Abstract
The growing field of miniaturized diagnostics is hindered by a lack of pre-analysis treatments that are capable of processing small sample volumes for the detection of low concentration analytes in a high-throughput manner. This letter presents a novel, highly efficient method for the extraction of low-molecular weight (LMW) proteins from biological fluids, represented by a mixture of standard proteins, using integrated microfluidic systems. We bound a polydimethylsiloxane layer patterned with a microfluidic channel onto a well-defined nanoporous silica substrate. Using rapid, pressure-driven fractionation steps, this system utilizes the size-exclusion properties of the silica nanopores to remove high molecular weight proteins while simultaneously isolating and enriching LMW proteins present in the biological sample. The introduction of the microfluidic component offers important advantages such as high reproducibility, a simple user interface, controlled environment, the ability to process small sample volumes, and precise quantification. This solution streamlines high-throughput proteomics research on many fronts and may find broad acceptance and application in clinical diagnostics and point of care detection.
Despite advances in the analysis of blood metabolites, the histochemical evaluation of tissue biopsies, and improvements in imaging approaches, early detection and diagnosis of human disease still suffers from severe limitations. A promising strategy to overcome this conundrum is the detection of molecular signatures from readily available body fluids.1 However, the onset of most diseases cannot be unequivocally identified on the basis of a single biomarker.2, 3, 4 Considerable attention has been devoted to the development of proteomic methods for the simultaneous, quantitative detection of multiple protein and peptide biomarkers (signature profiles) using mass spectrometry (MS).5, 6, 7 A critical aspect of the development of MS-based proteomics and peptidomics is the extraordinarily broad assortment of molecular species in blood, with concentrations ranging over more than ten orders of magnitude.8 This dynamic complexity is a significant barrier to the detection of disease-related peptides, many of which are present in trace amounts and are hidden within a background of highly abundant, nonrelevant proteins. Several strategies for sample treatment prior to MS analysis have been developed to address these limitations,9, 10 including conventional two-dimensional gel electrophoresis,11 prefractionation processes,12, 13 depletion of high-abundance proteins,14, 15 coated magnetic bead-based extractions,16 and beads equalization.17, 18 Despite these substantial advances, many of the listed techniques are prone to experimental variability, limited reproducibility, unwieldy handling procedures, protein instability during sample processing, and generation of misleading artifacts as a consequence of unreliable experimental procedures. These issues complicate the efficient detection of low-molecular weight (LMW) proteins and hinder the translation of LMW proteomic diagnostics to the clinic.19 We previously reported the development of mesoporous silica (MPS) substrates for the rapid on-chip fractionation of body fluids.6, 20, 21, 22 The ability of these chips to enrich previously undetectable LMW proteins opens the door to identification and analysis of these low abundance proteins, using matrix-assisted laser desorption∕ionization time-of-flight mass spectrometry (MALDI-TOF MS), whose spectra would otherwise be dominated by larger, more abundant serum proteins. The on-chip fractionation procedure, as well as the sample spectra demonstrating the ability of MPS chips to remove large proteins and enrich LMW proteins, is shown in Fig. S.1.23 Furthermore, our tests have demonstrated that the LMW species trapped inside the nanopores are protected from degradation through either size exclusion of proteases (e.g., trypsin) or through steric inhibition of their proteolytic activity in the confined space of the nanopores [see Fig. S.2 (Ref. 23)]. In this study, we couple a microfluidic system with our well established and characterized MPS platform for the purpose of achieving increased preconcentration of analytes, improved automation and compactness, and enhanced operational simplicity. This integrated system also overcomes the human error and variability associated with manual pipetting, thus ensuring better comparability between duplicates and among different samples.24, 25, 26, 27
Since no direct amplification process (such as polymerase chain reaction for nucleic acids) can be performed in proteomics, the sensitivity and dynamic range of a microanalytical system are of paramount importance.28, 29 Microanalysis technologies such as microfluidics offer promising features to address these needs, specifically as they pertain to proteomics research.30 Microfluidics has miniaturized biological procedures, allowing the creation of culture systems on a length scale similar to that of single cells or even molecule’s in vivo environment. This has resulted in a clear enhancement of kinetics and development rates with decreased degeneration as compared to traditional procedures. Microfluidic systems are typically created by microfabrication technologies using conventional lithography and etching techniques. These systems offer the ability to realize on-chip microchannel networks that can be computer-controlled to transport, separate, and detect chemical species using pressure flow of solution through their chambers. Once integrated with biological detection, these technologies have spurred the development of application-specific microstructured channels for proteomic and chemical analysis and have served as capillary mimics for tissue phantoms. In microfluidics, the surface-to-volume ratio is generally greatly increased. This is a key parameter for our application as the size-dependent capture processes operated by our chips take place on the sensor’s high surface area. Microfluidics technology allows precise control over local fluidic environments at submicron length scales over well-defined periods of time.31 With respect to proteomic nanochips, this level of control was tapped to provide greater control over the fluid environment at the MPS-fluid interface, further improving the sensitivity and efficiency of harvesting LMW proteins from complex biological samples.
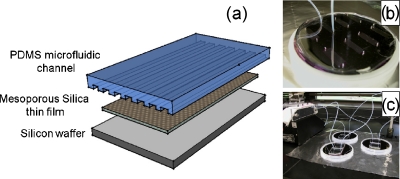
A typical procedure for the fabrication of MPS chips using triblock copolymer (Pluoronic F127) as a synthetic template is described in the supporting information. The microfluidic channels we fabricated exhibit rectangular cross sections (75 μm wide×20 μm deep). The device was fabricated from polydimethylsiloxane (PDMS) using the rapid prototyping technique. Figure 1a shows a schematic of the microfluidic device on porous silica substrate. The PDMS was replicated from SU8 masters created on Si wafers. SU8 photoresist was patterned on Si using a conventional photolithographic process. Channels with feature size of 75 μm width and 20 μm depth were created on SU8 master molds. The PDMS with curing agent (1:10 mixture ratio) was poured on the master mold and cured. The cured PDMS was then peeled off the mold to create the microchannel structure, which is then bonded to the porous silica substrate using oxygen plasma.
Figure 1.
Integrated microfluidic-MPS system. (a) Schematic of PDMS microfluidic channel on mesoporous silica substrate. (b) Image of single chip with input and output tubing. (c) Image of parallel experimental procedures performed over three devices at the same time.
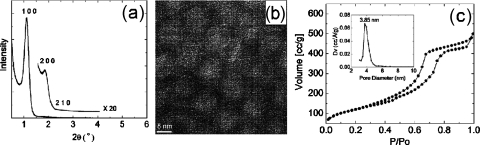
Characterization of the mesoporous silica thin films was carried out using several techniques. A variable angle spectroscopic ellipsometer (J. A. Woollam Co.) used in conjunction with WVASE32 modeling software was used to assess MPS thin film thickness (832.75±4.31 nm) and porosity (54.91%±0.22%), which were measured, respectively, using the Cauchy model and effective medium approximation model. Figure 2a presents an x-ray diffraction (XRD) pattern that shows three diffraction peaks, each of which corresponds to a different d-spacing measurement. These peaks, a sharp peak (d-spacing=8.22 nm) and two low intensity peaks (d-spacing=4.1 and 3.1 nm), can also be indexed as the 100, 200, and 210 planes characteristic of hexagonally arranged mesostructures. The pore structure can be verified through transmission electron microscope (TEM) imaging [Fig. 2b]. To further elucidate the physical pore parameters and morphologies, nitrogen adsorption∕desorption curves were plotted using a Brunauer–Emmett–Teller (BET) sorptometer [Fig. 2c] and the pore size distribution (3.85 nm) was calculated using the Barrett–Joyner–Halenda method [Fig. 2c inset]. The adsorption∕desorption isotherms show a type IV isotherm with an H2 hysteresis loop (identified by its sloped adsorption branch and nearly vertical desorption branch), which is indicative of an interconnected nanoporous structure. The inflection points at 0.40≤P∕P0≤0.75 indicate the presence of ink-bottle shaped nanopores [Fig. 2c].
Figure 2.
Characterizations of mesoporous silica thin films prepared with Pluoronic F127. (a) Small angle XRD patterns (0.2°–6°). (b) TEM imaging. (c) N2 adsorption∕desorption analysis isotherms and pore size distribution (inset).
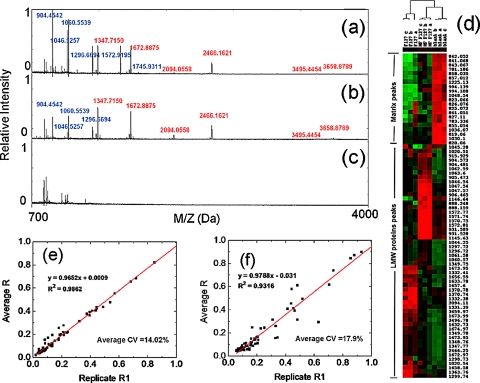
In order to emulate the complexity of a biological sample and to evaluate the improvement in protein enrichment by the integrated on-chip microfluidic system, we selected and assembled a standard mixture of proteins and peptides containing 26 different species with a broad range of molecular weights (900–66 500 Da), pIs (4.0–10.2), and concentrations (0.5–8 pmol∕μl) [see protein list in Fig. S.3 (Ref. 23)].We performed three experiments in parallel using (a) microfluidic integrated MPS chip, (b) MPS chip alone, and (c) microfluidic integrated with bare silicon (Si) substrate. While performing experiments with the integrated microfluidic system, we worked with 10 μl sample sizes of the aforementioned protein mixture. The solution was introduced to the microfluidic channels at a flow rate of 10 μl∕h and incubated for a period of 30 min. Following incubation, the protein solution was removed and 10 μl of de-ionized water was passed into the chamber to remove the residual, uncaptured proteins present in the microfluidic system. 10 μl of elution solution (1:1 volume ratio of acetonitrile: 0.1% trifluoroacetic acid) was then passed into the channels at a flow rate of 10 μl∕h. As the solution passed through the integrated microfluidic-porous silica structure, the LMW proteins were extracted. A similar experiment was performed on a bare Si with integrated microfluidic channels. The protocol for sample fractionation on the standard, uncoupled MPS chips is described in supporting information. The extracted solutions were then assessed using MALDI-TOF MS (Voyager-DE Pro). The spectra were collected in reflectron positive mode using standard instrument parameters.32 Figures 3a, 3b show the detection signals of the standard peptides (800–4000 Da) after on-chip fractionation by both the coupled microfluidic-MPS system and the MPS chip alone, respectively. All of the high molecular weight proteins [highlighted with green color in Fig. S.3 (Ref. 23)] in the mixture were successfully removed and no signal for large proteins was detected at high mass range from 3000 to 70 000 Da [see Fig. S.4 (Ref. 23)]. After normalizing the peak intensities, we were able to visually compare the differences in protein enrichment efficacy among the tested setups. The mass-to-charge ratio (m∕z) was labeled for each protein captured (12 markers from the microfluidic system and 10 markers from the MPS chip alone). All of the detected signals from the microfluidic-MPS chips [Fig. 3a] are either higher than (m∕z in blue) or equal to (m∕z in red) those from MPS chips [Fig. 3b]. Experiments using microfluidics coupled with bare Si [Fig. 3c] indicated no signal corresponding to the selected proteins in the mixture. This can be attributed to the absence of MPS to trap small peptides; the washing step effectively removed any residual proteins. The results obtained from this negative control experiment confirm that the MPS coating plays a critical role in the harvesting of LMW proteins from the biological complexes. The two-way hierarchical clustering presented in Fig. 3d shows enrichment patterns for LMW protein standards obtained using the microfluidic-MPS chip and corresponding control systems. Coupling of the existing MPS platform with a microfluidic system allows pressure control, which plays an important role in driving the desired LMW proteins into the silica pores. That is, both platforms are capable of capturing proteins within the defined range of 700–3000 Da. Differences in the clusters can be attributed to additional factors such as tertiary structure, which can affect their ability to enter the pores in slight but nonetheless noticeable ways. These differences are accentuated by the improved fractionation ability of the microfluidic system. As the microfluidic-MPS spectra is normalized, certain weak but present peaks are reduced in the presence of larger, more highly concentrated peaks.
Figure 3.
MALDI MS profiles and statistic analysis. (a) MS spectra of LMW proteins recovery from microfluidic-MPS device. (b) MPS chip and (c) microfluidic channel on bare silicon wafer. (d) Unsupervised hierarchical clustering analysis testing the ability of different settings to enrich LMW proteins (F127: MPS chip; MF F127: microfluidic-MPS device; blank: microfluidic channel on the bare Si wafer). Red indicates peak intensity higher than the median value, green indicates peak intensity lower than the median value, and black represents peak intensity equal to the median values. Each row represents an individual MALDI MS mass peak and each column represents a type of experimental setting. The reproducibility of LMW protein enrichment from (e) microfluidic-MPS chip and (f) MPS chip is investigated by regression curves.
The combined microfluidic-MPS system, due to the controlled flow environment, displayed superior enrichment of the LMW proteins present in the initial biological complex. It also avoided the mechanical damage to the microfluidic system at high flow rates. Using the microfluidic device, elution solution was flown in 5 s pulsed time intervals. The use of a second or even a third pulse was necessary to achieve complete elution. A total of ten pulses were performed to remove the adsorbed proteins onto the porous silica layer.
Reliable and reproducible fractionation is a key to the development of future proteomic and peptidomic screening techniques for clinical applications.33, 34 To assess the consistency of our on-chip fractionation strategy, we screened ten replicates with ten aliquots of the same standard proteins, testing both with microfluidic-MPS chips and MPS chips alone. Based on an unsupervised learning analysis, the regression curve and the equation comparing the peak intensities recovered from the replicates on the microfluidic-MPS chips are illustrated in Fig. 3e and exhibit a coefficient of regression R2=0.9862 and a lower average coefficient of variation (CV) (14.06%). This represents an improvement over the regression coefficient (0.9325) and an average CV at 17.9% obtained from MPS chips alone [see Fig. 3f]. The referenced CV curves and original MS spectra used for the reproducibility test are shown in Figs. S.5 and S.6, respectively.23 These results suggest that the automated operation of the microfluidic system eliminates man-made factors inherent to conventional pipetting methods and positions the described system as a platform capable of reliably profiling LMW and low abundance proteins in complex biological mixture for future clinical studies.
The human serum proteome has been extensively studied in order to identify and quantify protein and peptide biomarkers. As shown in Fig. S.7,23 we investigated the enrichment of LMW proteins from human serum sample (Sigma-Aldrich Co.) carried out using a MPS chip alone and a microfluidic-MPS chip. Figure S.7a displays the MALDI-TOF MS profile for LMW proteins from crude human serum, demonstrating that, without pretreatment, the signal for most of the peptides in the low mass range is suppressed by high-abundance large proteins. After sample fractionation with the MPS chip or the microfluidic-MPS system, as shown in Figs. S.7b and S.7c, the LMW serum proteins are significantly enriched. Linear regression analysis of the average intensities of detected MS peak for crude serum, MPS chip alone, and microfluidic-MPS chip is exhibited in Figs. S.7d–S.7f. The R2 (0.987) and CV (13.9%) carried by the microfluidic-MPS based on supervised statistical analysis are notably superior to the results from the process with MPS chip alone, indicating that the integration of the microfluidic system improves the overall reliability of the platform. However, more studies using human serum are needed in order to better assess the clinical possibilities that can be with this novel system.
In this letter, we presented a new fractionation device that presents features desirable for the exploratory screening and discovery of biomarkers. With the assistance of microfluidic modifications to our mesoporous silica substrates, LMW and low abundance proteins can be efficiently isolated and enriched with high reproducibility and high throughput compared to conventional methods. The results also confirmed that the on-chip sample fractionation integrated with microfluidic channels provided superior reproducibility and demonstrated the reliability of our MPS chips in the pretreatment of complex biological samples compared to other methods reported in the literature.35 As this approach evolves along with contemporary proteomic isolation and identification techniques, systems-mediated identification of disease-specific protein signatures may soon play a large role in the selection of personalized therapeutics, specifically pertaining to the real-time assessment of efficacy and toxicity, and in the rational modulation of therapy based on changes in the proteome associated with the prognosis of the disease.
Acknowledgments
The authors thank the Microelectronic Research Center (MRC) at the University of Texas at Austin. We thank Dr. Maria Person at the ICMB Protein and Metabolite Analysis Facility at the University of Texas at Austin. We also thank Professor Lidong Qin at the University of Texas Health Science Center at Houston for his advice for this letter. These studies were supported by the following grants issued in the U.S.: State of Texas’s Emerging Technology Fund, National Institute of Health (NIH) (Grant Nos. R21∕R33CA122864 and R01CA128797), National Aeronautics and Department of Defense (DoD) Breast Cancer Research Program Innovator Award (Grant No. W81XWH-09-1-0212) to M.F., and NSF CAREER Award (Grant No. 0846313) and DARPA Young Faculty Award (Grant No. YFA N66001-10-1-4049) to X.Z. The authors would like to recognize the contributions and support from the Alliance for NanoHealth (ANH).
References
- Hu Y., Fine D. H., Tasciotti E., Bouamrani A., and Ferrari M., Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology, March 2010. [DOI] [PMC free article] [PubMed]
- Etzioni R., Urban N., Ramsey S., McIntosh M., Schwartz S., Reid B., Radich J., Anderson G., and Hartwell L., Nat. Rev. Cancer 3, 243 (2003). 10.1038/nrc1041 [DOI] [PubMed] [Google Scholar]
- Papadopoulos M. C., Abel P. M., Agranoff D., Stich A., Tarelli E., Bell B. A., Planche T., Loosemore A., Saadoun S., Wilkins P., and Krishna S., Lancet 363, 1358 (2004). 10.1016/S0140-6736(04)16046-7 [DOI] [PubMed] [Google Scholar]
- Hye A., Lynham S., Thambisetty M., Causevic M., Campbell J., Byers H. L., Hooper C., Rijsdijk F., Tabrizi S. J., Banner S., Shaw C. E., Foy C., Poppe M., Archer N., Hamilton G., Powell J., Brown R. G., Sham P., Ward M., and Lovestone S., Brain 129, 3042 (2006). 10.1093/brain/awl279 [DOI] [PubMed] [Google Scholar]
- Liotta L. A., Ferrari M., and Petricoin E., Nature (London) 425, 905 (2003). 10.1038/425905a [DOI] [PubMed] [Google Scholar]
- Gaspari M., Ming-Cheng Cheng M., Terracciano R., Liu X., Nijdam A. J., Vaccari L., di Fabrizio E., Petricoin E. F., Liotta L. A., Cuda G., Venuta S., and Ferrari M., J. Proteome Res. 5, 1261 (2006). 10.1021/pr050417+ [DOI] [PubMed] [Google Scholar]
- Villanueva J., Shaffer D. R., Philip J., Chaparro C. A., Erdjument-Bromage H., Olshen A. B., Fleisher M., Lilja H., Brogi E., Boyd J., Sanchez-Carbayo M., Holland E. C., Cordon-Cardo C., Scher H. I., and Tempst P., J. Clin. Invest. 116, 271 (2006). 10.1172/JCI26022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson N. L. and Anderson N. G., Mol. Cell Proteomics 1, 845 (2002). 10.1074/mcp.R200007-MCP200 [DOI] [PubMed] [Google Scholar]
- Shores K. S. and Knapp D. R., J. Proteome Res. 6, 3739 (2007). 10.1021/pr070293w [DOI] [PubMed] [Google Scholar]
- Tirumalai R. S., Chan K. C., Prieto D. A., Issaq H. J., Conrads T. P., and Veenstra T. D., Mol. Cell Proteomics 2, 1096 (2003). 10.1074/mcp.M300031-MCP200 [DOI] [PubMed] [Google Scholar]
- Rabilloud T., Proteomics 2, 3 (2002). [DOI] [PubMed] [Google Scholar]
- Georgiou H. M., Rice G. E., and Baker M. S., Proteomics 1, 1503 (2001). [DOI] [PubMed] [Google Scholar]
- Pernemalm M., Orre L. M., Lengqvist J., Wikstrom P., Lewensohn R., and Lehtio J., J. Proteome Res. 7, 2712 (2008). 10.1021/pr700821k [DOI] [PubMed] [Google Scholar]
- Dekker L. J., Bosman J., Burgers P. C., van Rijswijk A., Freije R., Luider T., and Bischoff R., J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 847, 65 (2007). 10.1016/j.jchromb.2006.09.038 [DOI] [PubMed] [Google Scholar]
- Sahab Z. J., Iczkowski K. A., and Sang Q. X., Anal. Biochem. 368, 24 (2007). 10.1016/j.ab.2007.06.004 [DOI] [PubMed] [Google Scholar]
- Chang S. Y., Zheng N. -Y., Chen C. -S., Chen C. -D., Chen Y. -Y., and Wang C. R. C., J. Am. Soc. Mass Spectrom. 18, 910 (2007). 10.1016/j.jasms.2007.01.011 [DOI] [PubMed] [Google Scholar]
- Righetti P. G. and Boschetti E., Mass Spectrom. Rev. 27, 596 (2008). 10.1002/mas.20178 [DOI] [PubMed] [Google Scholar]
- Roux-Dalvai F., Gonzalez de Peredo A., Simo C., Guerrier L., Bouyssie D., Zanella A., Citterio A., Burlet-Schiltz O., Boschetti E., Righetti P. G., and Monsarrat B., Mol. Cell Proteomics 7, 2254 (2008). 10.1074/mcp.M800037-MCP200 [DOI] [PubMed] [Google Scholar]
- Davis M. T., Auger P. L., and Patterson S. D., Clin. Chem. 56, 244 (2010). 10.1373/clinchem.2009.127951 [DOI] [PubMed] [Google Scholar]
- Hu Y., Bouamrani A., Tasciotti E., Li L., Liu X., and Ferrari M., ACS Nano 4, 439 (2010). 10.1021/nn901322d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouamrani A., Hu Y., Tasciotti E., Li L., Chiappini C., Liu X., and Ferrari M., Proteomics 10, 496 (2010). 10.1002/pmic.200900346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geho D., Ming-Cheng Cheng M., Killian K., Lowenthal M., Ross S., Frogale K., Nijdam J., Lahar N., Johann D., Herrmann P., Whiteley G., Ferrari M., Petricoin E., and Liotta L., Bioconjugate Chem. 17, 654 (2006). 10.1021/bc0503364 [DOI] [PubMed] [Google Scholar]
- See supplementary material at http://dx.doi.org/10.1063/1.3528237 for the fabrication protocol of mesoporous silica thin films, the description of on-chip sample fractionation procedure, the scheme for on-chip fractionation and MALDI-TOF MS profiles for LMW proteins before and after enrichment, the MALDI-TOF MS to profile the effect of nanopores on inhibiting the proteolysis of LMW proteins, standard protein list, MALDI-TOF MS profiles of high molecular weight range from 3000 to 70 000 Da for the standard protein mixture, CV curves comparison (microfluidic-MPS chip versus MPS chip alone), the raw MALDI-TOF MS profilings for reproducibility study, and the comparison of LMW protein enrichment from human serum by different devices.
- Anderson J. R., Chiu D. T., Jackman R. J., Cherniavskaya O., McDonald J. C., Wu H., Whitesides S. H., and Whitesides G. M., Anal. Chem. 72, 3158 (2000). 10.1021/ac9912294 [DOI] [PubMed] [Google Scholar]
- McDonald J. C., Duffy D. C., Anderson J. R., Chiu D. T., Wu H., Schueller O. J. A., and Whitesides G. M., Electrophoresis 21, 27 (2000). [DOI] [PubMed] [Google Scholar]
- Rajaram N., Gopal A., Zhang X., and Tunnell J. W., Lasers Surg. Med. 42, 680 (2010). 10.1002/lsm.20933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy A. B., Tom W. J., Gopal A., Zhang X., and Dunn A. K., Opt. Express 16, 1975 (2008). 10.1364/OE.16.001975 [DOI] [PubMed] [Google Scholar]
- Lion N., Rohner T. C., Dayon L., Arnaud I. L., Damoc E., Youhnovski N., Wu Z. Y., Roussel C., Josserand J., Jensen H., Rossier J. S., Przybylski M., and Girault H. H., Electrophoresis 24, 3533 (2003). 10.1002/elps.200305629 [DOI] [PubMed] [Google Scholar]
- Figeys D. and Pinto D., Electrophoresis 22, 208 (2001). [DOI] [PubMed] [Google Scholar]
- Beebe D. J., Mensing G. A., and Walker G. M., Annu. Rev. Biomed. Eng. 4, 261 (2002). 10.1146/annurev.bioeng.4.112601.125916 [DOI] [PubMed] [Google Scholar]
- Brody J. P., Yager P., Goldstein R. E., and Austin R. H., Biophys. J. 71, 3430 (1996). 10.1016/S0006-3495(96)79538-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Person M. D., Shen J., Traner A., Hensley S. C., Lo H. H., Abbruzzese J. L., and Li D., J. Biomol. Tech. 17, 145 (2006). [PMC free article] [PubMed] [Google Scholar]
- Findeisen P., Sismanidis D., Riedl M., Costina V., and Neumaier M., Clin. Chem. 51, 2409 (2005). 10.1373/clinchem.2005.054585 [DOI] [PubMed] [Google Scholar]
- Seam N., Gonzales D. A., Kern S. J., Hortin G. L., Hoehn G. T., and Suffredini A. F., Clin. Chem. 53, 1915 (2007). 10.1373/clinchem.2007.091736 [DOI] [PubMed] [Google Scholar]
- De Bock M., de Seny D., Meuwis M. -A., Servais A. -C., Minh T. Q., Closset J., Chapelle J. -P., Louis E., Malaise M., Merville M. -P., and Fillet M., Talanta 82, 245 (2010). 10.1016/j.talanta.2010.04.029 [DOI] [PubMed] [Google Scholar]