hypocotyl lengths (+se; n ≥ 12) of 7-d-old wild-type, ech2-1, ibr1-2, ech2-1 ibr1-2, ibr3-1, ech2-1 ibr3-1, ibr10-1, ech2-1 ibr10-1, ibr1-2 ibr3-1 ibr10-1, and ech2-1 ibr1-2 ibr3-1 ibr10-1 pregerminated and then grown in the dark on medium supplemented with ethanol (0 μM IBA) or 20 to 140 μM IBA.
(I) ech2 enhances IBA resistance of ibr mutant roots. Mean root lengths (+se; n ≥ 12) of 10-d-old lines listed in (H) that were pregerminated and then grown in the light on medium supplemented with ethanol (0 μM IBA) or 10 to 80 μM IBA.
Wild-type Arabidopsis seedlings metabolize fatty acids stored in seeds by peroxisomal β-oxidation to fuel growth prior to photosynthesis (Hayashi et al., 1998). Mutants defective in IBR1, IBR3, or IBR10 display IBA resistance without dependence on exogenous fixed carbon sources for postgerminative growth (Zolman et al., 2000, 2007, 2008), consistent with the possibility that these enzymes are required for IBA-to-IAA conversion but not for fatty acid β-oxidation. Mutants defective in PXA1 or PED1, however, display both IBA resistance and dependence on sucrose to fuel growth following germination (Hayashi et al., 1998; Zolman et al., 2001), suggesting roles in both IBA and fatty acid β-oxidation. Dark-grown ech2-1 hypocotyls, like those of the wild type and the ibr mutants, elongated normally with or without sucrose (Figures 2E and 2F), suggesting that seed storage fatty acids are metabolized normally in ech2. Together, the IBA resistance and sucrose independence of ech2-1 suggest that ECH2 functions in IBA-to-IAA conversion (Figure 2G).
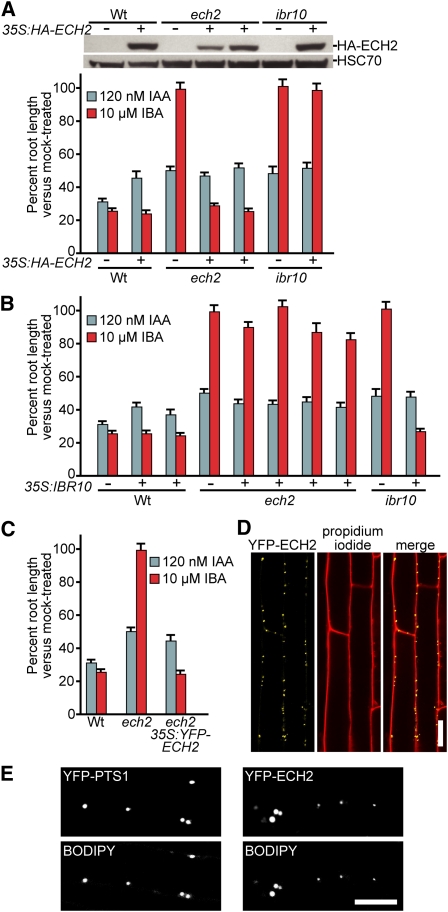
To test whether the ech2-1 lesion caused the observed IBA-response defects, we transformed ech2-1 with constructs driving N-terminally tagged versions of an ECH2 cDNA from the strong 35S promoter from cauliflower mosaic virus. ech2-1 was rescued by both 35S:HA-ECH2 and 35S:YFP-ECH2 constructs (Figures 3A and 3C), indicating that the IBA response defects resulted from reduced ECH2 function and that the N-terminal tags did not interfere with ECH2 function. Yellow fluorescent protein (YFP)-ECH2 localization in the ech2 35S:YFP-ECH2 line exhibited subcellular punctate fluorescence resembling the size and shape of peroxisomes (Figure 3D) and colocalized with a BODIPY probe (Figure 3E) that stains peroxisomes (Landrum et al., 2010). This localization is consistent with previous reports of peroxisomal localization of YFP-ECH2 in transiently transfected onion epidermal cells (Goepfert et al., 2006; Reumann et al., 2007) and identification of ECH2 in the proteome of peroxisomes purified from Arabidopsis leaves (Reumann et al., 2007) and cell cultures (Eubel et al., 2008).
Figure 3.
ech2 Is Rescued by ECH2 but Not IBR10.
(A) Overexpression of HA-tagged ECH2 restores IBA sensitivity to ech2-1 but not ibr10-1. Top: Immunoblot analysis of (left to right) 5-d-old light-grown wild type (Wt), wild type carrying 35S:HA-ECH2, ech2-1, two independent ech2-1 lines carrying 35S:HA-ECH2, ibr10-1, and ibr10-1 carrying 35S:HA-ECH2. Anti-HA and anti-HSC70 antibodies were used to detect HA-ECH2 and HSC70 (loading control), respectively. Bottom: Mean normalized root lengths (+se; n ≥ 11) of 8-d-old light-grown lines shown in the immunoblot above the graph.
(B) Overexpression of IBR10 restores IBA sensitivity to ibr10-1 but not ech2-1. Mean normalized root lengths (+se; n ≥ 9) of 8-d-old light-grown wild type, two independent wild-type lines carrying 35S:IBR10, ech2-1, four independent ech2-1 lines carrying 35S:IBR10, ibr10-1, and ibr10-1 carrying 35S:IBR10.
(C) YFP-tagged ECH2 rescues ech2-1. Mean normalized root lengths (+se; n ≥ 11) of 8-d-old light-grown wild type, ech2-1, and ech2-1 carrying 35S:YFP-ECH2.
(D) YFP-ECH2 localizes to punctate structures in Arabidopsis cells. Confocal images of root epidermal cells from 4-d-old ech2-1 expressing YFP-ECH2 counterstained with propidium iodide to visualize cell walls. Bar = 20 μm.
(E) YFP-ECH2 localizes to peroxisomes in Arabidopsis cells. Confocal images of root epidermal cells from 4-d-old wild type expressing a peroxisomally targeted YFP derivative (YFP-PTS1; left panels) (px-yk; Nelson et al., 2007) and ech2-1 expressing YFP-ECH2 (right panels). The top panel of each pair shows the fusion protein fluorescence and the bottom panel of each pair shows fluorescence from 8-(4-nitrophenyl)-BODIPY, which allows visualization of peroxisomes (Landrum et al., 2010). Bar = 10 μm.
Because ECH2 is an enoyl-CoA hydratase (Goepfert et al., 2006) and IBR10 resembles enoyl-CoA hydratases (Zolman et al., 2008), we examined whether ECH2 and IBR10 acted redundantly by comparing the effects of overexpressing ECH2 or IBR10 in both ech2-1 and ibr10-1. As previously demonstrated (Zolman et al., 2008), overexpressing IBR10 complemented ibr10-1 (Figure 3B). However, IBA responsiveness was not restored in ibr10 accumulating hemagglutinin (HA)-ECH2 to levels similar to the levels that complemented ech2-1 (Figure 3A; see Supplemental Figure 2 online), indicating that additional ECH2 did not compensate for decreased IBR10 activity. Likewise, overexpressing IBR10 failed to restore IBA responsiveness to the ech2-1 mutant (Figure 3B; see Supplemental Figure 2 online). These data suggest that ECH2 and IBR10, although members of the same protein superfamily (Figure 1D), are unlikely to act redundantly at the same step of IBA-to-IAA conversion (Figure 2G).
ech2-1 Enhances Effects of Mutants with Reduced IBA-to-IAA Conversion
We examined ech2-1 in combination with the previously characterized ibr1-2 (Zolman et al., 2008), ibr3-1 (Zolman et al., 2007), ibr10-1 (Zolman et al., 2008), and ibr1-2 ibr3-1 ibr10-1 (Zolman et al., 2008) mutants. ibr1-2 and ibr3-1 were mildly resistant to low IBA concentrations in dark-grown hypocotyl elongation assays (Figure 2H) and light-grown root elongation assays (Figure 2I), whereas ech2-1 and ibr10-1 were more strongly resistant (Figures 2H and 2I). ech2-1 resistance to the inhibitory effects of IBA on dark-grown hypocotyl and light-grown root elongation were slightly enhanced when combined with ibr1 or ibr3 and greatly enhanced when combined with ibr10 (Figures 2H and 2I). Similarly, ech2-1 2,4-DB resistance was enhanced when combined with ibr10 (see Supplemental Figure 1A online). Combining ech2-1 with ibr mutants did not confer sucrose dependence to dark-grown hypocotyls (see Supplemental Figure 1B online), suggesting that the encoded enzymes do not function redundantly in fatty acid β-oxidation. From here on, we will refer to ech2-1 as ech2. The ech2 ibr1 ibr3 ibr10 quadruple mutant displayed only slightly greater IBA resistance in hypocotyl elongation than the ech2 ibr10 double mutant (Figure 2H). The strong IBA resistance of ech2 ibr10 and ech2 ibr1 ibr3 ibr10 prompted us to examine these mutants for the developmental consequences of impeding IBA-to-IAA conversion.
Mutants Deficient in IBA-to-IAA Conversion Have Cell Expansion Defects
Several lines of evidence suggest that IBA-derived IAA contributes to root hair and cotyledon cell expansion. Mutants defective in the ABCG36/PDR8/PEN3 and ABCG37/PDR9/PIS1 ABC transporters, which probably function as IBA effluxers (reviewed in Strader and Bartel, 2011), display lengthened root hairs (Strader et al., 2008b; Strader and Bartel, 2009; Růžička et al., 2010), suggesting that increased IBA accumulation in root hairs can increase auxin levels. Additionally, abcg36 mutants display enlarged cotyledons, a second high-auxin phenotype (Strader and Bartel, 2009). Both of these abcg36 developmental phenotypes are suppressed when combined with the ibr1, ibr3, and ibr10 mutations, and the ibr1 ibr3 ibr10 triple mutant displays short root hairs and small cotyledons (Strader et al., 2010).
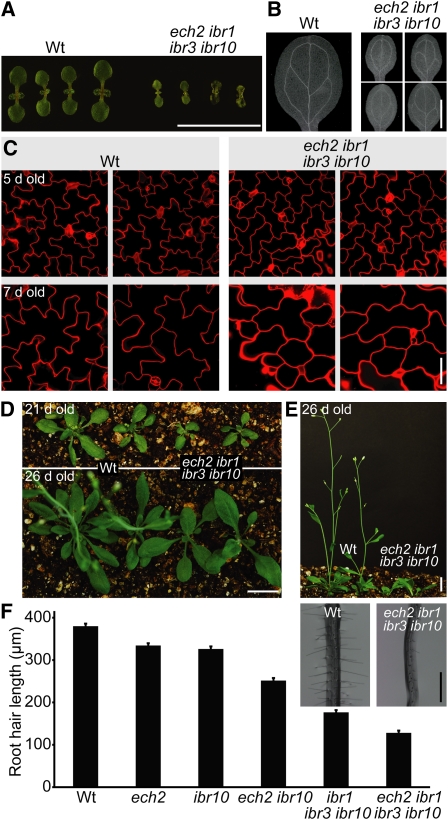
Like ibr1 ibr3 ibr10 (Strader et al., 2010), ech2 ibr1 ibr3 ibr10 seedlings exhibited smaller cotyledons than the wild type (Figure 4A). Moreover, the ech2 ibr1 ibr3 ibr10 quadruple mutant exhibited defects in cotyledon vascular patterning (Figure 4B). Because Arabidopsis cotyledons grow by cell expansion without cell division after germination (Mansfield and Briarty, 1996), this small size likely results from decreased cell expansion. Indeed, ech2 ibr1 ibr3 ibr10 cotyledon epidermal cells were smaller than those of the wild type and displayed aberrant shape with reduced interdigitation (Figure 4C), suggesting a role for IBA-derived IAA in shaping cotyledon pavement cells. The smaller size of ech2 ibr1 ibr3 ibr10 persisted in the first several true leaves, but older plants eventually produced wild-type-sized rosette leaves (Figure 4D). The smaller early leaves of the quadruple mutant were accompanied by slower development and a delay in the time to flowering (Figure 4E), but ech2 ibr1 ibr3 ibr10 plants did not display marked morphological defects at maturity (see Supplemental Figure 3 online).
Figure 4.
ech2 Enhances ibr1 ibr3 ibr10 Cell Expansion Defects.
(A) Seven-day-old light-grown ech2-1 ibr1-2 ibr3-1 ibr10-1 seedlings display decreased cotyledon size compared with wild-type (Wt) seedlings. Bar = 1 cm.
(B) Eight-day-old light-grown ech2-1 ibr1-2 ibr3-1 ibr10-1 seedlings display aberrant vascular patterning. Bar = 1 mm.
(C) Five-day-old (top panels) and 7-d-old (bottom panels) ech2-1 ibr1-2 ibr3-1 ibr10-1 seedlings display small cotyledon epidermal cells. Confocal images of propidium iodide–stained cells are shown. Bar = 50 μm.
(D) Soil-grown ech2-1 ibr1-2 ibr3-1 ibr10-1 plants are smaller than the wild type at 21 d (top panel) but begin to recover by 26 d (bottom panel). Two wild-type (left) and quadruple mutant (right) plants are shown. Bar = 1 cm.
(E) Soil-grown ech2-1 ibr1-2 ibr3-1 ibr10-1 plants flower later than the wild type. Profile of plants shown in (D). Bar = 1 cm.
(F) ech2 mutants display short root hairs. Mean root hair lengths (+se; n = 500) of 5-d-old wild-type, ech2-1, ibr10-1, ech2-1 ibr10-1, ibr1-2 ibr3-1 ibr10-1, and ech2-1 ibr1-2 ibr3-1 ibr10-1 light-grown seedlings. Inset: Root hairs from 5-d-old wild-type and ech2-1 ibr1-2 ibr3-1 ibr10-1 light-grown seedlings. Bar = 500 μm.
Root hairs are long, tubular outgrowths from certain root epidermal cell files that increase root surface area to aid in water and nutrient uptake. Because root hair expansion is auxin regulated, root hairs provide a sensitive single-cell model to study auxin levels and response (reviewed in Grierson and Schiefelbein, 2002). The ibr1 ibr3 ibr10 triple mutant exhibits short root hairs (Figure 4E) (Strader et al., 2010) that can be rescued by applying auxin (Strader et al., 2010). We found that ech2 displayed slightly shorter root hairs than the wild type and enhanced the short-root hair phenotype of ibr10 and ibr1 ibr3 ibr10 (Figure 4F; see Supplemental Figure 4 online), suggesting that ECH2 loss may decrease auxin levels in root hair cells.
ech2 ibr10 Displays Reduced Auxin-Responsive Transcription
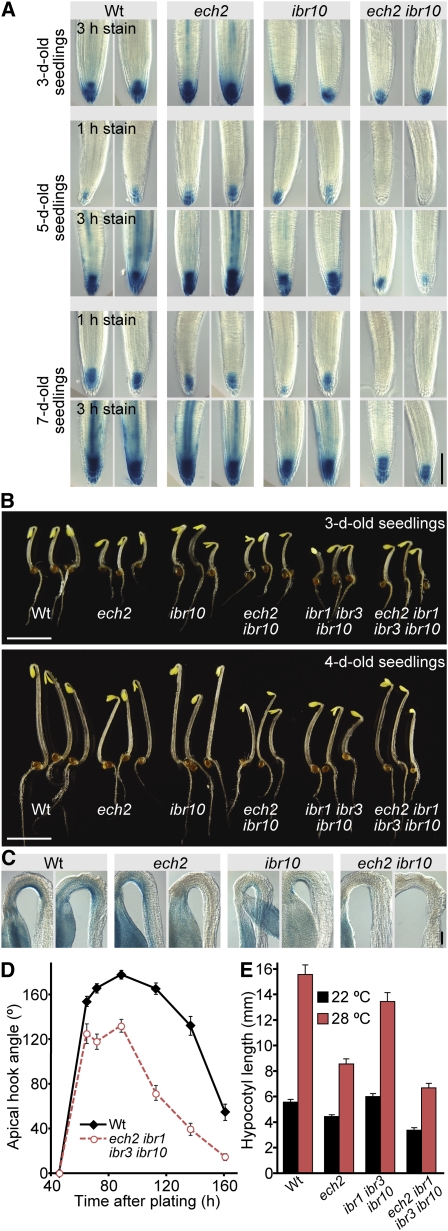
To assess local effects of ech2 and ibr10 on auxin-responsive transcription, we examined ech2, ibr10, and ech2 ibr10 carrying DR5-GUS, a reporter driving β-glucuronidase (GUS) expression from a synthetic auxin-responsive promoter (Ulmasov et al., 1997). Auxin levels (Petersson et al., 2009) and DR5-GUS activity (Sabatini et al., 1999) are high in the root meristem. As previously reported (Zolman et al., 2008), DR5-GUS activity was similar in ibr10 and wild-type root tips (Figure 5A). Similarly, ech2 did not display notably altered DR5-GUS activity (Figure 5A). However, DR5-GUS activity in ech2 ibr10 was undetectable in roots stained for the optimal time to visualize GUS activity in the wild type and was only observed after longer staining periods (Figure 5A). The differences in DR5-GUS activity between the wild type and ech2 ibr10 became more pronounced as seedlings matured; the small differences in 3-d-old seedlings were more apparent in 7-d-old seedlings (Figure 5A). The reduced DR5-GUS activity suggested that active auxin levels were decreased in ech2 ibr10 root meristems.
Figure 5.
ech2 ibr10 Displays Decreased Auxin Reporter Activity, Apical Hook Formation, and High-Temperature Hypocotyl Elongation.
(A) ech2 ibr10 displays decreased DR5-GUS activity in root tips. Three-, five-, and seven-day-old light-grown wild-type (Wt), ech2-1, ibr10-1, and ech2-1 ibr10-1 seedlings carrying the DR5-GUS construct (Ulmasov et al., 1997; Zolman et al., 2008) were stained for GUS activity for 1 or 3 h. Bar = 100 μm.
(B) ech2 mutants display decreased apical hook formation. Three-day-old (top panel) or 4-d-old (bottom panel) wild-type, ech2-1, ibr10-1, ech2-1 ibr10-1, ibr1-2 ibr3-1 ibr10-1, and ech2-1 ibr1-2 ibr3-1 ibr10-1 dark-grown seedlings are shown. Bar = 0.5 cm.
(C) ech2 ibr10 displays decreased DR5-GUS reporter activity in apical hooks. Three-day-old dark-grown wild-type, ech2-1, ibr10-1, and ech2-1 ibr10-1 seedlings carrying the DR5-GUS construct were stained for GUS activity for 4 h. Bar = 100 μm.
(D) ech2 ibr1 ibr3 ibr10 displays decreased apical hook formation and maintenance. Mean apical hook angles (±se; n ≥ 20) of wild-type and ech2-1 ibr1-2 ibr3-1 ibr10-1 dark-grown seedlings are shown.
(E) ech2 mutants display decreased high temperature–induced hypocotyl elongation. Mean hypocotyl lengths (+se; n = 16) of wild-type, ech2-1, ibr1-2 ibr3-1 ibr10-1, and ech2-1 ibr1-2 ibr3-1 ibr10-1 seedlings grown for 8 d at 22 or 28°C under yellow-filtered light.
Mutants Deficient in IBA-to-IAA Conversion Have Hypocotyl Defects
The decreased DR5-GUS activity and short root hairs suggested that auxin levels were low in ech2 ibr10 roots. To determine if ECH2 also impacted hypocotyl development, we examined two auxin-regulated processes in ech2 mutants: apical hook curvature and high temperature–induced hypocotyl lengthening. Dark-grown dicotyledonous seedlings, such as Arabidopsis seedlings, exhibit closed cotyledons and form an apical hook to protect the shoot apical meristem from damage when emerging from the soil (Goeschl et al., 1966; Guzmán and Ecker, 1990). Auxin response is necessary for proper apical hook formation and maintenance, and auxin-resistant and auxin-deficient mutants often are defective in apical hook curvature (Tian and Reed, 1999; Harper et al., 2000; Friml et al., 2002; Stepanova et al., 2008; Vandenbussche et al., 2010; Žádníková et al., 2010).
We found that apical hook curvature was slightly decreased in dark-grown ech2 and ibr10 single mutants and that ech2 ibr10, ibr1 ibr3 ibr10, and ech2 ibr1 ibr3 ibr10 displayed even less apical hook curvature in both 3-d-old and 4-d-old seedlings, with ech2 ibr1 ibr3 ibr10 exhibiting the greatest curvature defect (Figures 5B and 5D). The ech2 ibr1 ibr3 ibr10 mutants were defective in both apical hook formation, reaching a maximal angle of only 130° (versus 180° in the wild type), and apical hook maintenance, beginning to open when wild-type hooks were still largely closed (Figure 5D). Auxin promotes both hook formation and hook maintenance, and as previously observed (Friml et al., 2002; Vandenbussche et al., 2010; Žádníková et al., 2010), DR5-GUS activity was concentrated in the inner side of the apical hook in the wild type (Figure 5C). ech2 and ibr10 displayed decreased DR5-GUS activity in the inner face of the apical hook, and DR5-GUS activity in ech2 ibr10 was barely detectable in this region (Figure 5C), suggesting that the apical hook curvature defects in ech2 mutants were caused by decreased auxin response in this region.
Growth of seedlings at high temperature results in increased auxin levels and hypocotyl elongation (Gray et al., 1998). We found that ech2 and ech2 ibr1 ibr3 ibr10 displayed shorter hypocotyls than the wild type when grown at elevated temperatures (Figure 5E). Because the increased IAA at high temperature is thought to result from increased IAA synthesis (Gray et al., 1998), the high-temperature short-hypocotyl phenotype of ech2 is consistent with the possibility that ECH2 promotes IAA synthesis from IBA.
Mutants Deficient in IBA-to-IAA Conversion Have Decreased Lateral Root Production and Smaller Root Meristems
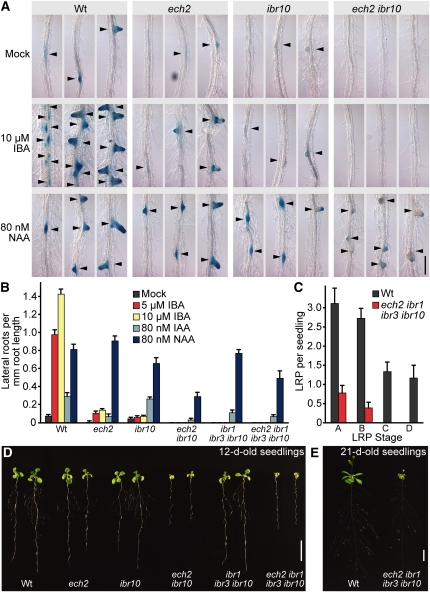
Auxin is a critical regulator of both lateral root initiation (reviewed in Casimiro et al., 2003) and lateral root emergence (Swarup et al., 2008). We examined lateral root initiation in the ech2 and ibr10 lines carrying the DR5-GUS reporter, which facilitates detection of lateral root primordia (LRP). In the absence of treatment, we did not detect any lateral roots or LRP in 8-d-old ech2 ibr10 seedlings (Figures 6A and 6B). Intriguingly, the LRP found in the ibr10 mutant displayed decreased DR5-GUS activity (Figure 6A), suggesting that auxin levels in these LRPs were sufficient to generate the lateral root but insufficient to highly activate the DR5-GUS reporter. Wild-type seedlings responded to IBA treatment with increased lateral root production (Figures 6A and 6B). Consistent with roles for ECH2 and IBR10 in IBA-to-IAA conversion, ech2, ibr10, and ech2 ibr10 were less responsive to IBA promotion of lateral root initiation (Figures 6A and 6B). Treatment with the synthetic auxin 1-naphthaleneacetic acid (NAA) increased lateral root production in all the tested lines (Figures 6A and 6B), indicating that the lack of LRPs in the ech2 ibr10 double mutant was not caused by an inability to make LRPs but by decreased active auxin in these tissues. Interestingly, the lateral roots and LRPs produced by ech2 ibr10 following NAA treatment displayed reduced DR5-GUS activity (Figure 6A), suggesting that even with NAA treatment, auxin levels in these LRP were sufficient to generate the lateral root but insufficient to highly activate the DR5-GUS reporter.
Figure 6.
ech2 ibr10 and ech2 ibr1 ibr3 ibr10 Are Defective in Lateral Root Production.
(A) ech2 ibr10 displays decreased DR5-GUS activity in lateral roots. Four-day-old light-grown wild-type (Wt), ech2-1, ibr10-1, and ech2-1 ibr10-1 seedlings carrying DR5-GUS were transferred to medium supplemented with ethanol (mock) or the indicated auxin and grown for an additional 4 d under yellow-filtered light at 22°C prior to staining for GUS activity for 1 h. Lateral roots and LRPs are highlighted with arrowheads. Bar = 200 μm.
(B) ech2 mutants produce fewer lateral roots than the wild type. Emerged lateral roots of wild-type, ech2-1, ibr10-1, ech2-1 ibr10-1, ibr1-2 ibr3-1 ibr10-1, and ech2-1 ibr1-2 ibr3-1 ibr10-1 were counted 4 d after transfer of 4-d-old seedlings to medium supplemented with either ethanol (mock) or the indicated auxins (mean + se, n ≥ 12).
(C) ech2 ibr1 ibr3 ibr10 displays fewer LRPs. Eight-day-old wild-type and ech2-1 ibr1-2 ibr3-1 ibr10-1 seedlings were cleared, and the number and stage of LRPs recorded (+se, n = 18). Stage A spans of the first anticlinal division of a pericycle cell to an LRP with three cell layers. Stage B includes unemerged lateral roots with more than three cell layers. Stage C includes emerged lateral roots shorter than 0.5 mm. Stage D consists of emerged lateral roots longer than 0.5 mm.
(D) Twelve-day-old wild-type, ech2-1, ibr10-1, ech2-1 ibr10-1, ibr1-2 ibr3-1 ibr10-1, and ech2-1 ibr1-2 ibr3-1 ibr10-1 light-grown seedlings. Bar = 1 cm.
(E) Twenty-one-day-old wild-type and ech2-1 ibr1-2 ibr3-1 ibr10-1 light-grown plants. Bar = 1 cm.
When we quantified lateral root emergence in response to auxins, we were surprised to find that ech2 ibr10 and ech2 ibr1 ibr3 ibr10 produced fewer lateral roots than the wild type not only in response to IBA but also in response to IAA and NAA (Figure 6B), suggesting that ECH2 and IBR10, and by implication IBA-to-IAA conversion, are necessary for full response not only to IBA but also to active auxins. We quantified LRPs in untreated roots and found that 8-d-old ech2 ibr1 ibr3 ibr10 seedlings had no emerged lateral roots and very few LRPs (Figure 6C). Despite this deficiency at 8 d, ech2 ibr1 ibr3 ibr10 did display emerged lateral roots after 12 (Figure 6D) and 21 d (Figure 6E). However, emerged lateral roots in ech2 ibr1 ibr3 ibr10 did not reach wild-type numbers even after 21 d, resulting in a reduced root system in the quadruple mutant (Figures 6D and 6E).
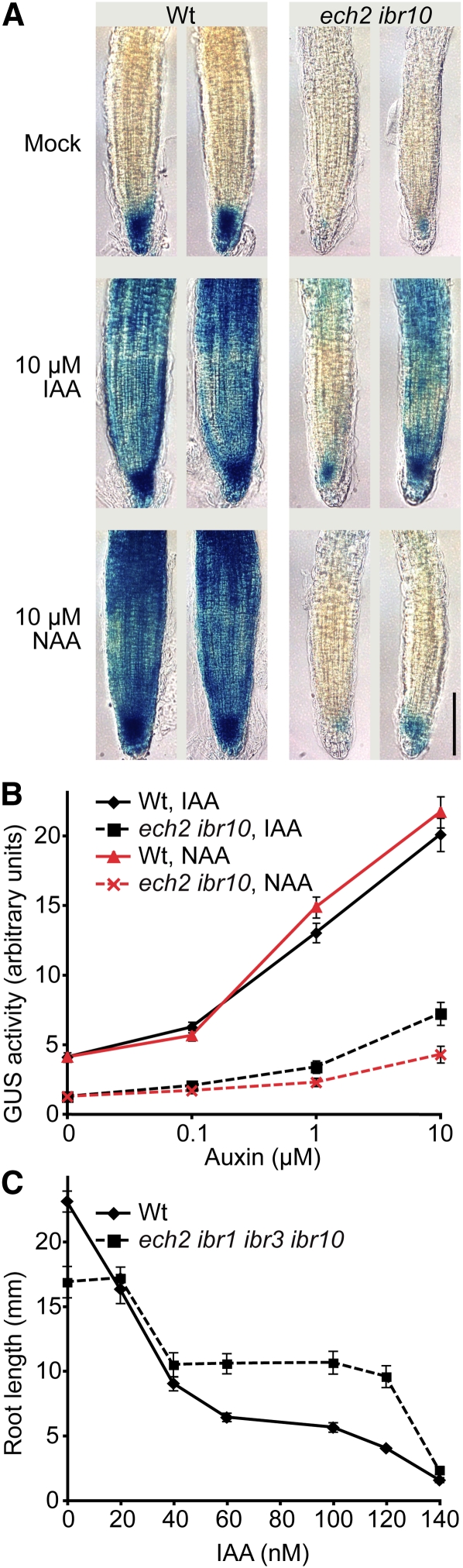
Because ECH2 and IBR10 were necessary for full response not only to IBA, but also to active auxins in 4-d-old lateral root formation assays (Figures 6A and 6B), we examined DR5-GUS in ech2 ibr10 following a shorter exposure to active auxins to determine if ECH2 and IBR10 also contribute to more rapid auxin responses. We found that ech2 ibr10 displayed dramatically decreased DR5-GUS activity in response to short (2-h) exposures to either NAA or IAA, both in root meristems (Figure 7A) and in whole seedlings (Figure 7B). Furthermore, we found that ech2 ibr1 ibr3 ibr10 displayed mild resistance to IAA in root elongation assays (Figure 7C), confirming that ECH2 and IBR10 are needed for full response to active auxins.
Figure 7.
ech2 ibr10 Displays Decreased Auxin Reporter Activity in Response to Active Auxins.
(A) Seven-day-old light-grown wild-type (Wt) and ech2-1 ibr10-1 seedlings carrying DR5-GUS were transferred to medium supplemented with ethanol (mock) or the indicated auxin and were incubated for 2 h at 22°C prior to staining for GUS activity for 1.5 h. Bar = 100 μm.
(B) Mean GUS activity (±se; n = 16) of 7-d-old light-grown wild-type and ech2-1 ibr10-1 seedlings carrying DR5-GUS treated for 2 h with ethanol (mock treatment; 0 μM auxin) or the indicated auxin.
(C) Mean root lengths (±se; n = 13) of 8-d-old light-grown wild-type and ech2-1 ibr1-2 ibr3-1 ibr10-1 seedlings grown on medium supplemented with ethanol (mock treatment; 0 nM IAA) or various concentrations of IAA.
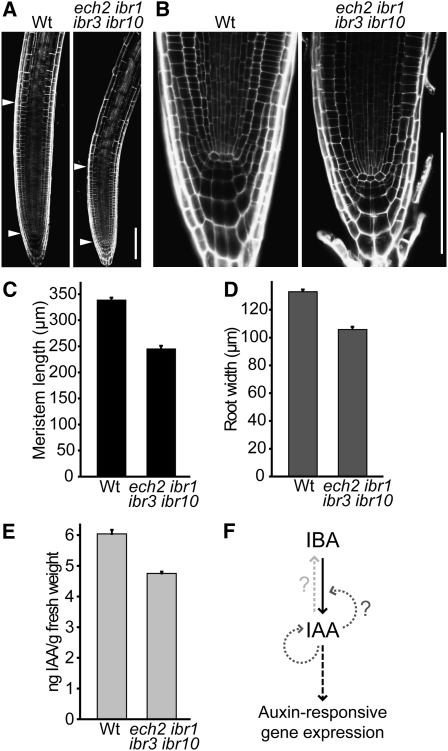
The root apical meristem is a region near the root tip that undergoes indeterminate growth, generating new root tissue through balanced cell division and differentiation. Establishment of an auxin maximum in the root tip is necessary to maintain the quiescent center, a small group of cells within the meristem that undergo only occasional cell divisions, and an auxin gradient is necessary to maintain root meristematic activity (reviewed in Iyer-Pascuzzi and Benfey, 2009). Consistent with reduced DR5-GUS activity in ech2 ibr10 root tips (Figure 5A), the root meristem, measured as the distance between the quiescent center and the first elongating cell of the root, was reduced by ~25% compared with the wild type in the ech2 ibr1 ibr3 ibr10 mutant (Figures 8A and 8C). Additionally, root width, measured in the cell elongation zone, was reduced by ~20% in ech2 ibr1 ibr3 ibr10 (Figure 8D). In spite of the reduced size of the root apical meristem in the quadruple mutant, we did not observe any obvious aberrations in meristem organization (Figure 8B).
Figure 8.
ech2 ibr1 ibr3 ibr10 Displays Meristem Defects and Decreased Auxin Levels.
(A) Confocal images of propidium iodide–stained root tips from 7-d-old light-grown wild-type (Wt) and ech2-1 ibr1-2 ibr3-1 ibr10-1 seedlings. Arrowheads delineate the top and bottom of the root meristem. Bar = 100 μm.
(B) Confocal images of propidium iodide–stained root tips from 7-d-old light-grown wild-type and ech2-1 ibr1-2 ibr3-1 ibr10-1 seedlings. Bar = 100 μm.
(C) Mean meristem lengths (+se; n ≥ 47) of 8-d-old light-grown wild-type and ech2-1 ibr1-2 ibr3-1 ibr10-1 seedlings.
(D) Mean root widths (+se; n ≥ 47) in the elongation zone of 8-d-old light-grown wild-type and ech2-1 ibr1-2 ibr3-1 ibr10-1 seedlings.
(E) Mean IAA levels (+se; n ≥ 3) in 5-mm root tips from light-grown wild-type and ech2-1 ibr1-2 ibr3-1 ibr10-1 seedlings.
(F) A model for the effects of IBA-to-IAA conversion on the auxin pool. IBA is converted to IAA in a process similar to peroxisomal fatty acid β-oxidation (solid black arrow). IAA also can be converted to IBA (dashed gray arrow; reviewed in Ludwig-Müller, 2000). Reduced DR5-GUS activity and decreased lateral root formation in ech2 ibr10 mutants treated with active auxins suggests that IBA-to-IAA conversion is necessary for full response to active auxins, either because auxin stimulates IBA-to-IAA conversion (positive feedback loops; dotted gray arrows) or because additional auxin is required to overcome the auxin deficit that results from blocking IBA-to-IAA conversion. Ultimately, increased auxin levels promote auxin signaling (dashed black arrow).
The ech2 ibr1 ibr3 ibr10 Mutant Has Reduced Auxin Levels
Although ibr1 ibr3 ibr10 seedling phenotypes are suggestive of low auxin levels, overall IAA levels are unchanged in ibr1 ibr3 ibr10 seedlings (Strader et al., 2010), indicating that any such reductions are minor and/or local. Because the reduced DR5-GUS activity in untreated ech2 ibr10 seedlings (Figures 5A and 7) along with the various ech2 ibr10 and ech2 ibr1 ibr3 ibr10 developmental phenotypes (Figures 4 to 6 and 8) suggested that auxin levels were further reduced, we used gas chromatography–mass spectrometry to measure free IAA levels in the root tips of ech2 ibr1 ibr3 ibr10 and the wild type. We found a 20% reduction of free IAA in the quadruple mutant compared with the wild type (Figure 8E). This reduction indicates that IBA-to-IAA conversion normally provides a significant input into the free IAA pool in developing seedlings.
DISCUSSION
Peroxisomal Enzymes Implicated in IBA-to-IAA Conversion
Screens for IBA resistance have revealed an IBA-to-IAA conversion pathway involving peroxisomal enzymes resembling fatty acid β-oxidation enzymes, suggesting that IBA undergoes β-oxidation to free IAA and acetyl-CoA (Figure 2G). IBR3 resembles acyl-CoA dehydrogenases/oxidases and may oxidize IBA-CoA to the α,β-unsaturated thioester (Zolman et al., 2007). Both IBR10 (Zolman et al., 2008) and ECH2 appear to be enoyl-CoA hydratases and may produce an IBA hydroxylacyl-CoA thioester intermediate. IBR1, a 3-hydroxylacyl-CoA-dehydrogenase-like protein, may perform the 3-hydroxyacyl-CoA dehydrogenase step in IBA-to-IAA conversion (Zolman et al., 2008). Removal of acetyl-CoA to give IAA-CoA may be catalyzed by the thiolase PED1 (Zolman et al., 2000), which acts in the analogous step in fatty acid β-oxidation (Hayashi et al., 1998). No genes annotated as encoding acyl-CoA synthetases or thioesterases have emerged from IBA response screens; however, either ECH2 or IBR10 may perform the thioesterase step, as discussed below.
Multiple ibr1 (Zolman et al., 2008) and ibr3 (Zolman et al., 2007) alleles have been isolated in IBA response screens, and some of these mutations result in premature stop codons. Moreover, T-DNA insertion alleles disrupting IBR1 and IBR3 have been characterized (Zolman et al., 2007, 2008; Wiszniewski et al., 2009). By contrast, no T-DNA insertions have been reported in IBR10 or ECH2, and only single alleles have emerged from IBA response screens: ibr10-1, which results in a 26–amino acid in-frame deletion (Zolman et al., 2008), and ech2-1, a missense allele (Figure 1), consistent with the possibility that null alleles of ibr10 and ech2 may confer more severe defects than the alleles that have been recovered to date. Unlike the 2,4-DB resistance of ech2-1 (Figure 1A; see Supplemental Figure 1 online), ech2 RNAi lines retain normal 2,4-DB responsiveness (Goepfert et al., 2006), suggesting that lines eliminating ECH2 function were not recovered in this study. Regardless of whether ibr10-1 or ech2-1 is a null allele, the observation that each confers greater IBA resistance than likely ibr1 or ibr3 null alleles suggests that the former mutations more effectively reduce IBA-to-IAA conversion than the latter.
If ibr1-2 and ibr3-1 are null alleles defective in dedicated IBA-to-IAA conversion enzymes, then why is IBA resistance enhanced when these alleles are combined with the ech2-1 missense allele? It is likely that alternate enzymes can catalyze the same IBA-to-IAA conversion steps as IBR3 and IBR1. For example, IBA-CoA oxidization may be catalyzed not only by IBR3 (Zolman et al., 2007), but also by ACX3 (Adham et al., 2005), a peroxisomal protein that also β-oxidizes fatty acyl-CoA esters (Froman et al., 2000). Because the ibr3 acx3 double mutant displays enhanced IBA resistance compared with either parent (Zolman et al., 2007), both IBR3 and ACX3 may oxidize IBA-CoA. Similarly, mutants in the multifunctional enzyme ABNORMAL INFLORESCENCE MERISTEM1 (AIM1), required for fatty acid β-oxidation (Richmond and Bleecker, 1999), also are IBA (Zolman et al., 2000) and 2,4-DB (Richmond and Bleecker, 1999) resistant. AIM1 can carry out the hydration and dehydrogenation steps of fatty acid β-oxidation (Richmond and Bleecker, 1999), whereas the combined activities of IBR1 (Zolman et al., 2008), IBR10 (Zolman et al., 2008), and/or ECH2 may perform the hydration and dehydrogenation steps of IBA β-oxidation. Because aim1 is IBA resistant (Zolman et al., 2000), it is possible that AIM1 may provide some IBA β-oxidation activity in addition to that provided by IBR1, IBR10, and ECH2. Interestingly, the enoyl-CoA domains of AIM1, IBR10, and ECH2 are dissimilar in primary sequence (Figure 1D).
ECH2 is conserved throughout the plant kingdom and does not share extensive sequence similarity with IBR10. In fact, ECH2 is more similar (~51% identical) to the enoyl-CoA hydratase domain from metazoan MULTIFUNCTIONAL ENZYME (MFE2) than it is the enoyl-CoA hydratase domains from IBR10 (11% identical) or plant multifunctional enzymes such as AIM1 (~8% identical) (Figure 1D). Unlike IBR10 and the plant multifunctional enzymes, members of the ECH2 family and the metazoan MFE2 family contain hot dog domains (see Supplemental Figure 5 online), a motif associated with both enoyl-CoA hydratases and thioesterases (Pidugu et al., 2009). ECH2 has bidirectional enoyl-CoA hydratase activity when heterologously expressed in Saccharomyces cerevisiae (Goepfert et al., 2006). That both ECH2 and IBR10 appear to encode enoyl-CoA hydratases (Figure 1D) and that mutation of either leads to strong IBA resistance (Figure 2) present a paradox: Are both of these enzymes catalyzing the enoyl-CoA hydratase step in IBA-to-IAA conversion? Our demonstration that overexpression of one cannot compensate for loss of the other (Figure 3) suggests that ECH2 and IBR10 catalyze different reactions. Given the mechanistic and sequence similarity between hydrolases and hydratases (Pidugu et al., 2009), it is tempting to speculate that one produces the IBA hydroxylacyl-CoA thioester intermediate and the other hydrolyzes IAA-CoA to release free IAA. Resolution of this question awaits the availability of postulated substrates needed for the biochemical characterization of these enzymes.
IBA-Derived IAA Plays Critical Roles in Seedling Development
IBA-derived IAA is important for seedling cell expansion; mutants defective in IBA efflux exhibit expanded root hairs and cotyledons, whereas mutants defective in IBA-to-IAA conversion display smaller root hairs and cotyledons (Strader and Bartel, 2008; Strader et al., 2010). IBA-derived IAA also plays a role in stamen filament elongation and lateral root production; pxa1 mutants display delayed filament elongation that is rescued by NAA application (Footitt et al., 2007) and decreased lateral rooting that is restored by IAA application (Zolman et al., 2001). The pex5-1 pex7-1 double mutant displays variably fused cotyledons (Woodward and Bartel, 2005a), a phenotype often associated with low auxin levels in embryogenesis (reviewed in Möller and Weijers, 2009). These indications of specific developmental roles for IBA-derived IAA are supported and extended by the wide-ranging auxin-related developmental defects in ech2 ibr1 ibr3 ibr10 seedlings, which included not only defects in lateral root production and cotyledon and root hair cell expansion, but also reduced cotyledon vasculature, delayed development, reduced apical hook curvature, reduced temperature-induced hypocotyl lengthening, reduced root width, smaller root meristems, and shorter roots. Together, the diverse low-auxin phenotypes displayed by ech2 ibr1 ibr3 ibr10 seedlings, our demonstration that auxin application at least partially rescues many of these defects, and the high-auxin phenotypes displayed by mutants with defects in IBA efflux suggest that IBA acts primarily or exclusively as an IAA precursor in Arabidopsis and that IBA-derived IAA contributes to developmental processes in multiple seedling tissues.
Previously characterized IAA biosynthetic mutants with low auxin levels include higher-order mutants in the YUCCA and TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1) families. Combinations of yuc1, yuc2, yuc4, and yuc6 exhibit various developmental defects, including altered plant stature, floral organ patterning, and vascular development (Cheng et al., 2006). Additionally, the yuc1 yuc4 yuc10 yuc11 quadruple mutant lacks a hypocotyl and a root meristem, indicating that YUCCA-synthesized IAA is critical during embryogenesis (Cheng et al., 2007). Mutants of TAA1 were isolated for altered shade avoidance (Tao et al., 2008), reduced ethylene responses (Stepanova et al., 2008), and reduced auxin transport inhibitor responses (Yamada et al., 2009). Like the yucca family, combining taa1 with tar2 or tar1 and tar2 alters plant stature, gravity response, vascular development, floral organ patterning, and embryo development (Stepanova et al., 2008). IBA-derived IAA appears to contribute to auxin-related developmental events primarily during seedling development; we find no evidence in ech2 ibr1 ibr3 ibr10 of the altered plant stature, floral organ patterning, gravity response, or embryo development found in higher-order yucca and taa1 family mutants, suggesting that the ECH2 pathway is distinct from the YUCCA and TAA1 pathways. However, because ech2 ibr1 ibr3 ibr10 mutants still respond to very high IBA levels (Figure 2), we cannot discount the possibility that residual IBA-to-IAA conversion occurs in the quadruple mutant and that phenotypes similar to those found in higher-order yucca and taa1 family mutants will be uncovered with a more complete block in IBA-to-IAA conversion. Indeed, a recent report of developmental alterations in mature plants resulting from increased IBA glucosylation suggests that IBA contributions are not limited to the seedling stage (Tognetti et al., 2010).
Is IBA an Auxin Storage Form or Biosynthetic Intermediate?
Previous data showing that IBA can be synthesized from IAA (reviewed in Ludwig-Müller, 2000), together with demonstrations that IBA is β-oxidized to IAA (Strader et al., 2010) suggest that IBA is an auxin storage form. However, no genes have been implicated in IAA-to-IBA conversion, and the observation that IBA is present at even lower levels than IAA in Arabidopsis seedlings (Ludwig-Müller and Epstein, 1993; Strader et al., 2010; Tognetti et al., 2010) is difficult to reconcile with the important roles of IBA-derived IAA in seedling development revealed here. It is tempting to speculate that IBA may function as an intermediate in a de novo IAA synthesis pathway in which IBA-to-IAA β-oxidation enzymes catalyze the final steps. Plants synthesize IAA using several pathways, none of which have been demonstrated at a level of detail such that all relevant enzymes and intermediates have been identified (reviewed in Strader and Bartel, 2008). Some pathways, such as the Trp-independent pathway, are particularly ill defined, with no identified intermediates between indole and IAA. Whether IBA serves as an IAA biosynthetic precursor, storage form, or both, the deep conservation of ECH2 and IBR10 homologs in the plant lineage (Figure 1D) suggests that IBA-to-IAA conversion is an ancient input to the free IAA pool.
Positive Feedback in Auxin Homeostasis
Positive feedback loops reinforcing auxin maxima are recurring themes in plant development, controlling root meristem maintenance, lateral root formation, vascular strand formation, and apical dominance maintenance (reviewed in Petrášek and Friml, 2009). Auxin canalization models to explain these maxima generally rely on the positive reinforcement provided by regulating polar auxin transport; however, temporal and spatial regulation of auxin synthesis also contributes to these maxima (reviewed in Zhao, 2010). Combining ech2 with ibr10 resulted in decreased activity of the DR5-GUS auxin-responsive reporter not only following IBA treatment (Figure 6A) but also in untreated seedlings (Figures 5 to 7), consistent with the reduced IAA levels we find in ech2 ibr1 ibr3 ibr10 root tips (Figure 8E). Surprisingly, DR5-GUS activity also was reduced in ech2 ibr10 seedlings following short- and long-term treatment with active auxins (Figures 6A and 7). Our observation that ech2 ibr10 mutants have reduced response not only to IBA, but also to IAA, suggests that auxin response, even to exogenous auxin, is dampened when endogenous IAA levels are reduced. Indeed, when release of IAA from amino acid conjugates is impaired, Arabidopsis seedlings show reduced IAA levels and less responsiveness to exogenous IAA (Rampey et al., 2004), consistent with the reduced auxin responsiveness that we find when IBA-to-IAA conversion is reduced. The molecular mechanisms underlying these reinforcing effects remain to be elucidated. Perhaps the IAA response machinery does not operate efficiently below some threshold IAA level, perhaps IAA response is impeded by the IBA that remains when IBA-to-IAA conversion is impaired, or perhaps IAA promotes IBA-to-IAA conversion, providing an additional positive feedback (Figure 8F). Given the importance of IBA-derived IAA in seedling development uncovered here, fully understanding the various inputs to the active auxin pool will require integration of IBA contributions into models of IAA homeostasis.
METHODS
Growth Conditions and Phenotypic Assays
Arabidopsis thaliana mutants were in the Colombia (Col-0) background, which was used as the wild type. Surface-sterilized seeds (Last and Fink, 1988) were plated on plant nutrient (PN) medium (Haughn and Somerville, 1986) supplemented with 0.5% (w/v) sucrose (PNS) solidified with 0.6% (w/v) agar and grown at 22°C under continuous illumination, unless otherwise noted.
To examine auxin-responsive elongation, seeds were plated on PNS supplemented with auxin and grown for 8 d under yellow-2208 (3-mm thick; A and C Plastics) long-pass (454-nm cutoff) plexiglass filters to slow indolic compound breakdown (Stasinopoulos and Hangarter, 1990) to monitor light-grown root and hypocotyl elongation or for 1 d under yellow long-pass filters followed by 4 d in the dark to monitor dark-grown hypocotyl elongation. To promote uniform germination on high IBA concentrations (Figures 2H and 2I), surface-sterilized seeds were imbibed in 0.1% agar for 3 d at 4°C and then incubated for 2 d under white light in liquid PN at 22°C prior to plating on medium supplemented with the indicated IBA concentrations and incubating for 8 d under yellow-filtered light (root elongation) or for 1 d under yellow-filtered light followed by an additional 4 d in darkness (hypocotyl elongation).
To examine sucrose dependence, seeds were plated on unsupplemented PN or on PNS and grown for 1 d under white light before growth for 4 d in the dark.
To examine cotyledon size, seedlings were grown for 7 d under white light on PNS before cotyledons were removed, mounted, and imaged. Cotyledon blade area was measured using NIH Image software.
To examine cotyledon vasculature, seedlings were grown for 8 d under white light on PNS, cleared with an ethanol series followed by 1 week in a chloral hydrate solution (80 g chloral hydrate, 20 mL glycerol, and 10 mL water), mounted in 50% glycerol, and imaged using a dissecting microscope.
To examine apical hooks, seeds were plated on PNS and grown for 1 d under white light before additional growth in the dark. Apical hooks were imaged, and apical hook angles were measured using NIH Image software.
To examine lateral roots, seedlings were grown for 4 d on PNS under yellow filters before transfer to PNS supplemented with the indicated auxin concentrations and grown for an additional 4 d. Emerged lateral roots were counted using a dissecting microscope.
To quantify LRPs, seedlings were grown for 8 d under white light on PNS, cleared by incubating for 1 week in chloral hydrate solution, mounted in 50% glycerol, and examined using a Zeiss Axioplan 2 microscope. Lateral roots and LRPs were counted and classified into four stages. Stage A includes LRP with three or fewer cell layers, corresponding to previously described Stages I to III (Malamy and Benfey, 1997), and Stage B includes unemerged LRPs with four or more cell layers, corresponding to previously described Stages IV to VIb (Malamy and Benfey, 1997). Stages C and D include emerged lateral roots less than and greater than 0.5 mm, respectively.
To examine root hair lengths, vertically grown 5-d-old seedlings grown under white light were imaged using a dissecting microscope, and root hairs lengths from the 4-mm root sections adjacent to the root-shoot junction were measured using NIH Image software.
To quantify meristem size and root width, 8-d-old seedlings grown under white light were fixed in ethanol:acetic acid (3:1) and mounted in chloral hydrate solution. Root tips were imaged using a Zeiss Axioplan 2 microscope, and the distance between the quiescent center and the first elongating cell was measured using NIH Image software.
Genetic Analyses
HR7 (in the Col-0 background) was outcrossed to Landsberg erecta for recombination mapping. DNA was isolated for mapping using markers (see Supplemental Table 2 online) from F2 individuals displaying long hypocotyls after 1 d in light and 4 d in darkness on 30 μM IBA. The ECH2 gene within the HR7 mapping interval was PCR amplified and sequenced from HR7 genomic DNA.
PCR analysis (see Supplemental Table 3 online) of segregating F2 plants was used to identify higher-order mutants.
Vector Construction and Plant Transformation
ECH2 was amplified from the U21373 cDNA obtained from the ABRC (Ohio State University) using Pfx Platinum Taq (Invitrogen) with 5′-CACCATGGCGACTAGCGATTCTGAATTCAATTC-3′ and 5′-TCATTACGCCAAGCAAACATCAAGAG-3′. The resulting PCR product was captured into the pENTR/D-TOPO vector (Invitrogen). The pENTR-ECH2 entry clone was cut with PvuII to linearize the vector to prevent transformation of Escherichia coli by the entry vector. ECH2 was recombined into the pEarleyGate 104 and pEarleyGate 201 plasmids (Earley et al., 2006) using LR Clonase (Invitrogen) to form pEarleyGate 104-ECH2 and pEarleyGate 201-ECH2, respectively, which express N-terminal YFP and HA fusions with ECH2 driven by the cauliflower mosaic virus 35S promoter. Recombinant plasmids were transformed into Agrobacterium tumefaciens strain GV3101 (Koncz and Schell, 1986), which was used to transform plants using the floral dip method (Clough and Bent, 1998). Transformants were selected on PNS plates supplemented with 7.5 μg/mL Basta (phosphinothricin), and lines homozygous for the transgene were selected in subsequent generations. To monitor HA-ECH2 levels, protein from 12 5-d-old seedlings was analyzed by immunoblotting as previously described (Strader et al., 2009), except that a 1:500 dilution of rat anti-HA antibody (3F10; Roche) was used.
Phylogenetic Analysis
Protein sequences corresponding to the hydratase domains of ECH2, IBR10, and relatives were aligned with Lasergene MegAlign (DNASTAR) using the ClustalW default settings with the Gonnet series protein weight matrix, then manually adjusted to optimize alignments. An unrooted phylogram was generated using PAUP 4.05b (Swofford, 2001) by performing the bootstrap method with 500 replicates with distance as the optimality criterion and all characters weighted equally.
GUS Assays
To histochemically examine GUS activity, seedlings carrying DR5-GUS (Ulmasov et al., 1997) were treated as indicated, fixed in 90% acetone for 20 min at −20°C, rinsed twice in GUS buffer (0.1 M NaPO4, pH 7.0, 0.5 mM K3[Fe(CN6)], 0.5 mM K4[Fe(CN6)], 10 mM EDTA, and 0.01% Triton X-100), and then incubated in GUS buffer supplemented with 0.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid at 37°C for the indicated time period. After staining, seedlings were fixed in 3:1 ethanol:acetic acid, mounted, and imaged using a Zeiss Axioplan 2 microscope.
For 4-methylumbelliferyl-β-d-glucuronide hydrate assays, 16 replicates of three 7-d-old seedlings were treated as indicated prior to GUS activity being monitored as previously described (Strader and Bartel, 2009).
Confocal Microscopy
Seedlings were counterstained in aqueous propidium iodide (10 μg/mL; Molecular Probes) to label cell walls or stained with 5 μM 8-(4-nitrophenyl)-BODIPY (diluted in water from a 5 mM stock in 90% acetone; Toronto Research Chemicals N503045) to label peroxisomes prior to mounting in water for imaging. Propidium iodide–stained YFP-ECH2 seedlings were imaged through a ×63 oil immersion lens on a Zeiss LSM 510 laser scanning confocal microscope equipped with a Meta detector. Samples were excited with the 488-nm laser line from an argon laser, and the resultant fluorescence was split through a 545-nm secondary dichroic beam splitter. Fluorescence <545 nm was further filtered through a 505-nm long-pass filter and false-colored yellow. Pixels resulting from fluorescence >545 nm were false-colored red.
Root apical meristem structure was imaged using propidium iodide–stained seedlings examined through a ×10 lens or a ×40 oil immersion lens on a Zeiss LSM 710 confocal laser scanning microscope. Samples were excited with the 543-nm laser, and resultant fluorescence between 566 and 719 nm was collected.
Seedlings stained with the peroxisome-associated 8-(4-nitrophenyl)-BODIPY fluorophore (Landrum et al., 2010) were imaged through a ×100 oil immersion lens on a Zeiss LSM 710 confocal laser scanning microscope. To detect BODIPY and YFP, samples were excited with the 488- and 514-nm lasers, and the resultant fluorescence between 493 and 516 nm, and 550 and 592 nm was collected, respectively. Images were converted and channels merged using NIH Image software.
Quantification of IAA
For IAA analysis, 150 μL of homogenization buffer (65% isopropanol and 35% 0.2 M imidazole buffer, pH 7.0) containing 10 ng [13C6]IAA internal standard (99 atom%; Cambridge Isotope Laboratories; Cohen et al., 1986) were added to each tissue sample. Samples were homogenized with two 3-mm tungsten carbide beads (Qiagen) in a Mixer-Mill (Qiagen) for 5 min at 25 Hz, then incubated on ice for 1 h to allow the internal standard to equilibrate with the endogenous IAA in the extract. After equilibration, debris was pelleted by centrifugation for 5 min at 10,000g, and 100 μL of supernatant was placed into a deep 96-well plate (Continental Lab Products). IAA extraction and solid phase purification was performed as described (Barkawi et al., 2008, 2010). The eluate, in 600 μL methanol, was methylated with 900 μL ethereal diazomethane (Cohen, 1984) in 1.5-mL screw-capped glass vials. Samples were then dried under N2 in a 55°C sand bath and resuspended in 30 μL ethyl acetate before being injected into an Agilent 6890 GC/5973 MS (Agilent Technologies) run in EI mode at 70 eV and equipped with a fused silica capillary column (HP-5MS, 30 m × 0.25-mm ID, 0.25-μm film). The injector temperature was 280°C, and the GC oven temperature was programmed to ramp from 70 to 280°C at 12°C/min. Helium was used as the carrier gas at a flow rate of 1 mL/min. Samples were analyzed in the selective ion monitoring mode. IAA levels were calculated by monitoring ions at mass-to-charge ratios (m/z) of 130 and 189 for endogenous IAA and m/z 136 and 195 for the [13C6]IAA standard, with quantities calculated using standard isotope dilution equations (Cohen et al., 1986).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession number At1g76150 (ECH2). Additional accession numbers are provided in Supplemental Table 1 online (loci corresponding to mutants used) and in Supplemental Figures 5 and 6 online (sequences used for alignments and phylogenetic analysis).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. ech2 ibr Mutants Are 2,4-DB Resistant (but Not Sucrose Dependent).
Supplemental Figure 2. ech2 Rescue Experiments in Hypocotyl Elongation.
Supplemental Figure 3. ech2 ibr Mutants Lack Morphological Defects as Adult Plants.
Supplemental Figure 4. ech2 ibr Mutants Have Short Root Hairs.
Supplemental Figure 5. Alignment of ECH2 and MFE2 Family Members.
Supplemental Figure 6. Alignment of ECH2, IBR10, MFE2, MFP2, and AIM1 Family Members.
Supplemental Table 1. Mutant Alleles Used in This Study.
Supplemental Table 2. New Markers Used in HR7 Mapping.
Supplemental Table 3. PCR Analysis of Mutant Genotypes.
Supplemental Data Set 1. Text File of the Sequences and Alignment Used for the Phylogenetic Analysis in Figure 1.
Supplementary Material
Acknowledgments
We thank Xing Liu (University of Minnesota) for assistance with mass spectrometry, the ABRC for providing the ECH2 cDNA and YFP-PTS1 (px-yk), and Wendell Fleming and Jerrad Stoddard for critical comments on the manuscript. This research was supported by the National Science Foundation (MCB-0745122 to B.B. and MCB-0725149 and IOS-0923960 to J.D.C.), the Robert A. Welch Foundation (C-1309 to B.B.), a Howard Hughes Medical Institute Professors Grant (to B.B.), the National Institutes of Health (1K99-GM089987 to L.C.S.), and the Gordon and Margaret Bailey Endowment for Environmental Horticulture (to J.D.C.). Confocal microscopy was performed on equipment obtained through a Shared Instrumentation Grant from the National Institutes of Health (S10RR026399-01).
References
- Adham A.R., Zolman B.K., Millius A., Bartel B. (2005). Mutations in Arabidopsis acyl-CoA oxidase genes reveal distinct and overlapping roles in β-oxidation. Plant J. 41: 859–874 [DOI] [PubMed] [Google Scholar]
- Barkawi L.S., Tam Y.Y., Tillman J.A., Normanly J., Cohen J.D. (2010). A high-throughput method for the quantitative analysis of auxins. Nat. Protoc. 5: 1609–1618 [DOI] [PubMed] [Google Scholar]
- Barkawi L.S., Tam Y.Y., Tillman J.A., Pederson B., Calio J., Al-Amier H., Emerick M., Normanly J., Cohen J.D. (2008). A high-throughput method for the quantitative analysis of indole-3-acetic acid and other auxins from plant tissue. Anal. Biochem. 372: 177–188 [DOI] [PubMed] [Google Scholar]
- Casimiro I., Beeckman T., Graham N., Bhalerao R., Zhang H., Casero P., Sandberg G., Bennett M.J. (2003). Dissecting Arabidopsis lateral root development. Trends Plant Sci. 8: 165–171 [DOI] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20: 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2007). Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cohen J.D. (1984). Convenient apparatus for the generation of small amounts of diazomethane. J. Chromatogr. A 303: 193–196 [Google Scholar]
- Cohen J.D., Baldi B.G., Slovin J.P. (1986). C(6)-[benzene ring]-indole-3-acetic acid: A new internal standard for quantitative mass spectral analysis of indole-3-acetic acid in plants. Plant Physiol. 80: 14–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein E., Ludwig-Müller J. (1993). Indole-3-butyric acid in plants: Occurrence, synthesis, metabolism, and transport. Physiol. Plant. 88: 382–389 [Google Scholar]
- Estelle M.A., Somerville C. (1987). Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol. Gen. Genet. 206: 200–206 [Google Scholar]
- Eubel H., Meyer E.H., Taylor N.L., Bussell J.D., O’Toole N., Heazlewood J.L., Castleden I., Small I.D., Smith S.M., Millar A.H. (2008). Novel proteins, putative membrane transporters, and an integrated metabolic network are revealed by quantitative proteomic analysis of Arabidopsis cell culture peroxisomes. Plant Physiol. 148: 1809–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett C.H., Wain R.L., Wightman F. (1960). The metabolism of 3-indolylalkanecarboxylic acids, and their amides, nitriles and methyl esters in plant tissues. Proc. R. Soc. Lond. B Biol. Sci. 152: 231–254 [DOI] [PubMed] [Google Scholar]
- Footitt S., Dietrich D., Fait A., Fernie A.R., Holdsworth M.J., Baker A., Theodoulou F.L. (2007). The COMATOSE ATP-binding cassette transporter is required for full fertility in Arabidopsis. Plant Physiol. 144: 1467–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J., Wiśniewska J., Benková E., Mendgen K., Palme K. (2002). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Froman B.E., Edwards P.C., Bursch A.G., Dehesh K. (2000). ACX3, a novel medium-chain acyl-coenzyme A oxidase from Arabidopsis. Plant Physiol. 123: 733–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goepfert S., Hiltunen J.K., Poirier Y. (2006). Identification and functional characterization of a monofunctional peroxisomal enoyl-CoA hydratase 2 that participates in the degradation of even cis-unsaturated fatty acids in Arabidopsis thaliana. J. Biol. Chem. 281: 35894–35903 [DOI] [PubMed] [Google Scholar]
- Goeschl J.D., Rappaport L., Pratt H.K. (1966). Ethylene as a factor regulating the growth of pea epicotyls subjected to physical stress. Plant Physiol. 41: 877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W.M., Östin A., Sandberg G., Romano C.P., Estelle M. (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson C., Schiefelbein J. (2002). Root hairs. In The Arabidopsis Book 1: e0061, doi/10.1199/tab.0060 [DOI] [PMC free article] [PubMed]
- Guzmán P., Ecker J.R. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper R.M., Stowe-Evans E.L., Luesse D.R., Muto H., Tatematsu K., Watahiki M.K., Yamamoto K., Liscum E. (2000). The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12: 757–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn G.W., Somerville C. (1986). Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol. Gen. Genet. 204: 430–434 [Google Scholar]
- Hayashi M., Toriyama K., Kondo M., Nishimura M. (1998). 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell 10: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer-Pascuzzi A.S., Benfey P.N. (2009). Transcriptional networks in root cell fate specification. Biochim. Biophys. Acta 1789: 315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Schell J. (1986). The promoter of the TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204: 383–396 [Google Scholar]
- Landrum M., Smertenko A., Edwards R., Hussey P.J., Steel P.G. (2010). BODIPY probes to study peroxisome dynamics in vivo. Plant J. 62: 529–538 [DOI] [PubMed] [Google Scholar]
- Last R.L., Fink G.R. (1988). Tryptophan-requiring mutants of the plant Arabidopsis thaliana. Science 240: 305–310 [DOI] [PubMed] [Google Scholar]
- Leyser H.M.O., Lincoln C.A., Timpte C., Lammer D., Turner J., Estelle M. (1993). Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364: 161–164 [DOI] [PubMed] [Google Scholar]
- Lingard M.J., Bartel B. (2009). Arabidopsis LON2 is necessary for peroxisomal function and sustained matrix protein import. Plant Physiol. 151: 1354–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig-Müller J. (2000). Indole-3-butyric acid in plant growth and development. Plant Growth Regul. 32: 219–230 [Google Scholar]
- Ludwig-Müller J., Epstein E. (1993). Indole-3-butyric acid in Arabidopsis thaliana. II. In vivo metabolism. Plant Growth Regul. 13: 189–195 [Google Scholar]
- Malamy J.E., Benfey P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Mansfield S.G., Briarty L.G. (1996). The dynamics of seedling and cotyledon cell development in Arabidopsis thaliana during reserve mobilization. Int. J. Plant Sci. 157: 280–295 [Google Scholar]
- Möller B., Weijers D. (2009). Auxin control of embryo patterning. Cold Spring Harb. Perspect. Biol. 1: a001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Perrot-Rechenmann C. (2010). Cellular responses to auxin: Division versus expansion. Cold Spring Harb. Perspect. Biol. 2: a001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson S.V., Johansson A.I., Kowalczyk M., Makoveychuk A., Wang J.Y., Moritz T., Grebe M., Benfey P.N., Sandberg G., Ljung K. (2009). An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 21: 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrášek J., Friml J. (2009). Auxin transport routes in plant development. Development 136: 2675–2688 [DOI] [PubMed] [Google Scholar]
- Pidugu L.S., Maity K., Ramaswamy K., Surolia N., Suguna K. (2009). Analysis of proteins with the ‘hot dog’ fold: Prediction of function and identification of catalytic residues of hypothetical proteins. BMC Struct. Biol. 9: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón N.M., Bartel B. (2010). Interdependence of the peroxisome-targeting receptors in Arabidopsis thaliana: PEX7 facilitates PEX5 accumulation and import of PTS1 cargo into peroxisomes. Mol. Biol. Cell 21: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampey R.A., LeClere S., Kowalczyk M., Ljung K., Sandberg G., Bartel B. (2004). A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 135: 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Babujee L., Ma C., Wienkoop S., Siemsen T., Antonicelli G.E., Rasche N., Lüder F., Weckwerth W., Jahn O. (2007). Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19: 3170–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond T.A., Bleecker A.B. (1999). A defect in β-oxidation causes abnormal inflorescence development in Arabidopsis. Plant Cell 11: 1911–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička K., et al. (2010). Arabidopsis PIS1 encodes the ABCG37 transporter of auxinic compounds including the auxin precursor indole-3-butyric acid. Proc. Natl. Acad. Sci. USA 107: 10749–10753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Scheres B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Stasinopoulos T.C., Hangarter R.P. (1990). Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol. 93: 1365–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A.N., Robertson-Hoyt J., Yun J., Benavente L.M., Xie D.Y., Dolezal K., Schlereth A., Jürgens G., Alonso J.M. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Strader L.C., Bartel B. (2008). A new path to auxin. Nat. Chem. Biol. 4: 337–339 [DOI] [PubMed] [Google Scholar]
- Strader L.C., Bartel B. (2009). The Arabidopsis PLEIOTROPIC DRUG RESISTANCE8/ABCG36 ATP binding cassette transporter modulates sensitivity to the auxin precursor indole-3-butyric acid. Plant Cell 21: 1992–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L.C., Bartel B. (February 28, 2011). Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol. Plant http://dx.doi.org/10.1093/mp/ssr006 [DOI] [PMC free article] [PubMed]
- Strader L.C., Beisner E.R., Bartel B. (2009). Silver ions increase auxin efflux independently of effects on ethylene response. Plant Cell 21: 3585–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L.C., Culler A.H., Cohen J.D., Bartel B. (2010). Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings. Plant Physiol. 153: 1577–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L.C., Monroe-Augustus M., Bartel B. (2008a). The IBR5 phosphatase promotes Arabidopsis auxin responses through a novel mechanism distinct from TIR1-mediated repressor degradation. BMC Plant Biol. 8: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L.C., Monroe-Augustus M., Rogers K.C., Lin G.L., Bartel B. (2008b). Arabidopsis iba response5 suppressors separate responses to various hormones. Genetics 180: 2019–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K., et al. (2008). The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10: 946–954 [DOI] [PubMed] [Google Scholar]
- Swofford D.L. (2001). PAUP*. Phylogenetic Analysis Using Parsimony (and Other Methods). (Sunderland, MA: Sinauer Associates; ). [Google Scholar]
- Tao Y., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q., Reed J.W. (1999). Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126: 711–721 [DOI] [PubMed] [Google Scholar]
- Tognetti V.B., et al. (2010). Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22: 2660–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F., Petrásek J., Zádníková P., Hoyerová K., Pešek B., Raz V., Swarup R., Bennett M., Zazímalová E., Benková E., Van Der Straeten D. (2010). The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 137: 597–606 [DOI] [PubMed] [Google Scholar]
- Wiszniewski A.A., Zhou W., Smith S.M., Bussell J.D. (2009). Identification of two Arabidopsis genes encoding a peroxisomal oxidoreductase-like protein and an acyl-CoA synthetase-like protein that are required for responses to pro-auxins. Plant Mol. Biol. 69: 503–515 [DOI] [PubMed] [Google Scholar]
- Woodward A.W., Bartel B. (2005a). The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol. Biol. Cell 16: 573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward A.W., Bartel B. (2005b). Auxin: Regulation, action, and interaction. Ann. Bot. (Lond.) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M., Greenham K., Prigge M.J., Jensen P.J., Estelle M. (2009). The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol. 151: 168–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Žádníková P., et al. (2010). Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137: 607–617 [DOI] [PubMed] [Google Scholar]
- Zhao Y. (2010). Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61: 49–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B.K., Bartel B. (2004). An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc. Natl. Acad. Sci. USA 101: 1786–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B.K., Martinez N., Millius A., Adham A.R., Bartel B. (2008). Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics 180: 237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B.K., Monroe-Augustus M., Silva I.D., Bartel B. (2005). Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell 17: 3422–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B.K., Nyberg M., Bartel B. (2007). IBR3, a novel peroxisomal acyl-CoA dehydrogenase-like protein required for indole-3-butyric acid response. Plant Mol. Biol. 64: 59–72 [DOI] [PubMed] [Google Scholar]
- Zolman B.K., Silva I.D., Bartel B. (2001). The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid β-oxidation. Plant Physiol. 127: 1266–1278 [PMC free article] [PubMed] [Google Scholar]
- Zolman B.K., Yoder A., Bartel B. (2000). Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156: 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.