Human POR gene transcription requires proximal sequences responsive to TR-β and Smad3/4. A common polymorphism at –152 bp reduces expression in liver and adrenal cells.
Abstract
P450 oxidoreductase (POR) is the flavoprotein that acts as the obligatory electron donor to all microsomal P450 enzymes, including those involved in hepatic drug metabolism as well as three steroidogenic P450 enzymes. The untranslated first exon of human POR was located recently, permitting analysis of human POR transcription. Expression of deletional mutants containing up to 3193 bp of the human POR promoter in human adrenal NCI-H295A and liver Hep-G2 cells located the proximal promoter at −325/−1 bp from the untranslated exon. Common human POR polymorphisms at −208 and −173 had little influence on transcription, but the polymorphism at −152 reduced transcription significantly in both cell lines. EMSA and supershift assays identified binding of Smad3/Smad4 between −249 and −261 and binding of thyroid hormone receptor-β (TRβ) at −240/−245. Chromatin immunoprecipitation showed that Smad3, Smad4, TRα, TRβ, and estrogen receptor-α were bound between −374 and −149. Cotransfection of vectors for these transcription factors and POR promoter-reporter constructs into both cell types followed by hormonal treatment showed that T3 exerts major tropic effects via TRβ, with TRα, estrogen receptor-α, Smad3, and Smad4 exerting lesser, modulatory effects. T3 also increased POR mRNA in both cell lines. Thyroid hormone also is essential for rat liver POR expression but acts via different transcription factor complexes. These are the first data on human POR gene transcription, establishing roles for TRβ and Smad3/4 in its expression and indicating that the common polymorphism at −152 may play a role in genetic variation in steroid biosynthesis and drug metabolism.
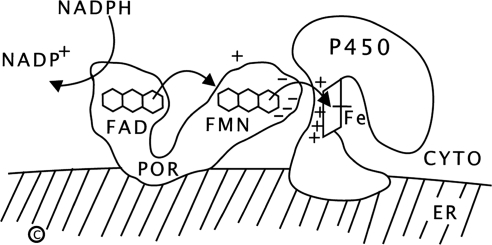
Human P450 oxidoreductase (POR) is a 77-kDa, 680-amino acid protein required for the activity of all microsomal (type 2) cytochrome P450 enzymes, including the steroidogenic enzymes P450c17 (17α-hydroxylase, 17,20 lyase), P450c21 (21-hydroxylase), and P450aro (aromatase), and all hepatic drug-metabolizing cytochrome P450 enzymes (reviewed in Ref. 1). POR contains two flavin moieties: a flavin adenine dinucleotide (FAD) and a flavin mononucleotide (FMN); each resides in a separate lobe connected by a flexible linker (2, 3). The FAD moiety of POR receives a pair of electrons from the reduced form of nicotinamide adenine dinucleotide phosphate; this elicits a conformational change, bringing the FAD and FMN moieties close together so that the electrons pass to the FMN domain; after reversion to its initial conformation, the FMN domain interacts with the redox-partner binding site of a microsomal P450 enzyme, donating the electrons and permitting P450-mediated catalysis (Fig. 1). POR also serves as a cofactor for several non-P450 enzymes, including squalene monoxygenase (4), fatty acid elongase (5), heme oxygenase (6), and cytochrome b5 (7).
Fig. 1.
Function of POR. Reduced nicotinamide adenine dinucleotide phosphate (NADPH) donates electrons (e-) to the FAD of POR, which is bound to the endoplasmic reticulum. Electron receipt elicits a conformational change, permitting the isoalloxazine rings of the FAD and FMN moieties to come close together, so that the electrons pass from the FAD to the FMN. POR then returns to its original orientation, permitting the FMN domain of POR to interact with the redox-partner binding site of the P450. Electrons from the FMN domain of POR reach the heme group to achieve catalysis. The interaction of POR and the P450 is coordinated by negatively charged acidic residues on the surface of the FMN domain of POR and positively charged basic residues in the redox-partner binding site of the P450. CYTO, Cytoplasm; ER, endoplasmic reticulum.
POR has attracted considerable recent interest following the discovery that POR mutations cause a broad spectrum of human disease, ranging from severe skeletal malformations associated with defective steroidogenesis (Antley-Bixler Syndrome), to phenotypically normal individuals with infertility (8–14). The human POR gene, located on chromosome 7:75382356–75454109, consists of 16 exons (15). The sequence of this gene in 842 normal persons from four ethnic groups revealed a high degree of polymorphism; most notably, the coding sequence variant A503V was found on approximately 28% of all alleles (16). That study identified 12 common single-nucleotide polymorphisms, having frequencies ranging from 3–80% of alleles in at least one of our four ethnic groups (see Table 4 in Ref. 16); three of these, 75382148 C→T (−208 C→T), 75382183 C→A (−173 C→A), and 75382204 C→A (−152 C→A), lie in the promoter region within 208 bp of the ATG translational start codon. Promoter polymorphisms contribute to genetic variation in hepatic drug metabolism mediated by some cytochrome P450 enzymes. For example, the −806 C→T polymorphism in the CYP2C19 promoter increases transcriptional activity (17), whereas −740 T→G and −730 C→T intron 1 polymorphisms in the CYP1A2 gene decreases binding of the Ets transcription factor and inducibility by dioxin (18). Similarly, ultrafast metabolism by CYP2D6 results from gene duplication (19). Thus variations in POR promoter activity might influence both steroidogenesis and drug metabolism.
The transcriptional regulation of the human POR gene has not been examined previously, but some studies have addressed the regulation of bovine and rat POR. In primary cultures of bovine adrenal cells, both ACTH and cAMP, but not phenobarbital, promoted accumulation of POR mRNA, roughly in parallel with the mRNAs for P450c17 and P450c21 (20). In intact rats, hypophysectomy reduced hepatic, and, to a lesser degree, adrenal POR activity, protein accumulation, and mRNA abundance; this reduction could not be reversed by treating the rats with ACTH, GH, or human chorionic gonadotropin, but was restored by treatment with T4 (21, 22). The discovery of the untranslated first exon of the rat POR gene showed that it lacked TATA or CCAAT boxes, but had several potential Sp1 sites (23). RNA polymerase run-on assays showed an increase in rat liver POR gene transcription in response to thyroid hormone, and analysis of the 5′-flanking DNA of the rat POR gene identified a thyroid-response region having the sequence AGGTGAgctgAGGCCA at bases −564 to −536 (24, 25). A proximal Egr-1/CACCC element at −206 was required, irrespective of thyroid status (26). With our identification of the untranslated first exon of the human POR gene (15), it is now possible to examine the transcriptional regulation of the human POR gene.
Results
Computational analysis of the POR promoter and design of promoter/reporter constructs
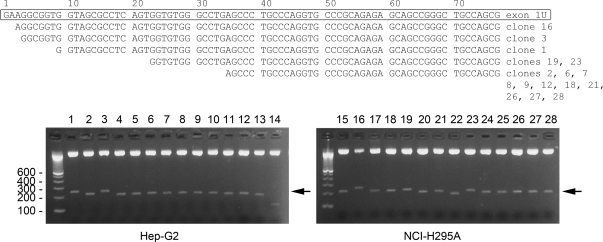
The identification of the untranslated upstream exon 1U was done by probing the human genome sequence with the 56-base sequence of the rat POR upstream untranslated exon (15), showing that the human POR gene is 71.8 kb long; however, the 5′-end of human POR mRNA was not determined experimentally. To determine whether the human POR gene contains additional untranslated exons upstream from exon 1U, we performed 5′-rapid amplification of cDNA ends (RACE) using homopolymeric tailed cDNAs prepared from Hep-G2 cells and NCI-H295A cells. The 5′-RACE products were cloned into pBluescript, 28 positive clones were isolated, and the eight longest 5′-RACE products from each cell line were sequenced (Fig. 2). The 5′-ends of these 16 longest clones were located within the first 36 nucleotides (nt) of exon 1U, and none of the clones extended upstream of exon 1U, indicating that the human POR gene does not contain exons that lie upstream of exon 1U.
Fig. 2.
Location of the POR transcriptional start site. Cloning the 5′-RACE products from Hep-G2 and NCI-H295A cells yielded 28 clones. The boxed sequence in the top panel shows the sequence of exon 1U as inferred (14), followed by the sequences of the eight longest 5′-RACE products from Hep-G2 cells (clones 1–3, 6–9, and 12 as indicated to the right of the sequence) and the eight longest 5′-RACE products from NCI-H295A cells (clones 16, 18, 19, 21, 23, and 26–28). The bottom panel shows the ethidium bromide-stained Tris-borate-EDTA-agarose gels of the 28 positive clones from Hep-G2 and NCI-H295A cells, digested with SalI-BamHI. Arrow indicates the POR 5′-RACE insert fragments.
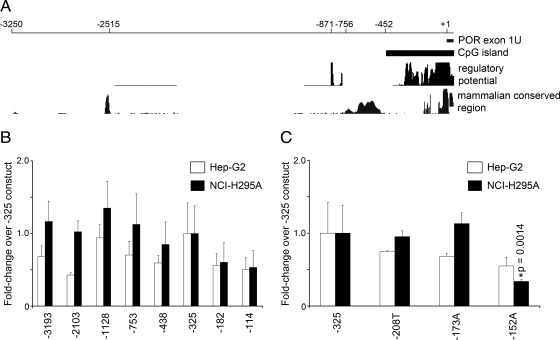
Using the UCSC Genome Browser (http://genome.ucsc.edu) and ECR browser (http://ecrbrowser.dcode.org/), we searched 10 kb of the POR 5′-flanking DNA sequence for upstream, putative regulatory elements that are conserved in mammals and sequences predicted to contain putative transcription factor-binding sites. The computational approaches identified an 810-base long CpG island extending from −454 to +356, with a 0.84 ratio of observed to expected CpG. Several regions that are highly conserved among mammals and several other regions that have sequences similar to transcription factor-binding sites were also identified (Fig. 3A). A series of eight promoter/reporter constructs was built based on this computational analysis and named according to the 5′-most base of the POR promoter: −3193 includes all the conserved regions identified; −2103 deletes the conserved mammalian region at −2515; −1128 deletes other apparently unimportant DNA; −753 deletes the two small clustered regions of apparent regulatory potential at −871 and −756; −438 deletes the broad region of conserved mammalian sequences between −756 and −452; −325 deletes to the beginning of the region rich in potential regulatory elements; −182 deletes to the beginning of the two proximal mammalian conserved regions; −114 deletes the upstream of these two mammalian conserved regions.
Fig. 3.
Analyses of human POR promoter activity in Hep-G2 and NCI-H295A cells. A, Computational mapping of the human POR promoter from nt −3250 to +78 (exon 1U). The regulatory potential (RP) compares frequencies of short alignment patterns between known regulatory elements and neutral DNA (61, 62). The mammalian conserved region was computed based on multiple sequence alignments of the POR genes from human, chimp, rhesus, mouse, rat, dog, and cow; the y-axis shows regions with greater than random nucleotide sequence identity. B, POR promoter-reporter deletional constructs −3193, −2103, −1128, −753, −438, −325, −182, and −114 were designed based on the analysis in panel A. The promoter segments were cloned into a luciferase reporter vector, transfected into Hep-G2 and NCI-H295A cells, and assayed for activities after 24 h. The data are expressed as fold-change over the −325 construct (mean ± sem) from three independent experiments, each done in triplicate. C, The wild-type −325 POR promoter-reporter construct and −325 construct carrying the polymorphisms −208 T, −173 A, and −152 A were transfected into Hep-G2 and NCI-H295A cells and assayed for assayed for luciferase activities. Asterisks indicate a significant difference between the wild-type −325 POR construct and the −152 A POR construct (P = 0.0014).
POR promoter-luciferase activity
Because POR is required by microsomal cytochrome P450 enzymes that are essential for adrenal steroidogenesis and hepatic drug metabolism, we analyzed the activities of the POR promoter constructs in human cell lines that model these two tissues: adrenal NCI-H295A cells and liver Hep-G2 cells. Compared to their activities in the adrenal cells, the activities of the constructs from −438 to −3193 were somewhat lower in Hep-G2 cells, and there was a consistently lower activity of the −2103 construct in Hep-G2 cells. However, there were no differences greater than about 2-fold among any of the constructs longer than −325, suggesting that the most important elements driving basal POR transcriptional activity in Hep-G2 and NCI-H295A cells lie within −325 bases upstream from the POR transcriptional start site (Fig. 3B). Therefore, we arbitrarily set the activity of the −325 construct as 100% for both cell lines, and compared the other constructs to the −325 construct. Deletion to −182 reduced activity in both cell lines to about half of the −325 level, and further deletion to −114 had no further effect. Thus the promoter/reporter data suggest that key elements required for the basal expression of the human POR gene lie between −182 and −325 bases from the transcriptional start site and would suggest that the region encompassing the polymorphism at −152 is not important for POR gene transcription (Fig. 3B). However, none of the apparent differences in Fig. 3B reached statistical significance.
By sequencing the proximal POR promoter in 702 normal persons of various ethnicities, we previously found three single-nucleotide polymorphisms, −208 C→T, −173 C→A and −152 C→A, in the 5′-flanking region at frequencies ranging from 3% to 13% (16). To examine the potential effects of the POR promoter polymorphisms at −208, −173, and −152 directly, we incorporated these polymorphisms into the −325 construct by site-directed mutagenesis. As shown in Fig. 3C, the −208T and −173A constructs had reporter activities comparable to the wild-type −325 construct, but the activity of the construct containing the −152 C→A polymorphism was reduced by about half in Hep-G2 cells (not significant) and to approximately 35% in NCI-H295A cells (P = 0.0014), suggesting that −152 is associated with an important regulatory element. However, because deletion to −114 did not reduce the activity compared with the −182 construct (Fig. 3B), the significance of the result with the −152 C→A change was unclear.
EMSA
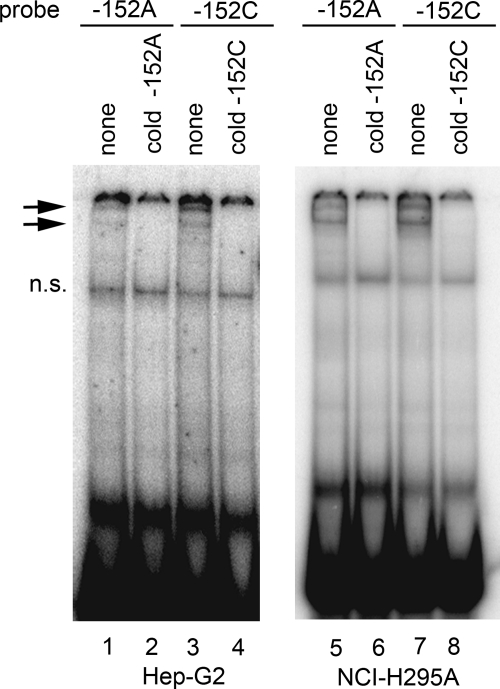
To determine whether the polymorphism at −152 affects binding of transcription factors, we performed EMSA using nuclear extracts from NCI-H295A or Hep-G2 cells and 32P-labeled double-stranded DNA probes encompassing bases −163 to −139 with either C or A at position −152 (Fig. 4). Both the −152A and −152C probes produced similar DNA-protein band-shift patterns (lanes 1, 3, 5, and 7) that could be competed by the corresponding unlabeled oligonucleotides (lanes 2, 4, 6, and 8), suggesting that both probes bound similar proteins from both NCI-H295A and Hep-G2 nuclear extracts. Thus the polymorphism at −152 did not visibly alter binding of nuclear proteins in vitro.
Fig. 4.
EMSA with wild-type −152C and mutant −152A probes. EMSA was done with nuclear extracts from Hep-G2 (lanes 1–4) and NCI-H295A (5–8) cells and 32P-labeled probes for −152A and −152C oligonucleotides in the absence (lanes 1 and 5) or presence (lanes 2–4 and 6–8) of unlabeled competitor DNA oligonucleotides as indicated above the lanes. Arrows indicate DNA-protein complexes. Both the A and C oligonucleotides bind the same pattern of complexes, and an excess of each cold oligonucleotide can compete for this binding. The nonspecific band is indicated by n.s.
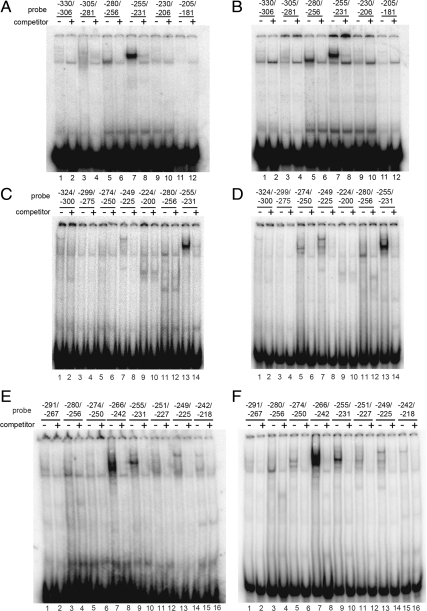
Because more basal transcriptional activity is contained within the −325 construct than in the −182 construct (Fig. 3B), we sought to identify the transcription factors that might bind to the promoter region from −325 to −182; to do this we scanned the region from −330 to −181 by EMSA using six oligonucleotides from the region −330/−306 to −205/−181. When incubated with nuclear extracts from either NCI-H295A or Hep-G2 cells, probes −280/−256 and −255/−231 produced band-shift patterns (in lanes 5 and 7, respectively) that could be competed by their corresponding unlabeled oligonucleotides (lanes 6 and 8, respectively) (Fig. 5, A and B). Next we repeated the experiment using a new set of probes that overlap the probes used in Fig. 5, A and B. As shown in Fig. 5, C and D, probes −324/−300 to −224/−200 produced weak band-shift patterns (lanes 1–10) compared with probes −280/−256 and −255/−231 in NCI-H295A and Hep-G2 extracts (lanes 11–14). To confirm that the POR promoter region −280 to −231 is important for binding transcription factors, we performed EMSA using probes that overlap one another from −291 to −218. As shown in Fig. 5, E and F, the strongest band-shift patterns were obtained using probes for −266/−242 (lane 7) and −255/−231 (lane 9). The probe −280/−256 produced weaker band-shift patterns with both NCI-H295A and Hep-G2 extracts (lane 3, Fig. 5, E and F). Thus our results indicate that the region −280 to −231 is required for binding transcription factors.
Fig. 5.
Scanning EMSA with probes from −330 to −181. EMSA was done with nuclear extracts from Hep-G2 (A, C, and E) and NCI-H295A (B, D, and F) cells and 32P-labeled probes corresponding to nucleotides −330/−306 (lanes 1 and 2), −305/−281 (lanes 3 and 4), −280/−256 (lanes 5 and 6), −255/−231 (lanes 7 and 8), −230/−206 (lanes 9 and 10), and −205/−181 (lanes 11 and 12) in the absence (−) or presence (+) of unlabeled competitor oligonucleotides. Hep-G2 (C) and NCI-H295A (D) nuclear extracts were used with 32P-labeled probes for regions −324/−300 (lanes 1 and 2), −299/−275 (lanes 3 and 4), −274/−250 (lanes 5 and 6), −249/−225 (lanes 7 and 8), −224/−200 (lanes 9 and 10), −280/−256 (lanes 11 and 12), and −255/−231 (lanes 13 and 14) in the absence (−) or presence (+) of unlabeled competitor oligonucleotides. Nuclear extracts from Hep-G2 (E) and NCI-H295A cells (F) were incubated with 32P-labeled probes for regions −291/−267 (lanes 1 and 2), −280/−256 (lanes 3 and 4), −274/−250 (lanes 5 and 6), −266/−242 (lanes 7 and 8), −255/−231 (lanes 9 and 10), −251/−227 (lanes 11 and 12), −249/−225 (lanes 13 and 14), and −242/−218 (lanes 15 and 16) in the absence or presence of unlabeled competitor oligonucleotides. In panels A and B, the −255/−231 probe binds better than the −280/−256 probe; in panels C and D, the −255/−231 binds better than the −274/−250 probe; in panels E and F the −266/−242 probe binds better than the −255/−231 probe.
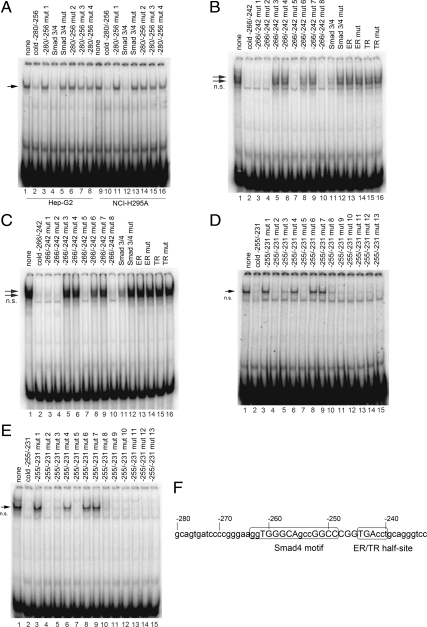
A search of the DNA from −280 to −231 using Transcription Element Search System (TESS; http://www.cbil.upenn.edu/cgi-bin/tess/tess) and rVISTA 2.0 (27) identified sequences potentially bound by heat shock factor 1, GAL4, c-Ets-2, Sp1, estrogen receptor (ER)α, GCF, steroidogenic factor 1, AP-1, thyroid hormone receptor (TR)α, PPARα, LVc, and Sma and Mad related protein 4 (Smad4). The EMSA band produced by the −280/−256 probe could be competed by excess unlabeled oligonucleotide (lanes 2 and 10) or by an oligonucleotide containing the Smad3/4 consensus sequence (lanes 4 and 12) (Fig. 6A). However, all four −280/−256 mutant oligonucleotides (lanes 3, 6–8, 11, 14–16) or the mutant of this Smad3/4 consensus (lanes 5 and 13) failed to compete with the −280/−256 probe for binding proteins from NCI-H295A and Hep-G2 nuclear extracts. To confirm that Smad3/4 binds to the −280/−256 region, we performed EMSA with a downstream overlapping probe −266/−242 using nuclear extracts from NCI-H295A and Hep-G2 cells. As shown in Fig. 6, B and C, the band shift patterns produced by the −266/−242 probe could be competed by unlabeled oligonucleotide (lane 2), oligonucleotide mutants 1, 2, 5, and 8 (lanes 3, 4, 7, and 10), and the Smad3/4 consensus (lane 11), but not by oligonucleotide mutants 3, 4, 6, and 7 (lanes 5, 6, 8, and 9), the Smad3/4 mutant (lane 12), or oligonucleotides containing the consensus sequences recognized by ER or TR, or their mutants (lanes 13–16). The sequence GGTGGGCAGCCGGCC in POR promoter region −263 to −249 matches the Smad4 motif (28). Taken together, the results in Fig. 6, A−C, suggest that Smad3/4 binds between bases −280 to −242 in the human POR promoter. As shown in Fig. 6, E and F, the −255/−231 probe produced a band that could be competed by excess unlabeled oligonucleotide (lane 2), but not by mutant −255/−231 oligonucleotides 4 (lane 6), and 6 and 7 (lanes 8 and 9). These mutant oligonucleotides carried mutations in a putative ER half-site, AGGTCA (29), which may prevent the binding of the nuclear receptors ER, retinoid X receptor (RXR), TR, vitamin D receptor (VDR), and RXR (Fig. 6, D and E). Thus nucleotides −261/−256 and −252/−247 in the Smad4 motif, −258/−256, and −246/−243 in the ER/TR half-site in POR promoter, are required for the binding of transcription factors (Fig. 6F).
Fig. 6.
EMSA scanning mutagenesis of the region between −231 and −280 identified in Fig. 5. A, EMSA was done with nuclear extracts from Hep-G2 cells (lanes 1–8) and NCI-H295A cells (lanes 9–16) and 32P-labeled −280/−256 probe in the absence (lanes 1 and 9) or presence (lanes 2–8 and 10–16) of unlabeled mutant competitor oligonucleotides (sequences shown in Table 1). A single DNA-protein complex is seen (arrow). B, EMSAs done with the −266/−242 probe and nuclear extracts from Hep-G2 cells in the absence (lane 1) or presence of cold competitors for −266/−242 mutants (lanes 2–10), and Smad3/4 consensus and mutant (lanes 11 and 12), ER consensus and mutant (lanes 13 and 14), and TR consensus and mutant (lanes 15 and 16). The nonspecific band is indicated by n.s. C, As in panel B except using nuclear extract from NCI-H295A cells. D, EMSA was done with the −255/−231 probe and nuclear extracts from Hep-G2 cells in the absence (lane 1) or presence of cold competitors for −255/−231 oligonucleotide (lane 2) and mutants (lanes 3–15). E, As in panel D except using nuclear extract from NCI-H295A cells. F, Sequence of the POR promoter between −280 to −231. A putative Smad4 motif is located at −263/−249, and an ER/TR half-site is located at −245/−240. The presence or absence of binding to the various mutant oligonucleotides used in panels A–E indicate that the bases shown in uppercase are required for the binding of transcription factors. mut, Mutant.
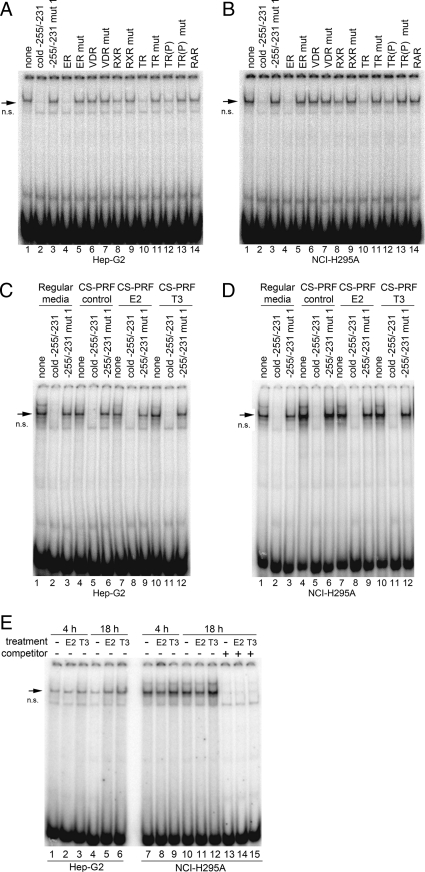
To determine whether ER, RXR, TR, VDR, or RXR bind to the −255/−231 region of the human POR promoter, we used their binding sequences as unlabeled competitors for the −255/−231 probe in EMSA. Unlabeled consensus oligonucleotides containing the sequences bound by ER, TR, and RXR were effective as competitors (lanes 4, 8, 10, and 12) using nuclear extracts from both Hep-G2 (Fig. 7A) and NCI-H295A (Fig. 7B), whereas oligonucleotides containing the mutant ER, TR, and RXR sequences or the consensus VDR and RXR sequences were ineffective as competitors. These data suggest that ER, TR, and RXR may act as transcription factors that bind to the −255/−231 region of POR.
Fig. 7.
EMSA with −255/−231 probes. EMSA was done with nuclear extracts from Hep-G2 (A) and NCI-H295A (B) cells and 32P-labeled −255/−231 probe in the presence of cold competitor oligonucleotides (lanes 2–16) as indicated above the lanes. The arrow indicates the DNA-protein complex. The oligonucleotides contain the following consensus sequences or their mutants (mut): ER, VDR, RXR, TR. The TR sequence contains two half-sites arranged as a direct repeat with four nucleotides spacing; in TR(P) the direct repeats are inverted. The nonspecific band is indicated by n.s. EMSA was also done with the −255/−231 probe and nuclear extracts from Hep-G2 (C) and NCI-H295A (D) cells cultured in regular growth media (lanes 1–3), or in hormone-free (charcoal-stripped, phenol red-free) (CS-PRF) media in the absence (lanes 4–6) or presence of 10 nm E2 (lanes 7–9) or 100 nm T3 (lanes 10–12). E, EMSA was done with 255/−231 probe and nuclear extracts from Hep-G2 (lanes 1–6) and NCI-H295A (lanes 7–15) cells cultured in charcoal-stripped media lacking phenol red in the absence (lanes 1, 4, 7, 10, and 13) or presence of 10 nm E2 (lanes 2, 5, 8, 11, and 14) or 100 nm T3 (lanes 3, 6, 9, 12, and 15) for 4 h (lanes 1–3 and 7–9) and 18 h (lanes 4–6, and 10–15).
Because TR, ER, and RXR appear to bind to the −255/−231 region of the human POR promoter, we determined whether estradiol (E2) or T3 affect the binding of transcription factors to this DNA. Because other agents can bind to ER and TR, we used nuclear extracts from Hep-G2 and NCI-H295A cells grown in media that contained no phenol red and used charcoal-stripped serum (hormone-free media). As shown in Fig. 7, C and D, the −255/−231 probe produced bands that can be competed by excess unlabeled oligonucleotide but not by the mutant oligonucleotide when incubated with nuclear extracts of cells grown in regular culture media (lanes 1–3) or in hormone-free media (lanes 4–6). Furthermore, the EMSA patterns were not altered by overnight treatments of Hep-G2 or NCI-H295A cells with 10 nm E2 (lanes 7–9) or 100 nm T3 (lanes 10–12). Similarly, the EMSA patterns were not altered when the cells were incubated with E2 or T3 for 4 h (Fig. 7E).
Antibody supershift assays
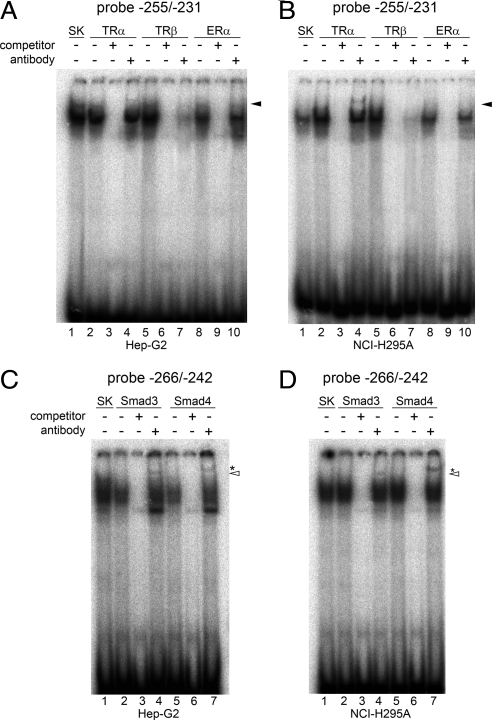
Antibody supershift experiments confirmed that TRα and Smad3/4 bind to the −255/−231 region of the human POR promoter. Hep-G2 and NCI-H295A cells were transfected with the expression plasmids for ERα, Smad3, Smad4, TRα, TRβ, or pBluescript SK vector only, and nuclear extracts were prepared after overnight treatments with 10 nm E2, 3 ng recombinant human TGFβ, or 100 nm T3 (Fig. 8). Supershift assays were done by preincubating nuclear extracts with the appropriate antibodies before adding 32P-labeled double-stranded DNA probes. Preliminary results suggested that the ligand-receptor complexes dissociated during the experiments; hence we added exogenous hormones to the nuclear extract-antibody reaction mixtures in the same concentrations used in the cellular incubations (100 nm T3 or 10 nm E2) to maintain the complexes during the supershift assays. The incubation of the −255/−231 probe with nuclear extracts from Hep-G2 and NCI-H295A cells expressing TRα produced a DNA-protein complex that was supershifted by the antibody against TRα (Fig. 8, A and B, lanes 2 vs. lane 4). Several antisera were tried; nevertheless, the supershifted bands were rather faint, even with long autoradiographic exposures. The complex formed by the −255/−231 probe and nuclear proteins from cells expressing TRβ was inhibited by the antibody to TRβ (Fig. 8, A and B, lane 5 vs. lane 7). In Fig. 8, C and D, antibodies against Smad3 and Smad4 supershifted (lanes 4 and 7) the DNA-protein complexes formed by the −266/−242 probe and nuclear extracts from cells expressing Smad3 and Smad4 (lanes 2 and 5). The available antisera to ERα failed to yield a supershift or detectable interference with complex formation (Fig. 8, A and B).
Fig. 8.
EMSA supershift assays. Hep-G2 and NCI-H295A cells were transfected with expression plasmids for either ERα, Smad3, Smad4, TRα, TRβ, or pBluescript SK vector and treated with either 10 nm E2, 3 ng recombinant human TGFβ, or 100 nm T3. EMSA supershift assays were done with 32P-labeled −255/−231 probe and nuclear extracts from the Hep-G2 and NCI-H295A cells transfected with ERα, TRα, TRβ, or pBluescript SK vector only (panels A and B) in the presence or absence of unlabeled oligonucleotide −255/−231 competitor or antibodies against either TRα, TRβ, and ERα (indicated above the lanes). For TR and ER supershift assays, 100 nm T3 or 10 nm E2 was added to the antibody-nuclear extract binding mixtures, respectively. The arrowhead in panel B indicates the antibody-supershifted band by TRα antibody. In panels C and D, supershift assays were done with −266/−242 probe and nuclear extracts from Hep-G2 and NCI-H295A cells transfected with Smad3, Smad4, or pBluescript SK vector only in the presence or absence of cold oligonucleotide −266/−242 competitor or antisera to either Smad3 or Smad4. The Smad4 supershift band migrates slower in the gel because Smad4 is a larger protein than Smad3 (552 aa vs. 425 aa) (54). The asterisks and open arrowheads indicate the supershift bands by Smad3 and Smad4 antibodies, respectively. Due to the weak intensities of the supershifted bands, long exposures of the autoradiograms are shown in panels A–D.
Chromatin immunoprecipitation (ChIP)
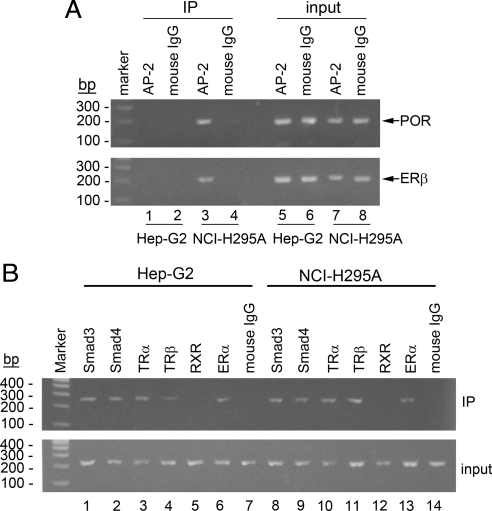
Searches of the POR promoter sequence from −325 to −1 using Transcription Element Search System (TESS; http://www.cbil.upenn.edu/cgi-bin/tess/tess) and rVISTA 2.0 (27) identified a potential activator protein 2 (AP-2 site) (GCGACCCAGCC) extending from −153 to −143, but identified no other potential transcription factors that might bind at the polymorphic base, −152. Hep-G2 cells do not express AP-2 (30); hence AP-2 was not a candidate to explain the impact of the −152 polymorphism in these cells, but it remained possible that AP-2 might be germane to this region's activity in NCI-H295A cells, which do express AP-2 (31). To determine whether AP-2 is recruited to this region of the POR promoter, we performed ChIP assays. DNA/protein complexes in HepG2 and NCI-H295A cells were cross-linked with formaldehyde, the chromatin was fragmented by sonication, and the cell lysates were immunoprecipitated with antibodies to AP-2. The DNA in the immunoprecipitated chromatin was extracted, and the POR promoter region from −222 to −36 was amplified by PCR. The presence of a PCR product indicates that the DNA was bound by AP-2. The data show that AP-2 is recruited to the POR promoter in NCI-H295A cells, but not in Hep-G2 cells (Fig. 9A).
Fig. 9.
ChIP. A, Proteins cross-linked to chromatin from Hep-G2 and NCI-H295A cells were precipitated with antiserum directed against AP-2, and the associated DNA sequences were amplified with PCR primers that amplify the POR promoter region between −222 to −36, spanning the putative AP-2 binding sites. PCR products from input chromatin before (input) and after immunoprecipitation (IP) are shown. PCR amplification with ERβ primers was used as a positive control for the AP-2 ChIP assay, and mouse IgG was used as a negative control. B, ChIP assay was done with antisera directed against ERα, Smad3, Smad4, TRα, TRβ, RXRα, and amplified with PCR primers that amplify the POR promoter region between −374 to −149, spanning the Smad3/4 and ER/TR binding sites.
Because the EMSA data indicated that Smad3/4, TR, ER, and possibly RXR bind to −280 to −231 of the POR promoter, we performed ChIP assays to determine whether these factors are recruited to this segment of DNA. Cell lysates from Hep-G2 and NCI-H295A cells were subjected to another ChIP assay with antibodies to Smad3, Smad4, TRα, TRβ, RXRα, and ERα, followed by PCR to amplify the POR promoter region from −374 to −149. As shown in Fig. 9B, Smad3 (lanes 1 and 8), Smad4 (lanes 2 and 9), TRα (lanes 3 and 10), TRβ (lanes 4 and 11), and ERα (lanes 6 and 13), but not RXRα (lanes 5 and 12), are recruited to the POR promoter in both Hep-G2 and NCI-H295A cells under basal conditions.
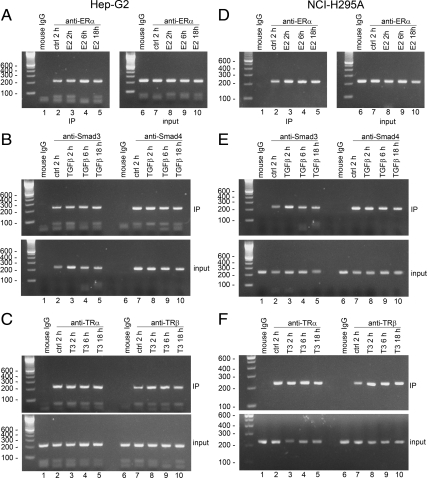
We next did ChIP assays to determine whether E2, TGFβ, or T3 affect the recruitment of ERα, Smad3, Smad4, TRα, and TRβ to the POR promoter. Hep-G2 and NCI-H295A cells were treated with these factors for various times, chromatin was immunoprecipitated with antibodies to ERα, Smad3, Smad4, TRα, and TRβ, and the POR promoter region from −374 to −149 was amplified by PCR. As shown in Fig. 10, compared with the untreated cells (lane 2), treatments with 10 nm E2, 3 ng TGFβ, or 100 nm T3 for 2 h, 6 h, or 18 h (lanes 3–5) did not alter the recruitment of ERα (Fig. 10, A and D), Smad3, Smad4 (Fig. 10, B and E), and TRα (Fig. 10, C and F) to the POR promoter in either Hep-G2 and NCI-H295A cells. However, the recruitment of TRβ to the endogenous POR promoter was increased after 2 h treatment with T3 (Fig. 10, C and F).
Fig. 10.
ChIP assays of the effects of E2, TGFβ, and T3 on recruitment of ERα, Smad3, Smad4, TRα, and TRβ to the POR promoter. Hep-G2 cells (A–C) and NCI-H295A cells (D–F) were treated for 2 h, 6 h, or 18 h in the absence (lanes 1 and 2) or presence of 10 nm E2, 3 ng TGFβ, or 100 nm T3 (lanes 3–5). Proteins cross-linked to chromatin were precipitated with antisera directed against ERα (A and D), Smad3, Smad4 (B and E), TRα and TRβ (C and F), and amplified with PCR primers that amplify the POR promoter region between −374 and −149. IP, Immunoprecipitation; ctrl, control.
T3 regulation of the POR promoter by TR
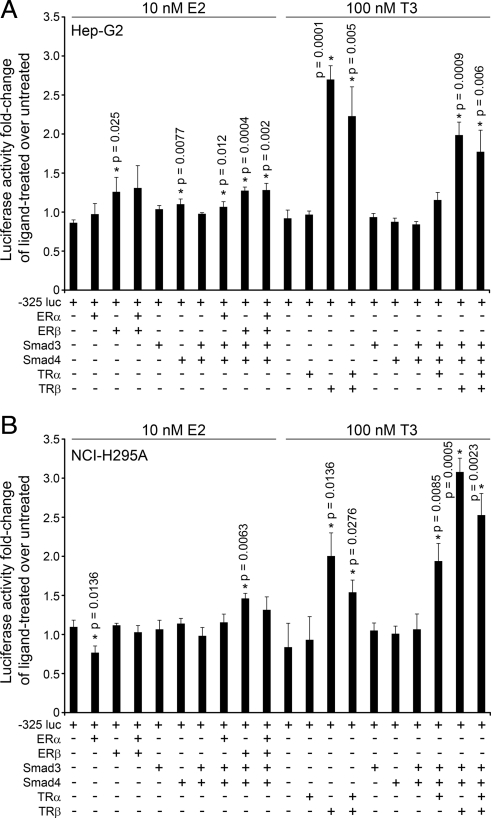
Because ERα, Smad3, Smad4, TRα, or TRβ are recruited to the POR promoter, we sought to determine whether these factors regulate POR promoter activity. Therefore, we cotransfected NCI-H295A and Hep-G2 cells with expression constructs for these transcription factors and for the −325 POR promoter-reporter construct and treated the cells with 10 nm E2 or 100 nm T3 for 18 h. As shown in Fig. 11, A and B, −325 POR promoter-reporter activity was not affected by ERα, ERβ, Smad3, or Smad4 alone, or the combination of ERα, ERβ, Smad3, and Smad4 in E2-treated or untreated cells. However, in the presence of T3, POR promoter activity in Hep-G2 cells was increased 2.8-fold by TRβ alone, 2.3-fold by TRα + TRβ, 2.1-fold by TRβ + Smad3 + Smad4, and 1.8-fold by TRα + TRβ + Smad3 + Smad4 (Fig. 11A). In NCI-H295A cells treated with T3, POR promoter activity increased 2.4-fold with TRβ alone, 1.8-fold with TRα + TRβ, 2.2-fold with TRα + Smad3 + Smad4, 3.7-fold with TRβ + Smad3 + Smad4, and 3.0-fold with TRα + TRβ + Smad3 + Smad4 (Fig. 11B). Thus, our data suggest that T3 regulation of POR promoter activity occurs predominantly by TRβ, although the combination of TRα, Smad3, and Smad4 may also contribute some activity.
Fig. 11.
Regulation of −325 POR promoter-reporter activity by T3. The −325 POR promoter-reporter construct was cotransfected into Hep-G2 (A) and NCI-H295A (B) cells (with a Renilla control) and expression plasmids for ERα, ERβ, Smad3, Smad4, TRα, and/or TRβ, as indicated. The cells were treated with 10 nm E2 or 100 nm T3 for 18 h and assayed for luciferase activity. Data are expressed as fold change of hormonally treated over untreated cells (mean ± sem) from three independent experiments, each done in triplicate. Statistically significant differences from control for each hormone and cell type are indicated by asterisks (*) and the corresponding P value.
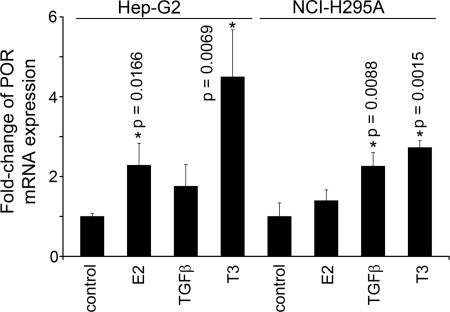
To determine whether E2, TGFβ, or T3 regulates the abundance of POR mRNA, Hep-G2 and NCI-H295A cells were treated for 18 h with 10 nm E2, 3 ng TGFβ, or 100 nm T3, and cDNAs were prepared for quantitative real-time PCR detection using the SYBR green fluorescent dye. In Hep-G2 cells, E2 increased POR expression by 2.2-fold, and T3 increased POR mRNA by 4.5-fold, but the apparent 1.9-fold induction by TGFβ was not statistically significant (Fig. 12). In NCI-H295A cells, E2 did not increase POR mRNA, and TGFβ and T3 increased POR mRNA 2.3-fold and 2.7-fold, respectively (Fig. 12).
Fig. 12.
Quantitative RT-PCR. Hep-G2 and NCI-H295A cells were treated for 18 h in the absence or presence of 10 nm E2, 3 ng TGFβ, or 100 nm T3, and cDNAs were prepared for qRT-PCR and SYBR green detection. The amount of POR mRNA was normalized to the amount of GAPDH mRNA, and the data are expressed as mean fold change of POR expression in treated cells over the untreated controls ± sd. Statistically significant differences from control for each cell type are indicated by asterisks (*) and the corresponding P value.
Discussion
POR is required for the activities of two adrenal P450 enzymes that participate in cortisol biosynthesis and for hepatic drug-metabolizing cytochrome P450 enzymes. Therefore, to study the transcriptional regulation of human POR, we used human adrenal NCI-H295A cells and human liver Hep-G2 cells as models of these two tissues, because our previous work permitted identification of adrenal-specific and liver-specific cis-acting transcriptional elements by this tactic (32). Deletional mutagenesis scanning 3193 bp upstream from the untranslated first exon showed that most basal transcriptional activity is mediated by sequences within the first 325 bp; thus, this DNA is the POR-proximal promoter. Our sequencing of this DNA in 701 normal persons identified nine rare sequence variants, present in less than 0.5% of the population, and three polymorphisms, present in more than 1% of the population (16). Among African-Americans (AA), Caucasian Americans (CA), Asian Americans (AS), and Mexican Americans (MA), the frequency of the C→T polymorphism at base −208 (genome position 75382148) was 6.2%, 10.8%, 3.8%, and 2.8%, respectively. For the C→A polymorphism at −173 (75382183), the frequency in the AA, CA, AS, and MA populations was 1.2%, 4.4%, 1.5%, and 3.1%, respectively. For the C→A polymorphism at −152 (75382204), the frequency in the AA, CA, AS, and MA populations was 2.6%, 13.0%, 7.8%, and 7.7%, respectively. The activities of the −325 construct containing the −208 and −173 polymorphisms were normal in NCI-H295A adrenal cells and in Hep-G2 liver cells, suggesting that these polymorphisms exert minimal effects on POR transcription. By contrast, activity of the −325 construct containing the −152 polymorphism was reduced to approximately 60% in Hep-G2 cells and to approximately 35% in NCI-H295A cells, suggesting that this polymorphism may be an important contributor to diminutions in the capacity to metabolize drugs or synthesize steroids. ChIP assays showed that the −152 POR promoter region binds AP-2 in NCI-H295A cells but not in Hep-G2 cells, which do not express AP-2 mRNA (30). Thus AP-2 is not the transcription factor responsible for the difference in POR transcription in the −152C vs. −152A alleles. The results suggest that the −152A sequence variant might recruit an inhibitory factor, but TESS and rVISTA searches using the mutant sequence to query the database did not identify a candidate factor.
We used EMSA to search for transcription factors that bind to sequences in the proximal promoter, showing that the region between −240 and −263 bound several factors, but we were unable to show protein binding to any of the polymorphic regions. Because the functional data clearly establish the importance of the region containing −152, it is likely that the factors binding to this DNA were either lost from our nuclear extract or bound with low affinity. Thus we sought to identify the factors binding between −240 and −263.
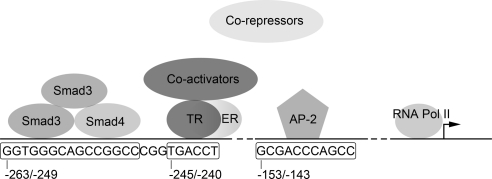
Current knowledge of the binding of transcription factors to the human POR promoter is summarized in Fig. 13. Smad3, Smad4, TRα, TRβ, and ERα are recruited to the proximal POR promoter between bases −240 and −263. Smad3 and Smad4 played a role in both NCI-H295A and Hep-G2 cells but Smad3 or Smad4 alone did not induce POR transcription. Smads bind DNA as a complex of one Smad4 and two Smad3 proteins (33); the motif at POR promoter region −263/−249 recruits this trimeric Smad complex. Thyroid hormone acts by recruiting a coactivator complex to replace a corepressor complex (34). TRα, TRβ, and ER could bind to the nuclear hormone half-site at −245/−240 (Fig. 13), but the effect of estradiol was minimal, and TRα may have slightly inhibited the induction by TRβ. The role of ER on POR regulation is unclear, although it is possible that TR may compete with ER for binding and interfere with transcription (35). In NCI-H295A cells, Smad3 and Smad4 appear to act synergistically with TRα to regulate POR expression in the presence of T3. These three factors could interact with one another to regulate POR promoter activity as another nuclear zinc-finger transcription factor, retinoic acid receptor (RAR)-γ, can interact with Smad3 (36). Smad3 signals through the TGFβ pathway (33). T3 treatment can increase TGFβ expression in Hep-G2 cells stably expressing TRα (37); this could be another mechanism by which TRα regulates POR expression (Fig. 13).
Fig. 13.
Binding of transcription factors to the human POR promoter. A heterotrimer of two Smad3 molecules and one Smad4 binds to the region between −263 and −249. Our in vitro data show that TR (TRα and/or TRβ) and ER can bind to the nuclear receptor half-site at −245/−240, but our functional data in transfected Hep-G2 and NCI-H295A cells indicate that the principal tropic regulator is TRβ. Binding of T3 to the TR recruits coactivators that displace corepressors and activate RNA polymerase II. The coactivators may interact with the Smad complex. AP-2 binds POR promoter region between −153 and −143 in NCI-H295A. The POR promoter probably binds additional factors, not yet identified. Pol II, Polymerase II.
In bovine adrenals (20) and in rat adrenals and liver (21, 22), ACTH via cAMP and T4 are the principal hormonal regulators of POR expression. In rat liver, a thyroid-responsive element at −564/−536 and an Egr-1 element at −206 are required to mediate this response (24–26). Our data show that the human POR promoter is also regulated by thyroid hormone and that the principal mediator of this regulation is TRβ, acting on a thyroid-response element at −245/−240. Consistent with this, T3 induced accumulation of POR mRNA in both NCI-H295A adrenal cells and Hep-G2 liver cells. In NCI-H295A cells, but not in Hep-G2 cells, TRα may also play a role. By contrast, the thyroid-responsive elements in the rat POR promoter are located at −564 to −536 and are bound by TRα-TRα and TRβ-TRβ homodimers and TRα-RXRα and TRβ-RXRα complexes (24). Thus, although transcription from both the rat and human POR promoters is induced by thyroid hormone, they achieve this with different DNA elements at different locations that bind different TR transcription complexes.
Because POR is the electron donor for type 2 microsomal P450 enzymes that function in hepatic drug metabolism and adrenal steroidogenesis, the effects of T3, E2, and TGFβ on POR and the P450 enzymes should be considered together. Our results show that POR gene transcription and mRNA accumulation are predominantly increased by T3 in Hep-G2 and NCI-H295A cells, with smaller effects of TGFβ and E2. By contrast, various microsomal P450 enzymes may have different responses to E2, TGFβ, or T3: for example, estrogen stimulates the 21-hydroxylase activity of CYP2C6 in a dose-related manner in primary cultures of rat hepatocytes (38); TGFβ inhibits the accumulation of P450c17 and ACTH receptor mRNA and protein in both bovine and human adrenal cells (39–41); TGFβ inhibits CYP2B6, CYP2C8, CYP2C9, CYP2C11, CYP2C19, and CYP3A4 expression in rat and human hepatocytes (42, 43); and thyroid hormone inhibits CYP2A1, CYP3A2, CYP3A4, and CYP4A2 but stimulated CYP2C7 and had no effect on CYP1A2, CYP2A2, CYP2C9, CYP2C11, CYP2C12, and CYP2E1, in rat and human hepatocytes (44–49). Thus, T3, E2 and TGFβ lead to complex effects on POR and microsomal P450 enzymes, so that any POR/CYP combination of interest will need to be studied individually.
Materials and Methods
RACE
RNA was prepared from Hep-G2 and NCI-H295A cells using the Trizol reagent (Invitrogen, Carlsbad, CA), and 4 μg was reverse transcribed in 20 mm Tris-HCl (pH 8.4), 50 mm KCl, 5 mm MgCl2, 10 mm dithiothreitol, 500 μm each dNTP, 2.5 pmol antisense oligonucleotide to POR (as329-rt, 5′-TGTTCCTCCCCGTTTTCTTCATCTTTT; nt 329–303), 40 U RNaseOUT (porcine liver Rnase A inhibitor; Invitrogen), and 200 U SuperScript III reverse transcriptase (Invitrogen) at 50 C for 50 min. The reaction was terminated at 85 C for 5 min, chilled on ice, and incubated with 2 U RNase H (Invitrogen) at 37 C for 20 min. The cDNA was purified through a QIAquick spin column (Qiagen, Chatsworth, CA), and a homopolymeric tail was added to the 3′-end of the cDNA in 10 mm Tris-HCl (pH 8.4), 25 mm KCl, 1.5 mm MgCl2, 200 μm dCTP, 20 U terminal deoxynucleotidyl transferase (New England Biolabs, Beverly, MA) at 37 C for 10 min. PCR was done with a sense adapter primer, s-anc2 5′-GGCCACGCGTCGACTAGTACGGGGGGGGGGGG and an antisense primer to POR as252-bam 5′-ATAGGATCCAACTCGGGGACTTCTTCTTTTT (nt 252–231), 5′-RACE products were digested with SalI/BamHI, and cloned into pBluescript SK vector. Clones containing the longest insert fragments were subjected to DNA sequencing on both strands.
Promoter-reporter plasmid construction
The 5′-flanking DNA of the human POR gene, extending from −1128 to +36 bp, was amplified by PCR of human genomic DNA using Advantage 2 polymerase (Clontech) and primer pairs −1128s and +36as (Table 1). Base +1 represents the transcriptional start site, i.e. the first nucleotide in the untranslated first exon of POR (15). The primers contain NheI and BglII sites at 5′- and 3′-ends, respectively, to permit cloning into NheI/BglII sites in the pGL3-basic vector (Promega Corp., Madison, WI), upstream from the firefly luciferase reporter gene. Deletion constructs comprising −753/+36, −438/+36, −325/+36, −182/+36, and −114/+36 were generated by PCR using Pfu Ultra II fusion HS DNA polymerase (Stratagene, La Jolla, CA) with the primer pairs −753s/+36as, −438s /+36as, −325s /+36as, −182s /+36as, and −114s /+36as (Table 1).
Table 1.
Sequences of oligonucleotides for PCR, site-directed mutagenesis (SDM), EMSA, ChIP, 5′-RACE, and qRT-PCR
| Type of assay | Sequences (5′ to 3′) | Restriction site |
|---|---|---|
| PCR | ||
| +36_as | GCACGTAGATCTTCAGGCCCACACCACTGAGG | BglII |
| −128_s | GAATTCGCTAGCGCGTGAGCCACCGTGCCTG | NheI |
| −753_s | GAATTCGCTAGCCGAGCTCTAGGCAGCGTGTG | NheI |
| −438_s | GAATTCGCTAGCTCAGGCTGGATGGAGGGAAC | NheI |
| −325_s | GAATTCGCTAGCGCCCGAAGGAGGAGGCTAGA | NheI |
| −182_s | GAATTCGCTAGCGGAACCACGCACTTTCATTTCTCT | NheI |
| −114_s | GAATTCGCTAGCGCCGTACCAAGAGCGCAAAT | NheI |
| −103_s | GAATTCGCTAGCGGTATTGTGCCCCACCCTGA | KpnI |
| −37_as | GCTGCCTAGAGCTCGCTTTTGAAGGC | None |
| −193_s | GAGGTACCGAAGAAGAGAAGCTTTCAGCTCGGTGTG | KpnI |
| −846_as | AGTCTCGCTATGTTGCCCA | None |
| SDM | ||
| −208-T_s | AGAAGCCGCAGCCGCTGTCTCCAGGCGACTC | None |
| −208-T_as | GAGTCGCCTGGAGACAGCGGCTGCGGCTTCT | None |
| −173-A_s | ACCCCCGGAACCACGAACTTTCATTTCTCTG | None |
| −173-A_as | CAGAGAAATGAAAGTTCGTGGTTCCGGGGGT | None |
| −152-A_s | CATTTCTCTGCCGGGAGACCCAGCCGAGCCG | None |
| −152-A_as | CGGCTCGGCTGGGTCTCCCGGCAGAGAAATG | None |
| EMSA | ||
| −330/−306 | CGGCGGCCCGAAGGAGGAGGCTAGA | None |
| −324/−300 | CCCGAAGGAGGAGGCTAGACCGGCG | None |
| −305/−281 | CCGGCGGGCGCACAGCCACAGTTCT | None |
| −299/−275 | GGCGCACAGCCACAGTTCTGCAGTG | None |
| −291/−267 | AGCCACAGTTCTGCAGTGATCCCCG | None |
| −280/−256 | GCAGTGATCCCCGGGAAGGTGGGCA | None |
| −274/−250 | ATCCCCGGGAAGGTGGGCAGCCGGC | None |
| −266/−242 | GGAAGGTGGGCAGCCGGCCCGGTGA | None |
| −255/−231 | GCCGGCCCGGTGACCTGCAGGGTCC | None |
| −251/−227 | GCCCGGTGACCTGCAGGGTCCGAGC | None |
| −249/−225 | CCGGTGACCTGCAGGGTCCGAGCTG | None |
| −242/−218 | CCTGCAGGGTCCGAGCTGTAGAAGC | None |
| −230/−206 | GAGCTGTAGAAGCCGCAGCCGCCGT | None |
| −224/−200 | TAGAAGCCGCAGCCGCCGTCTCCAG | None |
| −205/−181 | CTCCAGGCGACTCCGCCACCCCCGG | None |
| −163/−139 | TCTCTGCCGGGAGACCCAGCCGAGC | None |
| (−152A) | ||
| −163/−139 | TCTCTGCCGGGCGACCCAGCCGAGC | None |
| (−152C) | ||
| −280/−256 mutant 1 | GCAGTGATCCCCGGGAATTTGGGCA | None |
| −280/−256 mutant 2 | GCAGTGATCCCCGGGAAGTCGGGCA | None |
| −280/−256 mutant 3 | GCAGTGATCCCCGGGAAGGTTTGCA | None |
| −280/−256 mutant 4 | GCAGTGATCCCCGGGAAGGTGGTTC | None |
| −266/−242 mutant 1 | TTCAGGTGGGCAGCCGGCCCGGTGA | None |
| −266/−242 mutant 2 | GGACTTTGGGCAGCCGGCCCGGTGA | None |
| −266/−242 mutant 3 | GGAAGGCTTGCAGCCGGCCCGGTGA | None |
| −266/−242 mutant 4 | GGAAGGTGGTTCGCCGGCCCGGTGA | None |
| −266/−242 mutant 5 | GGAAGGTGGGCATTTGGCCCGGTGA | None |
| −266/−242 mutant 6 | GGAAGGTGGGCAGCCTTTCCGGTGA | None |
| −266/−242 mutant 7 | GGAAGGTGGGCAGCCGGCTTTGTGA | None |
| −266/−242 mutant 8 | GGAAGGTGGGCAGCCGGCCCGTCTA | None |
| −255/−231 mutant 1 | GCCGGCCCGGATTCCTGCAGGGTCC | None |
| −255/−231 mutant 2 | TTTGGCCCGGTGACCTGCAGGGTCC | None |
| −255/−231 mutant 3 | GCCTTCCCGGTGACCTGCAGGGTCC | None |
| −255/−231 mutant 4 | GCCGGTTCGGTGACCTGCAGGGTCC | None |
| −255/−231 mutant 5 | GCCGGCCTTGTGACCTGCAGGGTCC | None |
| −255/−231 mutant 6 | GCCGGCCCGTCGACCTGCAGGGTCC | None |
| −255/−231 mutant 7 | GCCGGCCCGGTTCCCTGCAGGGTCC | None |
| −255/−231 mutant 8 | GCCGGCCCGGTGATTTGCAGGGTCC | None |
| −255/−231 mutant 9 | GCCGGCCCGGTGACCCTCAGGGTCC | None |
| −255/−231 mutant 10 | GCCGGCCCGGTGACCTGTGGGGTCC | None |
| −255/−231 mutant 11 | GCCGGCCCGGTGACCTGCATTGTCC | None |
| −255/−231 mutant 12 | GCCGGCCCGGTGACCTGCAGGTCCC | None |
| −255/−231 mutant 13 | GCCGGCCCGGTGACCTGCAGGGTTT | None |
| SMAD3/4 consensus | TCGAGAGCCAGACAAAAAGCCAGACATTTAGCCAGACAC | None |
| SMAD3/4 mutant | TCGAGAGCTACATAAAAAGCTACATATTTAGCTACATAC | None |
| TR DR4 | AGCTTCAGGTCACAGGAGGTCAGAGAGCT | None |
| TR DR4 mutant | AGCTTCAGAACACAGGAGAACAGAGAGCT | None |
| TR palindrome [TR(P)] | GATCGTAAGATTCAGGTCATGACCTGAGGAGA | None |
| TR(P) mutant | GATCGTAAGATTCAGAACATGGACTGAGGAGA | None |
| RXR consensus | AGCTTCAGGTCAGAGGTCAGAGAGCT | None |
| RXR mutant | AGCTTCAGCACAGAGCACAGAGAGCT | None |
| ER consensus | GGATCTAGGTCACTGTGACCCCGGATC | None |
| ER mutant | GGATCTAGTACACTGTGTACCCGGATC | None |
| VDR consensus | AGCTTCAGGTCAAGGAGGTCAGAGAGCT | None |
| VDR mutant | AGCTTCAGAACAAGGAGAACAGAGAGCT | None |
| RAR consensus | TCGAGGGTAGGGTTCACCGAAAGTTCACTCG | None |
| ChIP | ||
| ERβ forward | CCACTATCCTTGTGGGTGGA | None |
| ERβ reverse | CAGCAGCTGGAGAAACTGAA | None |
| S222 | GAAGCCGCAGCCGCCGTCTC | None |
| AS36 | GCCGCCCCGCCCACTCG | None |
| S374 | GGAAACAGGGCCGTGGAGGAGACA | None |
| AS149 | TCGCCCGGCAGAGAAATGAAAGTG | None |
| 5′-RACE and RT-PCR | None | |
| as329-rt | TGTTCCTCCCCGTTTTCTTCATCTTTT | None |
| s-anc2 | GGCCACGCGTCGACTAGTACGGGGGGGGGGGG | SalI |
| as252-bam | ATAGGATCCAACTCGGGGACTTCTTCTTTTT | BamHI |
| s1622-or | ATGTTCGTGCGCAAGTCCCAGTTC | None |
| as2112-or | GCAGCCGTAGTACAGCAGCGTCTC | None |
| s702-gap | CGGGGCTCTCCAGAACATCATCC | None |
| as900-gap | CGACGCCTGCTTCACCACCTTCTT | None |
Restriction sites are underlined, and mutations are in boldface fonts. s, Sense; as, antisense.
To generate longer POR promoter-reporter constructs, the PCR product generated by primer pairs −1128s and +36as was first cloned into pcDNA3.1/V5-His-TOPO vector using pcDNA3.1/V5-His-TOPO TA cloning kit (Invitrogen), generating plasmid construct POR-1128-pcDNA3.1. The POR proximal promoter sequence from −2103 bp to −737 bp was amplified by PCR of human genomic DNA using Advantage 2 polymerase (Clontech Laboratories, Inc., Palo Alto, CA) and primer pairs −2103s and −737as; primer −2103s contains a KpnI site at its 5′-end. This PCR fragment was digested with KpnI and with EcoRI, which recognizes a site at −1042 bp, and then cloned into KpnI/EcoRI-digested POR-1128-pcDNA3.1, generating construct POR-2103-pcDNA3.1. The 5′-flanking DNA sequences of the POR gene from −2103 to +36 was excised from POR-2103-pcDNA3.1 with KpnI/BglII and inserted into KpnI/BglII-digested pGL3-basic vector (Promega). The proximal promoter sequence from −3193 to −1846 bp was similarly amplified by PCR using Advantage 2 polymerase and primer pairs −3193s, which contained a KpnI site at its 5′ end, and −1846as (Table 1). This PCR product was digested with KpnI and with BamHI, which which recognizes a site at −1892 bp, and then cloned into KpnI/BamHI-digested POR-1128-pcDNA3.1, generating POR-3193-pcDNA3.1. The 5′-flanking DNA from −3193 to +36 was excised from POR-3193-pcDNA3.1 with KpnI/BglII, and inserted into KpnI/BglII-digested pGL3-basic vector (Promega).
All PCR amplifications were performed under a touch-down cycling condition, starting at 95 C for 5 min, followed by 13 touch-down cycles of 95 C for 30 sec, 67 C to 61 C for 30 sec (the annealing temperature for each subsequent cycle was decreased by 0.5 C), and 72 C for 2 min 30 sec. This was followed by 35 amplification cycles at 95 C for 30 sec, 61 C for 30 sec, and 72 C for 2 min, 30 sec, a final extension held at 72 C for 7 min, after which the reaction was then stopped at 4 C. The sequences of all constructs were confirmed by sequencing on both DNA strands. POR promoter sequences were obtained from the University of California, Santa Cruz Genome Browser website (http://genome.ucsc.edu).
Plasmids containing the cDNAs for human estrogen receptor-α (ERα) (50) and -β (ERβ) (51) were kindly provided by Dr. Dale Leitman (University of California, San Francisco, CA). Plasmids containing the cDNAs for human TRα (52) and TRβ (53) were generous gifts from Dr. John Baxter (Methodist Hospital, Houston, TX). Plasmids containing the cDNAs for human Smad3 and Smad4 (54) cDNAs were generous gifts from Dr. Rik Derynck (University of California, San Francisco).
Cell culture
Human adrenal NCI-H295A cells (55–57) were cultured at 37 C and 5% CO2 in RPMI 1640, supplemented with 2% fetal calf serum, 5 μg/ml insulin, 5 μg/ml transferrin, 5 ng/ml selenium, and 50 μg/ml gentamycin. Human liver Hep-G2 cells (58) were cultured in DMEM with Eagle's basic salt solution medium, supplemented with 10% fetal calf serum, 1% nonessential amino acids, 1% sodium pyruvate, and 50 μg/ml gentamycin on 0.1% gelatin-treated plates. The cells were grown to 50%–80% confluence on 12-well tissue culture plates 24 h before trasfection. The POR promoter-reporter constructs or the empty vector pGL3-basic vector (300 ng each) were transfected with 2.4 μl of Enhancer (Qiagen) and 6 μl Effectene transfection reagent (Qiagen) into NCI-H295A or Hep-G2 cells. After 24 h, the cells were lysed and assayed for luciferase activity, normalized by Renilla luciferase activity expressed by cotransfected pRL-CMV (Promega).
To determine the effects of E2, TGFβ, or T3 stimulation on the recruitment of transcription factors, Hep-G2 and NCI-H295A cells were treated with either 10 nm E2, 100 nm T3 (Sigma), or 3 ng recombinant human TGFβ (R&D Systems, Minneapolis, MN) for various time points (2 h to 18 h) in serum-free phenol red-free media, and the cells were harvested for nuclear extracts or chromatin immunoprecipitation (ChIP) as described below.
Nuclear extract
NCI-H295A and Hep-G2 cells were grown to 50–80% confluence, harvested by centrifugation, and washed twice with 10 ml cold PBS. The cell pellets were resuspended in 10 mm HEPES (pH 7.8), 10 mm KCl, 1 mm phenylmethylsulfonyl fluoride and incubated on ice for 15 min; Nonidet P-40 was added to a final concentration of 0.58% to lyse the nuclei after which the suspension was vortexed for 15 sec and centrifuged at 8,000 × g rpm for 30 sec. The pellet was resuspended in 20 mm HEPES (pH 7.8), 50 mm KCl, 400 mm NaCl, 5.25% glycerol, 1 mm phenylmethylsulfonyl fluoride, vortexed vigorously for 1 h on a Fisher Vortexer Genie 2 (Fischer Scientific, Pittsburgh, PA), centrifuged at 16,000 × g for 5 min, and the supernatant containing nuclear proteins was collected.
EMSA and supershifts
Double-stranded EMSA oligonucleotides (Table 1) were end labeled with [γ-32P]ATP by T4 polynucleotide kinase (New England Biolabs, Beverly, MA), and purified on G-50 MicroSpin columns (GE Healthcare, Piscataway, NJ). Binding of nuclear proteins was done with 9 μg nuclear extract, 1 μg poly dI-dC (Sigma Chemical Co., St. Louis, MO), 50,000 cpm [γ-32P]ATP-labeled oligonucleotide probe in 10 mm Tris-HCl (pH 7.5), 100 mm KCl, 1 mm EDTA pH 8, 4% glycerol, 5 mm dithiothreitol, and 0.1 mg/ml BSA at room temperature for 20 min in the absence or presence of unlabeled oligonucleotide competitor (80 ng). For supershift assays, Hep-G2 and NCI-H295A cells were transfected with expression plasmids for either ERα, Smad3, Smad4, TRα, TRβ, or pBluescript SK vector only, treated with either 10 nm E2, 3 ng recombinant human TGFβ, or 100 nm T3; nuclear extract was preincubated with the appropriate antibody for 1 h at 4 C, and [γ-32P]ATP-labeled oligonucleotide probe was added to the reaction mixture and incubated at room temperature for 20 min. DNA-protein complexes were displayed by electrophoresis on 6% nondenaturing polyacrylamide gels in 1× Tris-borate-EDTA (pH 7.2) (50 mm Tris-HCl, 0.11 m boric acid, 1 mm EDTA); the gels were then dried and imaged with a Storm Phosphorimager (GE Healthcare). Each EMSA was done with at least three different preparations of nuclear extract, and representative autoradiograms are shown.
ChIP
ChIP assays were done similarly to our previous procedures (59). Hep-G2 and NCI-H295A cells were fixed in 1% formaldehyde solution, washed three times with PBS, collected, lysed on ice in 1% sodium dodecyl sulfate, 10 mm EDTA (pH 8), 50 mm Tris-HCl (pH 8) in the presence of protease inhibitors, and sonicated on ice with a Fisher Dismembrator 550 for a total of 24 sec (with cycles of 4 sec on pulses followed by 25 sec off), and chromatin was collected by centrifugation. Extracts were precleared with 20 μl of 50% protein A-agarose slurry containing 10 μg salmon sperm DNA, and 20 μg BSA. Before adding antibodies directed against AP-2α, Smad3 and Smad4, TRα, TRβ (Abcam, Inc., Cambridge, MA), RXRα (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), and ERα (Abcam), an aliquot (20 μl) of each sample was removed to use as an input for PCR. Immunoprecipitation was performed on a rocking platform at 4 C overnight, and immune complexes were captured by protein A-agarose beads (Invitrogen) and washed three times. Isolated chromatin was extracted with phenol, the DNA was precipitated with ethanol, and sequences of interest were amplified by PCR. For ERβ, the primer pairs were: 5′-CCACTATCCTTGTGGGTGGA (ERβ forward, bases −191 to −172) and 5′-CAGCAGCTGGAGAAACTGAA (ERβ reverse, bases 21 to 2), which span an AP-2 binding site in the ERβ promoter (60), thus serving as a positive control for AP-2 in the ChIP assay. For POR, the S222 sense primer was 5′-GAAGCCGCAGCCGCCGTCTC (nt −222 to −203), and the AS36 antisense primer was 5′-GCCGCCCCGCCCACTCG (−36 to −52), which span the putative AP-2 binding site; we also used POR S374 sense primer 5′-GGAAACAGGGCCGTGGAGGAGACA (nt −374 to −350) and AS149 antisense primer 5′-TCGCCCGGCAGAGAAATGAAAGTG (−149 to −173), which span the Smad3/4 and the ER/TR binding sites.
Quantitative RT-PCR (qRT-PCR)
QRT-PCR using the SYBR green fluorescent dye detection (Bio-Rad Laboratories, Inc., Hercules, CA) was done with cDNAs prepared from Hep-G2 and NCI-H295A cells treated for 18 h in absence or presence of 10 nm E2, 3 ng recombinant human TGFβ, or 100 nm T3 in serum-free phenol-red free media. The following primer pairs were used for qRT-PCR: POR, sense primer s1622-or 5′-ATGTTCGTGCGCAAGTCCCAGTTC (nt 1540–1563) and antisense primer as2112-or 5′-GCAGCCGTAGTACAGCAGCGTCTC (nt 1707–1684); glyceraldehyde-3-phosphate dehydrogenase (GAPDH), sense primer s702-gap 5′-CGGGGCTCTCCAGAACATCATCC (nt 702–724) and antisense primer as900-gap 5′-CGACGCCTGCTTCACCACCTTCTT (nt 900–877). The POR expression level was normalized to GAPDH level, both determined from the average threshold cycle (Ct) of triplicate samples.
Statistical analyses
Statistical analyses were performed using two-tailed unpaired t tests, and significance was accepted for tests where P < 0.05.
Acknowledgments
This work was supported by National Institutes of Health Grant GM073020 (to W.L.M.).
Disclosure Summary: The authors have nothing to disclose.
NURSA Molecule Pages:
Nuclear Receptors: TR-α | TR-β | ER-α | RXR-α;
Ligands: Thyroid hormone | 17β-estradiol.
Footnotes
- AP-2
- Activator protein 2
- ChIP
- chromatin immunoprecipitation
- E2
- estradiol
- ER
- estrogen receptor
- FAD
- flavin adenine dinucleotide
- FMN
- flavin mononucleotide
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- nt
- nucleotide
- POR
- P450 oxidoreductase
- qRT-PCR
- quantitative RT-PCR
- RACE
- 5′-rapid amplification of cDNA ends
- RAR
- retinoic acid receptor
- RXR
- retinoid-X receptor
- Smad
- Sma and Mad related protein
- TR
- thyroid hormone receptor
- VDR
- vitamin D receptor.
References
- 1. Miller WL, Auchus RJ. 2011. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr Rev 32:81–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang M, Roberts DL, Paschke R, Shea TM, Masters BS, Kim JJ. 1997. Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc Natl Acad Sci USA 94:8411–8416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ellis J, Gutierrez A, Barsukov IL, Huang WC, Grossmann JG, Roberts GC. 2009. Domain motion in cytochrome P450 reductase: conformational equilibria revealed by NMR and small-angle x-ray scattering. J Biol Chem 284:36628–36637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ono T, Bloch K. 1975. Solubilization and partial characterization of rat liver squalene epoxidase. J Biol Chem 250:1571–1579 [PubMed] [Google Scholar]
- 5. Ilan Z, Ilan R, Cinti DL. 1981. Evidence for a new physiological role of hepatic NADPH:ferricytochrome (P-450) oxidoreductase. Direct electron input to the microsomal fatty acid chain elongation system. J Biol Chem 256:10066–10072 [PubMed] [Google Scholar]
- 6. Wilks A, Black SM, Miller WL, Ortiz de Montellano PR. 1995. Expression and characterization of truncated human heme oxygenase (hHO-1) and a fusion protein of hHO-1 with human cytochrome P450 reductase. Biochemistry 34:4421–4427 [DOI] [PubMed] [Google Scholar]
- 7. Enoch HG, Strittmatter P. 1979. Cytochrome b5 reduction by NADPH-cytochrome P-450 reductase. J Biol Chem 254:8976–8981 [PubMed] [Google Scholar]
- 8. Flück CE, Tajima T, Pandey AV, Arlt W, Okuhara K, Verge CF, Jabs EW, Mendonçca BB, Fujieda K, Miller WL. 2004. Mutant P450 oxidoreductase causes disordered steroidogenesis with and without Antley-Bixler syndrome. Nat Genet 36:228–230 [DOI] [PubMed] [Google Scholar]
- 9. Adachi M, Tachibana K, Asakura Y, Yamamoto T, Hanaki K, Oka A. 2004. Compound heterozygous mutations of cytochrome P450 oxidoreductase gene (POR) in two patients with Antley-Bixler syndrome. Am J Med Genet A 128A:333–339 [DOI] [PubMed] [Google Scholar]
- 10. Arlt W, Walker EA, Draper N, Ivison HE, Ride JP, Hammer F, Chalder SM, Borucka-Mankiewicz M, Hauffa BP, Malunowicz EM, Stewart PM, Shackleton CH. 2004. Congenital adrenal hyperplasia caused by mutant P450 oxidoreductase and human androgen synthesis: analytical study. Lancet 363:2128–2135 [DOI] [PubMed] [Google Scholar]
- 11. Fukami M, Horikawa R, Nagai T, Tanaka T, Naiki Y, Sato N, Okuyama T, Nakai H, Soneda S, Tachibana K, Matsuo N, Sato S, Homma K, Nishimura G, Hasegawa T, Ogata T. 2005. Cytochrome P450 oxidoreductase gene mutations and Antley-Bixler syndrome with abnormal genitalia and/or impaired steroidogenesis: molecular and clinical studies in 10 patients. J Clin Endocrinol Metab 90:414–426 [DOI] [PubMed] [Google Scholar]
- 12. Huang N, Pandey AV, Agrawal V, Reardon W, Lapunzina PD, Mowat D, Jabs EW, Van Vliet G, Sack J, Flück CE, Miller WL. 2005. Diversity and function of mutations in P450 oxidoreductase in patients with Antley-Bixler syndrome and disordered steroidogenesis. Am J Hum Genet 76:729–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hershkovitz E, Parvari R, Wudy SA, Hartmann MF, Gomes LG, Loewental N, Miller WL. 2008. Homozygous mutation G539R in the gene for P450 oxidoreductase in a family previously diagnosed as having 17,20-lyase deficiency. J Clin Endocrinol Metab 93:3584–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sahakitrungruang T, Huang N, Tee MK, Agrawal V, Russell WE, Crock P, Murphy N, Migeon CJ, Miller WL. 2009. Clinical, genetic, and enzymatic characterization of P450 oxidoreductase deficiency in four patients. J Clin Endocrinol Metab 94:4992–5000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scott RR, Gomes LG, Huang N, Van Vliet G, Miller WL. 2007. Apparent manifesting heterozygosity in P450 oxidoreductase deficiency and its effect on coexisting 21-hydroxylase deficiency. J Clin Endocrinol Metab 92:2318–2322 [DOI] [PubMed] [Google Scholar]
- 16. Huang N, Agrawal V, Giacomini KM, Miller WL. 2008. Genetics of P450 oxidoreductase: sequence variation in 842 individuals of four ethnicities and activities of 15 missense mutations. Proc Natl Acad Sci USA 105:1733–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sim SC, Risinger C, Dahl ML, Aklillu E, Christensen M, Bertilsson L, Ingelman-Sundberg M. 2006. A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther 79:103–113 [DOI] [PubMed] [Google Scholar]
- 18. Aklillu E, Carrillo JA, Makonnen E, Hellman K, Pitarque M, Bertilsson L, Ingelman-Sundberg M. 2003. Genetic polymorphism of CYP1A2 in Ethiopians affecting induction and expression: characterization of novel haplotypes with single-nucleotide polymorphisms in intron 1. Mol Pharmacol 64:659–669 [DOI] [PubMed] [Google Scholar]
- 19. Johansson I, Lundqvist E, Bertilsson L, Dahl ML, Sjöqvist F, Ingelman-Sundberg M. 1993. Inherited amplification of an active gene in the cytochrome P450 CYP2D locus as a cause of ultrarapid metabolism of debrisoquine. Proc Natl Acad Sci USA 90:11825–11829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dee A, Carlson G, Smith C, Masters BS, Waterman MR. 1985. Regulation of synthesis and activity of bovine adrenocortical NADPH-cytochrome P-450 reductase by ACTH. Biochem Biophys Res Commun 128:650–656 [DOI] [PubMed] [Google Scholar]
- 21. Waxman DJ, Morrissey JJ, Leblanc GA. 1989. Hypophysectomy differentially alters P-450 protein levels and enzyme activities in rat liver: pituitary control of hepatic NADPH cytochrome P-450 reductase. Mol Pharmacol 35:519–525 [PubMed] [Google Scholar]
- 22. Ram PA, Waxman DJ. 1992. Thyroid hormone stimulation of NADPH P450 reductase expression in liver and extrahepatic tissues. Regulation by multiple mechanisms. J Biol Chem 267:3294–3301 [PubMed] [Google Scholar]
- 23. O'Leary KA, Beck TW, Kasper CB. 1994. NADPH cytochrome P-450 oxidoreductase gene: identification and characterization of the promoter region. Arch Biochem Biophys 310:452–459 [DOI] [PubMed] [Google Scholar]
- 24. O'Leary KA, Li HC, Ram PA, McQuiddy P, Waxman DJ, Kasper CB. 1997. Thyroid regulation of NADPH:cytochrome P450 oxidoreductase: identification of a thyroid-responsive element in the 5′-flank of the oxidoreductase gene. Mol Pharmacol 52:46–53 [DOI] [PubMed] [Google Scholar]
- 25. Li HC, Liu D, Waxman DJ. 2001. Transcriptional induction of hepatic NADPH: cytochrome P450 oxidoreductase by thyroid hormone. Mol Pharmacol 59:987–995 [DOI] [PubMed] [Google Scholar]
- 26. O'Leary KA, Kasper CB. 2000. Molecular basis for cell-specific regulation of the NADPH-cytochrome P450 oxidoreductase gene. Arch Biochem Biophys 379:97–108 [DOI] [PubMed] [Google Scholar]
- 27. Loots GG, Ovcharenko I. 2004. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res 32:W217–W221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Klock G, Strähle U, Schütz G. 1987. Oestrogen and glucocorticoid responsive elements are closely related but distinct. Nature 329:734–736 [DOI] [PubMed] [Google Scholar]
- 30. Williams T, Admon A, Lüscher B, Tjian R. 1988. Cloning and expression of AP-2, a cell-type-specific transcription factor that activates inducible enhancer elements. Genes Dev 2:1557–1569 [DOI] [PubMed] [Google Scholar]
- 31. Tee MK, Dong Q, Miller WL. 2008. Pathways leading to phosphorylation of P450c17 and to the posttranslational regulation of androgen biosynthesis. Endocrinology 149:2667–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tee MK, Thomson AA, Bristow J, Miller WL. 1995. Sequences promoting the transcription of the human XA gene overlapping P450c21A correctly predict the presence of a novel, adrenal-specific, truncated form of tenascin-X. Genomics 28:171–178 [DOI] [PubMed] [Google Scholar]
- 33. Söderberg SS, Karlsson G, Karlsson S. 2009. Complex and context dependent regulation of hematopoiesis by TGF-β superfamily signaling. Ann NY Acad Sci 1176:55–69 [DOI] [PubMed] [Google Scholar]
- 34. Harvey CB, Williams GR. 2002. Mechanism of thyroid hormone action. Thyroid 12:441–446 [DOI] [PubMed] [Google Scholar]
- 35. Vasudevan N, Davidkova G, Zhu YS, Koibuchi N, Chin WW, Pfaff D. 2001. Differential interaction of estrogen receptor and thyroid hormone receptor isoforms on the rat oxytocin receptor promoter leads to differences in transcriptional regulation. Neuroendocrinology 74:309–324 [DOI] [PubMed] [Google Scholar]
- 36. Pendaries V, Verrecchia F, Michel S, Mauviel A. 2003. Retinoic acid receptors interfere with the TGF-β/Smad signaling pathway in a ligand-specific manner. Oncogene 22:8212–8220 [DOI] [PubMed] [Google Scholar]
- 37. Lin KH, Chen CY, Chen SL, Yen CC, Huang YH, Shih CH, Shen JJ, Yang RC, Wang CS. 2004. Regulation of fibronectin by thyroid hormone receptors. J Mol Endocrinol 33:445–458 [DOI] [PubMed] [Google Scholar]
- 38. Endoh A, Natsume H, Igarashi Y. 1995. Dual regulation of 21-hydroxylase activity by sex steroid hormones in rat hepatocytes. J Steroid Biochem Mol Biol 54:163–165 [DOI] [PubMed] [Google Scholar]
- 39. Perrin A, Pascal O, Defaye G, Feige JJ, Chambaz EM. 1991. Transforming growth factor β1 is a negative regulator of steroid 17α-hydroxylase expression in bovine adrenocortical cells. Endocrinology 128:357–362 [DOI] [PubMed] [Google Scholar]
- 40. Le Roy C, Li JY, Stocco DM, Langlois D, Saez JM. 2000. Regulation by adrenocorticotropin (ACTH), angiotensin II, transforming growth factor-β, and insulin-like growth factor I of bovine adrenal cell steroidogenic capacity and expression of ACTH receptor, steroidogenic acute regulatory protein, cytochrome P450c17, and 3β-hydroxysteroid dehydrogenase. Endocrinology 141:1599–1607 [DOI] [PubMed] [Google Scholar]
- 41. Derebecka-Holysz N, Lehmann TP, Holysz M, Trzeciak WH. 2009. TGF-β inhibits CYP17 transcription in H295R cells acting via activin receptor-like kinase 5. Endocr Res 34:68–79 [DOI] [PubMed] [Google Scholar]
- 42. Iber H, Li-Masters T, Chen Q, Yu S, Morgan ET. 2001. Regulation of hepatic cytochrome P450 2C11 via cAMP: implications for down-regulation in diabetes, fasting, and inflammation. J Pharmacol Exp Ther 297:174–180 [PubMed] [Google Scholar]
- 43. Aitken AE, Morgan ET. 2007. Gene-specific effects of inflammatory cytokines on cytochrome P450 2C, 2B6 and 3A4 mRNA levels in human hepatocytes. Drug Metab Dispos 35:1687–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yamazoe Y, Ling X, Murayama N, Gong D, Nagata K, Kato R. 1990. Modulation of hepatic level of microsomal testosterone 7α-hydroxylase, P-450a (P450IIA), by thyroid hormone and growth hormone in rat liver. J Biochem 108:599–603 [DOI] [PubMed] [Google Scholar]
- 45. Ram PA, Waxman DJ. 1990. Pretranslational control by thyroid hormone of rat liver steroid 5α-reductase and comparison to the thyroid dependence of two growth hormone-regulated CYP2C mRNAs. J Biol Chem 265:19223–19229 [PubMed] [Google Scholar]
- 46. Waxman DJ, Ram PA, Notani G, LeBlanc GA, Alberta JA, Morrissey JJ, Sundseth SS. 1990. Pituitary regulation of the male-specific steroid 6 β-hydroxylase P-450 2a (gene product IIIA2) in adult rat liver. Suppressive influence of growth hormone and thyroxine acting at a pretranslational leve. Mol Endocrinol 4:447–454 [DOI] [PubMed] [Google Scholar]
- 47. Ram PA, Waxman DJ. 1991. Hepatic P450 expression in hypothyroid rats: differential responsiveness of male-specific P450 forms 2a (IIIA2), 2c (IIC11), and RLM2 (IIA2) to thyroid hormone. Mol Endocrinol 5:13–20 [DOI] [PubMed] [Google Scholar]
- 48. Sundseth SS, Waxman DJ. 1992. Sex-dependent expression and clofibrate inducibility of cytochrome P450 4A fatty acid ω-hydroxylases. Male specificity of liver and kidney CYP4A2 mRNA and tissue-specific regulation by growth hormone and testosterone. J Biol Chem 267:3915–3921 [PubMed] [Google Scholar]
- 49. Liddle C, Goodwin BJ, George J, Tapner M, Farrell GC. 1998. Separate and interactive regulation of cytochrome P450 3A4 by triiodothyronine, dexamethasone, and growth hormone in cultured hepatocytes. J Clin Endocrinol Metab 83:2411–2416 [DOI] [PubMed] [Google Scholar]
- 50. Green S, Walter P, Kumar V, Krust A, Bornert JM, Argos P, Chambon P. 1986. Human oestrogen receptor cDNA: sequence, expression and homology to v-erb-A. Nature 320:134–139 [DOI] [PubMed] [Google Scholar]
- 51. Kuiper GGJM, Gustafsson JA. 1997. The novel estrogen receptor-β subtype: potential role in the cell- and promoter-specific actions of estrogens and anti-estrogens. FEBS Lett 410:87–90 [DOI] [PubMed] [Google Scholar]
- 52. Sap J, Muñoz A, Damm K, Goldberg Y, Ghysdael J, Leutz A, Beug H, Vennström B. 1986. The c-erb-A protein is a high-affinity receptor for thyroid hormone. Nature 324:635–640 [DOI] [PubMed] [Google Scholar]
- 53. Weinberger C, Thompson CC, Ong ES, Lebo R, Gruol DJ, Evans RM. 1986. The c-erb-A gene encodes a thyroid hormone receptor. Nature 324:641–646 [DOI] [PubMed] [Google Scholar]
- 54. Zhang Y, Feng X, We R, Derynck R. 1996. Receptor-associated Mad homologues synergize as effectors of the TGF-β response. Nature 383:168–172 [DOI] [PubMed] [Google Scholar]
- 55. Gazdar AF, Oie HK, Shackleton CH, Chen TR, Triche TJ, Myers CE, Chrousos GP, Brennan MF, Stein CA, La Rocca RV. 1990. Establishment and characterization of a human adrenocortical carcinoma cell line that expresses multiple pathways of steroid biosynthesis. Cancer Res 50:5488–5496 [PubMed] [Google Scholar]
- 56. Staels B, Hum DW, Miller WL. 1993. Regulation of steroidogenesis in NCI-H295 cells: a cellular model of the human fetal adrenal. Mol Endocrinol 7:423–433 [DOI] [PubMed] [Google Scholar]
- 57. Rodriguez H, Hum DW, Staels B, Miller WL. 1997. Transcription of the human genes for cytochrome P450scc and P450c17 is regulated differently in human adrenal NCI-H295 cells than in mouse adrenal Y1 cells. J Clin Endocrinol Metab 82:365–371 [DOI] [PubMed] [Google Scholar]
- 58. Knowles BB, Howe CC, Aden DP. 1980. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science 209:497–499 [DOI] [PubMed] [Google Scholar]
- 59. Flück CE, Miller WL. 2004. GATA-4 and GATA-6 modulate tissue-specific transcription of the human gene for P450c17 by direct interaction with Sp1. Mol Endocrinol 18:1144–1157 [DOI] [PubMed] [Google Scholar]
- 60. Zhang X, Leung YK, Ho SM. 2007. AP-2 regulates the transcription of estrogen receptor (ER)-β by acting through a methylation hotspot of the 0N promoter in prostate cancer cells. Oncogene 26:7346–7354 [DOI] [PubMed] [Google Scholar]
- 61. Kolbe D, Taylor J, Elnitski L, Eswara P, Li J, Miller W, Hardison R, Chiaromonte F. 2004. Regulatory potential scores from genome-wide three-way alignments of human, mouse, and rat. Genome Res 14:700–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. King DC, Taylor J, Elnitski L, Chiaromonte F, Miller W, Hardison RC. 2005. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res 15:1051–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]