Abstract
Aberrant activation of the JAK–STAT pathway has been implicated in many human cancers. It has widely been assumed that the effects of STAT activation are mediated by direct transcriptional induction of STAT target genes. However, recent findings in Drosophila have identified a non-canonical mode of JAK–STAT signaling, which directly controls heterochromatin stability. This indicates that the JAK–STAT pathway also controls cellular epigenetic status, which affects expression of genes beyond those under direct STAT transcriptional control. Given the evolutionary conservation of the canonical pathway among different species, the non-canonical mode of JAK–STAT signaling might also operate in vertebrates. In this review, canonical versus non-canonical JAK–STAT signaling and the implications for gene regulation and cancer formation are discussed.
Introduction: a new twist to an old pathway
The Janus kinase (JAK)–signal transducer and activator of transcription (STAT) pathway was originally identified as an intracellular signaling pathway mediating cytokine signals in mammals [1-4]. Since its discovery in the early 1990s, much research has been done to decipher the functions of JAK–STAT signaling in animals and the mechanisms by which the activity of this pathway is regulated. Accumulating evidence indicates that dysregulation of the JAK–STAT pathway causes human diseases and cancers [5,6]. Studies of knockout mice have defined essential non-redundant physiological or developmental roles for each family member of mammalian STAT proteins [7]. However, despite the remarkable progress in understanding the JAK–STAT pathway, the precise molecular mechanisms by which JAK–STAT signaling functions during normal development and in pathogenesis remain incompletely understood.
Studies in the genetic model organism Drosophila melanogaster have been informative with regard to the mechanisms and functions of JAK–STAT signaling. The Drosophila genome contains single copies of the JAK and STAT genes, which constitute a canonical JAK–STAT pathway (Figure 1). The presence of single-copy genes simplifies interpretation of gene function based on gain- or loss-of-function mutations. Using Drosophila genetics, it has recently been demonstrated that the JAK–STAT pathway can engage in a non-canonical mode of signaling, which affects cellular epigenetic status by globally modulating heterochromatin stability [8,9]. Changes in the chromatin barrier to transcription-factor binding potentiate gene expression. Heterochromatin formation and remodeling are important for many cellular processes such as gene silencing, chromosomal packaging and segregation during mitosis, genome stability and cell differentiation [10-12], and these processes are all affected by JAK or STAT mutations [8,9] (S.-J. Yan et al., unpublished). This novel function of JAK–STAT signaling might lead to new mechanistic insight into the molecular process of tumor-igenesis that is induced by STAT over-activation. Here, canonical versus non-canonical JAK–STAT signaling is discussed, with the focus being on the latter.
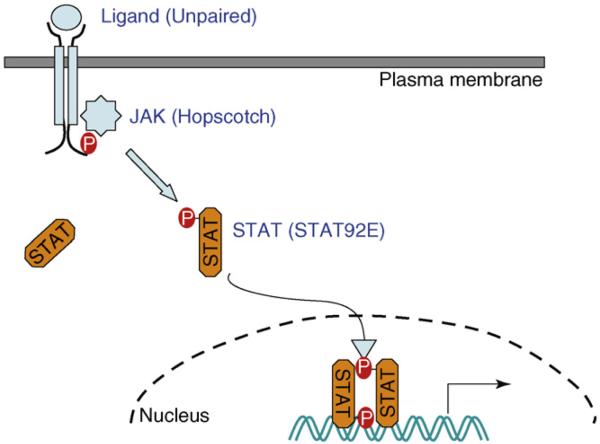
Figure 1.
Canonical JAK–STAT signaling. A simplified schematic representation of the Drosophila JAK–STAT pathway, which is typical of the canonical JAK–STAT pathway, in which unphosphorylated STAT resides in the cytoplasm. JAK activates STAT by phosphorylation, leading to nuclear translocation of dimerized phospho-STAT, which functions as a transcription factor. Names of the Drosophila homologs are in parentheses.
The canonical JAK–STAT pathway
There are seven STAT proteins (STAT1–4, 5A, 5B and 6) and four JAK kinases [JAK1–3 and tyrosine kinase 2 (TYK2)] in mammals. In the canonical mode of JAK–STAT signaling (Figure 1), activation of the pathway is initiated by binding of a peptide ligand (e.g. a cytokine) to transmembrane receptors. This leads to receptor dimerization and cross-activation of receptor-associated JAK kinases, which in turn phosphorylate tyrosine residues in the cytoplasmic tail of the receptor. These phospho-tyrosine residues function as docking sites for latent cytoplasmic STAT proteins, which are then phosphorylated by JAK on a crucial C-terminal tyrosine residue around the 700-amino-acid position. Phosphorylated STAT proteins dimerize via Src-homology 2 (SH2)-domain–phospho-tyrosine interactions and translocate to the nucleus, where they function as transcriptional activators, inducing expression of target genes [1,2].
Among the genes known to be regulated by mammalian STAT proteins are those encoding cell-survival factors, such as the B-cell lymphoma 2 (Bcl-2) family of proteins, those involved in cell proliferation, such as cyclin D1 and myc, and those implicated in angiogenesis or metastasis, such as vascular endothelial growth factor [5,6]. Conceivably, upregulation of genes that are important for cell proliferation and/or survival would promote cancer formation. Thus, it has been assumed that upregulation of these genes mediates the effects of STAT activation on cell behavior, which promotes cancer formation.
Genes regulated by the mammalian JAK–STAT pathway also include positive and negative regulators that modulate the magnitude and/or duration of signaling [2,13,14]. Activated STAT proteins drive their own expression, forming a positive-feedback loop or compensating for activation-induced STAT degradation. Several negative regulators of the pathway have been identified that are believed to be responsible for ‘turning off’ JAK–STAT signaling after activation. These include protein inhibitor of activated STAT (PIAS), suppressor of cytokine signaling (SOSC) and protein tyrosine phosphatases (PTPs) [2,13,14].
The Drosophila JAK–STAT pathway
The Drosophila genome contains a single JAK (Hopscotch or Hop) and a single STAT (STAT92E) protein, which are most homologous to JAK2 and STAT5, respectively [15,16]. Other components of this pathway identified by genetic studies in Drosophila include the secreted protein ligand Unpaired (Upd) [17], two Upd-related peptides, named Upd2 and Upd3 [18,19], and a transmembrane receptor (Domeless or Dom) [20,21], which are functionally similar to the mammalian cytokines and cytokine receptors upstream of JAK and STAT. In addition, as with the mammalian JAK–STAT pathway, the Drosophila pathway is autoregulatory by inducing positive and negative regulators [14]. As a positive regulator, STAT92E itself is transcriptionally induced by JAK–STAT signaling [22], similarly to mammalian STATs [2]. Some negative regulators of JAK–STAT signaling have been identified, including a PIAS [dPIAS or Su(var)2–10] [23], a SOCS (SOSC36E) [24-26] and a protein phosphatase PTP61F (a homolog of human phospo-Tyr phosphatase B1) [27,28]. Among these, at least SOCS36E and PTP61F are transcriptionally induced by STAT and then inhibit Hop and STAT92E activity by either hindering phosphorylation or by dephosphorylation [24-27].
Based on genetic and biochemical studies, the core components of the fly JAK–STAT pathway seem to function in a single linear manner that is typical of canonical JAK–STAT signaling (Figure 1). For example, after the finding that hop encodes a JAK kinase, STAT92E and Upd were genetically identified because their mutants exhibit the characteristic ‘hopscotch’ phenotypes – the larval cuticle is missing a few ventral denticle bands [15,17]. The fly pathway also shares biological functions with its mammalian counterpart, including regulating cell proliferation, differentiation, survival and migration [14,29]. The specific developmental processes that require JAK–STAT signaling in Drosophila include male-germline stem-cell maintenance [30,31], germ-cell function and migration [32,33], cell movement [34-36], hematopoiesis [37-39], immune responses [18,40,41], morphogenesis and patterning [42-45], imaginal tissue differentiation and proliferation [46-48] amongst others [14,26,29].
A Drosophila hematopoietic-tumor model: role of JAK–STAT
Studies of the Drosophila JAK–STAT pathway contributed to development of the initial hypothesis that human leukemia might be caused by JAK–STAT over-activation [37,38,49,50]. The Tumorous-lethal (Tum-l) mutation was first described in 1981 as causing a leukemia-like blood-cell over-proliferation. Tum-l turned out to be an allele of hop that encodes a temperature-sensitive hyper-active JAK kinase owing to a G341E substitution located in the N-terminal third of the protein, which is, presumably, a regulatory region [37,49]. Similarly to human leukemias, hopTum-l causes over-proliferation and clonal expansion of particular blood-cell types (plasmatocytes and lamellocytes; for a review of Drosophila-blood-cell development see Refs [51,52]) in heterozygous animals [37,39,49] (Figure 2). The hopTum-l mutation is associated with reduced animal viability and over-proliferation of blood cells that results in a high incidence of hematopoietic tumors, which are manifested as melanotic masses of aggregated blood cells in the larval or adult body cavity [37,38]. The idea that activating mutations in human JAK kinases would cause blood malignancies was later confirmed by the occurrences of the translocation-ets-leukemia (TEL)–JAK2 fusion oncoprotein and, more recently, the JAK2 V617F mutation in human leukemias [53-55].
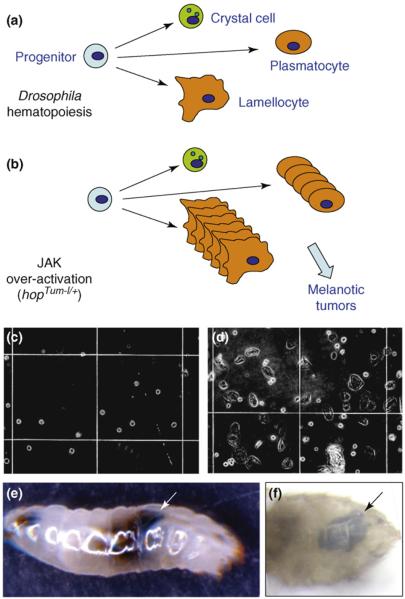
Figure 2.
Drosophila hemocyte development and blood-tumor formation. (a) The circulating hemocytes in the larval hemolymph (blood) consist of small crystal cells, intermediate phagocytic plasmatocytes and large, terminally differentiated phagocytic lamellocytes that are derived from a common stem cell or progenitor [51,52]. (b) The JAK-activating mutation hopTum-l causes overproliferation of plasmatocytes and lamellocytes, resulting in their aggregation and formation of melanotic tumors. (c,d) Micrographs show samples of larval blood from wild-type (c) and a hopTum-l+ (d) larva. Note the presence of large flat cells (lamellocytes) in (d). (e,f) Examples of melanotic tumors (arrows), which are visible without staining, in hopTum-l+ larvae (e) and adult flies (f).
JAK activation globally counteracts heterochromatin formation
Using the Drosophila hopTum-l hematopoietic-tumor model, genetic screens have been undertaken to identify genes important for tumor formation induced by over-activation of the JAK–STAT pathway [8,56]. A screen for suppressors of hopTum-l-associated animal lethality has identified a gain-of-function mutant Killer-of-prune (K-pn) allele of abnormal wing discs (awd), which encodes the Drosophila homolog of human-metastasis suppressor nm23-H1, a nucleoside diphosphate kinase [56]. The K-pn mutation exhibits no phenotype by itself, but causes lethality to the viable prune (pn) homozygous flies. A more recent screen for enhancers and suppressors of hopTum-l blood-tumor phenotype identified many predicted genes, including a loss-of-function mutation of awd as an enhancer of hopTum-l [8]. However, this screen also identified, unexpectedly, many modifier genes with chromatin-modifying functions. Among these are major heterochromatin components such as heterochromatin protein 1 (HP1), Su(var)3–9 (a histone methyltransferase) and Rpd3 (a histone deacetylase), and proteins that are important for heterochromatin formation such as RNA interference (RNAi) components Spn-E and Piwi [57]. These results indicate that heterochromatin formation normally antagonizes JAK-activation-induced hematopoietic tumorigenesis.
How does heterochromatin influence JAK-activation-induced tumorigenesis? Studies have shown that the heterochromatin-associated proteins do not modulate JAK–STAT signaling activity – they do not affect transcription of a STAT target reporter gene – but, rather conversely, it seems that JAK over-activation globally disrupts heterochromatin to cause blood-tumor formation [8]. Heterochromatin levels can be sensitively measured by position-effect variegation (PEV) (Box 1) and many heterochromatin components, including HP1 and Su(var)3–9, were initially isolated as dominant suppressor of variegation [Su(var)] mutations in Drosophila [10]. JAK gain- and loss-of-function mutations suppress and enhance, respectively, PEV at multiple independent chromosomal loci. Moreover, changes in levels of heterochromatin are also readily detectable in JAK gain- or loss-of-function mutant backgrounds by immunostaining with anti-HP1 or anti-histone H3 lys9 methylation (H3mK9) antibodies and by western blotting of total protein extracts [8].
Box 1. Position-effect variegation as a sensitive measurement of heterochromatin levels.
Position-effect variegation was first described as a Drosophila mutant by Nobel laureate Hermann Muller in 1938. By treating wild-type flies with X-ray, Muller found a mutant, which he named wm4 (white-mottled-4), in which the eye color was variegated (showing a red and white mosaic color). The red-eye pigmentation of wild-type Drosophila depends on expression levels of the white gene (w), which is located in euchromatin on the X chromosome. In the wm4 mutant, apparently, the white gene is expressed in some eye cells, but not in others. It was found that an X-ray induced inversion in the X chromosome caused the white gene to translocate next to pericentric heterochromatin. Owing to the loss of ‘heterochromatin boundary element’, heterochromatin formation during early development becomes metastable, resulting in mitotically heritable ‘on/off’ w+ expression patterns in the differentiated eye.
The overall w+ expression level (and red-eye pigmentation) in wm4 or other variegated mutant flies is inversely related to the level of heterochromatin. Indeed, genetic screens for mutations that suppress position-effect variegation have isolated >50 suppressors of variegation [Su(var)] mutations. Among the few Su(var) genes identified include those encoding integral heterochromatin components; for example, Su(var)205 encodes HP1 and Su(var)3–9 encodes a histone H3 lys9-specific histone methyltransferase (HMT) [10].
Consistent with the finding that JAK over-activation globally counteracts heterochromatin formation, a mutation of the negative regulator of JAK–STAT signaling, dPIAS, was originally isolated as Su(var)2–10, based on its ability to cause decreases in heterochromatin and, hence, to suppress PEV [58]. Further studies have shown that Su(var)2–10 is localized in the nucleus and regulates chromosome structure and function [59]. The PIAS family of proteins possesses small ubiquitin-related modifier (SUMO) E3-ligase activity [60]. Precisely how PIAS regulates chromosome structure and whether this is mediated by its ability to modulate STAT function remains unclear.
Furthermore, heterochromatin levels not only affect JAK-activation-induced hematopoietic tumorigenesis, but also significantly influence JAK-activation-induced cell over-proliferation during eye development [8]. Taken together, these results indicate that JAK over-activation globally disrupts heterochromatin, which would enable expression of genes not necessarily under direct STAT transcriptional regulation – a new mode of JAK signaling [61].
Unphosphorylated STAT stabilizes heterochromatin by binding to HP1
Given that STAT mediates JAK function in the canonical JAK–STAT pathway, the observation that Hop activation induces heterochromatin disruption raised the question of whether this effect is mediated by STAT92E. If STAT92E were required to positively mediate Hop function in this regard, reducing STAT92E levels would cause increased heterochromatin, just as reducing Hop levels causes increases in heterochromatin [8]. However, it was found that reducing Stat92E+ dosage had the opposite effect and strongly suppressed PEV [9]. The fact that loss of STAT92E had the same effect as Hop over-activation on PEV indicates that the effects of JAK–STAT signaling on heterochromatin do not occur simply via transcriptional regulation, at least, not via the canonical JAK–STAT pathway.
Further biochemical studies have shown that overexpression of Stat92E+ results in higher levels of heterochromatin (judging by increased H3mK9 and HP1 accumulation) and that, in Stat92E−/− cells, heterochromatin levels were much reduced [9]. Chromatin immunoprecipitation (ChIP) studies indicate that Stat92E RNAi knockdown reduced the association of HP1 with heterochromatin sequences [9]. Taken together, these results indicate that Stat92E is essential for HP1 localization and heterochromatin stability.
Because JAK activates STAT in the canonical JAK–STAT pathway [1,2], the observation that altering Stat92E levels has the opposite effects to hop gain- or loss-of-function on heterochromatin formation seems contradictory. However, a resolution to this paradox has been provided by the following findings.
First, Stat92E is found to be co-localized with HP1 in the nucleus in unstimulated Drosophila cells [9], in contrast to the notion that latent STAT proteins reside in the cytoplasm [1,2]. Moreover, Stat92E co-localization with HP1 in heterochromatic regions was also detected in S2 cells by ChIP or for a Stat92E–green fluorescent protein (GFP) transgene [9], demonstrating that at least a proportion of nuclear localized Stat92E was situated on heterochromatin. Staining of squashed polytene chromosomes from wild-type salivary glands indicated that Stat92E and HP1 co-localize in several regions of heterochromatin, including the chromocenter (in which the centromeres of polytene chromosomes attach to each other) and telomeres [9]. In contrast to the distribution of total Stat92E (which is detected by anti-Stat92E), phosphorylated Stat92E (which is detected by a phosphospecific antibody) is, more or less, uniformly distributed in the nucleus and does not co-localize with HP1 or heterochromatin [9]. These results indicate that some of the ‘inactive’ or unphosphorylated form of Stat92E, but not of phosphorylated Stat92E, is normally localized in the nucleus on heterochromatin.
Second, consistent with the co-localization results, Stat92E and HP1 physically interact. Stat92E contains a perfect and an imperfect HP1-binding sequence motif, Pro-Xaa-Val-Xaa-Leu [62]. Mutating both sites abolished Stat92E–HP1 interactions, indicating that either of the two sites might be sufficient to mediate Stat92E binding to HP1. Moreover, co-immunoprecipitation of Stat92E and HP1 decreases when levels of Stat92E phosphorylation increases (i.e. in hopTum-l/+ embryos or when Hop was overexpressed). This indicates that phosphorylation disrupts the association between Stat92E and HP1.
Third, examination of Stat92E–GFP and HP1 localization in ex vivo cultured salivary glands at different time points after the stimulation of Stat92E phosphorylation indicates that unphosphorylated Stat92E is associated with heterochromatin and that phosphorylation causes Stat92E dispersal and translocation to euchromatic regions, where it presumably binds to cognate promoters [9]. This result was corroborated by ChIP assays [9].
Finally, Stat92E phosphorylation-induced HP1 dispersal is not affected by blocking protein synthesis with cycloheximide (CHX) [9], indicating that STAT-activation-induced heterochromatin destabilization does not require new protein synthesis and, thus, cannot be mediated by induction of Stat92E transcriptional targets, including Stat92E itself.
Taken together, these results indicate that unphosphorylated Stat92E normally stabilizes HP1 localization and that activation of Stat92E by phosphorylation causes Stat92E dispersal from heterochromatin, thereby leading to HP1 displacement and heterochromatin destruction in a transcription-independent manner.
Non-canonical JAK-STAT signaling
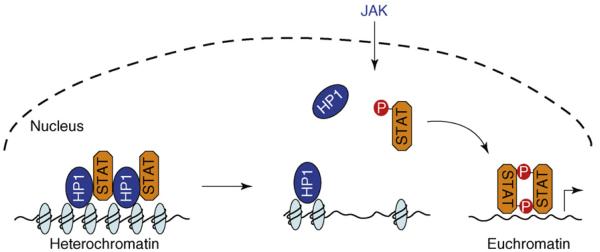
Studies in Drosophila have demonstrated a non-canonical mode of JAK–STAT signaling [8,9] (Figure 3). In contrast to the canonical mode of signaling, in which the latent STAT protein is localized in the cytoplasm, in the non-canonical mode, a portion of the unphosphorylated-STAT pool is localized in the nucleus on heterochromatin in association with HP1. The heterochromatin-associated unphosphorylated STAT is essential for maintaining HP1 localization and heterochromatin stability. Activation of STAT by phosphorylation causes STAT dispersal from heterochromatin, which in turn leads to HP1 displacement and heterochromatin destabilization. This process apparently does not require induction of STAT transcriptional target genes [9].
Figure 3.
Non-canonical JAK–STAT signaling. In the non-canonical mode of JAK–STAT signaling, unphosphorylated STAT is localized on heterochromatin in association with HP1 in the nucleus. Increasing STAT phosphorylation (by JAK or other tyrosine kinases) reduces the amount of unphosphorylated STAT localized on heterochromatin. This, in turn, leads to HP1 displacement from heterochromatin and heterochromatin instability. Dispersed phospho-STAT binds to cognitive sites in euchromatin to induce target-gene expression. Genes originally localized in heterochromatin are now accessible to STAT or other transcription factors.
Consistent with the findings in Drosophila, a predominantly nuclear localization of unphosphorylated mammalian-STAT proteins (STAT3 and STAT5A) has also been reported [63-66], although their subnuclear localization relative to heterochromatin has not been determined. Moreover, it has been reported that unphosphorylated STAT proteins are constantly shuttling between cytoplasmic and nuclear compartments [63-65]. Furthermore, unphosphorylated mammalian-STAT proteins have been shown to influence gene transcription by mechanisms distinct from those used by phosphorylated STAT [67,68].
It remains to be determined whether JAK enters the nucleus to phosphorylate STAT, or whether the redistribution of unphosphorylated nuclear STAT results from an altered equilibrium between nuclear and cytoplasmic or phosphorylated and unphosphorylated STAT proteins in response to JAK activation. Interestingly, JAK nuclear translocation has previously been reported for mammalian JAK2 [69].
Does STAT activation always cause heterochromatin destabilization under physiological conditions? This question is yet to be answered. The outcome of STAT activation might depend on the intensity of activation signals. Perhaps low levels of JAK–STAT activation sustain the expression of certain STAT target genes without affecting heterochromatin, whereas high levels of JAK–STAT activation cause heterochromatin disruption, resulting in global epigenetic changes in gene transcription.
Chromatin remodeling caused by JAK–STAT activation has been shown to occur in mammalian cells. For instance, transcriptional induction of human major histocompatibility complex (MHC) during immune response requires JAK–STAT signaling. It has been shown that JAK–STAT activation triggers higher-order chromatin remodeling of the entire MHC locus, resulting in chromosomal decondensation before transcriptional activation [70]. In another example, it has been shown recently that JAK3–STAT5 activation causes chromatin remodeling at the interferon-γ locus during T-helper-cell differentiation [71]. These findings indicate that non-canonical JAK–STAT signaling is present in mammals.
Epigenetic gene regulation in development and disease
Currently, it is not known if the changes in chromatin structure and gene expression caused by the non-canonical JAK–STAT pathway are transient or whether it can also induce inheritable (epigenetic) changes. Epigenetic regulation of gene expression refers to stably heritable repression or activation of gene expression via covalent modifications of DNA or histones, without changing the DNA sequence of the gene [12,72,73]. Epigenetic regulation has important roles at different stages of development, particularly during early embryonic development in which totipotent stem cells give rise to different cell lineages [74,75]. Differentiation is associated with permanent activation and repression of subsets of genes by stable histone or DNA modification.
There is increasing evidence that perturbation of epigenetic gene regulation has an important role in many human diseases [76,77]. For instance, local hypermethylation of tumor-suppressor genes and global hypomethylation of genomic DNA are both commonly found in human cancers and can have a causal role in tumorigenesis [76,77]. Histone modifications represent another type of epigenetic gene regulation. For instance, methylation of histone H3 at lysine 9 (H3mK9), catalyzed by histone methyltransferase Su(var)3–9 homologs, is associated with gene silencing, heterochromatin formation, and cellular senescence. H3mK9 provides binding sites for HP1, a key component of heterochromatin, which in turn recruits more Su(var)3–9 and associated proteins, thereby leading to heterochromatin assembly and spreading [10]. However, acetylation of histones, which is catalyzed by histone acetyltransferases (HATs), is associated with transcriptional activation [77]. Two well-studied HATs, p300 and CBP, function as co-factors for a large number of transcription factors or oncoproteins, such as p53 and AP1. Recruitment of HATs results in chromatin remodeling, thereby facilitating transcription [78]. Finally, non-canonical JAK–STAT signaling regulates heterochromatin stability, resulting in altered histone H3 methylation and/or chromatin remodeling [8,9,70,71].
Heterochromatin and tumor suppression
Heterochromatin, marked by histone H3 lys9 methylation (H3mK9), consists of highly condensed chromatin. Constitutive heterochromatin remains condensed throughout the cell cycle and is typically found in pericentromeric and telomeric regions of chromosomes. Facultative heterochromatin, however, represents chromosomal regions that can assume euchromatic or heterochromatic properties depending on the cellular context [10,11]. Heterochromatin, especially constitutive heterochromatin, has, until recently, been regarded as stable and static. However, studies of the exchange rate of HP1 indicate that constitutive heterochromatin can be quite dynamic and accessible, and is subject to remodeling by regulatory factors [79,80].
Heterochromatin formation has been implicated in tumor suppression [81,82]. Heterochromatin is associated with silencing of many oncogene-regulated promoters in quiescent cells [82]. Global disruption of heterochromatin conceivably enables de-repression of many ‘oncogenes’ and their targets. Indeed, the tumor suppressor Rb promotes heterochromatin formation at certain proto-oncogene promoters by recruiting HP1 [81,83]. By contrast, the oncoprotein Myc prevents heterochromatin formation by globally maintaining active chromatin structures [84]. Moreover, increased heterochromatin is a hallmark of cellular senescence, which functions as a barrier to cell proliferation and to oncogene-induced tumor formation in mammals [85,86]. In accordance with these data, in Drosophila, heterochromatin formation counteracts JAK–STAT activation-induced tumor formation and loss of heterochromatin promotes tumorigenesis [8]. Consistent with these results, in mammalian models, loss of heterochromatin or H3mK9 resulting either from loss of the key heterochromatin component HP1 or the Suv39h1 methyltransferase promotes cancer formation and/or progression [85,87,88]. These observations support the idea that heterochromatin is important for tumor suppression. It remains to be determined whether JAK–STAT over-activation disrupts heterochromatin to cause human leukemias.
Concluding remarks
Cell–cell communication via signal-transduction pathways has essential roles both in normal animal development and in physiological responses to the environment. Dysfunctional cell–cell communication, such as aberrant activation or inactivation of signal-transduction pathways, invariably causes diseases and cancers. Indeed, constitutive activation of STAT proteins, especially STAT3 and STAT5, has been detected in a variety of human cancers [5,6]. Furthermore, various oncoproteins have been shown to directly cause STAT activation [89,90]. These include the leukemogenic fusion oncoprotein tyrosine kinases such as breakpoint cluster region (Bcr)–Abelson (Abl), TEL–JAK2 and TEL–platelet-derived growth-factor-β receptor. Thus, it has been postulated that aberrant STAT activation might have a central role in cancer development. Whether heterochromatin disruption underlies any of these oncogenic events is yet to be determined but, in light of the recent studies in Drosophila, controlling heterochromatin stability by signaling pathways such as the JAK–STAT pathway could represent a new paradigm for understanding cancer development. A testable hypothesis is that heterochromatin formation constitutes a tumor suppression mechanism, and that heterochromatin destabilization is a crucial change in global chromatin structure that promotes oncogene-induced tumorigenesis.
Acknowledgements
I thank the Li laboratory members for discussion and comments. This work is supported by grants from the National Institutes of Health, an American Cancer Society Research Scholar Grant and a Leukemia and Lymphoma Society Research Scholar Grant.
Footnotes
Publisher's Disclaimer: This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research and education use, including for instruction at the authors institution and sharing with colleagues.
Other uses, including reproduction and distribution, or selling or licensing copies, or posting to personal, institutional or third party websites are prohibited.
Reference
- 1.Aaronson DS, Horvath CM. A road map for those who don't know JAK-STAT. Science. 2002;296:1653–1655. doi: 10.1126/science.1071545. [DOI] [PubMed] [Google Scholar]
- 2.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 3.Fu XY, et al. The proteins of ISGF-3, the interferon α-induced transcriptional activator, define a gene family involved in signal transduction. Proc. Natl. Acad. Sci. U.S.A. 1992;89:7840–7843. doi: 10.1073/pnas.89.16.7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schindler C, et al. Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. 1992;257:809–813. [PubMed] [Google Scholar]
- 5.Bromberg J. Stat proteins and oncogenesis. J. Clin. Invest. 2002;109:1139–1142. doi: 10.1172/JCI15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu H, Jove R. The STATs of cancer – new molecular targets come of age. Nat. Rev. Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 7.Ihle JN. The Stat family in cytokine signaling. Curr. Opin. Cell Biol. 2001;13:211–217. doi: 10.1016/s0955-0674(00)00199-x. [DOI] [PubMed] [Google Scholar]
- 8.Shi S, et al. JAK signaling globally counteracts heterochromatic gene silencing. Nat. Genet. 2006;38:1071–1076. doi: 10.1038/ng1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi S, et al. Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat. Cell Biol. 2008;10:489–496. doi: 10.1038/ncb1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grewal SI, Elgin SC. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 2002;12:178–187. doi: 10.1016/s0959-437x(02)00284-8. [DOI] [PubMed] [Google Scholar]
- 11.Grewal SI, Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 12.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 13.Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat. Rev. Immunol. 2003;3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 14.Arbouzova NI, Zeidler MP. JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development. 2006;133:2605–2616. doi: 10.1242/dev.02411. [DOI] [PubMed] [Google Scholar]
- 15.Hou XS, et al. Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell. 1996;84:411–419. doi: 10.1016/s0092-8674(00)81286-6. [DOI] [PubMed] [Google Scholar]
- 16.Yan R, et al. Identification of a Stat gene that functions in Drosophila development. Cell. 1996;84:421–430. doi: 10.1016/s0092-8674(00)81287-8. [DOI] [PubMed] [Google Scholar]
- 17.Harrison DA, et al. Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. 1998;12:3252–3263. doi: 10.1101/gad.12.20.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agaisse H, et al. Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev. Cell. 2003;5:441–450. doi: 10.1016/s1534-5807(03)00244-2. [DOI] [PubMed] [Google Scholar]
- 19.Gilbert MM, et al. A novel functional activator of the Drosophila JAK/STAT pathway, unpaired2, is revealed by an in vivo reporter of pathway activation. Mech. Dev. 2005;122:939–948. doi: 10.1016/j.mod.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Brown S, et al. Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr. Biol. 2001;11:1700–1705. doi: 10.1016/s0960-9822(01)00524-3. [DOI] [PubMed] [Google Scholar]
- 21.Chen HW, et al. mom identifies a receptor for the Drosophila JAK/STAT signal transduction pathway and encodes a protein distantly related to the mammalian cytokine receptor family. Genes Dev. 2002;16:388–398. doi: 10.1101/gad.955202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xi R, et al. A gradient of JAK pathway activity patterns the anterior-posterior axis of the follicular epithelium. Dev. Cell. 2003;4:167–177. doi: 10.1016/s1534-5807(02)00412-4. [DOI] [PubMed] [Google Scholar]
- 23.Betz A, et al. A Drosophila PIAS homologue negatively regulates stat92E. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9563–9568. doi: 10.1073/pnas.171302098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Callus BA, Mathey-Prevot B. SOCS36E, a novel Drosophila SOCS protein, suppresses JAK/STAT and EGF-R signalling in the imaginal wing disc. Oncogene. 2002;21:4812–4821. doi: 10.1038/sj.onc.1205618. [DOI] [PubMed] [Google Scholar]
- 25.Karsten P, et al. Cloning and expression of Drosophila SOCS36E and its potential regulation by the JAK/STAT pathway. Mech. Dev. 2002;117:343–346. doi: 10.1016/s0925-4773(02)00216-2. [DOI] [PubMed] [Google Scholar]
- 26.Rawlings JS, et al. Two Drosophila suppressors of cytokine signaling (SOCS) differentially regulate JAK and EGFR pathway activities. BMC Cell Biol. 2004;5:38. doi: 10.1186/1471-2121-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baeg GH, et al. Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes Dev. 2005;19:1861–1870. doi: 10.1101/gad.1320705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller P, et al. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature. 2005;436:871–875. doi: 10.1038/nature03869. [DOI] [PubMed] [Google Scholar]
- 29.Hou SX, et al. The JAK/STAT pathway in model organisms. Emerging roles in cell movement. Dev. Cell. 2002;3:765–778. doi: 10.1016/s1534-5807(02)00376-3. [DOI] [PubMed] [Google Scholar]
- 30.Kiger AA, et al. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 2001;294:2542–2545. doi: 10.1126/science.1066707. [DOI] [PubMed] [Google Scholar]
- 31.Tulina N, Matunis E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science. 2001;294:2546–2549. doi: 10.1126/science.1066700. [DOI] [PubMed] [Google Scholar]
- 32.Li J, et al. Coactivation of STAT and Ras is required for germ cell proliferation and invasive migration in Drosophila. Dev. Cell. 2003;5:787–798. doi: 10.1016/s1534-5807(03)00328-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown S, et al. JAK/STAT signalling in Drosophila controls cell motility during germ cell migration. Dev. Dyn. 2006;235:958–966. doi: 10.1002/dvdy.20709. [DOI] [PubMed] [Google Scholar]
- 34.Silver DL, Montell DJ. Paracrine signaling through the JAK/STAT pathway activates invasive behavior of ovarian epithelial cells in Drosophila. Cell. 2001;107:831–841. doi: 10.1016/s0092-8674(01)00607-9. [DOI] [PubMed] [Google Scholar]
- 35.Beccari S, et al. The JAK/STAT pathway is required for border cell migration during Drosophila oogenesis. Mech. Dev. 2002;111:115–123. doi: 10.1016/s0925-4773(01)00615-3. [DOI] [PubMed] [Google Scholar]
- 36.Johansen KA, et al. Localized JAK/STAT signaling is required for oriented cell rearrangement in a tubular epithelium. Development. 2003;130:135–145. doi: 10.1242/dev.00202. [DOI] [PubMed] [Google Scholar]
- 37.Hanratty WP, Dearolf CR. The Drosophila Tumorouslethal hematopoietic oncogene is a dominant mutation in the hopscotch locus. Mol. Gen. Genet. 1993;238:33–37. doi: 10.1007/BF00279527. [DOI] [PubMed] [Google Scholar]
- 38.Harrison DA, et al. Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J. 1995;14:2857–2865. doi: 10.1002/j.1460-2075.1995.tb07285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zettervall CJ, et al. A directed screen for genes involved in Drosophila blood cell activation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14192–14197. doi: 10.1073/pnas.0403789101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boutros M, et al. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev. Cell. 2002;3:711–722. doi: 10.1016/s1534-5807(02)00325-8. [DOI] [PubMed] [Google Scholar]
- 41.Agaisse H, Perrimon N. The roles of JAK/STAT signaling in Drosophila immune responses. Immunol. Rev. 2004;198:72–82. doi: 10.1111/j.0105-2896.2004.0133.x. [DOI] [PubMed] [Google Scholar]
- 42.Li J, et al. Patterns and functions of STAT activation during Drosophila embryogenesis. Mech. Dev. 2003;120:1455–1468. doi: 10.1016/j.mod.2003.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Josten F, et al. Cooperation of JAK/STAT and Notch signaling in the Drosophila foregut. Dev. Biol. 2004;267:181–189. doi: 10.1016/j.ydbio.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 44.Assa-Kunik E, et al. Drosophila follicle cells are patterned by multiple levels of Notch signaling and antagonism between the Notch and JAK/STAT pathways. Development. 2007;134:1161–1169. doi: 10.1242/dev.02800. [DOI] [PubMed] [Google Scholar]
- 45.Ayala-Camargo A, et al. The JAK/STAT pathway regulates proximo-distal patterning in Drosophila. Dev. Dyn. 2007;236:2721–2730. doi: 10.1002/dvdy.21230. [DOI] [PubMed] [Google Scholar]
- 46.Bach EA, et al. A sensitized genetic screen to identify novel regulators and components of the Drosophila janus kinase/signal transducer and activator of transcription pathway. Genetics. 2003;165:1149–1166. doi: 10.1093/genetics/165.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsai YC, Sun YH. Long-range effect of upd, a ligand for Jak/STAT pathway, on cell cycle in Drosophila eye development. Genesis. 2004;39:141–153. doi: 10.1002/gene.20035. [DOI] [PubMed] [Google Scholar]
- 48.Mukherjee T, et al. Opposing roles for Drosophila JAK/STAT signalling during cellular proliferation. Oncogene. 2005;24:2503–2511. doi: 10.1038/sj.onc.1208487. [DOI] [PubMed] [Google Scholar]
- 49.Luo H, et al. An amino acid substitution in the Drosophila hopTum-l Jak kinase causes leukemia-like hematopoietic defects. EMBO J. 1995;14:1412–1420. doi: 10.1002/j.1460-2075.1995.tb07127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dearolf CR. Fruit fly “leukemia”. Biochim. Biophys. Acta. 1998;1377:M13–M23. doi: 10.1016/s0304-419x(97)00031-0. [DOI] [PubMed] [Google Scholar]
- 51.Evans CJ, et al. Thicker than blood: conserved mechanisms in Drosophila and vertebrate hematopoiesis. Dev. Cell. 2003;5:673–690. doi: 10.1016/s1534-5807(03)00335-6. [DOI] [PubMed] [Google Scholar]
- 52.Meister M, Lagueux M. Drosophila blood cells. Cell. Microbiol. 2003;5:573–580. doi: 10.1046/j.1462-5822.2003.00302.x. [DOI] [PubMed] [Google Scholar]
- 53.Peeters P, et al. Fusion of TEL, the ETS-variant gene 6 (ETV6), to the receptor-associated kinase JAK2 as a result of t(9;12) in a lymphoid and t(9;15;12) in a myeloid leukemia. Blood. 1997;90:2535–2540. [PubMed] [Google Scholar]
- 54.James C, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 55.Levine RL, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 56.Zinyk DL, et al. Drosophila awdK-pn, a homologue of the metastasis suppressor gene nm23, suppresses the Tum-1 haematopoietic oncogene. Nat. Genet. 1993;4:195–201. doi: 10.1038/ng0693-195. [DOI] [PubMed] [Google Scholar]
- 57.Pal-Bhadra M, et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 58.Wustmann G, et al. The genetics of position-effect variegation modifying loci in Drosophila melanogaster. Mol. Gen. Genet. 1989;217:520–527. doi: 10.1007/BF02464926. [DOI] [PubMed] [Google Scholar]
- 59.Hari KL, et al. The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev. 2001;15:1334–1348. doi: 10.1101/gad.877901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharrocks AD. PIAS proteins and transcriptional regulation–more than just SUMO E3 ligases? Genes Dev. 2006;20:754–758. doi: 10.1101/gad.1421006. [DOI] [PubMed] [Google Scholar]
- 61.Betz A, Darnell JE., Jr A Hopscotch-chromatin connection. Nat. Genet. 2006;38:977–979. doi: 10.1038/ng0906-977. [DOI] [PubMed] [Google Scholar]
- 62.Thiru A, et al. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 2004;23:489–499. doi: 10.1038/sj.emboj.7600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reich NC, Liu L. Tracking STAT nuclear traffic. Nat. Rev. Immunol. 2006;6:602–612. doi: 10.1038/nri1885. [DOI] [PubMed] [Google Scholar]
- 64.Vinkemeier U. Getting the message across, STAT! Design principles of a molecular signaling circuit. J. Cell Biol. 2004;167:197–201. doi: 10.1083/jcb.200407163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu L, et al. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-α3. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8150–8155. doi: 10.1073/pnas.0501643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iyer J, Reich NC. Constitutive nuclear import of latent and activated STAT5a by its coiled coil domain. FASEB J. 2008;22:391–400. doi: 10.1096/fj.07-8965com. [DOI] [PubMed] [Google Scholar]
- 67.Yang J, et al. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFκB. Genes Dev. 2007;21:1396–1408. doi: 10.1101/gad.1553707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang J, Stark GR. Roles of unphosphorylated STATs in signaling. Cell Res. 2008;18:443–451. doi: 10.1038/cr.2008.41. [DOI] [PubMed] [Google Scholar]
- 69.Ram PA, Waxman DJ. Interaction of growth hormone-activated STATs with SH2-containing phosphotyrosine phosphatase SHP-1 and nuclear JAK2 tyrosine kinase. J. Biol. Chem. 1997;272:17694–17702. doi: 10.1074/jbc.272.28.17694. [DOI] [PubMed] [Google Scholar]
- 70.Christova R, et al. P-STAT1 mediates higher-order chromatin remodelling of the human MHC in response to IFNγ. J. Cell Sci. 2007;120:3262–3270. doi: 10.1242/jcs.012328. [DOI] [PubMed] [Google Scholar]
- 71.Shi M, et al. Janus-kinase-3-dependent signals induce chromatin remodeling at the Ifng locus during T helper 1 cell differentiation. Immunity. 2008;28:763–773. doi: 10.1016/j.immuni.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 73.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 74.Boyer LA, et al. Molecular control of pluripotency. Curr. Opin. Genet. Dev. 2006;16:455–462. doi: 10.1016/j.gde.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 75.Hochedlinger K, Jaenisch R. Nuclear reprogramming and pluripotency. Nature. 2006;441:1061–1067. doi: 10.1038/nature04955. [DOI] [PubMed] [Google Scholar]
- 76.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat. Rev. Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 77.Lund AH, van Lohuizen M. Epigenetics and cancer. Genes Dev. 2004;18:2315–2335. doi: 10.1101/gad.1232504. [DOI] [PubMed] [Google Scholar]
- 78.Imhof A, et al. Acetylation of general transcription factors by histone acetyltransferases. Curr. Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 79.Cheutin T, et al. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- 80.Festenstein R, et al. Modulation of heterochromatin protein 1 dynamics in primary mammalian cells. Science. 2003;299:719–721. doi: 10.1126/science.1078694. [DOI] [PubMed] [Google Scholar]
- 81.Narita M, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 82.Ogawa H, et al. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- 83.Nielsen SJ, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- 84.Knoepfler PS, et al. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Braig M, et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- 86.Mathon NF, Lloyd AC. Cell senescence and cancer. Nat. Rev. Cancer. 2001;1:203–213. doi: 10.1038/35106045. [DOI] [PubMed] [Google Scholar]
- 87.Norwood LE, et al. A requirement for dimerization of HP1Hsα in suppression of breast cancer invasion. J. Biol. Chem. 2006;281:18668–18676. doi: 10.1074/jbc.M512454200. [DOI] [PubMed] [Google Scholar]
- 88.Cloos PA, et al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- 89.Benekli M, et al. Signal transducer and activator of transcription proteins in leukemias. Blood. 2003;101:2940–2954. doi: 10.1182/blood-2002-04-1204. [DOI] [PubMed] [Google Scholar]
- 90.Sternberg DW, Gilliland DG. The role of signal transducer and activator of transcription factors in leukemogenesis. J. Clin. Oncol. 2004;22:361–371. doi: 10.1200/JCO.2004.10.124. [DOI] [PubMed] [Google Scholar]