Abstract
Axon path-finding plays an important role in normal and pathogenic brain development as well as in neurological regenerative medicine. In both scenarios, axonal growth is influenced by the microenvironment including the soluble molecules and contact-mediated signaling from guiding cells and cellular matrix. Microfluidic devices are a powerful tool for creating a microenvironment at the single cell level. In this paper, an asymmetrical-channel-based biochip, which can be later incorporated into microfluidic devices for neuronal network study, was developed to investigate geometric as well as supporting cell control of polarized axonal growth in forming a defined neuronal circuitry. A laser cell deposition system was used to place single cells, including neuron-glia pairs, into specific microwells of the device, enabling axonal growth without the influence of cytophilic∕phobic surface patterns. Phase microscopy showed that a novel “snag” channel structure influenced axonal growth in the intended direction 4:1 over the opposite direction. In heterotypic experiments, glial cell influence over the axonal growth path was observed with time-lapse microscopy. Thus, it is shown that single cell and heterotypic neuronal path-finding models can be developed in laser patterned biochips.
INTRODUCTION
Axon path-finding plays an important role in normal and pathogenic brain development as well as in neurological regenerative medicine. The list of known disorders caused by genetic mutations of axon development pathways is growing.1 With the emergence of functional magnetic resonance imaging (fMRI) tractography, many developmental disorders such as autism have been shown to be associated with white matter aberrations.2, 3 Recent genetic studies of autism have implicated axon path-finding pathways.4 Furthermore, a large portion of nerve regeneration technologies is based on directing axonal growth across lesions with chemical and physical guides. Consequently, there is much to be gained by understanding and manipulating the mechanisms involved in axon path-finding.
However, the human brain’s complex architecture is comprised of billions of cells and quadrillions of synapses. Elucidating the mechanisms by which the nervous system develops is therefore a daunting task. To address this challenge, assays that provide high spatial and temporal resolutions and a simplified environment to study the influences of various factors to the path-finding of single neuronal processes are needed. This paper details a polydimethylsiloxane (PDMS) biochip assay that enables single-cell-resolution assessment of polarized axonal growth in the presence of geometric and glial cell influence.
Topographical guidance cues were shown to influence axonal growth as early as 1987.5 More recently, the geometry of substrate microfeatures has been shown to influence the turning of axonal growths.6 Francisco and colleagues7 were able to regulate axonal guidance by varying the angle of turns in open microchannels. Similar results have been shown for axons navigating the turns of cytophilic surface patterns.8 While the geometry of microtopologies and surface patterns has been proven to influence axonal outgrowth, exploitation of these phenomenon to define the polarity of a circuit has not been shown. Yet there is a demand for techniques that define polarity of neuronal networks.9, 10
While in vitro experiments have probed and demonstrated physical and chemical guidance cues, in vivo glial cells guide neuron migration during development and thus are critical for brain architecture.11 If neurons and glia do not arrive at the correct location so as to receive the proper support signals, they will be eliminated through apoptotic processes known as pruning. Consequently, having the ability to control the locations of neurons and glial cells is critical for the investigation of in vivo relevant axon path-finding in an in vitro system.
After cell patterning, cell polarity is established when one neurite (one of the first two neurites to extend in pyramidal neurons) develops into the axon.12 In unipolar neurons, the other neurites develop into dendrites. Pioneer axons are guided over intermediate distances by diffusible factors released by glia and over short distances by contact with glia that act as guidepost cells and diffusible cues from targets and astrocytes. Pioneer axons are the first to extend through an axon-free environment, laying the groundwork for follower axons.13 Glial cells also extend cellular processes that have matrices upon which axons extend. These glial processes provide contact guidance and the initial scaffold on which extending pioneer axons grow.11
The research interest in the role of glia in the nervous system, and astrocytes in particular, is rapidly expanding due to their emerging function in development and normal maintenance.11 Conversely, when glial cells go awry, they contribute to disease and neurodegeneration.14 Because of their importance to axon path-finding and implications in neural pathologies, glial cells should not be overlooked in axonal growth research and the associated technologies.
The nervous system is actually a nonpermissive environment for neurons and glia, when the cells do not receive the appropriate paracrine and autocrine survival signals.15, 16 By reducing culture volumes, and therefore, the diffusive loss of excreted factors, microfluidic cell culture devices are a powerful tool for culturing and studying single, isolated neurons.17 The use of effective cell culture volumes on the same scale as cells themselves has a dual benefit: cells can regulate their own microenvironment that is more physiologically relevant, and the reduced volume allows cells to concentrate trophic survival factors. Unfortunately, decreasing cell culture volume also amplifies the impact of toxins and cell waste. The modified PDMS recipe and solvent leaching cleaning protocols described herein address the presence of cytotoxic oligomers in PDMS. Working down in size from the classic campenot18 chamber used to isolate neurites from their cell bodies, microscale devices allow for a more refined isolation. Jeon’s group demonstrated several microfluidic devices, which are basically microcampenot chambers used for isolating axons19 from neuron cell bodies and for axonal injury.20 Besides isolating for study, microfluidics can be used to define connectivity of neuronal networks. Morin and colleagues21 did so, but with high density neuronal populations in relatively large (600 μm2) microwells. Dworak and Wheeler’s group22 employed a similar approach using microtunnels in a PDMS membrane to guide the neurites of large neuron cultures over a set of electrodes. Toward the single neuron manipulation, Claverol-Tinture and colleagues23 were able to guide the axon of a single invertebrate neuron over a series of electrodes using similar elastomeric membranes.
The small volumes inside microfluidic channels can aid in culturing neurons at very low densities. Millet and co-worker17 successfully cultured neurons in simple microfluidic channels at densities not possible otherwise. The channels were also used to coat the substrate with poly-D-lysine and laminin and to slowly flow the media over the neurons which improved viability.
In the previously reported elastomeric membrane-based neuronal networks, placement of individual cells to specific wells or electrodes was performed by contact manipulation with a micropipette. This process involves cumbersome micromanipulators that usually necessitate an open environment prone to contamination. Furthermore, the restricted access angles of a micropipette and micromanipulator may limit the types of three-dimensional (3D) structure into which a cell may be placed. While placement of single cells into specific places on a substrate has been demonstrated using microfluidic devices, the nontrivial process would be further complicated when combined with the microfluidic guidance devices described above.
Previous research has demonstrated that physical confinement and restrictive guidance, imposed by 3D constructs, is a simple and highly effective method for patterning cells and controlling axonal growth.6, 7, 24 Yet, there has been only one example of single-cell-resolution neuronal circuits to date, which employed dynamic etching of channels and microwells in agar.25 In this work, we investigated axonal growth in a PDMS biochip with a novel microchannel designed to influence the polarity of axon growth. Aided by the use of the unique laser cell deposition system (LCDS), experiments were performed with single neurons. The influence of glial cells on axonal growth was also observed by placing single neuron-glial cell pairs into the biochip.
MATERIALS AND METHODS
Biochip design
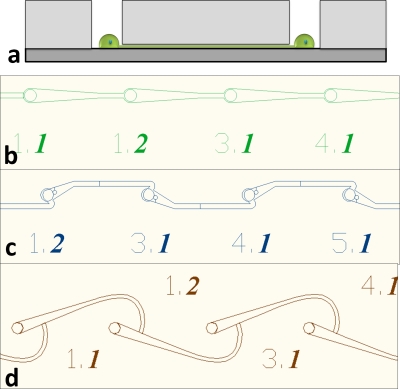
The biochip design was based on a simple system of microwells connected by microtunnels. The microwells were used to confine the cell body and the microtunnels were designed to guide axonal growth along a defined path to an adjacent microwell with a target neuron. The microwells and microtunnels were created when an elastomeric membrane with through-holes and shallow channels was aligned and attached to a glass substrate. Prior work has shown that neurites navigate acute angles more slowly than obtuse turns.6, 24 This phenomenon of controlling the direction of neurite growth by geometric guidance was exploited in the channel design of our biochip. For the photolithography process we used laser-photoplotted transparencies, limiting the feature size to 8–10 μm. Channels of this width would have been incapable of excluding the neuronal cell bodies that were about 8 μm in diameter. Therefore, to prevent migration of neurons from the microwells into the microchannels, channels∕tunnels were fabricated with very low aspect ratios (height of <2.5 μm), as shown in Fig. 1a. These features allowed the isolation of cell bodies and confinement of axons to define interneuron connectivity and enable clear observation of axonal behavior. In previous work,26 we used purely tapered microtunnel designs [Fig. 1b]. These showed no influence on initial neurite outgrowth direction and subsequent axonal growth control (data not shown). In this work, we analyze the influence of an asymmetric, angled “snag” microtunnel design [Fig. 1c] on the direction of axonal growth. We also demonstrate the creation of single neuron-glia pairs in a “hook” microtunnel design [Fig. 1d] as a platform for studying heterotypic axon path-finding and glial cell migration. With consideration for future applications in neuronal network electrophysiology, both designs have microwells that are 200 μm apart and 30 μm in diameter. These are the dimensions of a standard commercially available microelectrode array (MEA) from multichannel systems. MEA results are beyond the scope of this paper and will be reported in future publications.
Figure 1.
(a) Elastomeric membrane concept. The microwells confine the neurons while the short microtunnels allow only the neurites to pass through. (b) Directed microstructure design. (c) Snag microstructure design. (d) Hook microstructure design.
Biochip fabrication
Elastomeric membranes (35 μm thick) were fabricated in PDMS using standard soft lithography and replica molding methods. Microscale patterns were first created using computer-aided design (CAD) and then laser-photoplotted by CAD∕Art Services, Inc. The transparencies were used as the masks for photolithographic microfabrication using a Karl-Suss MJB-3 aligner 200 W lamp house. The multiheight molds were created by successive layering of SU-8 (2000.5, 2005, 2025, and 2050) negative photoresist (Microchem, Newton, MA). With these materials it was possible to create molds with small (<2 μm) channel features and strong 30–60 μm tall posts. The silicon molds were silanized with 1H,1H,2H,2H-perfluorooctyltrichlorosilane to aid in the removal of elastomeric membranes.
PDMS was obtained as a two-part elastomer, Sylgard™ 184, from World Precision Instruments. The base was mixed with the curing agent in 9:1 (rather than the recommended 10:1) as a higher curing agent content has been shown to be more biocompatible to in vitro cell cultures.27 To this mixture we added 10% xylene to decrease viscosity, allowing more uniform breakthrough of the through-hole-forming pillars. The uncured PDMS solution was spin-coated onto the silicon molds at speeds sufficient to reduce the PDMS thickness to just below the 40 μm posts. The PDMS was then cured by baking the wafer on a hotplate at 125 °C for 1–3 min. The elastomeric membranes were then baked in a vacuum oven for 2 h to ensure maximum crosslinking before a three solvent oligomer extraction cleaning was used to improve cell survival.17 The membranes were then vacuum-baked for at least 2 h to make sure solvents were removed. The assembled microdevices were then cleaned, sterilized, and activated by oxygen plasma. The membranes were then attached to a coverglass and heated at 50 °C for 2 h to create a permanent bond.
Surface modification
To improve the biocompatibility of the biochip, the surface was modified by physiosorption of a cationic polymer to improve cell attachment and neurite outgrowth on the underlying glass substrate. Immediately following a 10 min oxygen plasma treatment, the biochip was immersed in a solution of polyethylimine diluted to 0.05% w∕v in 8.5 pH borate buffer. After 24 h the microchip was removed and rinsed three times over 48 h in deionized (DI) water.
Cell culture
Two primary cell types were used in our experiments. Chick forebrain neurons were harvested from day 7 embryonic white leghorn chicks as described by Heidemann.28 Astrocyte cultures were derived from day 14 chick embryonic cerebral hemispheres (E15CH) based on the procedure of Kentroti.29 High purity astrocyte cultures were obtained by passage 3.30 Once established, astrocyte cultures were kept for four more passages (one doubling per passage) for culture use and creating conditioned media and then discarded as their ability to produce astrocytic factors became questionable.31 Both the neurons and astrocytes were maintained in a 37 °C, 5% CO2 incubator.
To promote the survival of single neurons grown in the biochip, the neural support medium was conditioned by mature astrocyte cultures. Prior to conditioning by astrocytes, the medium was comprised of Neurobasal™ medium (without l-glutamine or phenol red) supplemented with 1× GlutaMAX™ (Gibco), 1% antibiotic∕antimycotic (10 000 units∕mL penicillin G sodium, 10 000 μg∕mL streptomycin sulfate), 50 μg∕mL gentamicin, and 2.5 μg∕mL amphotericin B. Astrocyte conditioned medium (ACM) was created by using the unconditioned to support a confluent astrocyte culture for 24 h. A T150 flask of confluent neurons was used to condition three 20 mL volumes of media over 3 days before it was passaged. Conditioned media (20 mL) was removed after 24 h and added to a 250 mL bottle of frozen conditioned medium and returned to the freezer. When a bottle was filled, it was thawed and filtered with a 0.22 μm filter and aliquoted into 50 mL tubes and refrozen. As needed, 50 mL tubes were thawed and 2% B27 and 100 ng∕mL nerve growth factor (NGF) 7s were added to create a supplemented ACM. The final neuronal support medium contained Neurobasal™ medium, 1× GlutaMAX™, 1% antibiotic∕antimycotic, 50 μg∕mL gentamicin, 2.5 μg∕mL amphotericin B, astrocytic trophic factors (from conditioning), 2% B27, and 100 ng∕mL NGF 7s. In preliminary studies, this supplemented ACM supported neuron survival at densities as low as 10 cells∕cm2. The other medium used in our experiments, referred to as glial media, was comprised of Media 199 (without l-glutamine or phenol red) with 10% fetal bovine serum, 1× GlutaMAX™ (Gibco), 1% antibiotic∕antimycotic, 50 μg∕mL gentamicin, and 2.5 μg∕mL amphotericin B. This medium was used to plate and grow astrocyte cultures to confluency.
Laser cell patterning
Placement of single cells of different types into specific features (e.g., microwells) of the biochip at different times was critical to this work. The required high temporal and spatial positioning of multiple cell types was achieved by using the LCDS we recently developed at Clemson University as previously described.32 The LCDS used a micropump system to inject nanoliter volumes of a cell suspension into a 1 mm(z)×1 cm2 cell deposition chamber with the biochip as the chamber bottom. Using live video microscopy, an individual cell would be selected from an injection of cell suspension and guided to specific points on the substrate (e.g., microwells) by the optical force generated from a continuous wave laser beam (800 nm, 100 mW) with a Gaussian profile, which was weakly focused into the chamber. This process was repeated for every cell deposited to the biochip. Heterogeneous cell patterns were formed by first depositing neurons in one laser cell patterning session and then depositing glial cells beside those neurons in a second session 24 h later.
Biochip construction
Investigation of the geometric guidance of axonal growth was performed in eight experiments. Single neurons were laser patterned into the microwells of a biochip, each with up to 59 microwells. Not every microwell was filled with a functional neuron, due to biochip flaws (poorly formed features, nonthrough holes), obstruction of microwells by debris, and an imposed 1 h limit to the cell patterning process. Glial cell guidance of axonal growth was also investigated in a similar experiment where neurons were initially patterned into the microwells of a biochip and cultured for 24 h. After this initial culture period, glial cells were then copatterned into those same microwells for observing glial-neuron interactions in microchannels at the single cell level.
Generally, cell suspensions were prepared with 33×104 cells∕ml in serum free, phenol-red free Media 199. A volume of 25–100 nL cell suspension was injected into the chamber by the microinjection system to a region near (Z<1000 μm, XY<2000 μm) but not directly above the desired deposition section. This was done because, on occasion, some clumping occurred while cells were sitting in the syringe and injection fiber and these clumps could not be allowed to fall into the desired deposition section. Single, round, dark cells (due to the illumination system, the cell looks dark) were trapped by the laser beam from the injected suspension and moved at about 150 μm∕s horizontally and 25 μm∕s vertically toward the target microwell on the biochip. Rows were patterned at once, from the closest to the farthest from the injection region. The microinjection fiber was inserted or withdrawn from the chamber every 2–3 rows to minimize the distance (and time) between the microinjection region and the target microwells on the biochip. It took 30 to 45 s to guide each cell into its target microwell. However, injection and finding healthy-looking single cells added to the overall patterning time. The overall average time to pattern a cell was closer to 90 s.
Immediately after patterning, the biochip was removed from the cell deposition chamber in a sterile hood and transferred to a 35 mm Petri dish prefilled with 2 mL of culture medium. The patterned biochip was then incubated at 37 °C and 5% CO2.
Viability observation
Neurons in both control cultures and microwells were observed to assess viability via counting frequency of neurite outgrowth. Control cultures were prepared with the same cell suspension used for the patterning experiment, after setting in a sealed (non-CO2-buffered) vial for the duration of the patterning process (1 h). Neurons in the control vial were seeded at a low density of 104 cells∕cm2 on 35 mm glass-bottom fluorodishes (World Precision Instruments) with the same surface modifications and culture medium as neurons grown in the biochips.
The dishes were fixed after 24 h in culture. Five different micrographs with the same 10× size field-of-view were taken for each dish. Using the NIH IMAGEJ image processing application33 with the cell counter plug-in (Kurt De Vos) every cell or cell-sized particle was counted. The number of neurite outgrowths was also counted. If a cell had more than one outgrowth, each was counted. If a neurite connected two cells, the interconnecting neurite was counted only once. The total number of neurite outgrowths was counted and divided by the total number of neuron-sized (6–12 μm) round bodies.
Immunocytochemistry and microscopy
To verify the purity of the primary astrocyte lines, a sample of first passage astrocyte culture was fixed with paraformaldehyde∕glutaraldehyde and stained with a primary antibody for glial fibrillary acidic protein (GFAP; MAB360, Millipore). In neuronal cultures, axons (afferent neurites) were identified by staining with an antibody for neurofilaments (MAB1621, Millipore). Dendrites (efferent neurites) were identified by staining with an anti-microtubule-associated protein 2 (MAP2) antibody (IHCR1004-6, Millipore). Alexa Fluor 488 anti-mouse and Alex Fluor 594 anti-rabbit were used as secondary antibody fluorescent markers. Live-cell phase microscopy was performed on a microscope equipped with an onstage incubator maintaining a 37 °C and 5% CO2 environment.
RESULTS AND DISCUSSIONS
Biochip compatibility
The biochip, comprised of microwells and microchannels, was designed to facilitate different axon path-finding experiments, including future incorporation of a microfluidic system to study the effects of different soluble factors. The biochip was designed to study neurite outgrowth without the influence∕use of surface patterning, necessitating the use of some other cell placement method. The LCDS was chosen because of its ability to deposit single cells with high spatial and temporal resolution. Not only did the LCDS process prove to be compatible with the biochip design, the LCDS was also capable of propelling cells onto the bottom of the 35 μm deep microwells, resulting in a cell pattern resistant to washout from media change. Furthermore, the LCDS could pattern both neuronal and glial cells without selection via adhesive surface modification. The use of the LCDS enabled the unique biochip design to be implemented.
Viability and occurrence of axonal growth (initially in the form of neurite outgrowth) of single neurons growing in microwells of the biochip was compared with that of randomly seeded neurons in the control cultures. For this comparison, a total of 33 cells were deposited with the LCDS into unobstructed microwells. After 24 h in culture, 28 of these 33 cells survived (retained a round, unblebbed structure) and 14 had extended neurites. The percentage of patterned cells that survived and extended neurites was 48%, better than that in the control culture of 104 cells∕cm2, where the neurite-to-cell ratio was 36% with a standard error of 0.03%. We believe that the slightly higher ratio is due to the selection procedure by the laser beam, in which visually healthier neurons were chosen and placed into the microwells.
These results are comparable to those seen by Heidemann for neuronal cultures of 104 cells∕cm2.28 Furthermore, Heidemann and colleagues found little to no neurite outgrowth from chick forebrain neurons plated below 5×103 cells∕cm2. At 104 cells∕cm2 only about 50% of the plated cells survived,28 whereas 85% of the neurons laser patterned into microwells in our biochip survived.
Geometric axonal guidance
Previous research has evaluated polarized axonal growth10, 34 by immunocytochemical staining of neurofilaments (abundant in the axon but not in the dendrites) and MAP2 (abundant in the dendrites but not in the axon). Following these examples, we stained neuronal circuits grown for 72 h in our laser patterned biochip with standard fluorescent antibodies for neurofilaments and MAP2. Unlike in previous research, where axons and dendrites occupied mutually exclusive areas of the substrate, our method of defining polarity allowed both axons and dendrites to occupy the same space. This allowed for a large overlap of each stain within the microchannels so that once fully connected, the polarity of a circuit could not be discerned, as shown in Fig. 2a. However, by time-lapse imaging the time history of axonal growth, we could validate the polarized axonal growth when the circuits did not fully connect, as shown in Fig. 2b. Consequently, we proposed an alternative method to characterize a neurons polarity.
Figure 2.
Fluorescence microscope images of neurons 72 h after deposition into snag microstructures: Red indicates MAP2 abundant in dendrites, green indicates neurofilaments abundant in axons, and yellow areas indicate an overlap of the two stains. White arrows point to neuron cell bodies and striped arrows point to axons. (a) A circuit connecting two neurons. (b) An incomplete circuit where an obscured neuron (left) extended its axon down the intended microtunnel direction toward a clearly identifiable neuron (right) extending an axon back in the unintended direction toward the first neuron.
Because of the documented tendency of the first neurite of a neuron to become the axon, identifiable by its length12 (instead of identifying the polarity by axon and dendrite specific markers), we used live-cell phase microscopy to observe the time history of axonal growth, before they fully connected to neighboring cells. The observed neurite outgrowth was classified as one of the four following scenarios: extension of a neurite (a) in the direction intended by the microchannel geometry (+); (b) in the opposite direction (−); (c) into both intended and unintended channels (both); or (d) neither microchannel (neither). Neurons that did not extend neurites at all were not included in the analysis. Only the elastomeric membranes with the snag microstructures were considered, as preliminary data showed that the simple tapered channels of the “directed” microstructures shown in Fig. 1b had no influence on the polarization of axonal growths. Table 1 shows the compiled results for 87 neurons with visible neurite outgrowth present in eight different laser patterned biochips. Figure 3 depicts the typical results for each scenario.
Table 1.
Neurite growth behavior. The four classifications (as depicted in Fig. 3) are neither direction (neither), both directions (both), the unintended direction (−), and the intended direction (+). The table shows the results for only the 87 neurons that exhibited neurite outgrowth after 24 h.
| Observed | Total (%) | |
|---|---|---|
| Neurite (+) | 43 | 49.4 |
| Neurite (−) | 13 | 14.9 |
| Neurites (both) | 9 | 10.3 |
| Neurites (neither) | 22 | 25.3 |
| Total | 87 | 100 |
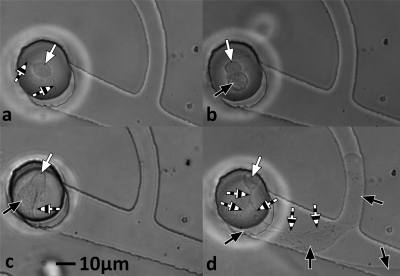
Figure 3.
Classifications of neurite growth. Phase contrast micrographs of neurons 24 h after deposition into snag microstructures. White arrows point to neuron cell bodies and striped arrows point to axons. (a) A typical neuron with a neurite (here split in two directions) that did not extend far enough down either microtunnel to be ascribed to either direction (neither). (b) A typical neuron with multiple neurites that extended significantly into both microtunnels (both). (c) A typical neuron extending a single neurite into the intended microtunnel∕direction (+). (d) A typical neuron extending a neurite into the unintended microtunnel∕direction (−).
By using live-cell phase microscopy to observe neurite outgrowth before complete axonal connection between neurons was achieved, we were able to analyze the polarity of neurite outgrowth for a majority of the cells. Each microwell was analyzed using this time-lapse approach, and the resulting data showed that 49% of neurons exhibiting neurite outgrowth extended those outgrowths in the desired direction, compared with only 15% directed in the opposing direction. This equates to over three times as many neurites being guided in the intended direction than in the opposite direction. Furthermore, only 10% of neurons that exhibited neurite outgrowth extended those neurites in both directions. Still, for fully defined circuits with control over polarity, the probability of having four neurons connected with the intended polarity would be approximately 5%. Even though the snag structure was successful, there were some design flaws that led to less-than-ideal control over polarity. We are currently investigating the geometric guidance of a new hook structure, including microstructures that have 90° intersections. We anticipate that these new structures will offer the unique ability to determine whether the presynaptic axon more frequently turns toward or away from a neuron when it is incident on that neuron’s axon at 90°. In addition, more high resolution versions of the snag biochip may be implemented to improve the effect.
Circuit connectivity
While the immunocytochemistry-stained circuits were not indicative of neuron and circuit polarity [Fig. 2a], they demonstrate that single neurons can produce axons that bridge the 200 μm channels defining an interneuronal circuit with the same scale as a standard MEA. Cells in the microwells took longer (5–6 days) to extend neurites than randomly seeded neurons in glass-bottom Petri dishes (2–3 days) prepared with the same surface modification procedures.
Heterotypic patterning: Glial migration and contact guidance
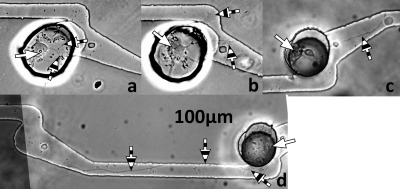
Heterotypic patterning of neurons and glial cells into a single microwell in the biochip with hook microtunnels was successfully demonstrated. Figure 4 shows several micrographs in time progression for one microwell as the two cell types were added. A neuron was added first and cultured for 23 h [panel (a)]. After this culture period, one glial cell was then patterned into the microwell adjacent to the previously deposited neuron [panel (b)], which had already extended a neurite. Figure 4, panels (c) and (d), shows how the coculture responded over the following 72 h.
Figure 4.
Time-lapse phase contrast micrographs of cells patterned into a hook microstructure. White arrows point to neuron cell bodies, striped arrows point to axons, and black arrows point to astrocytes. (a) Neuron 23 h after deposition with laser cell patterning system. Two neurites are visible. (b) Neuron and astrocyte 1 h after astrocyte deposition with laser patterning system (24 h after neuron deposition). Neurites are missing or obscured by the astrocyte. (c) Neuron and astrocyte 24 h after astrocyte deposition (48 h after neuron deposition). The astrocyte has changed morphology and a single neurite is visible. (d) Neuron and astrocyte 72 h after astrocyte deposition (96 h after neuron deposition). The astrocyte has proliferated and migrated down the microtunnel and into the narrow perpendicular tunnel. The neuron has extended a neurite down the channel but it is blocked by the astrocyte barrier and directed along the side of the astrocyte.
In previous work we had shown that the microtunnels successfully confine neurons to the microwells. We expected the same outcome for glial cells. The initial intent of creating neuron-glia pairs was to quantitatively assess the effect of glial-cell-contact on the viability and neurite outgrowth of single neurons. However, contrary to the results shown for neurons, the heterotypic patterning results show that glial cells can easily migrate beneath the 2 μm tall microtunnels. In fact, in this set of heterotypic patterning experiments, glial cells rarely stayed within the microwell, preferring to migrate into the short and narrow channels (panel d), and then proliferating once inside. This made a controlled assessment of the effect of glial-cell-contact impossible to perform. Still, the results are of interest. In Fig. 4d, the axon of the neuronal cell can clearly be distinguished from the glial cell body. The glial cell body formed a physical boundary, which guided the axon along its upper side to the perpendicular channel. The heterotypic hook observations offer in vitro evidence and provide an in vitro platform for investigation of the widespread and well documented35in vivo phenomenon of contact guidance and barrier activity between neurons and glial cells.
As shown in prior literature examples,7, 8 and again in this work by using the snag microstructure, axons tend to extend past sharp turns. However, in this research, glial cells tended to migrate into tight spaces, despite a sharp turn. The tendency of glia to turn into a tight space, and guide an axon, which might not normally turn that way, is something that should be investigated in more detail. Such investigations are well suited to this experimental design. The placement of a second cell type next to a developing cell to form a heterotypic cell pair was made possible by the high spatial and temporal resolution of the LCDS and demonstrates how laser trapping and other cell printing technologies can create cellular patterns and structures in nontraditional microenvironments such as microfluidic chips.
CONCLUSION
In this study, we assessed the ability of a PDMS biochip with microwells and microchannels to support survival and influence axonal growth polarity of individual neurons and heterotypic neuron-glia pairs. The results showed that some microchannel geometries (snag) were effective at influencing axonal growth polarity and that such microstructures can be used as a tool in defining the polarity of neuronal circuits. The laser cell pattering system and the microfabrication system were effective tools for placing two cells of different types in close contact within a closed, marked environment so that the cell-cell interactions could be easily tracked over time. Results from the heterotypic neuron∕glia pairs demonstrated glial contact guidance of the axonal growth. These results indicated that the biochip∕laser cell deposition system combination used in this work is a powerful research path for creation and study of heterotypic, single-cell-resolution assays for polarized axonal growth.
The future work will focus on creating neuronal circuits on electrode arrays to investigate electrophysiological characteristics of single-cell-resolution circuits, or to determine a minimum number of cells that reliably produce spontaneous or evoked action potentials and spike propagation. Asymmetrical channel geometries (i.e., snag) will be used to create and study neuronal networks with defined polarity as well as connectivity. Microchannels may also be used to investigate the migration of glial cells through different geometries and sizes as well as with different chemical attractants and repellants, whereby motility could be assessed by measuring the distance traveled down a long microchannel. In the case of the hook microstructure, the developed systems will allow the investigation of how an axon is guided, up or down the postsynaptic cells axon, when it is incident at 90°. Finally, we plan to extend the utility of the biochip to monitor the axonal growth in the presence of signaling molecules and neurotoxins by intersecting microfluidic channels with the axonal guidance channels.
ACKNOWLEDGMENTS
This work has been partially supported by NIH SC INBRE (Grant No. 2p20rr16461-05 and its supplementary grant 3P20RR016461-09S2), SC COBRE (Grant No. P20RR021949), and Career Award (Grant No. 1k25hl088262-01); NSF MRI (Grant No. CBET-0923311); SC GEAR program. B.Z.G. acknowledges the National Natural Science Foundation of China (Grant No. 31070847); R.K.P. would like to thank the National Research Council for his Research Associateship.
References
- Engle E. C., Cold Spring Harbor Perspectives in Biology, March 2010; 2 a001784. 10.1101/cshperspect.a001784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung C., Chua S. E., Cheung V., Khong P. L., Tai K. S., Wong T. K. W., Ho T. P., and McAlonan G. M., J. Child Psychol. Psychiatry 50, 1102 (2009). 10.1111/j.1469-7610.2009.02086.x [DOI] [PubMed] [Google Scholar]
- Keller T. A., Kana R. K., and Just M. A., NeuroReport 18, 23 (2007). 10.1097/01.wnr.0000239965.21685.99 [DOI] [PubMed] [Google Scholar]
- Sbacchi S., Acquadro F., Calo I., Cali F., and Romano V., Current Genomics 11, 136 (2010). 10.2174/138920210790886880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark P., Connolly P., Curtis A. S., Dow J. A., and Wilkinson C. D., Development 99, 439 (1987). [DOI] [PubMed] [Google Scholar]
- Mahoney M. J., Chen R. R., Tan J., and Mark Saltzman W., Biomaterials 26, 771 (2005). 10.1016/j.biomaterials.2004.03.015 [DOI] [PubMed] [Google Scholar]
- Francisco H., Yellen B. B., Halverson D. S., Friedman G., and Gallo G., Biomaterials 28, 3398 (2007). 10.1016/j.biomaterials.2007.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers G. S. and Conrad D., J. Neurobiol. 66, 1183 (2006). 10.1002/neu.20298 [DOI] [PubMed] [Google Scholar]
- Suzuki I., Sugio Y., Moriguchi H., Jimbo Y., and Yasuda K., J Nanobiotechnology 2, 7 (2004). 10.1186/1477-3155-2-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenger D. A., Hickman J. J., Bateman K. E., Ravenscroft M. S., Ma W., Pancrazio J. J., Shaffer K., Schaffner A. E., Cribbs D. H., and Cotman C. W., J. Neurosci. Methods 82, 167 (1998). 10.1016/S0165-0270(98)00047-8 [DOI] [PubMed] [Google Scholar]
- Allen N. J. and Barres B. A., Nature (London) 457, 675 (2009). 10.1038/457675a [DOI] [PubMed] [Google Scholar]
- Calderon de Anda F., Gärtner A., Tsai L. -H., and Dotti C. G., J. Cell. Sci. 121, 178 (2008). 10.1242/jcs.023143 [DOI] [PubMed] [Google Scholar]
- Hidalgo A., Biochem. Soc. Trans. 31, 50 (2003). 10.1042/BST0310050 [DOI] [PubMed] [Google Scholar]
- Miller G., Science 308, 778 (2005). 10.1126/science.308.5723.778 [DOI] [PubMed] [Google Scholar]
- Levi-Montalcini R., Science 237, 1154 (1987). 10.1126/science.3306916 [DOI] [PubMed] [Google Scholar]
- Fernandez P. -A., Tang D. G., Cheng L., Prochiantz A., Mudge A. W., and Raff M. C., Neuron 28, 81 (2000). 10.1016/S0896-6273(00)00087-8 [DOI] [PubMed] [Google Scholar]
- Millet L. J., Stewart M. E., Sweedler J. V., Nuzzo R. G., and Gillette M. U., Lab Chip 7, 987 (2007). 10.1039/b705266a [DOI] [PubMed] [Google Scholar]
- Campenot R. B., Independent Control of the Local Environment of Somas and Neurites, Cell Culture: Methods in Enzymology, Vol. 58, 1979, edited by Jacoby W. B. and Pastan I. H. (Academic Press, New York: ), p. 302. [DOI] [PubMed] [Google Scholar]
- Park J. W., Kim H. J., Byun J. H., Ryu H. R., and Li Jeon Professor N., J. Biotechnol. 4, 1573 (2009). 10.1002/biot.200900159 [DOI] [PubMed] [Google Scholar]
- Taylor A. M., Blurton-Jones M., Woo Rhee S., Cribbs D. H., Cotman C. W., and Li Jeon N., Nat. Methods 2, 599 (2005). 10.1038/nmeth777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin F., Nishimura N., Griscom L., LePioufle B., Fujita H., Takamura Y., and Tamiya E., Biosens. Bioelectron. 21, 1093 (2006). 10.1016/j.bios.2005.04.020 [DOI] [PubMed] [Google Scholar]
- Dworak B. J. and Wheeler B. C., Lab Chip 9, 404 (2009). 10.1039/b806689b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverol-Tinture E., Cabestany J., and Rosell X., IEEE Trans. Biomed. Eng. 54, 331 (2007). 10.1109/TBME.2006.880903 [DOI] [PubMed] [Google Scholar]
- Smeal R. M., Rabbitt R., Biran R., and Tresco P. A., Ann. Biomed. Eng. 33, 376 (2005). 10.1007/s10439-005-1740-z [DOI] [PubMed] [Google Scholar]
- Suzuki I., Sugio Y., Jimbo Y., and Yasuda K., Lab Chip 5, 241 (2005). 10.1039/b406885h [DOI] [PubMed] [Google Scholar]
- Pirlo R. K., Peng X., Yuan X., and Zhi Gao B., Optoelectronics Letters 4, 387 (2008). 10.1007/s11801-008-8059-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. N., Jiang X., Ryan D., and Whitesides G. M., Langmuir 20, 11684 (2004). 10.1021/la048562+ [DOI] [PubMed] [Google Scholar]
- Heidemann S. R., Reynolds M., Ngo K., and Lamoureux P., Methods Cell Biol. 71, 51 (2003). 10.1016/S0091-679X(03)01004-5 [DOI] [PubMed] [Google Scholar]
- Kentroti S. and Vernadakis A., J. Neurosci. Res. 47, 322 (1997). [DOI] [PubMed] [Google Scholar]
- Barnea A., Roberts J., Keller P., and Ann Word R., Brain Res. 896, 137 (2001). 10.1016/S0006-8993(01)02141-2 [DOI] [PubMed] [Google Scholar]
- Taylor A. R., Gifondorwa D. J., Newbern J. M., Robinson M. B., Strupe J. L., Prevette D., Oppenheim R. W., and Milligan C. E., J. Neurosci. 27, 634 (2007). 10.1523/JNEUROSCI.4947-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirlo R., Rev. Sci. Instrum. 82, 013708 (2011). 10.1063/1.3529919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramoff M. D., Magelhaes P. J., and Ram S. J., Biophotonics Int. 11, 36 (2004). [Google Scholar]
- Vogt A. K., Daniel Stefani F., Best A., Nelles G., Yasuda A., Knoll W., and Offenhäusser A., J. Neurosci. Methods 134, 191 (2004). 10.1016/j.jneumeth.2003.11.004 [DOI] [PubMed] [Google Scholar]
- Fitch M. T. and Silver J., Cell Tissue Res. 290, 379 (1997). 10.1007/s004410050944 [DOI] [PubMed] [Google Scholar]