Abstract
Vascular function, homeostasis, and pathological development are regulated by the endothelial cells that line blood vessels. Endothelial function is influenced by the integrated effects of multiple factors, including hemodynamic conditions, soluble and insoluble biochemical signals, and interactions with other cell types. Here, we present a membrane microfluidic device that recapitulates key components of the vascular microenvironment, including hemodynamic shear stress, circulating cytokines, extracellular matrix proteins, and multiple interacting cells. The utility of the device was demonstrated by measuring monocyte adhesion to and transmigration through a porcine aortic endothelial cell monolayer. Endothelial cells grown in the membrane microchannels and subjected to 20 dynes∕cm2 shear stress remained viable, attached, and confluent for several days. Consistent with the data from macroscale systems, 25 ng∕ml tumor necrosis factor (TNF)-α significantly increased RAW264.7 monocyte adhesion. Preconditioning endothelial cells for 24 h under static or 20 dynes∕cm2 shear stress conditions did not influence TNF-α-induced monocyte attachment. In contrast, simultaneous application of TNF-α and 20 dynes∕cm2 shear stress caused increased monocyte adhesion compared with endothelial cells treated with TNF-α under static conditions. THP-1 monocytic cells migrated across an activated endothelium, with increased diapedesis in response to monocyte chemoattractant protein (MCP)-1 in the lower channel of the device. This microfluidic platform can be used to study complex cell-matrix and cell-cell interactions in environments that mimic those in native and tissue engineered blood vessels, and offers the potential for parallelization and increased throughput over conventional macroscale systems.
INTRODUCTION
The vascular environment is a complex milieu in which local mechanical forces and biochemical signals combine to regulate biological responses. Endothelial cells (ECs) that line blood vessels are responsible for maintaining vascular homeostasis in response to circulating cytokines, hemodynamic shear stress, and inflammatory cells.1 These factors are interdependent, increasing the complexity of the system significantly. For example, hemodynamic shear stress can cause changes in EC morphology, metabolism, and gene expression, thereby altering the cytokines secreted by ECs, the expression of adhesion molecules by ECs, and the responsiveness of ECs to a variety of cytokines and growth factors.2 This, in turn, can affect interactions with circulating inflammatory cells.1
Systematic investigation of complicated biological systems in vitro requires model systems that capture in vivo interactions. Many key components of the in vivo vascular environment are relevant for vascular (patho)biology, tissue engineering, and regenerative medicine. These include hemodynamic shear stress, the extracellular matrix (ECM) of the vascular wall, and interactions between multiple cell types, including EC-vascular smooth muscle cell interactions that are important for vascular homeostasis and EC-inflammatory cell interactions, such as adhesion and transmigration, which are important in disease initiation. Fundamental insights into the integrated effects of these factors on vascular function require in vitro technologies that mimic components of the in vivo environment and permit efficient and systematic experimentation.
Macroscale parallel plate-based flow systems have incorporated some of these features,3, 4, 5 but are often too cumbersome, costly, and inefficient to use for higher throughput experimentation. Microfluidics provides a viable alternative, as the natural scale of microfluidic systems makes reagent use more economical and parallelization more feasible than with macroscale systems. Microfluidic technologies have been used effectively to mimic components of the liver,6 lung,7, 8 and heart,9 recapitulating organ functions and processes that were otherwise unattainable with conventional macroscale systems. Microfluidics has been used to study various aspects of the vasculature and circulation,10, 11, 12, 13, 14, 15 but a platform that mimics multiple features of the complex vascular microenvironment has yet to be realized.
Here, we describe a microfluidic platform for the study of long-term cell-cell interactions in physiologically relevant vascular microenvironments that include shear stress, extracellular matrix proteins, biochemical signals, and multiple interacting cell types. As an initial demonstration of the technology, we investigated (1) the independent and combined effects of hemodynamic shear stress and tumor necrosis factor-α (TNF-α; an inflammatory cytokine) on monocyte attachment to ECs and (2) monocyte transmigration across the endothelium in response to a chemoattractant. Monocyte attachment to the endothelium and subsequent transmigration are important initial steps in the pathogenesis of atherosclerosis. Regions of the vasculature that are prone to disease development in vivo correlate spatially with regions that experience low magnitude shear stress and disturbed or oscillatory flow patterns.16 These regions are also often laden with inflammatory cytokines and adherent monocytes.17 Therapeutic strategies to arrest atherosclerotic lesion development by reducing chronic inflammation are particularly promising.18 Endothelial-monocyte interactions have been investigated in vitro previously (see, e.g., Refs. 19, 20, 21, 22), but with macroscale technologies that are not amenable to high-throughput, multiparameter investigations. New technologies that enable the complex vascular environment to be studied in vitro in a systematic and efficient manner may provide novel insights into pathological mechanisms and identify promising drug candidates prior to preclinical testing.
MATERIALS AND METHODS
Device fabrication
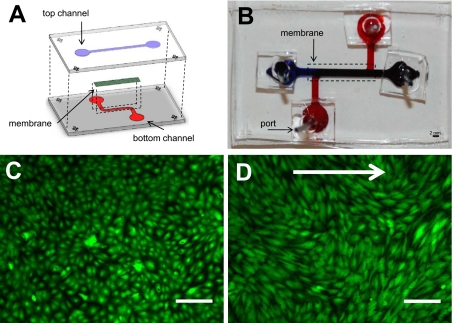
Polydimethylsiloxane (PDMS) (Sylgard 184; Dow Corning, Midland, MI) microchannels were fabricated using standard soft lithography. A polyethylene terephthalate (PET), track-etched porous membrane with 8 μm diameter pores was cut out from standard cell culture inserts (Becton Dickinson, Mississauga, ON, Canada) and sandwiched between two PDMS microchannels [Figs. 1a, 1b]. Others have detailed the process by which porous membranes are incorporated into microdevices7, 12, 23 and demonstrated their utility for biological studies.7, 8, 12, 24, 25 For the devices used in this study, the top straight channel and bottom S-shaped channel were 2000 μm wide by 300 μm high.
Figure 1.
Membrane microfluidic device and endothelial cell culture. (a) Illustration of the membrane microfluidic device. Two PDMS channels were used to sandwich a porous, PET membrane. (b) Photograph of the actual device (top view) with 2 mm scale bar. (c) Confluent monolayer of ECs grown in a membrane device and stained with calcien AM. (d) ECs stained with calcien AM after 24 h of flow in a membrane microfluidic device. Arrow indicates the direction of flow. Scale bar in panels (c) and (d) is equal to 200 μm.
Endothelial cell culture and seeding
Primary porcine aortic endothelial cells (PAECs) were provided by Lowell Langille (University of Toronto). PAECs were cultured in M199 (Wisent, St. Bruno, QC, Canada) supplemented with 5% fetal bovine serum (FBS) (Fisher Scientific, Ottawa, ON, Canada), 5% cosmic calf serum (CS) (Fisher Scientific), and 1% penicillin-streptomycin (P-S) (Sigma Aldrich, Oakville, ON, Canada) (PAEC media) and used between passages 4–8. The membrane in the top channel was coated with 100 μg∕ml bovine plasma fibronectin (FN) (Sigma-Aldrich) for 1 hour after devices were rinsed sequentially with 100% ethanol, 70% ethanol, and then phosphate-buffered saline (PBS). PAECs were seeded into the top channel at a cell density of 2.0×106 cells∕ml. Devices were maintained in an incubator (37 °C and 5% CO2) for 5–6 h to allow cell attachment. The top and bottom channels were replenished with 500 μl of media at this point and every 12–16 h thereafter.
Application of shear stress
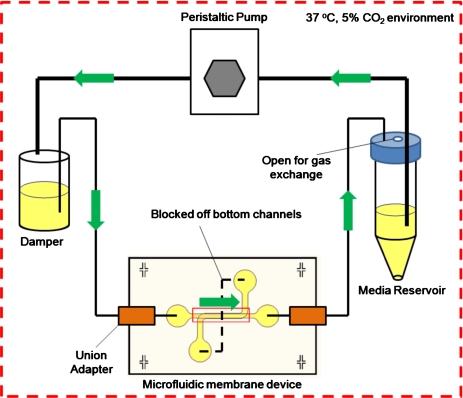
The membrane devices were connected to a closed-loop recirculatory flow system (Fig. 2). One day after the ECs reached confluence. A peristaltic pump (Cole Parmer, Vernon Hills, IL) forced media from a reservoir to a damper, which converted pulsatile flow coming from the pump into continuous, steady flow entering the top microchannel. The inlets to the bottom channel were sealed off by connecting the two ports to one another using polyethylene tubing (Becton Dickinson). Low pressure union adapters (Upchurch Scientific, Oak Harbor, WA) were used at the inlets and outlets of the microfluidic device to prevent leakage during shear flow. The entire setup was placed in an incubator at 37 °C and 5% CO2. PAECs in the top channel were subjected to either 0 (static) or 20 dynes∕cm2 flow-induced shear stress for 24 h to several days, depending on the experiment. To demonstrate cell viability, in some experiments endothelial cells were fluorescently labeled with 2 μM calcien AM (Invitrogen), a vital dye, for 30 min, then washed with PBS, and replenished with medium for 30 min.
Figure 2.
Recirculatory flow loop. A peristaltic pump was used to force media from the reservoir into the damper that converted the flow from pulsatile to continuous and steady. Union adapters were used to connect tubing from the damper to the inlet port of the top channel. Tubing connected to the outlet of the top channel was fed back into the reservoir completing the recirculatory flow loop. Both ports of the bottom channel were connected to each other using a small piece of tubing to maintain static conditions in the lower compartment. The entire setup was placed in an incubator (37 °C and 5% CO2).
TNF-α treatment
ECs in microchannels were treated with or without 25 ng∕ml TNF-α (Invitrogen, Burlington, ON, Canada) in PAEC media. In some experiments, ECs were subjected to static or shear stress conditions for 24 h and then treated with or without TNF-α for 4 h under static or shear conditions, respectively. In other experiments, ECs were subjected to 28 h of static or shear stress conditions and simultaneously treated with or without TNF-α. After treatment, monocytes were injected into the top channel.
Monocyte adhesion assay
RAW264.7 cells (ATCC, Manassas, VA) were cultured in Dulbecco’s modified Eagle’s medium (Wisent) supplemented with 10% FBS and 1% P-S (RAW medium) and used between passage 2 and 6. One hour before seeding into microchannels, RAW264.7 cells were fluorescently labeled using 2 μM calcien AM (Invitrogen) for 20 min, then washed with PBS, and replenished with RAW medium for 40 min. RAW264.7 cells were detached from the surface using a cell scraper and suspended at a cell density of 106–107 cells∕ml. Cells were injected into the top channel using a volume of 200–300 μl and allowed to adhere for 1 h. A multisyringe pump (Cole Parmer) was then used to wash away any unbound cells at a shear stress of 1 dyne∕cm2 for 3 min.
Microchannels containing fluorescently labeled RAW264.7 cells were imaged using an inverted fluorescent microscope (IX-71; Olympus, Markham, ON, Canada). Three representative images were taken for each device after washing. The number of adherent cells in each location was quantified using IMAGEJ software (NIH) and averaged to give a mean count for each microchannel device. Data are presented as mean±standard deviation. Differences in monocyte adhesion between conditions were tested with Student’s t-tests.
Monocyte transmigration assay
PAECs were seeded at 5×105 cells∕ml and allowed to grow to confluence in the upper channel. At confluence, ECs were treated with 25 ng∕ml TNF-α for 4 h under static conditions. The THP-1 human monocytic cell line was used to demonstrate transmigration. THP-1 cells were cultured in RPMI 1640 medium (Gibco, Burlington, ON, Canada) supplemented with 10% FBS (THP-1 medium). THP-1 nuclei were labeled with Hoechst 33342 (2 μg∕ml) (Invitrogen, Burlington, ON, Canada) for 30 min, and then the cells were centrifuged and resuspended in THP-1 medium for 30 min. Labeled THP-1 cells were injected into the top channel at 1×106 cells∕ml in the PAEC medium. The lower channel contained THP-1 medium with or without 500 ng∕ml monocyte chemoattractant protein-1 (MCP-1) (Becton Dickonson). After overnight incubation, five random locations were imaged in the lower channel of a device under each condition.
RESULTS AND DISCUSSION
A membrane microfluidic device was successfully built with multiple features that mimic the structural and functional components of the vascular wall [Figs. 1a, 1b]. The lumen-like top channel contains a membrane that can be coated with ECM proteins of interest and seeded with ECs. Pores in the membrane permit the transport of soluble factors and circulatory cells (similar to the basement membrane) between the top and bottom channels. The bottom channel can contain soluble factors and mural cells (e.g., smooth muscle cells) either as a two-dimensional layer or in a three-dimensional gel protected from flow. Labeled cells in both top and bottom channels can be imaged in real-time using epifluorescence or confocal microscopy. Cells in the upper and lower channels can be isolated independently for assaying by, for example, polymerase chain reaction or immunoblotting. The integration of the channel with a recirculatory flow loop allows long-term circulation of media and stimulation with shear stress. More complex flow profiles can be applied by modifying the flow loop using a programmable syringe pump. Despite the high shear stresses and associated pressures, leakage or failure of the devices and recirculatory loop was not observed, demonstrating the robustness of the system.
ECs were successfully cultured to confluence in the membrane microfluidic device and remained viable for up to at least 1 week, as observed by calcien AM staining [Fig. 1c]. After 24 h of shear stress at 20 dynes∕cm2, ECs remained adhered to the membrane, maintained confluence, and elongated without any preferential alignment over this time period [Fig. 1d]. We have observed viability and maintenance of adhesion and confluence after several days of shear stress application. To our knowledge, this is the first report of physiological shear stress being applied long-term to ECs cultured in a membrane microfluidic chip.
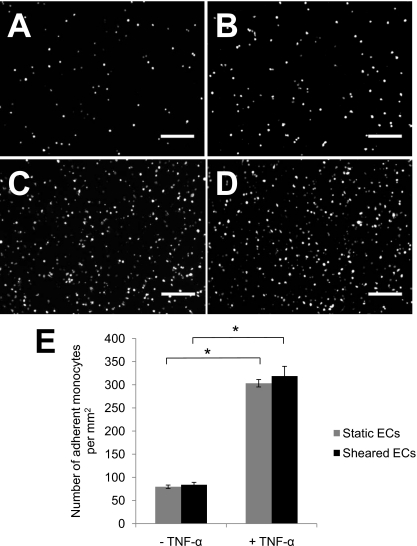
To validate the use of this device to investigate monocyte-EC interactions, ECs were first stimulated with TNF-α and monocyte adhesion was compared between stimulated and nonstimulated EC monolayers under static (no flow) conditions. TNF-α has been shown to increase the expression of surface adhesion proteins such as vascular cell adhesion molecule-1 (VCAM-1) and E-selectin that bind monocytes and other inflammatory cells.26 Thus, in macroscale culture systems, TNF-α increases monocyte adhesion to the endothelium.19, 22, 26 Similarly, in the microdevice monocytes adhered to ECs within 1 h of seeding in both TNF-α-stimulated and nonstimulated conditions, with significantly more adhesion to TNF-α-stimulated ECs (p<0.05; Fig. 3).
Figure 3.
Monocyte adhesion to preconditioned or static ECs with and without TNF-α stimulation. Representative images of fluorescently labeled RAW264.7 monocyte attachment to ECs under (a) static and (b) shear conditions without TNF-α treatment and under (c) static and (d) shear conditions with TNF-α treatment. Initial monocyte cell density was 2×106 cells∕ml in these experiments. Scale bar=200 μm. (e) The number of monocytes attached per unit area was significantly greater with TNF-α treatment under both static and sheared conditions. Data presented as mean±standard deviation: *p<0.05.
We next tested whether preconditioning the ECs with 24 h of 20 dynes∕cm2 flow-induced shear stress influenced TNF-α-induced monocyte adhesion. After 24 h of shear stress preconditioning, ECs were stimulated with TNF-α for an additional 4 h with the flow maintained. Similar to the static case, monocyte adhesion to ECs preconditioned with shear stress was significantly enhanced by TNF-α stimulation (p<0.05; Fig. 3). ECs preconditioned by flow-induced shear stress showed no significant difference in monocyte adhesion compared to static ECs with or without TNF-α stimulation (Fig. 3). This result is consistent with the observations made by Gonzales and Wick19 using a macroscale system in which monocyte adhesion to ECs preconditioned with high magnitude shear stress (30 dynes∕cm2) was not significantly different from static conditions. Notably, Gonzales and Wick19 found that monocyte adhesion to ECs preconditioned with low magnitudes of shear stress (<10 dynes∕cm2) was three- to fourfold greater than in static conditions, emphasizing the sensitivity of the monocyte adhesion and EC activation to shear stress conditions.
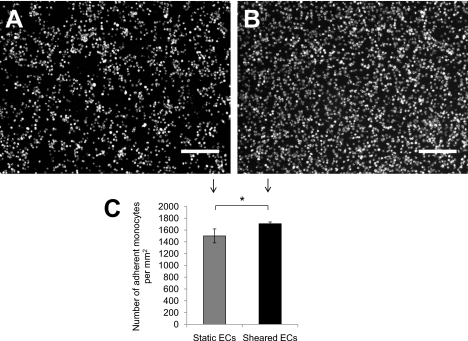
We also tested the effect of simultaneous shear stress and TNF-α stimulation for 28 h to match the total duration of the preconditioning experiments. In contrast to the preconditioned response, monocyte adhesion was significantly greater to sheared ECs than static ECs when TNF-α was applied simultaneously with flow (p<0.05) (Fig. 4). Fluid shear stress has been shown in macroscale systems to increase the endothelial adhesiveness to monocytes through upregulation of VCAM-1 and intercellular adhesion molecule-1 (ICAM-1).19, 20, 27 This effect is temporally dynamic; for example, ICAM-1 expression increases significantly after 8 h of shear stress stimulation and continues to increase up until 48 h.20 Thus, differences in TNF-α-induced monocyte adhesion to ECs preconditioned versus simultaneously simulated with shear stress may reflect differences in the temporal expression of adhesion molecules.
Figure 4.
Monocyte adhesion to ECs under static or shear stress conditions and simultaneously stimulated with TNF-α. Monocyte attachment to ECs under (a) static and (b) sheared conditions and simultaneously stimulated with TNF-α. The initial monocyte cell density was 10×106 cells∕ml. (c) The number of monocytes attached per unit area was significantly greater on the ECs subjected to shear stress and TNF-α for 28 h. Data presented as mean±standard deviation: *p<0.05.
We observed a proadhesive effect of shear stress19, 20, 27 only in the presence of TNF-α (Fig. 4), suggesting that the effects of shear stress and TNF-α were additive (and therefore more readily detectable) or that shear stress modulated the effect of TNF-α on EC adhesion molecule expression. In contrast, other authors reported that laminar shear stress decreases inflammatory cell adhesion to ECs.21, 22, 28 The discrepancies between studies are likely attributable to the differences in multiple experimental parameters including cell types; type and duration of flow stimulation; and type, concentration, and duration of biochemical stimuli, emphasizing the sensitivity of fundamental endothelial responses to the combined effects of multiple environmental cues. Because of miniaturization and parallelization, systematic investigation of the integrated response to multiple cues is more efficient with microfluidic platforms than with macroscale systems. The microfluidic device reported here could be readily extended to include multiple parallel membrane channels on a single chip as an enabling technology for combinatorial studies.
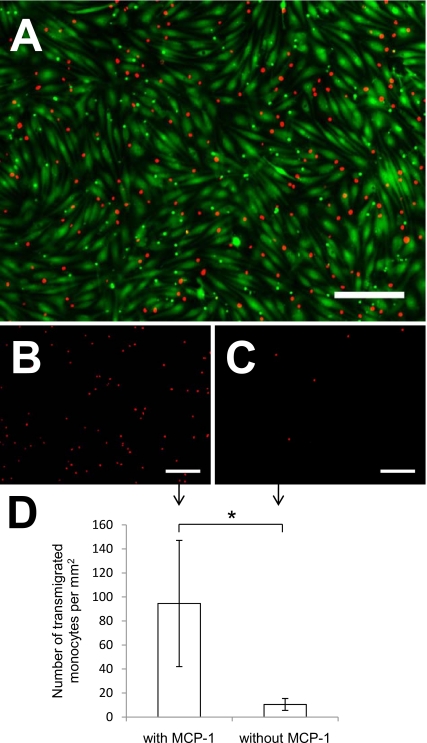
To exhibit the unique capabilities of a bilayer microfluidic membrane device, we demonstrated transmigration of THP-1 monocytic cells. Prelabeled monocytes adhered to an activated endothelium in the top channel [Fig. 5a] and then transmigrated across the EC-membrane barrier into the underlying channel. The addition of MCP-1 (a chemoattractant) in the lower channel caused greater transmigration [Figs. 5b, 5d] compared to a control containing no MCP-1 [Figs. 5c, 5d].
Figure 5.
Monocyte adhesion and transmigration. (a) Nuclear-stained (red) THP-1 monocyte adhesion to a confluent monolayer of calcien AM-stained (green) ECs. (b) Representative image of transmigrated monocytes in response to MCP-1, in the bottom channel of the device. (c) Representative image of transmigrated monocytes without the addition of MCP-1. (d) Addition of MCP-1 in the lower channel increased monocyte transmigration. Scale bar=200 μm. Data presented as mean±standard deviation: *p<0.05.
CONCLUSIONS
A membrane microfluidic device has been developed to mimic important physiologically relevant biomechanical and biochemical factors that influence vascular cell interactions and functions. As an initial demonstration of the utility of the device, the complex interactions between shear stress and TNF-α stimulation on monocyte adhesion to ECs and preferential transmigration of monocytes in response to a chemoattractant were investigated. The microfluidic platform is readily scalable and designed to enable study of other important phenomena, including drug and macromolecule permeability of ECs, EC-mural cell paracrine interactions, and the interaction of multiple cells types with ECM proteins, all under hemodynamic flow. The utility of this platform will be its potential to investigate phenomena under physiologically relevant conditions that are difficult to replicate using conventional macroscale systems.
ACKNOWLEDGMENTS
This study was supported by a Collaborative Health Research Project from the Natural Science and Engineering Research Council (NSERC) of Canada and the Canadian Institutes of Health Research (Grant No. CHRP 385956-2010), an Ontario Graduate Scholarship and a scholarship from the NSERC CREATE graduate training program in Microfluidic Applications and Training in Cardiovascular Health (to S.S.), an NSERC Undergraduate Student Research Award (to C.L.), and the Canada Research Chairs in Bioanalytical Chemistry (to A.R.W.) and Mechanobiology (to C.A.S.).
References
- Traub O. and Berk B. C., Arterioscler., Thromb., Vasc. Biol. 18, 677 (1998). [DOI] [PubMed] [Google Scholar]
- Davies P. F., Physiol. Rev. 75, 519 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangos J. A., McIntire L. V., and Eskin S. G., Biotechnol. Bioeng. 32, 1053 (1988). 10.1002/bit.260320812 [DOI] [PubMed] [Google Scholar]
- Sill H. W., Chang Y. S., Artman J. R., Frangos J. A., Hollis T. M., and Tarbell J. M., Am. J. Physiol. Heart Circ. Physiol. 268, H535 (1995). [DOI] [PubMed] [Google Scholar]
- Saunders K. B. and D’Amore P. A., In Vitro Cell. Dev. Biol.: Anim. 28, 521 (1992). 10.1007/BF02634136 [DOI] [Google Scholar]
- Powers M. J., Domansky K., Kaazempur-Mofrad M. R., Kalezi A., Capitano A., Upadhyaya A., Kurzawski P., Wack K. E., Beer Stolz D., Kamm R., and Griffith L. G., Biotechnol. Bioeng. 78, 257 (2002). 10.1002/bit.10143 [DOI] [PubMed] [Google Scholar]
- Huh D., Fujioka H., Tung Y. -C., Futai N., R.PaineIII, Grotberg J. B., and Takayama S., Proc. Natl. Acad. Sci. U.S.A. 104, 18886 (2007). 10.1073/pnas.0610868104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D., Matthews B. D., Mammoto A., Montoya-Zavala M., Hsin H. Y., and Ingber D. E., Science 328, 1662 (2010). 10.1126/science.1188302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camelliti P., Gallagher J. O., Kohl P., and McCulloch A. D., Nat. Protoc. 1, 1379 (2006). 10.1038/nprot.2006.203 [DOI] [PubMed] [Google Scholar]
- Tan W. and Desai T. A., J. Biomed. Mater. Res. Part A 72A, 146 (2005). 10.1002/jbm.a.30182 [DOI] [PubMed] [Google Scholar]
- Vickerman V., Blundo J., Chung S., and Kamm R., Lab Chip 8, 1468 (2008). 10.1039/b802395f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young E. W. K., Watson M. W. L., Srigunapalan S., Wheeler A. R., and Simmons C. A., Anal. Chem. 82, 808 (2010). 10.1021/ac901560w [DOI] [PubMed] [Google Scholar]
- Schaff U. Y., Xing M. M. Q., Lin K. K., Pan N., Jeon N. L., and Simon S. I., Lab Chip 7, 448 (2007). 10.1039/b617915k [DOI] [PubMed] [Google Scholar]
- Genes L. I., Tolan N. V., Hulvey M. K., Scott Martin R., and Spence D. M., Lab Chip 7, 1256 (2007). 10.1039/b712619k [DOI] [PubMed] [Google Scholar]
- Neeves K. B. and Diamond S. L., Lab Chip 8, 701 (2008). 10.1039/b717824g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C., Tempel D., van Haperen R., van der Baan A., Grosveld F., Daemen M. J. A. P., Krams R., and de Crom R., Circulation 113, 2744 (2006). 10.1161/CIRCULATIONAHA.105.590018 [DOI] [PubMed] [Google Scholar]
- Weber C., Zernecke A., and Libby P., Nat. Rev. Immun. 8, 802 (2008). 10.1038/nri2415 [DOI] [PubMed] [Google Scholar]
- Libby P., Ridker P. M., and Hansson G. K., J. Am. Coll. Cardiol. 54, 2129 (2009). 10.1016/j.jacc.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales R. S. and Wick T. M., Ann. Biomed. Eng. 24, 382 (1996). 10.1007/BF02660887 [DOI] [PubMed] [Google Scholar]
- Nagel T., Resnick N., Atkinson W. J., C. F.Dewey, Jr., and M. A.Gimbrone, Jr., J. Clin. Invest. 94, 885 (1994). 10.1172/JCI117410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicha I., Beronov K., Lopez Ramirez E., Osterode K., Goppelt-Struebe M., Raaz D., Yilmaz A., Daniel W. G., and Garlichs C. D., Atherosclerosis 207, 93 (2009). 10.1016/j.atherosclerosis.2009.04.034 [DOI] [PubMed] [Google Scholar]
- Sheikh S., Rainger G. E., Gale Z., Rahman M., and Nash G. B., Blood 102, 2828 (2003). 10.1182/blood-2003-01-0080 [DOI] [PubMed] [Google Scholar]
- Chueh B. -H., Huh D., Kyrtsos C. R., Houssin T., Futai N., and Takayama S., Anal. Chem. 79, 3504 (2007). 10.1021/ac062118p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismagilov R. F., Ng J. M. K., Kenis P. J. A., and Whitesides G. M., Anal. Chem. 73, 5207 (2001). 10.1021/ac010502a [DOI] [PubMed] [Google Scholar]
- Song J. W., Cavnar S. P., Walker A. C., Luker K. E., Gupta M., Tung Y. C., Luker G. D., and Takayama S., PLoS ONE 4, e5756 (2009). 10.1371/journal.pone.0005756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlos T., Kovach N., Schwartz B., Rosa M., Newman B., Wayner E., Benjamin C., Osborn L., Lobb R., and Harlan J., Blood 77, 2266 (1991). [PubMed] [Google Scholar]
- Morigi M., Zoja C., Figliuzzi M., Foppolo M., Micheletti G., Bontempelli M., Saronni M., Remuzzi G., and Remuzzi A., Blood 85, 1696 (1995). [PubMed] [Google Scholar]
- Thin Luu N., Rahman M., Stone P. C., Rainger G. E., and Nash G. B., J. Vasc. Res. 47, 451 (2010). 10.1159/000302613 [DOI] [PubMed] [Google Scholar]