Abstract
Microfluidic techniques have been recently developed for cell-based assays. In microfluidic systems, the objective is for these microenvironments to mimic in vivo surroundings. With advantageous characteristics such as optical transparency and the capability for automating protocols, different types of cells can be cultured, screened, and monitored in real time to systematically investigate their morphology and functions under well-controlled microenvironments in response to various stimuli. Recently, the study of stem cells using microfluidic platforms has attracted considerable interest. Even though stem cells have been studied extensively using bench-top systems, an understanding of their behavior in in vivo-like microenvironments which stimulate cell proliferation and differentiation is still lacking. In this paper, recent cell studies using microfluidic systems are first introduced. The various miniature systems for cell culture, sorting and isolation, and stimulation are then systematically reviewed. The main focus of this review is on papers published in recent years studying stem cells by using microfluidic technology. This review aims to provide experts in microfluidics an overview of various microfluidic systems for stem cell research.
INTRODUCTION
Microfluidic platforms for cell-based assays have provided a promising means to observe cellular behavior in a mimetic microenvironment. They provide more in vivo-like microenvironments than the existing culture dishes since they enable perfusion-based culture systems. Furthermore, more physical and chemical factors for cell stimulation can be simultaneously supplied by using an on-chip culture device. Moreover, microfluidic devices offer the ability to precisely control these factors, i.e., well-controlled magnitudes and concentrations, which is especially important for stem cell culture and differentiation. In this review, recent papers reporting on microfluidic systems for cell study will be first introduced, followed by thorough discussions on their applications on stem cells. This review aims to provide experts in microfluidics an overview of microfluidic systems for stem cell research.
Microfluidics
Microfluidic technology developed at the beginning of the 1980s is a functional extension of microelectromechanical-systems technology, and has enabled the realization of a “lab-on-a-chip (LOC)” or a “micrototal-analysis-system (μTAS).” These miniaturized systems can be used to realize entire protocols performed in traditional laboratories with advantages of a smaller size, less reaction∕analysis time, less sample∕reagent consumption, reduced risk of contamination, higher sensitivity, higher reliability, and, most importantly, the ability to run multiple processes sequentially or in a parallel in an integrated system.1, 2 This technology has attracted scientists and researchers from different fields to study the behavior and manipulation of fluids in microchannels ranging from 0.1 to 1000 μm.3
Microfluidic systems can be fabricated by using a variety of microfabrication process by using materials such as polymers,4 glass,5 silicon,6 and metals,7 depending on their specific requirements and specifications. Figure 1 shows a schematic illustration of the microfabrication procedures for a polydimethylsiloxane (PDMS)-based microfluidic device with several useful characteristics such as elasticity, durability, biocompatibility, optical transparency, gas permeability, no toxicity, and low electrical conductivity.8, 9 PDMS is replicated against a mold usually made from a soft-lithography technique because of its advantages including low cost, ease to fabricate, and rapid processing.10, 11, 12 A variety of microfluidic devices have been fabricated based on this microfabrication process, such as micropumps,13 microvalves,14 microseparators,15 microconcentrators,16 and micromixers.17 Furthermore, these PDMS-based microfluidic devices can even be integrated to perform a specific assay for various applications such as molecular diagnosis,18 protein analysis,19 and cell studies.20 However, there are still potential limitations for PDMS-based microfluidic systems. For example, a PDMS surface is prone to nonspecific protein adsorption.21 Nonspecific protein absorption onto PDMS may limit the cell-culture applications, which may consume or change protein levels in the medium and hence affect the cellular behavior. The solution to this issue is to make the PDMS surface hydrophilic either by the use of an oxygen plasma treatment22 or surface treatment with poly(ethylene oxide) (PEO) (Ref. 23) to lower the levels of protein absorption. Besides, the bubbles formed in microfluidic devices may block the flow of liquids and cause cell damage attributed to the lack of nutrition.24 Similarly, the dead volume is another crucial issue that may cause serious contamination issues when cells are cultured.25 Typically, a streamline-shaped design in microfluidic devices has been adopted so that fluids flow smoothly to address this problem.26 In this review, recent advancements in cell studies, especially for stem cells, in microfluidic systems are reviewed. Advances in microfluidic systems for stem cell study will be summarized and discussed.
Figure 1.
Schematic illustration of the simplified fabrication process for a cell-culture device by using a SU-8 mold and PDMS replication process. (a) Lithography process to define the SU-8 mold pattern. (b) PDMS casting process to pour PDMS onto the SU-8 mold and then to thermally cure the polymer. (c) PDMS replication process to mechanically separate the SU-8 mold and the PDMS replica. (d) Bonding process to bond the PDMS layers and a glass substrate to form a complete microfluidic chip by using an oxygen plasma treatment prior to assembly. [Reprinted with permission from C. W. Huang and G. B. Lee, J. Micromech. Microeng. 17, 1266 (2007). © 2007, Institute of Physics.]
Stem cells
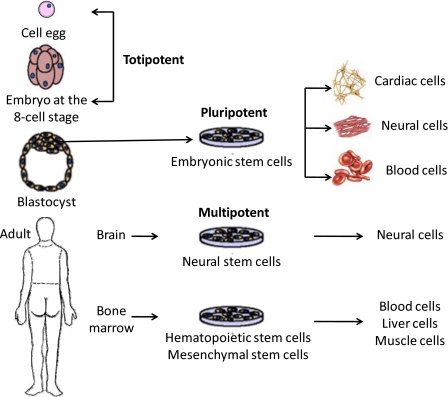
It is well recognized that stem cells have the ability to genetically differentiate into a specific cell type for promising applications such as tissue engineering or cell therapy. For embryonic stem cells (ESCs) and adult stem cells (mesenchymal, hematopoietic, and other tissue specific stem cells), they are also known for their self-renewal capability when undergoing multilineage differentiation with plasticity.27 There are three potencies for stem cells including totipotent, pluripotent, and multipotent, which have been summarized in Table 1. Briefly, totipotent stem cells are able to differentiate into any cell type. Pluripotent stem cells are the descendants of totipotent cells and have the ability to differentiate into nearly all cell types, while multipotent stem cells can only differentiate into a closely related family of cells.28, 29 Table 2 lists the differentiators between the characteristics of ESCs and those of adult stem cells.30 Briefly, major differences between ESCs and adult stem cells are their abilities in differentiating in terms of cell numbers and cell types. ESCs can become all cell types found in the body because they are pluripotent. Adult stem cells are limited in differentiating into different cell types from their original tissues (multipotent). Sections 1B1, 1B2 will describe both of these stem cells in detail.
Table 1.
Comparison of the three potencies of stem cells (totipotent, pluripotent, and multipotent) (Refs. 28, 29).
| Potency | Description | Examples |
|---|---|---|
| Totipotent | Each cell is able to develop into any cell types. | Cells from early (1–3 days) embryos |
| Pluripotent | Cells can form most of the cell types except placental tissues. | Some cells of blastocyst (5–14 days) |
| Multipotent | Cells differentiated, but can make a range of other particular tissues. | Fetal tissue, cord blood, and adult stem cells |
Table 2.
The differentiations between the characteristics of embryonic stem cells and those of adult stem cells.
| Cell type | Human adult stem cella | Human embryonic stem cellsa |
|---|---|---|
| Ease of isolation | Difficult | Easy, however, blastocyst may be destroyed |
| Potency | Multipotent (lose pluripotent ability) | Pluripotent (infinite life span theoretically) |
| Telomerase level | Low | High |
| Chromosomes | Tend to be shortened with aging | Maintain through serial passages |
| Apoptosis | Early | Late |
| Teratoma risk | No | Yes |
| Possibility of immune rejection | No | Yes |
| Ethical issues | No | Yes |
Reference 30.
Embryonic stem cells
ESCs are isolated from the inner cell mass and blastocysts which have the pluripotent ability to form all embryonic germ layer derivatives except extracellular tissues. The potential of ESCs to differentiate into almost all cell types not only provides an unlimited number of cells but also has attracted great interest in medical applications such as pharmaceutical31 and cell therapy.32 For example, pluripotent stem cells derived from ESCs can be used in drug development, drug screening, and cell toxicology.33, 34 ESC lines with genetic disease markers have been generated through nuclear transfer technology to study the underlying cause of diseases.13 However, ESCs would rapidly differentiate if optimal culture conditions or genetic manipulation were not assured.35Recently, ESCs have been extensively explored for cell therapy.32 Nonetheless, there still exist some difficulties in avoiding transplant rejection when ESCs are induced into usable cells. Furthermore, ethical issues also need to be properly addressed when using ESCs.
Adult stem cells
Stem cells found in adult organisms such as mesenchymal stem cells (MSCs), hematopoietic stem cells (HSCs), and neural stem cells (NSCs) are considered multipotent since they can produce mature cell types of one or more lineages but cannot reconstitute the organism as a whole.36 A great deal of adult stem cell research has focused on investigating their capacity to divide or self-renew indefinitely and their differentiation potential. Adult stem cells can be found in the brain, bone marrow, peripheral blood, blood vessels, skeletal muscle, skin, teeth, heart, liver, ovaries, and the epithelium. They are usually easily derived from peripheral tissues without ethical concerns when compared with human embryonic research.37, 38
Many types of cells can be differentiated from MSCs from adults, including osteoblasts, adipocytes, chondrocytes, astrocytes, and myoblasts.39 It is evident that MSCs derived from adult marrows,40 cord blood,41 placentas,42 and aminotic fluids43 also have the ability to differentiate into cells with ectoderm, mesoderm, and endoderm characteristics. Alternatively, it has been reported that MSCs can be also derived from fetal livers and adult or fetal peripheral bloods.44, 45, 46
In addition, human HSCs are defined by their ability to replenish all blood cell types and their ability to self-renew, containing both multipotent and lineage committed progenitors which give rise to all the blood cell types.47 The presence of HSCs in cord blood is an attractive cell source that is readily available and can be collected noninvasively with a lower occurrence of cytomegalovirus infection.48 Moreover, HSCs extracted from cord blood for transplantation have been widely used, and are capable of decreasing human leukocyte antigen (HLA) mismatches from a related family donor.49 Therefore, there exists a great need to isolate the HSCs from cord blood. HSCs cannot be easily observed directly and, therefore, their behavior needs to be inferred indirectly. Some clonal studies are the closest method for observing a single HSC in vivo.50 The clonal expansion of this stem cell can then be observed over time by monitoring the percent donor-type cells in the blood as the host is reconstituted.
The properties of NSCs are similar to HSCs, that is, they are able to differentiate into various cell types found in the immune system when injected into blood.51 Typically, NSCs can self-renew and generate three principal cell types commonly found in the central nervous system (CNS)—neurons, astrocytes, and oligodendrocytes.52 They can be propagated for extended periods of time and differentiated into both neuronal and glia cells, and therefore behave as stem cells. NSCs have a therapeutic effect in the treatment of some neurodevelopmental and neurodegenerative diseases, such as Parkinson’s and Alzheimer’s diseases.53 This behavior has been studied in some research which has proposed that the progeny of stem cell division undergoes strictly limited replication cycles in vivo.54 Moreover, NSCs are not able to behave in the same manner as stem cells when transplanted back into the brain.55 Stem cells, including totipotent stem cells, pluripotent stem cells, multipotent stem cells, with different characteristics that can be extracted from the human body and are capable of differentiating into various cells, are classified and illustrated in Fig. 2. After fertilization and union of the sperm with an egg, the created zygote is a totipotent cell. Pluripotent stem cells can be derived from the primitive germ line stem cells that exist from the blastocyst stage, which is able to differentiate into many cells such as cardiac cells, neural cells, and blood cells. Multipotent stem cells are derived from adult bodies, which can give rise to a limited number of other particular types of cells. For example, neural stem cells from the brain can give rise to nerve cells.
Figure 2.
Schematic illustration of stem cells with different characteristics obtained from a human body capable of differentiating into various cell types.
Cancer stem cells (CSCs) are cancer cells found in tumors or hematological cancers which possess similar characteristics to normal stem cells, specifically the ability to give rise to all cell types found in a particular cancer sample.56 For instance, CSCs may generate tumors through the stem cell processes of self-renewal and differentiation into multiple cell types. Research into CSCs holds hope for improvement of survival rates and the quality of life of cancer patients by developing specific therapies targeted at CSCs, especially for metastatic diseases.
Recently, a significant advancement has been reported that induced pluripotent stem cells (iPSCs) can be obtained from a reverse process using a direct reprograming method on somatic cells.57 Pluripotent stem cells created by this strategy may help researchers reprogram cells to repair damaged human tissues. It has opened up a new route for stem cell study since 2003.58
Microfluidic systems for cell studies are first reviewed. We mainly focus on the behaviors of different types of stem cells that can be explored using microfluidic systems. Several critical issues related to the isolation, culture, and differentiation of stem cells will be discussed and examined.
CELLS IN MICROFLUIDICS
Cell culture in microfluidics
It is well recognized that the preliminary development of most biomedical applications such as tissue engineering, pharmacology, and cell biology is via cell-culture experiments.59 However, for the conventional in vitro cell-culture process, this usually requires a large number of cell-culture areas, a large volume of culture media, expensive and automated equipment, and experienced personnel. Besides, a culture dish provides a macroscale environment that is totally different from that in an in vivo biological system.60, 61 Therefore, it is necessary to build up a new method for an in vivo cell culture. Recently, biomicroelectromechanical-system (bio-MEMS) techniques have been demonstrated as an enabling technology for cell culture.62 Many advantages include small device size, high surface-area-to-volume ratio, high throughput, continuous-perfusion cell-culture environments, and automation can be achieved by using this technique. Microfluidic-based cell-culture systems are developed to provide an in vivo-like microenvironment which can be more easily maintained than that in macroscale dishes. It also represents a new culture vessel to control the cellular environment and provides the capacity of running a multiple-channel assay process.14 Typically, these microfluidic systems for cell culture can be classified into two types: either as two-dimensional (2D) or three-dimensional (3D) platforms.
2D cell culture
Most adherent human cells are cultured using a 2D platform, where cells are seeded on a planar surface. With this approach, it is easy to manipulate large quantities of cells and simplify the analysis of their cellular behavior. Moreover, a fluorescence detection method can be used for the direct detection of the cellular behavior.63, 64 Many 2D approaches to culture cells in a microfluidic system have been reported in the literature. For example, photo- and soft-lithography-based patterning techniques have been widely used for a variety of applications including confining cells in specific regions and detecting cellular responses.9, 61, 62 2D cell cultures were then performed to verify cell viability. Alternatively, a dry etching technique has also been used to pattern the substrate by defining cell-adhesive and nonadhesive areas in which different types of cells can be cultured two-dimensionally inside microfluidic chips.65 The laminar flow of liquids in capillary systems has also been used to pattern the cells so that selected cells can be controlled and cultured by tuning the characteristics of the surface. Besides, PDMS structures have been widely used for 2D cell culture because of the simple fabrication process, high flexibility, optical transparency, and good biocompatibility.66, 67
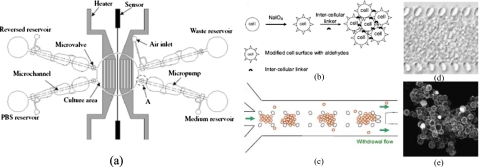
Most of the 2D cell-culture studies in microfluidic systems have adopted a static culture strategy, which used a fixed amount of culture media over a period of time.14 Although static culture systems are simple to operate and easy to maintain, the culture environment in such a system may fluctuate due to the periodic medium replacement processes.68 Alternatively, a continuous-perfusion cell culture has been recently presented with advantages that include a continuous supply of medium and transport of waste way in a sustainable microenvironment. For example, a microfluidic-based artificial liver inspired by an endothelial-like barrier capable of mass transport properties similar to the liver acinus was reported.69 A continuous, perfusion-based microfluidic cell-culture array has also been reported70 which contained a 10×10 array to perform 100 cell-based experiments in parallel for the functions including repeated cell growth∕passage cycles, reagent introduction, and real-time optical analysis. Moreover, a microfluidic chip with integrated micropumps, microvalves, and a microheater has been demonstrated to supply and transfer medium, buffer, and waste automatically14 [Fig. 3a]. A suitable thermal environment for cell growth has also been provided to replace the traditional manual process.
Figure 3.
(a) Schematic illustration of a 2D, automatic, cell-culture system including a cell-culture area, four micropumps, four microcheck valves, microchannels, reservoirs, two heaters, and a microtemperature sensor (Ref. 14). [(b)–(e)] A 3D, gel-free microfluidic device. (b) Cell surfaces modified by sodium periodate (NaIO4) have aldehyde groups which react with hydrazides on the intercellular linker to form multicellular aggregates. (c) Cells are suspended in a cell-culture medium with a dissolved intercellular linker and are seeded into the microfluidic channel with an exit flow at the outlet. (d) Transmission image of the cellular construct in a gel-free 3D-mFCCS after seeding. (e) Confocal microscopic image of cells aggregated with fluorescent intercellular linkers. Additional details regarding the fabrication of devices and experimental procedures can be found in Ref. 74. [Reprinted with permission from S. M. Ong et al., Biomaterials 29, 3237 (2008). © 2008, Elsevier.]
3D cell culture
Since it has been found that cells lose their phenotypes when cultured in 2D environments, a 3D culture offers a more biologically relevant model to conduct microscale, mammalian, cell-based research and development.71 There are many studies about cells cultured in 3D environments with compatible polymer (such as PDMS) fabrication of bioreactors, which can prevent damage to cells.12 A micro-3D cell-culture platform with perfusion functionality using microfluidic technologies can be achieved in high-throughput cell-culture assays, and anticancer drug testing was successfully demonstrated using the proposed culture platform.72, 73 A gel-free 3D-microfluidic cell-culture system has been presented using a transient intercellular polymeric linker with in situ microfabricated pillar arrays, which were released using an embedded hydrogel for cellular support74 [Figs. 3b, 3c, 3d, 3e]. Besides, 3D cell patterning methods capable of seeding cells in a confined region have been investigated extensively to identify polymer structure designs that can minimize the induced shear stress.75
Most of the cell-culture systems for mammalian cell cultures use 2D systems, where cells are cultured as a monolayer on a surface and do not reproduce the morphology that the cells possess in the original tissue.74 Compared with 2D culture systems, the cells grow more similarly to their in vivo phenotypes when cultured in 3D microenvironments through properly regulated cell-cell and cell-matrix interactions.76 Therefore, when studying mammalian cells, 3D culture platforms are preferable over 2D ones.
Cell sorting and isolation in microfluidics
The sorting and isolation of a concentrated cell mixture is also a crucial issue in many biological fields. There are several bio-MEMS approaches for cell sorting and isolation. Typical mechanisms include a combination of gravity and hydrodynamic forces,77 dielectrophoretic (DEP),78, 79, 80, 81, 82, 83, 84 electrokinetic,85 magnetic,86, 87 acoustic,88 laminar flow control,89, 90, 91, 92 and microfiltering approaches.93, 94 For example, a microfluidic sorter was presented that used a combination of gravity and hydrodynamic forces for continuous mass-dependent separation of cells.77 For a DEP force-based microfluidic cell sorter, when an inhomogeneous electric field exists, polarized cells will move due to the induced dipole such that cells with different sizes or different dielectric properties can be sorted. Obstacles such as ridges and wedges embedded in the microchannel can be also used to generate the inhomogeneous electric field required for the DEP approach to cell sorting.78, 79, 80, 81, 83 Alternatively, an optically induced dielectrophoresis (ODEP) device can be used to generate various types of virtual microelectrodes by projecting a light pattern onto photoconductive materials to manipulate beads and cells.82, 84 With this method, there is no need to fabricate microelectrodes to manipulate and sort cells. Besides, a microdevice using an inhomogeneous magnetic field perpendicular to the direction of the flow such that cells of different sizes can be separated was also reported.86, 87 However, such aforementioned systems may damage cell samples when external forces are applied.
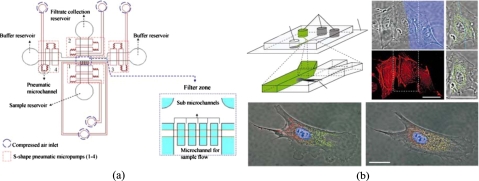
Alternatively, cell sorting and isolation can be also performed by hydrodynamic filtration based on flow control and appropriate microchannel design.92, 95 For instance, a microfluidic device composed of slanted obstacles was developed for continuous blood cell separation.95 Successful isolation of white blood cells (WBCs) with 210-fold enrichment has been demonstrated. Similarly, leukocytes were successfully enriched from blood.92 Furthermore, microfilters with different geometries have been inserted into the microchannels for isolation of red blood cells (RBCs) and plasma.93 Similarly, a cross-flow method in an array of microchannels was designed as a separator for isolating WBCs from human whole blood.96 The concentration of WBCs can be enriched about 4000 times. WBC separation was also performed by another silicon-based microfilter,97 which was designed with four different types of structures, including weirs, pillars, crossflows, and membranes, to compare their performance. The results indicated that the crossflow design has a better separation efficiency and could be employed for fully integrated genomic analysis on a chip. These filtration-based techniques used simple fabrication techniques to facilitate the experiments and to prevent cell damage during operation. However, the channel fouling problem is still a crucial issue. Recently, a size-tunable microfilter modulated by pneumatic pressure for cell separation has also been developed98 [Fig. 4a]. When the cell samples are sparse or the harvested specimen is limited, this device can be used to isolate cells from biosamples. The fouling issue has been solved by using a back-flush operating mode. The results showed that an excellent cell passage efficiency of 93% and a cell viability of 96% for the separation of chondrocytes can be achieved.
Figure 4.
(a) Schematic illustration of a pneumatically tunable microfilter and a close-up view of the filter zone. [Reprinted with permission from S. B. Huang et al., Sens. Actuators B 142, 389 (2009). © 2009, Elsevier.] (b) Differential manipulation of regions of a single bovine capillary endothelial cell using multiple laminar flows. [Reprinted with permission from S. Takayama et al., Nature (London) 411, 1016 (2001). © 2001, Macmillan.]
Recently, biomedical applications utilizing a magnetic bead-based immunoassay implemented in a microfluidic system have been reported.98, 99, 100, 101 The magnetic beads coated with antibodies specific to target pathogens can be used to isolate biosamples99 with the advantages of less labor-intensive operation, lower cost, and less sample∕reagent consumption in these miniature systems. However, how to detach the magnetic beads from the cell-magnetic-bead complexes for a cell culture needs special consideration.
Cell analysis in microfluidics
Since a microfluidic system has the ability to mimic the cellular microenvironment, biological pathways can be studied in a high-throughput manner on this platform. For example, microfluidic systems can be used to control the stimulation factor in an extracellular environment such that the cell phenotype and behavior can be explored. Typically, the impact of shear stress on cell growth, differentiation, and migration can be studied by controlling different flow rates.100 Chemical gradients established by the diffusive mixing properties of a flow capable of generating a complex range of concentrations and distributions of soluble factors to the culture area can also be integrated into the microfluidic chip.101 Compared with conventional pipetting methods, it is possible to simultaneously change the chemical gradients and observe the resulting cell behavior. For example, well-controlled concentration gradients of IL-8 created by a laminar flow were reported to promote neutrophil migration.101 A chemotaxis gradient experiment using the chemokine CXCL8 showed that increasing the flow rates significantly affected cell trajectories along a specific axis.105 Neural axon regeneration, extension,102 and neural cell differentiation20 have also been studied by using microfluidic gradient generators. Furthermore, a microfluidic device consisting of a membrane-covered source region and a large volume sink region, connected by a microfluidic channel, was reported to offer a steady gradient distribution that preserved autocrine and paracrine signaling.103
Besides, slow diffusive mixing in a flow environment results in a separate liquid-phase area that provides the capability to spatially and temporally deliver specific chemical dosages by using a patterned laminar fluid flow. For instance, a flow-focusing system consisting of three inlet channels converging into a cell-culture microchannel was demonstrated104 [Fig. 4b]. It was capable of maintaining selective labeling of different regions of the cell to study the movement of subcellular mitochondria and changes in the cytoskeletal structure. The results indicated that using laminar flows was an effective way to deliver small molecules to selected domains inside single mammalian cells. Similarly, the study about the effect of temperature on an embryo of Drosophila has also been developed.105 Laminar flows were used to create temperature differences by flowing two converging aqueous streams around an embryo. It was confirmed that the warmer half of the embryo develops more rapidly. Those approaches to localized stimulation using microfluidics may solve some obstacles encountered in conventional methods, such as screening the cell behavior in a microenvironment simultaneously since microfluidics provides a high-throughput approach to allow the simultaneous changing of these stimuli.
Alternatively, segmented flow microfluidic systems have been used for high-throughput cellular analysis.106, 107, 108, 109, 110, 111 For instance, a direct cell analysis approach has been reported using microfluidics, which was based on analysis of interphoton burst recurrence times from a single particle for discrimination between different cell populations, allowing species discrimination in a real-time fashion with the potential for cell counting and single-cell sizing.106 Besides, a microfluidic stochastic confinement approach utilizing wells and droplets to separate rare single cells from a bulk mixture was reported.107 It allows detection signals to be locally accumulated to a higher concentration around a single cell, which can be used for the detection and analysis of single cell. Another method has also been proposed for cancer cell sorting and multimodal typing using columns of magnetic beads self-assembled in a microfluidic channel in the form of an array of magnetic traps, allowing high-resolution and quantitative immunophenotyping and morphological analysis.109
Furthermore, droplet-based microfluidic systems have also been used for cell analysis.110, 111 For instance, simultaneous determination of gene expression and enzymatic activity in individual bacterial cells has been performed in microdroplets utilizing a microfluidic device capable of storing picoliter droplets containing single bacteria with constant volumes.110 Furthermore, similar techniques have even been adopted for investigation of stem cells.111 Cells labeled with antibodies and fluorescent dyes have been explored by using droplet-based microfluidic systems to physically encapsulate and simultaneously characterize single cells, which may be useful for the enrichment of stem cells. Those methods provide a direct way to investigate the cellular behavior simultaneously and have potential for further medical applications.
STEM CELL TYPES AND APPLICATIONS IN MICROFLUIDICS
In recent years, there have emerged a variety of applications for stem cells in microfluidic systems. The microfluidic stem cell systems are categorized according to stem cell types and are presented in Table 3. Briefly, a comparison of the origins, differentiation pathways, and microfluidic applications between stem cell types including ESCs, HSCs, MSCs, NSCs, and CSCs is described. Details of representative works for each stem cell type are described in Secs. 3A, 3B, 3C, 3D.
Table 3.
The categorizations of microfluidic systems for stem cells based on stem cell types.
| Cell type | Derivation | Differentiation pathways | Microfluidic applications |
|---|---|---|---|
| Embryonic stem cells | Early stage embryo (Ref. 27), inner cell mass of the blastocyst (Ref. 27) | Any type of cells in the human body (Ref. 27) | Size tuning of EB (Refs. 115, 116, 117, 118, 119, 120, 121, 122, 123, 124), culture and differentiation (Refs. 115, 123, 125, 126, 127, 138), temperature stimulation (Ref. 111), physical stimulation (Refs. 132, 133, 134, 135, 136, 137, 138), chemical stimulation (Ref. 139), and signaling analysis (Refs. 130, 131, 171, 172, 173 |
| Hematopoietic stem cells | Bone marrow, umbilical cord blood, placenta, peripheral blood (Ref. 47) | All types of blood cells (Ref. 47) | Purification and separation (Refs. 145, 153, 154, 155, 156, 157, 158), signaling analysis (Refs. 159, 160, 161, 162, 163, 164), electrophoresis assay and microfluidic digital RT-PCR assay (Refs. 165, 166, 167) |
| Mesenchymal stem cells | Adult marrow (Ref. 40), cord blood (Ref. 41), placenta (Ref. 42), aminotic fluid (Ref. 43) | Osteocytes, adipocytes, chondrocytes, astrocytes, and myoblasts (Ref. 39) | Culture and differentiation (Refs. 129, 130, 131, 132, 133, 134, 137, 138, 139, 171), stimulation (Refs. 135, 136), separation (Ref. 173), and transfection (Ref. 140) |
| Neural stem cells | Adult brain (Ref. 52) | Neuron, astrocytes, oligodendrocytes (Ref. 52) | Signaling analysis (Refs. 20, 167), sorting (Ref. 168) and gene expression profiling (Ref. 169) |
| Cancer stem cells | Tumors, hematological cells (Ref. 56) | Tumor cells (Ref. 56) | None |
Embryonic stem cells
ESCs are pluripotent cells derived from the inner cell mass of blastocyst, and an early-stage embryo, which can be harvested from a donor mother animal.112 The proliferation and differentiation of the stem cells can be well controlled by manipulating the chemical environment in a microfluidic system. For example, the proliferation of mouse ESCs has been observed under a continuous medium-exchange condition.67 It is a common technique to use mouse cells and other animal cells to maintain the pluripotency of actively dividing stem cells. Differentiation of ESCs is often initiated by the generation of embryoid bodies (EBs), which are three-dimensional cell aggregates formed by culturing ESCs on a nonadherent substrate.113, 114
In this section, a variety of methods for studying the response of ESCs in microfluidic systems are introduced and reviewed, including microwells,115, 116, 117, 118, 119, 120, 121 membrane fabrication,122, 123, 124 feeder-layer coating,125, 126, 127 arrayed microreactors with microwells,128, 129, 130, 131 and physical and chemical stimulation111, 132, 133, 134, 135, 136, 137, 138, 139 to study the morphology of ESCs.
Size-controlled microwells
In conventional large-scale systems, it is difficult to regulate the EB size since large cell clusters tend to differentiate at the borders, and clusters with different sizes may lead to heterogeneous differentiation patterns within embryo bodies.115 Studies have shown that the initial number of cells in a self-assembled aggregate highly affects the preferential commitment of the EB toward a specific germ layer.116 Therefore, a new method to culture human ESCs with a controlled cluster size and number through a merging of biomaterials and microfabrication technologies has been demonstrated by using size-controlled microwells.115 Polymeric microwells were fabricated to control the size and uniformity of human embryonic stem cell (hESC) clusters when cocultured with murine embryonic fibroblasts (MEFs). Experimental data showed that hESCs could be cultured homogeneously while maintaining their undifferentiated state (Fig. 5). Similarly, a 3D microwell-based approach has been reported to maintain hESC for weeks without differentiating by using physical and extracellular matrix (ECM) patterns to control the colony growth.117 The results showed that over 90% of the hESCs cultured in the microwells were viable for 2–3 weeks.
Figure 5.
(a) Surfaces are treated with fibronectin and are seeded with MEF cells growing into a monolayer. [(b)–(g)] hES-MEF cocultures at various time intervals. MEFs are stained with CFSE (green) and hES cells with Vybrants DiD (red). (h) Confocal microscopic images of hES-MEF cocultures within a microwell on day 1. Additional details regarding the coating method can be found in Ref. 115. [Reprinted with permission from A. Khademhosseini et al., Biomaterials 27, 5968 (2006). © 2006, Elsevier.]
Creating uniform EBs from murine embryonic stem cells (mESCs) and hESCs still remains a challenge.118, 119 EB populations formed from conventional suspension culture are heterogeneous in size and morphology so that their epigenetic expression revealed that they were unsuitable for potential clinical applications.120 Therefore, it was reported that honeycomb microwells with a high-aspect ratio fabricated by a laser-jet printer and replica molding method can be used to produce uniform EBs of tunable sizes.121 This approach can be used to control the initial size of the cell aggregate and the concentration of the cell suspension by printing various microwell patterns onto prestressed polystyrene sheets to provide ESCs with a suitable lineage commitment. The size-controlled microwell method may simply confine uniform ESCs in a certain region and can be useful for stem cell study.
Membrane fabrication
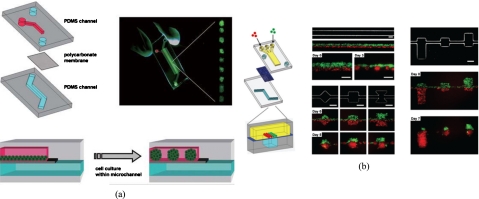
Another approach to producing EBs has been recently reported by using a semiporous membrane channel on a compartmentalized microfluidic device such that a suitable environment for the culture of EBs was provided [Fig. 6a].122 It was demonstrated that the size of EBs developed uniformly on the upper channel while the nonattached EBs were isolated in the lower semiporous membrane channel.126 The results showed that the size of the EBs was controlled by adjusting the cross-sectional size of microchannels or by changing their geometry. It has been also demonstrated that the EBs were extracted from microchannels without affecting their ability to differentiate into healthy neuron-like cells. Similarly, 3D coculture spheroid formations of various geometries and compositions of EB have also been reported in a microfluidic system [Fig. 6b].123 It allowed observation of a spheroid in a single step and multicell-type 3D features were spatially arranged by using a membrane. By simply changing the geometric pattern of the channel, experimental results showed that spheroids of HepG2 cells and mESCs were formed successfully. Furthermore, the regional differentiation of the mESCs was observed.
Figure 6.
(a) Schematic illustration of the microfluidic EB formation device. Two PDMS microchannels are separated by a semiporous polycarbonate membrane, and the cells self-aggregate and form EBs because channel surfaces are rendered resistant to cell adhesion. [Reprinted with permission from Y. S. Torisawa et al., Lab Chip 7, 770 (2007). © 2007, The Royal Society of Chemistry.] (b) Two types of cells, MDA-MB-231 cells (green) and COS7 cells (red), are juxtaposed in the top layer as fluid flow focuses them together into one channel in the bottom layer, and cellular patterning on the bottom channel with distinct geometric features for shape control. [Reprinted with permission from Y. S. Torisawa et al., Integr. Biol. 1, 649, (2009). © 2009, The Royal Society of Chemistry.]
Compared with coculture spheroids formed by the conventional method which randomly mixed the two cell types, this flow hydrodynamically focused cells in the Y-shape microchannel allowing different cell types to be patterned simultaneously in well-defined spatial arrangements. Furthermore, the EBs were loaded into a three-layer microfluidic chip with a uniform microenvironment, and then perfused with vitamin C (VC).124 The results indicated that EBs could extend in the in vivo-like microchip and tended to differentiate into cardiomyocytes. In summary, the membrane approach opens up a new route for manipulating the reactions between different cell types in 3D microenvironment and may be useful for stem cell study.
Feeder-layer coating
Another approach is to coat the surfaces of the microfluidic chip with a feeder layer to maintain the phenotype of ECSs and to analyze their primitive characteristics. For instance, a microfluidic platform was reported to coculture MEFs and mESCs, which used MEFs as a feeder layer to keep the ESCs in a primordial state.125 It was observed that primary MEFs and mESCs cultured in this microfluidic environment demonstrated similar viability and morphology when compared with those in a culture dish. In order to balance nutrient delivery and secretion removal to sustain MEF densities, an optimal flow rate was used for the self-renewal of mESCs. Similarly, an external membrane of a fertilized zebra fish egg was used as a feeder layer and combined with a microfluidic culture system has been demonstrated as a type of ECM for culturing mESCs.126 Mouse P19 embryonic carcinoma (EC) stem cells were xenotransplanted into this type of chorion biomaterial and their differentiation was studied. It was found that neural cells were located in the outer layer of the cell mass and the beating cardiomyocytes existed in the inner layer of the embryo body. Another approach was reported to combine conventional culture materials and microfabrication techniques to create a miniaturized 3D microenvironment on a silicon chip, which was coated with a protein∕ECM for cell attachment and long-term growth.127
Arrayed microreactors and microwells
Arrayed structures have been commonly reported to study stem cells. For instance, hESCs were induced to differentiate into endothelial cells in an array of 12 microfluidic bioreactors.128 This hybrid platform combined bioreactors and microfluidic devices and has advantages of smaller volumes, independent culture wells, high throughput, steady-state conditions, and enhanced mass transport. It was reported that a higher shear stress and a low seeding density in the culture chambers led to larger differentiated cells. Moreover, the operating requirements for hESC culture microsystems were further explored.129 The optimal criteria for selecting operating parameters and the configuration of the bioreactors were characterized, leading to control of the cellular environments for hESC differentiation.
The shape of retained EBs in microfabricated polyethylene glycol (PEG) microwells that exhibited a high viability has been explored recently.130 In order to raise the throughput, an approach using microarrays capable of providing a high cell-number spatial density was used for nanoliter-scale synthesis of biomaterials and characterization of their interactions with cells.131 It was shown that over 1700 hESC-material interactions were characterized simultaneously by the microarray, and a host of unexpected material effects that offer new levels of control over the behavior of hESC behavior was identified.
These approaches provide high performance throughput for the culture of ESCs, and have the ability to simultaneously analyze the ESCs.
Physical factors
Mechanical forces have played an important role on the differentiation and cellular behaviors of stem cells.132 Shear stress can be generated by controlling the flow rate in the microfluidic devices such that the behavior of the stem cells can be observed in real time. For example, hESC-derived endothelial cells can be induced to elongate and align in the direction of the flow by modulating gene expressions.133 Their transcriptional response to shear stress was found to be similar to the primary cells tested. A logarithmic range of flow rates in a microfluidic device has been used to analyze the mESCs.134 It was found that cell colonies can grow with healthy, rounded morphology at higher flow rates, while cells in slower flow rates did not proliferate.
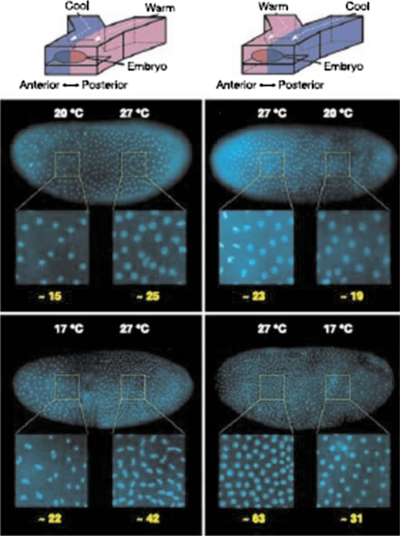
For most of studies on EB differentiation, they were only subjected to a single differentiation condition.135 Recently, a Drosophila embryo was placed in a Y-shaped microfluidic device with two fluid streams at different temperatures, allowing the observation of stem cells under multiple differentiation conditions.105 The laminar flow in microfluidic systems was used to create temperature differences by flowing two converging aqueous streams around an embryo to rapidly provide the supply and removal of heat (Fig. 7). This device was used to precisely control the temperature of different parts of the embryo both spatially and temporally so that the rate of development in the anterior and posterior halves of the embryo was differentially controlled. The results showed that a lower cell density and patterning was observed in the cooler half of the embryo’s body. Similarly, a platform based on a microfluidic Y-channel device with two inlets made up of biocompatible materials for two different culturing media was reported. This device was able to independently cultivate two halves of an EB in two separate media due to the laminar characteristics at low Reynolds number flow in a microchannel.136 Thus, cell differentiation was induced on half of the EB, while the other half of the EB was maintained in an uninduced state simultaneously. Similarly, by using a patterned laminar flow, it was also demonstrated that patterned delivery of reagents and dissociation enzymes to selected regions of individual hESC colonies was feasible.137 Similarly, maintenance of hESC self-renewal abilities and the ability of differentiating into three germ cell lineages with or without flow on a microchip have been demonstrated. This method may allow propagation of signals from one end of a colony to the other end which represents gradient development. Hence, treatment with subcellular spatial selectivity on target portions of hESC colonies can be realized and a single colony can serve as its own experimental control.
Figure 7.
The rate of development in each half of an embryo exposed to a T-step is affected by temperature. Higher nuclear density is observed in the warmer half of the embryo. [Reprinted with permission from E. M. Lucchetta et al., Nature (London) 434, 1134 (2005). © 2005, Nature Publishing Group. ]
In addition, a unidirectional perfusion microfluidic device has been developed to observe the effect of human embryonic germ (hEG) cells on SKOV3 cells. They were cocultured in the inlet and the outlet of the device to observe their changes. The result showed the hEG cells induced the apoptosis of SKOV3 cells, revealing that the stem cells can inhibit the growth of cancer cells (SKOV3 cells).138
Chemical factors
In addition to physical factors, the effect of chemical factors on stem cells has been recently investigated by using microfluidic systems. For instance, a computational model and experiments were recently presented to study mESCs. This study suggested that the removal of putative survival factors could affect stem cell survival in a culture. It was also experimentally confirmed that mESC viability is dependent on soluble factors secreted by mESCs, instead of being supplied from the cell-culture media. It also revealed that mESCs undergo an adaptive∕selective process in response to a lack of paracrine signaling (Fig. 8).139
Figure 8.
(a) Schematic diagram of a dual-layer microfluidic device. The fluidic layer is shown in red and the push-down valves are shown in blue. (b) Phase contrast image of single-cell colonies seeded inside the microfluidic device. (c) The single-cell colonies where DAPI indicates staining of dead cell (the arrows). (d) Phase contrast image of a small multicellular colony inside the microfluidic device. (e) DAPI image of a multicellular colony stained with CellTrackert. Additional details regarding the experimental procedures and results can be found in Ref. 139. [Reprinted with permission from D. Ellison et al., Mol. BioSyst. 5, 1004 (2009). © 2009, The Royal Society of Chemistry.]
Mesenchymal stem cells
Different methods for applications with MSC are described in this section, including microenvironments built for MSC culture and differentiation,26, 140, 141, 142, 143 stimulation of MSCs by chemical and physical factors in microfluidic systems,144, 145 cell growth by patterned substrates, and MSC coculture.146, 147, 148, 149, 150 Moreover, other applications for MSCs such as separation and transfection are also introduced and reviewed.151, 152
Microculture and differentiation environment
Multipotent MSCs are adult stem cells capable of differentiating into many cell types such as osteoblasts, adipocytes, chondrocytes, astrocytes, and myoblasts.39 There are several microfluidic platforms developed for the study of MSCs. For example, a microfluidic system was demonstrated to provide a microculture and differentiation environment for chondrogenic differentiation of MSCs and for investigating the role of mechanical signals.140 It was demonstrated that this device, actuated by an electromagnetic force, can generate a cyclic compression load (CCL) which enhanced the chondrogenic differentiation of rabbit MSCs. The application of micromechanical stimulation in this microfluidic system generated a much greater magnitude of hydrostatic and compression loads compared with conventional systems. Furthermore, multiple samples were cultured simultaneously under uniform conditions. Therefore, the behavior of the MSCs can be systematically investigated.
In most microfluidic-based culture systems, cell seeding density is controlled only by the concentration of the cell suspension. However, paracrine or juxtacrine signaling between cells affects their behavior because of broad distributions of cell numbers across the culture chambers, resulting in a higher variability in the local environment. Therefore, an automated cell-culture platform with 96 individual culture chambers made from PDMS material has been developed to solve this problem.141 In this device, the cell number can be precisely and independently controlled with a single-cell suspension concentration by using a sequential loading procedure. This performance of this versatile system was verified by measuring the influence of transient stimulation on the proliferation, osteogenic differentiation, and motility of human primary MSCs. It was observed that the reversibility and dynamics on cell motility were affected by osteogenic differentiation factors (Fig. 9).
Figure 9.
(a) Schematic diagram of the fluidic path in the chip. (b) A photograph of the chip with the channels filled with colored water to indicate different parts of the device. Additional details regarding the fabrication of devices and results can be found in Ref. 141. [Reprinted with permission from R. Gómez-Sjöberg et al., Anal. Chem. 79, 8557 (2007). © 2007, American Chemical Society.]
Even though these microfluidic devices can perform differentiation of stem cells, the long-term culture of these cells still remains an issue. Moreover, these devices were equipped with large and expensive syringe pumps which increased the complexity and cost of the experimental setup. When compared with a static culture device, a continuous medium flow was provided to keep the culture environment fresh. Therefore, long-term culture and differentiation of MSCs was performed more reliably with a continuous approach.142 The integration of the micropumps makes the system capable of performing these functions automatically. Various differentiation conditions can be also tested and observed by designing several culture areas in the microfluidic chip. Similarly, a micrototal cell-culture system composed of a pulsating pumping system, cell reservoir, cell-culture chamber, and decoding microswitches using PDMS material was reported for studying hMSCs.146 The results showed that the pulsatile pumping plays an important role on the culture and differentiation of two cell types [human breast adenocarcinoma cells (MCF-7) and hMSCs].
Culturing of mammalian cells is increasingly used in drug development and biological research applications. As mentioned previously, these cells exhibit 3D features and should be culture in 3D microenvironments. However, there are several limitations associated with the use of hydrogels in 3D cell cultures in microfluidic systems. For instance, hydrogels need to be spatially localized within the microchannel to avoid blocking the medium flow. Dense cells and poor ECM are hard to embed into hydrogels to form 3D features in vitro. Alternatively, a 3D, microfluidic, channel-based cell-culture system has been reported to precisely control 3D cell-to-cell and cell-to-matrix interactions.143 In order to immobilize and support cells with 3D cell-to-cell and cell-to-matrix interactions, an array of micropillars fabricated in the microchannel was used in this work instead of hydrogels. The laminar flow was also used to hydrodynamically focus the 3D matrix into the center compartment of the microchannel which facilitated a perfusion culture. The experimental results demonstrated that various types of immortalized (HepG2 and MCF7) and primary [hepatocytes and bone marrow mesenchymal stem cells (BMSCs)] mammalian cells were successfully cultured in the 3D-microfluidic culture device. Besides, a gel-free 3D-microfluidic culture system combined with transient intercellular polymeric linkers and micropillar arrays has also been reported for seeding and culturing of mammalian cells [two mammalian cell lines (A549 and C3A) and a primary mammalian cell (BMSCs)] without the use of hydrogels.74 Cell surfaces modified with sodium periodate (NaIO4) were successfully used to react with the hydrazides on the intercellular linker to form multicellular aggregates. In summary, culture and differentiation of MSCs in these microenvironments can offer a more in vivo-like surrounding for studying MSCs and still attract considerable research interest.
Chemical and physical factors
Similar to other stem cells, chemical and physical factors have been applied to study MSCs. For instance, an osmosis-driven pump capable of generating a concentration gradient of fetal bovine serum (FBS) was used to culture hMSCs.144 A very slow microchannel flow was used to provide the medium. The attachment, viability, and morphology of hMSCs were investigated. It was reported that the cell density in the higher FBS concentration area was twice as high as that in the pure culture medium. Similarly, a microfluidic chip which can provide physiologically relevant models of interstitial flow, as well as create a combined gradient of chemical and mechanical stimuli on living organisms, was reported.145 This work realized a culture platform capable of producing chemical and mechanical stimuli similar to realistic physiological conditions. The performance of the system was evaluated by monitoring L929 mouse fibroblast cells for a continuous and long-term culture, which can be applied to the differentiation of stem cells.
It is well recognized that cells respond dynamically to their microenvironments with biochemical and mechanical cues. Small convective flows can cause large changes in morphogenetic distributions even though they only generate very low shear stresses. Therefore, a very slow flow produced in a microchannel is capable of affecting the cellular phenotype and can be used to monitor their variations in the mimic microenvironment. Those studies demonstrated that MSCs can be very sensitive to chemical or physical factors during the culture and differentiation processes.
Patterning structures
Patterned substrates have been used for the culture and differentiation of MSCs. For instance, a method using direct-write CO2-laser micromachining to engrave PDMS microchannels to pattern and coculture neuron cells and MSCs was reported, and which allows the evaluation of trans-differentiated MSCs.146 It was observed that these patterned cells proliferated, extended, and migrated across the interval space between neurons and MSCs and came into contact with each other due to cell-to-cell interactions. Alternatively, a self-assembled monolayer (SAM) substrate was developed to create arrays of spatially distinct hMSC culture environments.147 A broad range of cell adhesion peptide densities of hMSCs adhered to patterned features was observed. Moreover, the peptide density significantly affects the formation, extension, and focusing the hMSC adhesion complex.
It is a great need to align ECM structures with native tissues because they lose their native organization and adopt random distributions that do not resemble the original physiological tissue architecture when being assessed outside the ECM. Therefore, aligned ECM matrices were used to support multilineage mesenchymal differentiation.148 It was reported that aligned collagen matrices structures are capable of guiding the differentiation of resident mesenchymal stem and progenitor cells into specific lineages. It was also shown that the reconstituted, aligned ECM structures could maintain multilineage (osteogenic∕adipogenic∕chondrogenic) differentiation of MSCs. Another interesting work reported that nanofiber-based extracellular matrix proteins can be used for patterning substrates of hMSCs.149 Various types of nanostructures can be engineered to mimic microenvironments using submicron sized synthetic nanofibers. It was observed that the cells on the AA-grafted nanofibers greatly promoted the hMSC adhesion, migration, and proliferation when compared to untreated nanofibers. In summary, patterned substrates are promising methods for evaluating the interactions and signaling between cells and will attract considerable interest in the near future.
MSC coculture
Coculture techniques have been demonstrated in microfluidic systems for studying MSCs. For instance, the dynamic migration process of bone marrow MSCs to the pancreatic islets has been investigated by using a coculture microfluidic chip.150 A tunnel was designed to connect BMSCs and pancreatic islets in two separate microchambers. The results demonstrated that the migration of BMSCs to pancreatic islets can be accelerated by the factors generated by pancreatic islets. Besides, coculturing with BMSCs provided pancreatic islets a better survival rate as well. This indicated that MSCs may be promising for use in clinical therapy in the future.
MSC separation and transfection
The separation of MSCs is an important issue and, therefore, has been extensively studied using microfluidic systems. For example, a cell separation chip was reported in which the MSC cells were successfully isolated by a combination of a T-junction focusing structure and louver-like structures embedded as a microfilter, which allowed the target cells to flow through and any unwanted larger cells to flow away from the tilted channel due to hydrodynamic forces.15 Therefore, the fouling issue is solved successfully and the cells remain intact due to the lack of a large applied external force during the separation process. Experimental results showed that a relatively high separation efficiency (90%) was achieved. This method can effectively prevent damage to the fragile cells, which is an important issue in the study of MSCs.
Gene transfection for the study of MSCs has also attracted considerable interest recently. It was reported that existing biological techniques which introduced foreign DNA into mammalian cells based on conventional procedures have specific limitations, such as a limit on the amount of DNA uptake and the site of genomic integration.151 These limitations have caused a serious concern that stem cells are unable to be genetically modified for therapeutic applications. Therefore, microfluidic systems have been demonstrated for MSC transfection. For example, a microfluidic device capable of single-cell electroporation was used to transfect cells with a high efficiency (>75%).152 It has been shown that electroporation of single mouse myoblastic C2C12 cells and single human MSC was successfully performed utilizing this microfluidic device. The development of a flow-through microfluidic chip capable of cell immobilization, electroporation, and transfection of genes in an individual stem cell may become an extremely valuable tool in this field.
Hematopoietic stem cells
Recently, microfluidic technology has been widely used for the study of HSCs. Many processing tools for HSCs are introduced and reviewed in this section including purification and separation,145, 154, 155, 156, 157, 158 signaling analysis,159, 160, 161, 162, 163, 164 microchip electrophoresis assays, and microfluidic-based, digital, reverse transcription-polymerase chain reaction (RT-PCR) assays.165, 166, 167
Purification and separation
HSCs from cord blood for transplantation have been widely used. They are capable of decreasing HLA mismatches from a related family donor. Enrichment and purification of hematopoietic stem and progenitor cells are important in transplantation therapies for hematologic disorders and in basic stem cell research such as cell transplantation for regenerative medicine.153 Therefore, there exists a great need to isolate the HSCs from cord blood. In order to isolate the CD34+ fraction, the mononuclear cells (MNCs) can be treated with fluorescent anti-CD34 conjugates and purified using a high-speed flow cytometer. Alternatively, the antibody can be conjugated to magnetic beads or iron-dextran nanoparticles (MiniMACS, CliniMACS) to capture the target cells and to isolate them by using a magnetic field. Although a pure population (>90%) of CD34+ hematopoietic stem and progenitor cells (HSPCs) can be achieved by using flow cytometry, the process is time-consuming, while immunomagnetic cell enrichment is much faster than the flow cytometry but the resulting purities are less consistent (from 18% to 99%).154 Recently, there has been a significant improvement in conventional methods by utilizing selectin-mediated enrichment to recover HSPCs from adult bone marrow (ABM) based on differences in rolling velocities which is independent of CD34 expression. However, HSC isolation still requires a relatively time-consuming and lengthy process, costly and bulky equipment, and experienced personnel to achieve reasonable separation efficiency.
Recently, microfluidic systems have been used for the purification and separation of HSCs. For example, a microfluidic technique using dielectrophoretic field-flow-fractionation (DEP-FFF) was reported to separate hematopoietic CD34+ stem cells from human breast cancer MDA-435 cells according to their dielectric properties.155 The DEP force was generated by an array of microelectrodes patterned on the bottom surface of a narrow chamber. It was demonstrated that CD34+ stem cells can be separated from human breast cancer MDA-435 cells by DEP-FFF with a purity of higher than 99.2%. Similarly, differences in average rolling velocities between cell populations can be used as a method for sorting and enriching hematopoietic stem and progenitor cells.156
The amount of HSCs available for transplantation in clinical usage has to be counted precisely for efficacy issues and to maximize the amount recovered from the cord blood. Therefore, an automatic microfluidic system that integrates the two major functions of sample pretreatment and a magnetic, bead-based flow cytometer for isolation, counting, and sorting of HSC from cord blood has been recently developed.157 By using an immunoassay technique, the antibody-conjugated magnetic beads labeled with fluorescence dyes can be used to identify a surface antigen specific for HSCs and bind with the target cells. Subsequently, the cell-magnetic-bead complexes were optically detected, counted, and finally sorted into the target collector through the feedback signal. Furthermore, an anti-CD34 antibody-immobilized, cell-rolling column has been developed with the ability to separate CD34+ cells by the CD34 density of the cell surfaces.158 This device consisted of a tubular column with a continuous surface for cell separation such that the anti-CD34 antibody was immobilized at a high density on the surface. Then the injected cells were rolled onto the inner surface of the column to sheer force of the medium flow. This validated the ability to isolate cells via a cell-rolling process using columns, based on surface marker density under labeling-free conditions. Note that although density gradient centrifugation in conventional methods is a relatively simple task to perform, the process is not specific for stem cells. The antibody-based strategy is much more effective and specific for the isolation of HSCs.
Microwells and microcavities
Technologies to detect and isolate specific cells of interest from heterogenic populations are in great demand in the field of cell analysis.159 Even though the traditional approach that seeds cells in a large dish is adequate in most cases, knowledge about the behavior of an individual cell is still a critical issue which cannot be solved by this existing method. For example, the phenotypes of stem cells are almost the same as their already partially differentiated progenitor cells. It is crucial to evaluate the changes in cell cycle kinetics, self-renewal, and differentiation. One of the simple platforms to analyze a single cell in a medium with a high throughput is an array of microwells. Microwell arrays consist of high-density, micron-sized cavities of a desirable geometry, which can be fabricated using standard microfabrication processes such as photolithography. For instance, PDMS microwell arrays were reported to monitor the dynamic behavior of single mouse HSCs and correlate proliferative behavior in vitro with in vivo functions.160 Similarly, a PEG hydrogel was also used to fabricate microwell arrays to track the molecular signaling of HSCs and progenitor cell populations. It was observed that individual HSCs possess prolonged cycle times compared to the progenitor cells. The change of single mouse HSCs in response to selected putative niche protein components was performed on a microfluidic device to clarify the characteristic of HSCs.161 The influence of spatial restrictions and adhesive interactions on human CD133+ HSCs has been recently investigated.162 The HSC behavior in fibronectin-coated microwell arrays was analyzed using live-cell microscopy combined with in vivo transplantation assays. Lower proliferation and less differentiation in morphology was observed when patterning single HSCs in the small microwells. Furthermore, the analysis of a single cell in the smaller cavities revealed higher levels of HSC marker expression and decreased DNA synthesis.
Moreover, 10 000 microcavities were reported to efficiently capture single CD34+ HSCs, followed by detection and isolation of CD34+ HSCs.163 Approximately 0.1% of the total population of peripheral blood mononuclear cells (PBMCs) was observed. Typically, a cell suspension is introduced into the chamber and trapped into the microcavities by applying negative pressure. Approximately 90% of the loaded cells were successfully captured. This technique presents a simple and promising tool to identify cells from human PBMCs using image-based gene expression analysis of single CD34+ cells.
Since hematopoietic cells are nonadherent and thus difficult to study over time using conventional cell-culture techniques, a microfluidic platform embedded with microcavities, which have been first reported by Di Carlo et al.,164 was adapted to study patient-derived, single CD34+ stem cells.165 These microcavities can replace the two-layer cup-shaped PDMS structures, which usually require a complicated fabrication process. A HSC disorder, chronic myeloid leukemia (CML), resulting from a reciprocal translocation between chromosomes was studied. CML research needs to examine the signal dynamics on the single-cell level in order to identify variations in cellular characteristics which protect certain cells against drug-induced cell death. The study investigated patient-derived normal and CML stem cell responses to the Tyrosine Kinase Inhibitor (TKI) dasatinib, a small molecule inhibitor for treatment of dasatinib resistant CML. Results showed that CML stem cells exhibit increased resistance to cell death upon exposure to dasatinib, while the drug reduced the mobility of CML cells within the microfluidic device (Fig. 10).
Figure 10.
Graphic representation of the microfluidic device concept. Cells can be trapped by hydrodynamic flow. [Reprinted with permission from D. Di Carlo et al., Anal. Chem. 78, 4925 (2006). © 2009, The Royal Society of Chemistry.]
Microelectrophoresis assay and digital RT-PCR assay
Allogeneic hematopoietic stem cell transplantation (allo-HSCT) has become one of the most promising treatment methods in a variety of malignant and nonmalignant diseases.166 Chimerism is a dynamic process which signifies the proportion of cells derived from the donor in the recipient’s blood, that is, a mixture of recipient and donor cells in the recipient hematopoietic system may occur after allo-HSCT.167 It has been reported that the determination of donor chimerism is feasible by using a microfluidic system. Engraftment status was quantitatively evaluated in post-allo-HSCT patients using variable numbers of tandem repeat (VNTR) markers and microelectrophoresis technology. Monitoring of post-allo-HSCT chimerism status using microelectrophoresis for both large- and small-scale post-allo-HSCT chimerism centers was also quantified. Besides, a microfluidic-based “digital RT-PCR” assay was demonstrated, which is a miniaturized version of PCR assays commonly used in molecular biology for nucleic acid amplification.168 The system was used to count template molecules in cDNA samples prepared from single cells with systematic, quantitative population analysis of transcription factor expression within developmental progenitors. These on-chip assays provide a simpler and faster way to analyze and quantify HSCs.
Neural stem cells
Recently, microfluidic technology has been extensively used for the study of NSCs. Many applications for NSCs are introduced and reviewed in this section including signaling analysis,20, 169 NSC sorting, and gene expression profiling.170, 171
Signaling analysis
NSCs are able to self-renew and differentiate into three principal cell types associated with the CNS.52 Recently, microfluidic techniques have been used for signaling analysis of NSCs as well so as to tackle the problems which cannot be properly addressed by using conventional culture technology. For instance, a recent study exposed NSCs to growth factors with a concentration gradient under a continuous flow to minimize autocrine and paracrine signaling of NSCs.20 This work optimized proliferation and differentiation conditions for NSCs using a gradient-generating microfluidic device. Proliferation and differentiation of NSCs into astrocytes were screened by time-lapsed microscopy and immunocytochemistry. The results showed that the NSCs remained healthy throughout the entire culture period. It was also experimentally verified that a proportional expression of cell proliferation and differentiation varied directly with the concentration of growth factor. Similarly, a hybrid microfluidic-vacuum device, which integrated a network of vacuum channels with a microfluidic gradient generator, has been developed for NSCs study.169 The system generated concentration gradients of soluble factors in a single chip with several advantages such as the capability to perform experiments on sensitive cells, such as neurons, which are difficult to culture inside a microfluidic environment, and high reproducibility and functionality.
NSC sorting and gene expression profiling
Recently, two types of neural cells were characterized on a microfluidic-based platform using electrical admittance spectroscopy.170 The difference in the electrical admittance of the cells was measured as they flew through. Hence, the cells were sorted and isolated according to their characteristic electrical signatures which could be useful for stem cell therapy.
Besides, to produce consecutive gene expression profiles from target cell populations, a large number of cells must be profiled to cover all the significant cellular stages. Microfluidic technology has been used for gene expression profiling of NSCs. For instance, a microfluidic device was reported for high-throughput, single-cell, gene expression profiling.171 It was observed that cDNA obtained from 50 individual cells was amplified within 3 h and can be used for whole genome microarray analysis. In summary, NSCs are extremely sensitive and require special attention when they are studied in a microfluidic environment.
Integrated microsystems for stem cell applications
Recently, with the advances in microfluidic technology, integrated microfluidic systems with more functionality have been used for cell analysis and genetic analysis.172, 173, 174 For example, an integrated microfluidic system with six individual cell-culture chambers was reported.172 It allows for reproducible, quantitative, parallel hESC culture and multiparameter analyses in sequence. Several pluripotency markers for immunoassays were performed to confirm the pluripotent ability of hESC colonies. It was reported that hESC can be cultured and analyzed in a single microfluidic device, which is capable of parallel culture, real-time screening, and automated examination of self-renewal and differentiation. A significantly improved efficacy of the response of mESCs to bone morphogenesis protein (BMP) signaling on an integrated microfluidic platform was also reported.173 This work demonstrated an efficient method for directing the differentiation of ESCs using the microfluidic system to minimize paracrine and autocrine effects on ESCs in a controlled manner. Besides, the gene expression of hESCs was performed on an integrated microfluidic system, which extracted total mRNA from individual single cells and then synthesized cDNA with high mRNA-to-cDNA efficiency.174 The results showed that the relative expression levels used in gene expression studies were very different between measurements from cDNA of a cell population and a single-cell cDNA. In summary, integrated microfluidic systems with more functionalities can assure automatic operations for the study of stem cells. Consequently, the integrated microfluidic system provided a fast, compact, and reliable approach for stem cell analysis.
CONCLUSIONS
Microfluidic systems offer cells an in vivo-like microenvironment that is challenging to be realized in conventional large-scale systems. The main advantages of microfluidic systems that include low sample and reagent consumption, fast operation time, high throughput, less labor-intensiveness, and high levels of integration make them promising assay platforms for cell study. Moreover, the capability to perform perfusion allows the infusion of assay reagents while the high optical transparency enables real-time imaging and analysis of the cellular response. These characteristics significantly improve the ability to culture and analyze stem cells, which are important in the medical fields. Consequently, there have been a great number of precision microfluidic platforms developed recently for studies of different types of stem cells (ESCs, MSCs, HSCs, NSCs, etc.) with many promising capabilities such as isolation, culture and differentiation, stimulation, and signaling analysis. Monitoring the real-time reaction response from stem cells offers a literal, straightforward result because they tend to change their morphology easily after a long experimental process. This paper reviews recent works on stem cells on microfluidic systems. It shows that these miniature systems are promising in many aspects. However, there are still several unresolved issues with the use of microfluidic systems for studying stem cells. For example, the efficient separation of stem cells still needs to be greatly improved. Currently, throughput is limited by the slow flow rates in microfluidics. A multichannel array may resolve this bottleneck if they can be properly addressed. The effect of drugs on stem cell differentiation can be screened and monitored in this useful tool. These types of devices can be useful for drug development since the drug-screening throughput can be greatly enhanced. Furthermore, stem cell therapy in clinical applications requires rapid isolation and precise counting of these cells, which has not yet been properly addressed in microfluidic systems. A fundamental understanding of stem cells using microfluidic systems is still under extensive investigation. By controlling multiple parameters that affect the metabolic rates and viability, any cell types including stem cells can be studied and characterized in these systems more efficiently. Therefore, microfluidic systems may become a powerful platform for both fundamental stem cell research and commercial applications175 in the near future.
ACKNOWLEDGMENTS
The authors gratefully acknowledge the financial support provided to this study by the National Science Council in Taiwan (Grant No. NSC 98-2120-M-006-001).
NOMENCLATURE AND ABBREVIATIONS
- 2D
two-dimensional
- 3D
three-dimensional
- ABM
adult bone marrow
- BMSCs
bone marrow mesenchymal stem cells
- allo-HSCT
allogeneic hematopoietic stem cell transplantation
- bio-MEMS
biomicroelectromechanical-system
- BMP
bone morphogenesis protein
- CCL
cyclic compression load
- CSCs
cancer stem cells
- CML
chronic myeloid leukemia
- CNS
central nervous system
- DEP-FFF
dielectrophoretic field-flow-fractionation
- DEP
dielectrophoretic
- EBs
embryoid bodies
- ECM
extracellular matrix
- EC
embryonic carcinoma
- ESCs
embryonic stem cells
- FBS
fetal bovine serum
- FACS
flow cytometry
- HSCs
hematopoietic stem cells
- HLA
human leukocyte antigen
- HSPCs
hematopoietic stem and progenitor cells
- hEG
human embryonic germ
- hESCs
human embryonic stem cells
- hMSCs
human mesenchymal stem cells
- iPSCs
induced pluripotent stem cells
- LOC
lab-on-a-chip
- MEFs
murine embryonic fibroblasts
- mESCs
murine embryonic stem cells
- MSCs
mesenchymal stem cells
- MNCs
mononuclear cells
- NaIO4
sodium periodate
- NSCs
neural stem cells
- ODEP
optically induced dielectrophoresis
- PDMS
polydimethylsiloxane
- PEG
polyethylene glycol
- PEO
poly(ethylene oxide)
- PBMCs
peripheral blood mononuclear cells
- RT-PCR
reverse transcription-polymerase chain reaction
- RBCs
red blood cells
- SAM
self-assembled monolayer
- VC
vitamin C
- VNTR
variable number of tandem repeat
- WBCs
white blood cells
- μTAS
micrototal-analysis-system
References
- McDonald J. C., Duffy D. C., Anderson J. R., Chiu D. T., Wu H. K., Schueller O. J. A., and Whitesides G. M., Electrophoresis 21, 27 (2000). [DOI] [PubMed] [Google Scholar]
- Park T. H. and Shuler M. L., Biotechnol. Prog. 19, 243 (2003). 10.1021/bp020143k [DOI] [PubMed] [Google Scholar]
- Whitesides G. M., Nature (London) 442, 368 (2006). 10.1038/nature05058 [DOI] [PubMed] [Google Scholar]
- Xiao F., Wu T. H., and Chiou P. Y., Appl. Phys. Lett. 97, 031112 (2010). 10.1063/1.3459977 [DOI] [Google Scholar]
- Hirano-Iwata A., Oshima A., Nasu T., Taira T., Kimura Y., and Niwano M., Supramol. Chem. 22, 406 (2010). 10.1080/10610278.2010.487564 [DOI] [Google Scholar]
- van Honschoten J. W., Berenschot J. W., Ondarcuhu T., Sanders R. G. P., Sundaram J., Elwenspoek M., and Tas N. R., Appl. Phys. Lett. 97, 014103 (2010). 10.1063/1.3462302 [DOI] [Google Scholar]
- Matović J. and Jaksic Z., Microelectron. Eng. 87, 1487 (2010). 10.1016/j.mee.2009.11.074 [DOI] [Google Scholar]
- Randall G. C. and Doyle P. S., Proc. Natl. Acad. Sci. U.S.A. 102, 10813 (2005). 10.1073/pnas.0503287102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibel D. B., Garstecki P., and Whitesides G. M., Curr. Opin. Neurobiol. 15, 560 (2005). 10.1016/j.conb.2005.08.013 [DOI] [PubMed] [Google Scholar]
- Xia Y. N. and Whitesides G. M., Annu. Rev. Mater. Sci. 28, 153 (1998). 10.1146/annurev.matsci.28.1.153 [DOI] [Google Scholar]
- Martin R. S., Root P. D., and Spence D. M., Analyst (Cambridge, U.K.) 131, 1197 (2006). 10.1039/b611041j [DOI] [PubMed] [Google Scholar]
- Baudoin R., Corlu A., Griscom L., Legallais C., and Leclerc E., Toxicol. In Vitro 21, 535 (2007). 10.1016/j.tiv.2006.11.004 [DOI] [PubMed] [Google Scholar]
- Huang S. B., Wu M. H., Cui Z. F., Cui Z., and Lee G. B., J. Micromech. Microeng. 18, 12 (2008). [Google Scholar]
- Huang C. W. and Lee G. B., J. Micromech. Microeng. 17, 1266 (2007). 10.1088/0960-1317/17/7/008 [DOI] [Google Scholar]
- Wu H. W., Lin X. Z., Hwang S. M., and Lee G. B., Biomed. Microdevices 11, 1297 (2009). 10.1007/s10544-009-9349-x [DOI] [PubMed] [Google Scholar]
- Lee H. Y. and Voldman J., Anal. Chem. 79, 1833 (2007). 10.1021/ac061647q [DOI] [PubMed] [Google Scholar]
- Yang S. Y., Lin J. L., and Lee G. B., J. Micromech. Microeng. 19, 9 (2009). [Google Scholar]
- Chang H. J., Huang M. Y., Yeh C. S., Chen C. C., Yang M. J., Sun C. S., Lee C. K., and Lin S. R., Clin. Microbiol. Infect. 16, 1090 (2010). 10.1111/j.1469-0691.2009.03045.x [DOI] [PubMed] [Google Scholar]
- Dai W., Zheng Y. Z., Luo K. Q., and Wu H. K., Biomicrofluidics 4, 024101 (2010). 10.1063/1.3398319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung B. G., Flanagan L. A., Rhee S. W., Schwartz P. H., Lee A. P., Monuki E. S., and Jeon N. L., Lab Chip 5, 401 (2005). 10.1039/b417651k [DOI] [PubMed] [Google Scholar]
- Huang B., Wu H. K., Kim S., Kobilka B. K., and Zare R. N., Lab Chip 6, 369 (2006). 10.1039/b515840k [DOI] [PubMed] [Google Scholar]
- Hillborg H., Ankner J. F., Gedde U. W., Smith G. D., Yasuda H. K., and Wikstrom K., Polymer 41, 6851 (2000). 10.1016/S0032-3861(00)00039-2 [DOI] [Google Scholar]
- Hawk G. L., Dufault L. B., and Cameron J. A., Prep. Biochem. Biotechnol. 2, 193 (1972). 10.1080/00327487208061469 [DOI] [PubMed] [Google Scholar]
- Zheng W., Wang Z., Zhang W., and Jiang X., Lab Chip 10, 2906 (2010). 10.1039/c005274d [DOI] [PubMed] [Google Scholar]
- Sabourin D., Snakenborg D., and Dufva M., Microfluid. Nanofluid. 9, 87 (2010). 10.1007/s10404-009-0520-8 [DOI] [Google Scholar]
- Wu H. W., Lin X. Z., Hwang S. M., and Lee G. B., Biomed. Microdevices 11, 869 (2009). 10.1007/s10544-009-9304-x [DOI] [PubMed] [Google Scholar]
- Hwang N. S., Varghese S., and Elisseeff J., Adv. Drug Delivery Rev. 60, 199 (2008). 10.1016/j.addr.2007.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitalipov S. and Wolf D., Eng. Stem Cells 114, 185 (2009). 10.1007/10_2008_45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulloa-Montoya F., Verfaillie C. M., and Hu W. S., J. Biosci. Bioeng. 100, 12 (2005). 10.1263/jbb.100.12 [DOI] [PubMed] [Google Scholar]
- National Institutes of Health, Stem Cells: Scientific Progress and Future Research Directions (University Press of the Pacific, Honolulu, 2001). [Google Scholar]
- Phillips B. W. and Crook J. M., Biodrugs 24, 99 (2010). 10.2165/11532270-000000000-00000 [DOI] [PubMed] [Google Scholar]
- Brignier A. C. and Gewirtz A. M., J. Allergy Clin. Immunol. 125, S336 (2010). 10.1016/j.jaci.2009.09.032 [DOI] [PubMed] [Google Scholar]
- Rohwedel J., Guan K., Hegert C., and Wobus A. M., Toxicol. In Vitro 15, 741 (2001). 10.1016/S0887-2333(01)00074-1 [DOI] [PubMed] [Google Scholar]
- Menendez P., Bueno C., and Wang L., Cytotherapy 8, 530 (2006). 10.1080/14653240601026654 [DOI] [PubMed] [Google Scholar]
- Di Giorgio F. P., Carrasco M. A., Siao M. C., Maniatis T., and Eggan K., Nat. Neurosci. 10, 608 (2007). 10.1038/nn1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston S. L., Alison M. R., Forbes S. J., Direkze N. C., Poulsom R., and Wright N. A., Mol. Pathol. 56, 86 (2003). 10.1136/mp.56.2.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- in‘tAnker P. S., Scherjon S. A., Kleijburg-van der Keur C., Noort W. A., Claas F. H. J., Willemze R., Fibbe W. E., and Kanhai H. H. H., Blood 102, 1548 (2003). 10.1182/blood-2003-04-1291 [DOI] [PubMed] [Google Scholar]
- Kakishita K., Nakao N., Sakuragawa N., and Itakura T., Brain Res. 980, 48 (2003). 10.1016/S0006-8993(03)02875-0 [DOI] [PubMed] [Google Scholar]
- Cipriani S., Bonini D., Marchina E., Balgkouranidou I., Caimi L., Zucconi G. G., and Barlati S., Cell Biol. Int. 31, 845 (2007). 10.1016/j.cellbi.2007.01.037 [DOI] [PubMed] [Google Scholar]
- Jiang Y. H., Jahagirdar B. N., Reinhardt R. L., Schwartz R. E., Keene C. D., Ortiz-Gonzalez X. R., Reyes M., Lenvik T., Lund T., Blackstad M., Du J. B., Aldrich S., Lisberg A., Low W. C., Largaespada D. A., and Verfaillie C. M., Nature (London) 447, 880 (2007). 10.1038/nature05812 [DOI] [Google Scholar]
- Lee O. K., Kuo T. K., Chen W. M., Lee K. D., Hsieh S. L., and Chen T. H., Blood 103, 1669 (2004). 10.1182/blood-2003-05-1670 [DOI] [PubMed] [Google Scholar]
- Yen B. L., Huang H. I., Chien C. C., Jui H. Y., Ko B. S., Yao M., Shun C. T., Yen M. L., Lee M. C., and Chen Y. C., Stem Cells 23, 3 (2005). 10.1634/stemcells.2004-0098 [DOI] [PubMed] [Google Scholar]
- Tsai M. S., Hwang S. M., Tsai Y. L., Cheng F. C., Lee J. L., and Chang Y. J., Biol. Reprod. 74, 545 (2006). 10.1095/biolreprod.105.046029 [DOI] [PubMed] [Google Scholar]
- Campagnoli C., Roberts I. A. G., Kumar S., Bennett P. R., Bellantuono I., and Fisk N. M., Blood 98, 2396 (2001). 10.1182/blood.V98.8.2396 [DOI] [PubMed] [Google Scholar]
- Hu Y., Liao L. M., Wang L. Y., Ma L., Ma G. J., Jiang X. Y., and Zhao R. C., J. Lab. Clin. Med. 141, 342 (2003). 10.1016/S0022-2143(03)00022-2 [DOI] [PubMed] [Google Scholar]
- Romanov Y. A., Svintsitskaya V. A., and Smirnov V. N., Stem Cells 21, 105 (2003). 10.1634/stemcells.21-1-105 [DOI] [PubMed] [Google Scholar]
- Dhot P. S., Nair V., Swarup D., Sirohi D., and Ganguli P., Indian J. Pediatr. 70, 989 (2003). 10.1007/BF02723826 [DOI] [PubMed] [Google Scholar]
- Mossad S., Int. J. Infect. Dis. 14, e329 (2010). 10.1016/j.ijid.2010.02.222220594890 [DOI] [Google Scholar]
- Tomonari A., Takahashi S., Takasugi K., Ooi J., Tsukada N., Konuma T., Iseki T., Tojo A., and Asano S., Int. J. Hematol. 84, 438 (2006). 10.1532/IJH97.06119 [DOI] [PubMed] [Google Scholar]
- Smith L. G., Weissman I. L., and Heimfeld S., Proc. Natl. Acad. Sci. U.S.A. 88, 2788 (1991). 10.1073/pnas.88.7.2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornson C. R. R., Rietze R. L., Reynolds B. A., Magli M. C., and Vescovi A. L., Science 283, 534 (1999). 10.1126/science.283.5401.534 [DOI] [PubMed] [Google Scholar]
- Gregg C. T., Chojnacki A. K., and Weiss S., J. Neurosci. Res. 69, 708 (2002). 10.1002/jnr.10340 [DOI] [PubMed] [Google Scholar]
- Kennea N. L. and Mehmet H., J. Pathol. 197, 536 (2002). 10.1002/path.1189 [DOI] [PubMed] [Google Scholar]
- Doetsch F., Petreanu L., Caille I., Garcia-Verdugo J. M., and Alvarez-Buylla A., Neuron 36, 1021 (2002). 10.1016/S0896-6273(02)01133-9 [DOI] [PubMed] [Google Scholar]
- Marshall G. P., Laywell E. D., Zheng T., Steindler D. A., and Scott E. W., Stem Cells 24, 731 (2006). 10.1634/stemcells.2005-0245 [DOI] [PubMed] [Google Scholar]
- Park C. H., Bergsage D. E., and McCulloc E. A., J. Natl. Cancer Inst. 46, 411 (1971). [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., and Yamanaka S., Cell 131, 861 (2007). 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Byrne J. A., Simonsson S., Western P. S., and Gurdon J. B., Curr. Biol. 13, 1206 (2003). 10.1016/S0960-9822(03)00462-7 [DOI] [PubMed] [Google Scholar]
- Yoo S. K., Lee J. H., Yun S. S., and Gu M. B., Biosens. Bioelectron. 22, 1586 (2007). 10.1016/j.bios.2006.07.014 [DOI] [PubMed] [Google Scholar]
- Andersson H. and van den Berg A., Sens. Actuators B 92, 315 (2003). 10.1016/S0925-4005(03)00266-1 [DOI] [Google Scholar]
- Zhang S. G., Nat. Biotechnol. 22, 151 (2004). 10.1038/nbt0204-151 [DOI] [PubMed] [Google Scholar]
- Saltzman W. M. and Olbricht W. L., Nat. Rev. Drug Discovery 1, 177 (2002). 10.1038/nrd744 [DOI] [PubMed] [Google Scholar]
- Matsubara Y., Murakami Y., Kobayashi M., Morita Y., and Tamiya E., Biosens. Bioelectron. 19, 741 (2004). 10.1016/j.bios.2003.08.001 [DOI] [PubMed] [Google Scholar]
- Meyvantsson I. and Beebe D. J., Annu. Rev. Anal. Chem. 1, 423 (2008). 10.1146/annurev.anchem.1.031207.113042 [DOI] [PubMed] [Google Scholar]
- Rhee S. W., Taylor A. M., Tu C. H., Cribbs D. H., Cotman C. W., and Jeon N. L., Lab Chip 5, 102 (2005). 10.1039/b403091e [DOI] [PubMed] [Google Scholar]
- Takayama S., McDonald J. C., Ostuni E., Liang M. N., Kenis P. J. A., Ismagilov R. F., and Whitesides G. M., Proc. Natl. Acad. Sci. U.S.A. 96, 5545 (1999). 10.1073/pnas.96.10.5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch A. and Toner M., Annu. Rev. Biomed. Eng. 2, 227 (2000). 10.1146/annurev.bioeng.2.1.227 [DOI] [PubMed] [Google Scholar]
- Sung K. E., Su G., Pehlke C., Trier S. M., Eliceiri K. W., Keely P. J., Friedl A., and Beebe D. J., Biomaterials 30, 4833 (2009). 10.1016/j.biomaterials.2009.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. J., Hung P. J., and Lee L. P., Biotechnol. Bioeng. 97, 1340 (2007). 10.1002/bit.21360 [DOI] [PubMed] [Google Scholar]
- Hung P. J., Lee P. J., Sabounchi P., Lin R., and Lee L. P., Biotechnol. Bioeng. 89, 1 (2005). 10.1002/bit.20289 [DOI] [PubMed] [Google Scholar]
- Albrecht D. R., Underhill G. H., Wassermann T. B., Sah R. L., and Bhatia S. N., Nat. Methods 3, 369 (2006). 10.1038/nmeth873 [DOI] [PubMed] [Google Scholar]
- Wu M. H., Huang S. B., Cui Z. F., Cui Z., and Lee G. B., Sens. Actuators B 129, 231 (2008). 10.1016/j.snb.2007.07.145 [DOI] [Google Scholar]
- Wu M. H., Huang S. B., Cui Z. F., Cui Z., and Lee G. B., Biomed. Microdevices 10, 309 (2008). 10.1007/s10544-007-9138-3 [DOI] [PubMed] [Google Scholar]
- Ong S. M., Zhang C., Toh Y. C., Kim S. H., Foo H. L., Tan C. H., van Noort D., Park S., and Yu H., Biomaterials 29, 3237 (2008). 10.1016/j.biomaterials.2008.04.022 [DOI] [PubMed] [Google Scholar]
- Khademhosseini A., Langer R., Borenstein J., and Vacanti J. P., Proc. Natl. Acad. Sci. U.S.A. 103, 2480 (2006). 10.1073/pnas.0507681102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukierman E., Pankov R., Stevens D. R., and Yamada K. M., Science 294, 1708 (2001). 10.1126/science.1064829 [DOI] [PubMed] [Google Scholar]
- Huh D., Bahng J. H., Ling Y. B., Wei H. H., Kripfgans O. D., Fowlkes J. B., Grotberg J. B., and Takayama S., Anal. Chem. 79, 1369 (2007). 10.1021/ac061542n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbulovic-Nad I., Xuan X. C., Lee J. S. H., and Li D. Q., Lab Chip 6, 274 (2006). 10.1039/b513183a [DOI] [PubMed] [Google Scholar]
- Barrett L. M., Skulan A. J., Singh A. K., Cummings E. B., and Fiechtner G. J., Anal. Chem. 77, 6798 (2005). 10.1021/ac0507791 [DOI] [PubMed] [Google Scholar]
- Gascoyne P. R. C., Huang Y., Pethig R., Vykoukal J., and Becker F. F., Meas. Sci. Technol. 3, 439 (1992). 10.1088/0957-0233/3/5/001 [DOI] [Google Scholar]
- Han K. H. and Frazier A. B., Lab Chip 8, 1079 (2008). 10.1039/b802321b [DOI] [PubMed] [Google Scholar]
- Lin Y. H. and Lee G. B., Biosens. Bioelectron. 24, 572 (2008). 10.1016/j.bios.2008.06.008 [DOI] [PubMed] [Google Scholar]
- Pommer M. S., Zhang Y. T., Keerthi N., Chen D., Thomson J. A., Meinhart C. D., and Soh H. T., Electrophoresis 29, 1213 (2008). 10.1002/elps.200700607 [DOI] [PubMed] [Google Scholar]
- Ohta A. T., Chiou P. Y., Han T. H., Liao J. C., Bhardwaj U., McCabe E. R. B., Yu F. Q., Sun R., and Wu M. C., J. Microelectromech. Syst. 16, 491 (2007). 10.1109/JMEMS.2007.896717 [DOI] [Google Scholar]
- Chien Y. S., Lin C. H., Kao F. J., and Ko C. W., Mater. Sci. Forum 505–507, 643 (2006). 10.4028/www.scientific.net/MSF.505-507.643 [DOI] [Google Scholar]
- Lin Y. A., Wong T. S., Bhardwaj U., Chen J. M., McCabe E., and Ho C. M., J. Micromech. Microeng. 17, 1299 (2007). 10.1088/0960-1317/17/7/012 [DOI] [Google Scholar]
- Rong R., Choi J. W., and Ahn C. H., J. Micromech. Microeng. 16, 2783 (2006). 10.1088/0960-1317/16/12/035 [DOI] [Google Scholar]
- Laurell T., Petersson F., and Nilsson A., Chem. Soc. Rev. 36, 492 (2007). 10.1039/b601326k [DOI] [PubMed] [Google Scholar]
- Sai Y., Yamada M., Yasuda M., and Seki M., J. Chromatogr. A 1127, 214 (2006). 10.1016/j.chroma.2006.05.020 [DOI] [PubMed] [Google Scholar]
- Wu Z. G., Liu A. Q., and Hjort K., J. Micromech. Microeng. 17, 1992 (2007). 10.1088/0960-1317/17/10/010 [DOI] [Google Scholar]
- Yamada M., Nakashima M., and Seki M., Anal. Chem. 76, 5465 (2004). 10.1021/ac049863r [DOI] [PubMed] [Google Scholar]
- Yamada M. and Seki M., Lab Chip 5, 1233 (2005). 10.1039/b509386d [DOI] [PubMed] [Google Scholar]
- Sethu P., Sin A., and Toner M., Lab Chip 6, 83 (2006). 10.1039/b512049g [DOI] [PubMed] [Google Scholar]
- Wilding P., Kricka L. J., Cheng J., Hvichia G., Shoffner M. A., and Fortina P., Anal. Biochem. 257, 95 (1998). 10.1006/abio.1997.2530 [DOI] [PubMed] [Google Scholar]
- Choi S., Song S., Choi C., and Park J. K., Lab Chip 7, 1532 (2007). 10.1039/b705203k [DOI] [PubMed] [Google Scholar]
- VanDelinder V. and Groisman A., Anal. Chem. 79, 2023 (2007). 10.1021/ac061659b [DOI] [PubMed] [Google Scholar]
- Ji H. M., Samper V., Chen Y., Heng C. K., Lim T. M., and Yobas L., Biomed. Microdevices 10, 251 (2008). 10.1007/s10544-007-9131-x [DOI] [PubMed] [Google Scholar]
- Huang S. B., Wu M. H., and Lee G. B., Sens. Actuators B 142, 389 (2009). 10.1016/j.snb.2009.07.046 [DOI] [Google Scholar]
- Lien K. Y., Lin J. L., Liu C. Y., Lei H. Y., and Lee G. B., Lab Chip 7, 868 (2007). 10.1039/b700516d [DOI] [PubMed] [Google Scholar]
- Gray B. L., Lieu D. K., Collins S. D., Smith R. L., and Barakat A. I., Biomed. Microdevices 4, 9 (2002). 10.1023/A:1014211627166 [DOI] [Google Scholar]
- Jeon N. L., Baskaran H., Dertinger S. K. W., Whitesides G. M., Van de Water L., and Toner M., Nat. Biotechnol. 20, 826 (2002). [DOI] [PubMed] [Google Scholar]
- Taylor A. M., Blurton-Jones M., Rhee S. W., Cribbs D. H., Cotman C. W., and Jeon N. L., Nat. Methods 2, 599 (2005). 10.1038/nmeth777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abhyankar V. V., Lokuta M. A., Huttenlocher A., and Beebe D. J., Lab Chip 6, 389 (2006). 10.1039/b514133h [DOI] [PubMed] [Google Scholar]
- Takayama S., Ostuni E., LeDuc P., Naruse K., Ingber D. E., and Whitesides G. M., Nature (London) 411, 1016 (2001). 10.1038/35082637 [DOI] [PubMed] [Google Scholar]
- Lucchetta E. M., Lee J. H., Fu L. A., Patel N. H., and Ismagilov R. F., Nature (London) 434, 1134 (2005). 10.1038/nature03509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edel J. B. and Demello A. J., Appl. Phys. Lett. 90, 053904 (2007). 10.1063/1.2435327 [DOI] [Google Scholar]
- Vincent M. E., Liu W. S., Haney E. B., and Ismagilov R. F., Chem. Soc. Rev. 39, 974 (2010). 10.1039/b917851a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini T. E., Hannezo E., Trepat X., Fredberg J. J., and Weitz D. A., Phys. Rev. Lett. 104, 168104 (2010). 10.1103/PhysRevLett.104.168104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba A. E., Saias L., Psychari E., Minc N., Simon D., Bidard F. C., Mathiot C., Pierga J. Y., Fraisier V., Salamero J., Saada V., Farace F., Vielh P., Malaquin L., and Viovy J. L., Proc. Natl. Acad. Sci. U.S.A. 107, 14524 (2010). 10.1073/pnas.1001515107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J. U., Olguin L. F., Whyte G., Scott D., Babtie A., Abell C., Huck W. T. S., and Hollfelder F., J. Am. Chem. Soc. 131, 15251 (2009). 10.1021/ja904823z [DOI] [PubMed] [Google Scholar]
- Srisa-Art M., Bonzani I. C., Williams A., Stevens M. M., deMello A. J., and Edel J. B., Analyst (Cambridge, U.K.) 134, 2239 (2009). 10.1039/b910472k [DOI] [PubMed] [Google Scholar]
- Thomson J. A., Science 282, 1145 (1998). 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- Desbaillets I., Ziegler U., Groscurth P., and Gassmann M., Exp. Physiol. 85, 645 (2000). 10.1017/S0958067000021047 [DOI] [PubMed] [Google Scholar]
- Itskovitz-Eldor J., Schuldiner M., Karsenti D., Eden A., Yanuka O., Amit M., Soreq H., and Benvenisty N., Mol. Med. 6, 88 (2000). [PMC free article] [PubMed] [Google Scholar]
- Khademhosseini A., Ferreira L., Blumling J., Yeh J., Karp J. M., Fukuda J., and Langer R., Biomaterials 27, 5968 (2006). 10.1016/j.biomaterials.2006.06.035 [DOI] [PubMed] [Google Scholar]
- Falconnet D., Csucs G., Grandin H. M., and Textor M., Biomaterials 27, 3044 (2006). 10.1016/j.biomaterials.2005.12.024 [DOI] [PubMed] [Google Scholar]
- Mohr J. C., de Pablo J. J., and Palecek S. P., Biomaterials 27, 6032 (2006). 10.1016/j.biomaterials.2006.07.012 [DOI] [PubMed] [Google Scholar]
- Ungrin M. D., Joshi C., Nica A., Bauwens C., and Zandstra P. W., PLoS ONE 3, e1565 (2008). 10.1371/journal.pone.0001565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang S. M., Gerecht-Nir S., Chen J., Itskovitz-Eldor J., and Zandstra P. W., Stem Cells 22, 275 (2004). 10.1634/stemcells.22-3-275 [DOI] [PubMed] [Google Scholar]
- Bratt-Leal A. M., Carpenedo R. L., and McDevitt T. C., Biotechnol. Prog. 25, 43 (2009). 10.1002/btpr.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D., Sa S., Pegan J. D., Rich B., Xiang G. X., McCloskey K. E., Manilay J. O., and Khine M., Lab Chip 9, 3338 (2009). 10.1039/b914091c [DOI] [PubMed] [Google Scholar]
- Torisawa Y. S., Chueh B. H., Huh D., Ramamurthy P., Roth T. M., Barald K. F., and Takayama S., Lab Chip 7, 770 (2007). 10.1039/b618439a [DOI] [PubMed] [Google Scholar]
- Torisawa Y. S., Mosadegh B., Luker G. D., Morell M., O’Shea K. S., and Takayama S., Integr. Biol. 1, 649 (2009). 10.1039/b915965g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai X., Liang Q., Wang Y., and Luo G., IFPT’6: Progress on Post-Genome Technologies, 2009, p. 154.
- Villa M., Pope S., Conover J., and Fan T. H., Biomed. Microdevices 12, 253 (2010). 10.1007/s10544-009-9380-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. W., Na D. S., Kang J. Y., Lee S. H., and Ju B. K., Biosci., Biotechnol., Biochem. 70, 1325 (2006). 10.1271/bbb.50609 [DOI] [PubMed] [Google Scholar]
- Ramadan Q., Yu C., Miao J. H., Cheong H. W., Oh S., Choo A., and Liang T. H., Sens. Actuators B 126, 351 (2007). 10.1016/j.snb.2007.03.024 [DOI] [Google Scholar]
- Figallo E., Cannizzaro C., Gerecht S., Burdick J. A., Langer R., Elvassore N., and Vunjak-Novakovic G., Lab Chip 7, 710 (2007). 10.1039/b700063d [DOI] [PubMed] [Google Scholar]
- Cimetta E., Figallo E., Cannizzaro C., Elvassore N., and Vunjak-Novakovic G., Methods 47, 81 (2009). 10.1016/j.ymeth.2008.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp J. M., Yeh J., Eng G., Fukuda J., Blumling J., Suh K. Y., Cheng J., Mahdavi A., Borenstein J., Langer R., and Khademhosseini A., Lab Chip 7, 786 (2007). 10.1039/b705085m [DOI] [PubMed] [Google Scholar]
- Anderson D. G., Levenberg S., and Langer R., Nat. Biotechnol. 22, 863 (2004). 10.1038/nbt981 [DOI] [PubMed] [Google Scholar]
- Lu H., Koo L. Y., Wang W. C. M., Lauffenburger D. A., Griffith L. G., and Jensen K. F., Anal. Chem. 76, 5257 (2004). 10.1021/ac049837t [DOI] [PubMed] [Google Scholar]
- Metallo C. M., Vodyanik M. A., de Pablo J. J., Slukvin I. I., and Palecek S. P., Biotechnol. Bioeng. 100, 830 (2008). 10.1002/bit.21809 [DOI] [PubMed] [Google Scholar]
- Kim L., Vahey M. D., Lee H. Y., and Voldman J., Lab Chip 6, 394 (2006). 10.1039/b511718f [DOI] [PubMed] [Google Scholar]
- Wichterle H., Lieberam I., Porter J. A., and Jessell T. M., Cell 110, 385 (2002). 10.1016/S0092-8674(02)00835-8 [DOI] [PubMed] [Google Scholar]
- Fung W. -T., Beyzavi A., Abgrall P., Nguyen N. -T., and Li H. -Y., Lab Chip 9, 2591 (2009). 10.1039/b903753e [DOI] [PubMed] [Google Scholar]
- Villa-Diaz L. G., Torisawa Y. S., Uchida T., Ding J., Nogueira-De-Souza N. C., O’Shea K. S., Takayama S., and Smith G. D., Lab Chip 9, 1749 (2009). 10.1039/b820380f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X. Y., Kong B. H., and Li D., Biotechnol. Lett. 30, 1537 (2008). 10.1007/s10529-008-9725-2 [DOI] [PubMed] [Google Scholar]
- Ellison D., Munden A., and Levchenko A., Mol. BioSyst. 5, 1004 (2009). 10.1039/b905602e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. H., Sim W. Y., Park S. W., Yang S. S., Choi B. H., Park S. R., Park K., and Min B. H., Tissue Eng. 12, 3107 (2006). 10.1089/ten.2006.12.3107 [DOI] [PubMed] [Google Scholar]
- Gómez-Sjöberg R., Leyrat A. A., Pirone D. M., Chen C. S., and Quake S. R., Anal. Chem. 79, 8557 (2007). 10.1021/ac071311w [DOI] [PubMed] [Google Scholar]
- Kim J. Y., Park H., Kwon K. H., Park J. Y., Baek J. Y., Lee T. S., Song H. R., Park Y. D., and Lee S. H., Biomed. Microdevices 10, 11 (2008). 10.1007/s10544-007-9105-z [DOI] [PubMed] [Google Scholar]
- Toh Y. C., Zhang C., Zhang J., Khong Y. M., Chang S., Samper V. D., van Noort D., Hutmacher D. W., and Yu H. R., Lab Chip 7, 302 (2007). 10.1039/b614872g [DOI] [PubMed] [Google Scholar]
- Park J. Y., Hwang C. M., and Lee S. H., Lab Chip 7, 1673 (2007). 10.1039/b710777c [DOI] [PubMed] [Google Scholar]
- Park J. Y., Yoo S. J., Hwang C. M., and Lee S. H., Lab Chip 9, 2194 (2009). 10.1039/b822006a [DOI] [PubMed] [Google Scholar]
- Wang D. Y. and Huang Y. Y., Biomed. Eng. Appl. Basis Commun. 20, 231 (2008). 10.4015/S1016237208000829 [DOI] [Google Scholar]
- Koepsel J. T. and Murphy W. L., Langmuir 25, 12825 (2009). 10.1021/la901938e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfer B., Seib F. P., Freudenberg U., Stamov D., Bley T., Bornhauser M., and Werner C., Biomaterials 30, 5950 (2009). 10.1016/j.biomaterials.2009.07.039 [DOI] [PubMed] [Google Scholar]
- Lee K. H., Kwon G. H., Shin S. J., Baek J. Y., Han D. K., Park Y., and Lee S. H., J. Biomed. Mater. Res. Part A 90A, 619 (2009). 10.1002/jbm.a.32059 [DOI] [PubMed] [Google Scholar]
- Lin P., Chen L., Li D., Yang N. L., Sun Y., and Xu Y. X., Neuroendocrinol. Lett. 30, 204 (2009). [PubMed] [Google Scholar]
- Wang J., Zhan Y. H., Ugaz V. M., and Lu C., Lab Chip 10, 2057 (2010). 10.1039/c004472e [DOI] [PubMed] [Google Scholar]
- Valero A., Post J. N., van Nieuwkasteele J. W., ter Braak P. M., Kruijer W., and van den Berg A., Lab Chip 8, 62 (2008). 10.1039/b713420g [DOI] [PubMed] [Google Scholar]
- Narasipura S. D., Wojciechowski J. C., Charles N., Liesveld J. L., and King M. R., Clin. Chem. 54, 77 (2008). 10.1373/clinchem.2007.089896 [DOI] [PubMed] [Google Scholar]
- Charles N., Liesveld J. L., and King M. R., Biotechnol. Prog. 23, 1463 (2007). 10.1021/bp0702222 [DOI] [PubMed] [Google Scholar]
- Huang Y., Yang J., Wang X. B., Becker F. F., and Gascoyne P. R. C., J. Hematother. Stem Cell Res. 8, 481 (1999). 10.1089/152581699319939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg A. W. and Hammer D. A., Biotechnol. Bioeng. 73, 111 (2001). 10.1002/bit.1043 [DOI] [PubMed] [Google Scholar]
- Wu H. W., Hsu R. C., Lin C. C., Hwang S. M., and Lee G. B., Biomicrofluidics 4, 024112 (2010). 10.1063/1.3454767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahara A. and Yamaoka T., Biotechnol. Prog. 26, 441 (2010). [DOI] [PubMed] [Google Scholar]
- Nagrath S., Sequist L. V., Maheswaran S., Bell D. W., Irimia D., Ulkus L., Smith M. R., Kwak E. L., Digumarthy S., Muzikansky A., Ryan P., Balis U. J., Tompkins R. G., Haber D. A., and Toner M., Nature (London) 450, 1235 (2007). 10.1038/nature06385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykstra B., Ramunas J., Kent D., McCaffrey L., Szumsky E., Kelly L., Farn K., Blaylock A., Eaves C., and Jervis E., Proc. Natl. Acad. Sci. U.S.A. 103, 8185 (2006). 10.1073/pnas.0602548103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutolf M. P., Doyonnas R., Havenstrite K., Koleckar K., and Blau H. M., Integr. Biol. 1, 59 (2009). 10.1039/b815718a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth I., Franke K., Pompe T., Bornhauser M., and Werner C., Integr. Biol. 1, 427 (2009). 10.1039/b903711j [DOI] [PubMed] [Google Scholar]
- Hosokawa M., Arakaki A., Takahashi M., Mori T., Takeyama H., and Matsunaga T., Anal. Chem. 81, 5308 (2009). 10.1021/ac900535h [DOI] [PubMed] [Google Scholar]
- Di Carlo D., Aghdam N., and Lee L. P., Anal. Chem. 78, 4925 (2006). 10.1021/ac060541s [DOI] [PubMed] [Google Scholar]
- Faley S. L., Copland M., Wlodkowic D., Kolch W., Seale K. T., Wikswo J. P., and Cooper J. M., Lab Chip 9, 2659 (2009). 10.1039/b902083g [DOI] [PubMed] [Google Scholar]
- Myers K. C. and Davies S. M., Biol. Blood Marrow Transplant 15, 279 (2009). 10.1016/j.bbmt.2008.11.037 [DOI] [PubMed] [Google Scholar]
- Daud S. S., Ibrahim K., Choong S. S., Vengidasan L., Chong L. A., and Ariffin H., Anal. Biochem. 397, 181 (2010). 10.1016/j.ab.2009.10.008 [DOI] [PubMed] [Google Scholar]
- Warren L., Bryder D., Weissman I. L., and Quake S. R., Proc. Natl. Acad. Sci. U.S.A. 103, 17807 (2006). 10.1073/pnas.0608512103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung B. G., Park J. W., Hu J. S., Huang C., Monuki E. S., and Jeon N. L., BMC Biotechnol. 7, 60 (2007). 10.1186/1472-6750-7-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J., Flanagan L., Jeon N., Monuki E., Schwartz P. H., and Lee A. P., in Micro Total Analysis Systems 2004, 8th International Conference on Miniaturized Systems for Chemistry and Life Science, Malmo, Sweden, 25–30 September 2004, Vol. 1, 363 (2005).
- Chen Y. and Zhong J. F., Methods Mol. Biol. 438, 293 (2008). 10.1007/978-1-59745-133-8_22 [DOI] [PubMed] [Google Scholar]
- Kamei K. I., Guo S. L., Yu Z. T. F., Takahashi H., Gschweng E., Suh C., Wang X. P., Tang J. G., McLaughlin J., Witte O. N., Lee K. B., and Tseng H. R., Lab Chip 9, 555 (2009). 10.1039/b809105f [DOI] [PubMed] [Google Scholar]
- Luong M. N., Sun A. X., Shin H. S., Tian L., Jeon N. L., and Cho K., Dev. Biol. 331, 501 (2009). 10.1016/j.ydbio.2009.05.429 [DOI] [Google Scholar]
- Zhong J. F., Chen Y., Marcus J. S., Scherer A., Quake S. R., Taylor C. R., and Weiner L. P., Lab Chip 8, 68 (2008). 10.1039/b712116d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. J., Botchwey E. A., Levine E. M., Pollack S. R., and Laurencin C. T., Proc. Natl. Acad. Sci. U.S.A. 101, 11203 (2004). 10.1073/pnas.0402532101 [DOI] [PMC free article] [PubMed] [Google Scholar]