Abstract
Cell movement is highly sensitive to stimuli from the extracellular matrix and media. Receptors on the plasma membrane in cells can activate signal transduction pathways that change the mechanical behavior of a cell by reorganizing motion-related organelles. Cancer cells change their migration mechanisms in response to different environments more robustly than noncancer cells. Therefore, therapeutic approaches to immobilize cancer cells via inhibition of the related signal transduction pathways rely on a better understanding of cell migration mechanisms. In recent years, engineers have been working with biologists to apply microfluidics technology to study cell migration. As opposed to conventional cultures on dishes, microfluidics deals with the manipulation of fluids that are geometrically constrained to a submillimeter scale. Such small scales offer a number of advantages including cost effectiveness, low consumption of reagents, high sensitivity, high spatiotemporal resolution, and laminar flow. Therefore, microfluidics has a potential as a new platform to study cell migration. In this review, we summarized recent progress on the application of microfluidics in cancer and other cell migration researches. These studies have enhanced our understanding of cell migration and cancer invasion as well as their responses to subtle variations in their microenvironment. We hope that this review will serve as an interdisciplinary guidance for both biologists and engineers as they further develop the microfluidic toolbox toward applications in cancer research.
INTRODUCTION
In the past decade, microfluidics has become a great tool in cancer research and other biological studies.1 Microfluidics typically deals with the manipulation of fluids that are geometrically constrained to a submillimeter scale. Such small scales offer a number of advantages including cost effectiveness, low consumption of reagents, high sensitivity, high resolution, and other less obvious features such as laminar flow.2 Its application to biological systems is compelling because it allows manipulation at the single or even subcellular level. However, microfluidic technology’s marriage to biology is at its infancy stage and there is still a gap between the engineers’ focus and the biologists’ needs. Therefore, there has been a push toward applying microfluidic tools to specific biological research areas so that development of these engineering approaches can be better guided.3
Microfluidic devices allow for a lab-on-a-chip array to simplify single cell analysis by providing a microenvironment that is of micrometer dimension and containing nanomoles of reagent∕media. Furthermore, microfluidic devices allow for controlled placement of cells and precise delivery of factors.4 Therefore, microfluidic technology has features of reliable, efficient, and cost-effective single cell selection and navigation. In this review, we will discuss the recent progress in using microfluidics in cell migration and its application to cancer and cancer stem cell models.
CANCER CELL MIGRATION
Cell movement is highly sensitive to stimuli from the extracellular matrix (ECM) and media. Receptors on the plasma membrane in cells can activate signal transduction pathways that change the mechanical behavior of a cell by reorganizing motion-related organelles.5 Cancer cells change their migration mechanisms in response to different environments more robustly than noncancer cells.6 For instance, they exhibit transitions to modes of migration that can lower dependence to the suppressed cellular pathway and increase cell motility in response to protease inhibitors, adhesion inhibitors, etc.6 Therefore, the therapeutic approaches to immobilize cancer cell via inhibition of the related signal transduction pathways rely on a better understanding of cell migration mechanisms. The migration modes of cancer cells can be categorized into three groups: mesenchymal, amoeboid, and collective cell migration modes with the former two at single cell level. As illustrated in Fig. 1, the three groups are distinguished by their morphology and migration characteristics in response to stimuli (usually, a combination of soluble chemokines, ECM, and surface-bound cues5). Here, we briefly summarize the migration processes in each mode by highlighting some of the essential elements that interact with the microenvironment.
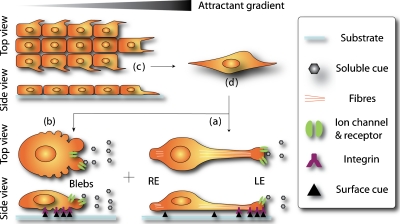
Figure 1.
Three modes of cancer cell movement. Similar to that of other cells, the migration of cancer cells is categorized into three modes: (a)–(c). Binding of extracellular cues to receptors and integrins transduces changes of the intercellular organization and morphology of the cell. A mesenchymal single cancer cell (a) forms a protrusion at the LE toward high chemoattractant levels and traction at the rear edge. The LE contains focal complexes that firmly adhere to stimuli through membrane receptors and integrins. An amoeboid single cancer cell (b) has dynamic focal complexes and high deformability. It migrates through blebbing movement induced by ECM and other stimuli. Collective cancer cells (c) migrate in a manner similar to mesenchymal cell, except that the intercellular connection is maintained during migration. Collective cell migration can change to single cell migration via epithelial-mesenchymal transition (C→D→A+B). Single cells can also switch from one mode to another.
Mesenchymal single cell migration
Mesenchymal single cell migration is the most studied mode.5 It is composed of four overlapping processes: cell polarization, protrusion, traction, and disassembly [Fig. 1a]. In response to extracellular cues, mesenchymal cells acquire an elongated morphology and strong polarity by rearrangement of proteins and membrane lipids in the cell. Cells form protrusions at their leading edge (LE), which is near to a high concentration of chemoattractants, and traction at their rear edge. Located at the LE are binding complexes of chemokine receptors and integrins that create strong adhesion of the cell membrane to the substrate. These adhesions are called focal adhesions. The corresponding larger macromolecular assembly is referred to as a focal complex. The formation of focal complexes triggers the activation of signal transduction cascades in the cell. Polymerized actins and myosin fibers act together to generate motor force at the LE.5 In this manner, cells adhesively crawl forward with continuous assembly and disassembly of LE.
Amoeboid single cell migration
Other single cancer cells, as exemplified by Walker carcinosarcoma cells7 and mammalian tumor cells,8 migrate in an amoeboid mode that carries little intrinsic polarity but high deformability. In this mode [Fig. 1b], cells also form focal complexes in response to stimuli, but the generated adhesion is too weak to exert forces sufficient for movement.9 Instead, cell motility mainly relies on three-dimensional (3D) blebbing as the driving force for locomotion. There are four stages in bleb formation: initiation, expansion, contractile cortex assembly, and retraction. Loose contact of the plasma membrane with the ECM or small space transduces damage to the membrane by either local breakage of actin cortex10 or detachment of the membrane from the actin cortex.11 The outward hydrostatic pressure pushes against the undermined membrane portion, resulting in bleb formation. The space in the bleb eventually is filled with cytosol and actin-related proteins until the local hydrostatic pressure is balanced and the cortex is repaired. This will lead to retraction and disappearance of blebs. It is believed that the formation of membrane blebs causes the transmission of traction forces and hydrostatic force to move the cell.9 This nonadhesive motility mode is particularly attractive to cancer study12 because cells migrate extremely fast in this mode and is related to the highly invasive phases of cancer cells and the therapy resistance of tumors.13
Collective cell movement and transition between migration modes
Collective migration is also important to cancer research as it is involved in the dissemination of some invasive tumor cells such as epithelial originated carcinomas.6, 14 In this mode [Fig. 1c], bulk cancer cells migrate as a sheet with cell-cell contact (usually through membrane receptors), which is maintained during migration.15 Individual cells behave similar to those in the mesenchymal migration mode in that they polarize and generate protrusions toward the migration direction in response to focal adhesion formation. A strong polarity is usually observed in the leading cells that have edges free from cell-cell contact. Under certain circumstances, collective migration can change into mesenchymal migration through dissolution of cell-cell contact [Fig. 1a], a process referred to as epithelial-mesenchymal transition.16 In this process, dissociated single cancer cells lose their internal polarity and are capable of converting into the mesenchymal or amoeboid mode, depending on how much cell polarity is induced. Cancer cells can also make the transition between mesenchymal and amoeboid mode at the single cell level in response to different environmental cues, as observed in metastasis.17 This mode switching has to be attributed to their high-motility and putative escaping mechanism.17
MICROFLUIDIC APPLICATION FOR MIGRATION STUDIES
One conventional system that is commonly used as a model to study cell migration is the transwell Boyden chamber, in which a porous membrane with pore size of 5–10 μm is placed between cells and chemoattractant so that cells are attracted to move across the membrane.18 However, lack of the capability of visual inspection and geometrical control has limited its application to quantification of overall cell population migration, although a few modifications have attempted to improve on this.19 On the other hand, microfluidic devices are capable of producing a microenvironment that is suitable for single cell study and have intrinsic characteristics that make them good candidates for studying cancer migration. Moreover, microfluidic devices usually use glass coverslips, polydimethylsiloxane (PDMS) membranes, or other polymer substrates, whose high transparency and thinness make them compatible with biological microscopy. In this review, we collectively demonstrate recent progress in the usage of microfluidic devices to study cell migration.
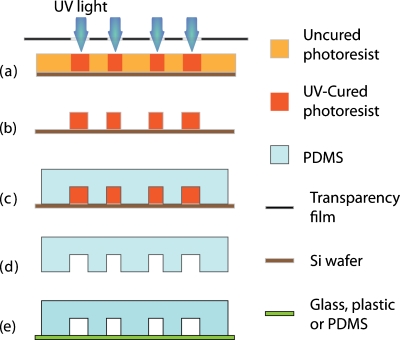
Advances in microtechnology have made microfluidic devices easy to design and construct. Typically, PDMS stamps are molded off through soft lithography and other rapid prototyping techniques, as described by Xia and Whitesides.20 As briefly illustrated in Fig. 2, a mask with the desired channel patterns is printed through computer-aided design and the patterns are projected on a master coated with photosensitive resist via optical lithography. Once a master is made, a negative in bas-relief is made by casting a mixture of PDMS with a crosslinking reagent and allowing it to harden. The resulting PDMS mold is then sealed by another substrate, often a glass or other PDMS, to form channels. The sealing can be either irreversible through oxygen plasma bonding or reversible via the naturally adhesive surface of PDMS. Refinements in the fabrication process, such as e-beam lithography, makes it possible to fabricate channels on the submicron or even nanoscale, which in theory would be able to constrain a fluid volume down to a femtoliter (billionth of microliter) range.21 Another important technique is the fabrication of multicompartment devices,22 which align several layers of patterns during the master fabrication process. These technical advances give engineers more flexibility, allowing them to construct complicated channel structures to meet specific biological needs. Here, we highlight some of the recent microfluidic approaches in studying cellular migration.
Figure 2.
Schematic of typical PDMS microfluidic device via soft lithography. (a) A mask with pattern designs printed on a transparency film is projected on a master coated with negative photoresist via optical lithography. [(b) and (c)] After the resulting master is made, a negative in bas-relief is made by casting a mixture of PDMS with a crosslinking reagent and allowing it to harden. [(d) and (e)] The resulting PDMS mold is released from the master and is sealed by another substrate, often a glass or other PDMS, to form channels. The sealing can be either irreversible through oxygen plasma bonding or reversible via the naturally adhesive surface of PDMS.
Interaction with mechanical and 3D environment
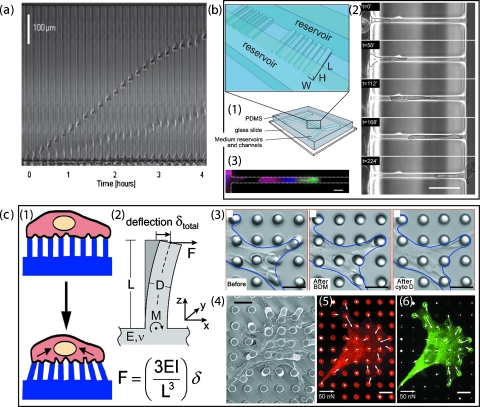
Spreading of cancer cells often follow the path of lymphatic vessels, nerves, white matter tracts, or other heterogeneous structures in tissues. It has been shown that interference of the mechanical properties of the local environment plays an important role in cancer migration, especially to low-adhesive and high-deformative amoeboid migration.23 However, visualizing the migration process while quantifying the migration speed is too difficult to be practical for conventional method (such as the transwell Boyden chamber). PDMS-made microchannels with dimensions of 3–12 μm in height and 6–100 μm in width, bonded on a glass coverslip and then coated or filled with ECM, provide a mechanically constrained environment for cancer cells to migrate through spontaneously [Fig. 3a].24 These systems predefine the cell migration path within the microsized channels (similar in size to in vivo conditions) such that migration speed and direction can be precisely quantified via time-lapse images. Migration properties of cancer cells have been characterized with a variety of origins from lung, breast, prostate, glioblastma, and golorectal adenocarcinoma. They mostly exhibit a much stronger motility and persistence as compared to noncancerous migratory cells. Cancer cell migration speed also varies between cell lines, with lung cancer and prostate cancer as the fastest movers.24 Figure 3a illustrates a dramatic case in which an amoeboid cell migrates almost three times as fast as a mesenchymal cell.
Figure 3.
Schematic of cell interaction with mechanical environment. (a) Sequential frames over 4 h show the displacement of cells, one with amoeboid (faster) and one with mesenchymal migration mode, inside the same channel. Cell migration speed can be quantified. Reproduced and adapted with permission from D. Irimia and M. Toner, Integrative Biology 1, 506 (2009). Copyright © 2009 by Royal Society of Chemistry Publishing group. (b) Cell spontaneously “squeezes” through microchannel. (b1) The dual-layered microfluidic device is fabricated with PDMS such that microchannels bridge between two parallel cell culture channels. (b2) Images of a time-lapse with a SPC-treated Panc-1 cell, migrating from left to right through a micron channel (7×11×150 μm), exhibit cell deformation in five stages. (b3) The corresponding localizations of cytokeratin (in green), filamentous actin (magenta), and the nuclei (blue) are visualized by staining. Scale bar: 50 μm. Reproduced and adapted with permission from C. G. Rolli et al., PLoS ONE 5, e8726 (2010). Copyright © 2010 by Public Library of Science. (c) Arrays of microposts can be used for measurement of cell adhesion during migration process. (c1) Under proper geometric constraints of postheight and width, cell exerting traction forces would deflect the elastomeric posts. The corresponding force can be quantified by measuring the displacement of micropoles (c2). (c3) Differential interference contrast micrographs of a smooth muscle cell cultured for 2 h on a postarray. These demonstrate loss of traction forces in response to the treatment of 2,3-butanedione monoxime or cytochalasin D, inhibitors to myosin contractility. [(c4)–(c6)] Phase contrast and immunofluorescence images of a smooth muscle cells on the microposts. Localization of fibronectin (in red) and focal adhesion protein vinculin (in green) indicates a strong correlation between direction and magnitude of traction force and focal adhesion area. Reproduced and adapted with permission from J. L. Tan et al., Proc. Natl. Acad. Sci. U.S.A. 100, 1484 (2003). Copyright © 2003 by National Academy of Sciences; I. Schoen et al., Nano Lett. 10, 1823 (2010). Copyright © 2010 by American Chemistry Society Publications.
Invasiveness of cancer cells is related to their high deformability that enables them to move through 3D-confined spaces. For example, human pancreatic adenocarcinoma cells in macroscale culture were reported to undergo drastic cell and nuclear deformation during migration when their proteolytical activity is inhibited.25 Such deformative migration of cancer cells, accompanied with substantial reorganization of cytoskeleton and other organelles, can accelerate by 1.5-fold upon treatment of sphingosylphosphorylcholine (SPC), a bioactive phospholipid.26 In order to investigate the underlying organization of cytoskeleton, Rolli et al.27 used a bilayered microfluidic device that has microchannel bridging between two larger chambers [Fig. 3b]. Tumor cells seeded on one chamber side of the microchannels tend to squeeze and permeate to the other side through microchannels (∼7 μm). After the cells enter the microchannels, their migration speeds increase by threefold. Unlike the ones in macroculture, the migration speed of cells inside the microchannels is not promoted further by SPC treatment. Instead, the overall proportion of permeative cells increases dramatically from 7% to 33% with SPC treatment. Such a drastic accelerating motility mentioned earlier is shown to be mostly adhesion-independent in the study of integrin-ablated cell models.28 Instead, cells migrate by the sole force of actin-network expansion, suggesting a high-motility phase of mesenchymal mode, which promotes protrusive flowing of the leading edge. The underlying mechanism is proposed to be mainly powered by actin polymerization at the cell membrane, which strongly relies on geometric confinement and cell deformability.29
Cell migration within 3D matrices is another important process in tumor spreading and invasion. Unlike two-dimensional surface movement, 3D migration through ECM involves highly regulated and reciprocal interaction between the ECM and the cells, often including reorganization of ECM microstructures by cellular behavior.30 Such behavior is contractility-dependent and is also relevant to cell alignment31 to collagen stripes made through soft lithography. A recent work by Beebe and colleagues32 has established a microfluidics-based system for making arrays of type-I collagen, an important ECM component, allowing further investigation of cell-ECM interaction (also see Sec. 3B).
As we described earlier, mesenchymal cells migrate primarily through traction forces at focal adhesions, which involves tremendous rearrangements of ECM ligand-integrin interaction and cytoskeleton structure, via the consequential reorganization of mechanic pulling and contraction force to the substrate. How these forces are coordinated spatiotemporally in response to stimuli is another central question to the migration study of cancer cell. Microfluidic devices that contain nano- or microsized pillars can be used to study the mechanical interaction between the migrating cell and the underlying substrate. Arrays of micropillars, selectively or uniformly coated with adhesion molecules, are patterned as adhesive substrate for cell migration.33 One can measure adhesion force-induced deformations of the pillars and then calculate the corresponding force values according to the elastic properties of the pillars [Fig. 3c].34 A number of studies demonstrate a strong connection between contraction force and cell morphology: migratory cells exert much higher contraction forces than stationary cells in protrusion segments, and the more spreading these segments are, the more sensitive the cell will respond to chemoattractant and surface stimuli.35 A similar approach uses silicon nanowires instead of PDMS pillars and shows that cancer cells exhibits a larger traction force than normal cells by 20% for Hela cell line and 50% for L929 cell line.36 When cells were subjected to multiple force actuations, by magnetic manipulation of the micropillars, focal adhesion recruitment is further enhanced.37
Gradient generator
Under physiological conditions, tumor cells rarely encounter a uniform environment. Instead, they often have to deal with a complicated chemical environment where chemotaxis is guided by concentration gradients of growth factors, chemokines, and surface ligands. The gradients can have soluble, surface-bound, or ECM-adsorbed forms. Cells adjust their behavior accordingly, sensing the subtle variation in chemical concentration across an area as small as a single cell.38 Conventional chambers that are used for gradient generation either lack sufficient resolution or are unable to sustain a steady gradient over time. Recent advances in microfluidic device design allow for the establishment of a steady gradient profile, which can be engineered and controlled. These designs can be divided into three groups: flow-based gradient generator, diffusion-based gradient generator, and surface micropatterning. They simulate stimulus gradient that are in soluble, surface-bound, and ECM-adsorbed forms, respectively.
Flow-based
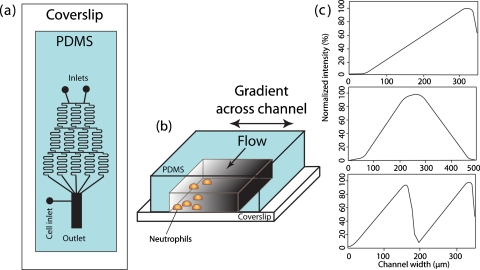
When fluid dimensions are miniaturized, its dynamic properties are also altered and unusual phenomena take place. Probably, the most unique and useful characteristic of the microfluidic flow is the extraordinarily low Reynolds number. The Reynolds number is the ratio of inertial forces that produce turbulence to the viscous forces that result in laminar flow in which the fluid flows in parallel layers with no disruption between layers. As the cross sectional area of a chamber becomes smaller, convection forces decrease and molecules move mainly by diffusion along the concentration gradients. Under these conditions, laminar flow dominates. Engineers have taken advantage of these flow conditions by manipulating geometrically patterned overturns to produce turbulent flow and define gradients.39 A good example of a flow-based gradient generator is the “Christmas tree,” which was pioneered by Jeon and colleagues. It was initially used for neutrophil chemotaxis40 (Fig. 4) and leukocyte trafficking.41 In this device, two concentrations of biomolecules separately flow into a network of microchannels, where spring-shaped mixers are patterned to convectively mix adjacent streamlines and form a stair-shaped chemical gradient. This chemical gradient in the laminar flow is then run through a cell chamber where diffusion will smoothen the gradient profile. Additional inlets and microchannel network manipulation will lead to controlled gradient profiles with defined intensity and shape [Fig. 4c].42 Stair-shaped gradient generators were also used to study EGF-induced motility in breast cancer cells.43 Furthermore, MDA-MB-231 breast cancer cells exhibit chemotaxis to a polynomial gradient of 0–50 ng∕ml of EGF but not to a linear gradient.44
Figure 4.
Schematic representation of the gradient generator and gradient characterization. (a) Top view of the device consisting of the gradient mixer and cell culture chamber. (b) 3D representation of the cell culture chamber where cells are exposed to gradients of chemoattractant. (c) Gradient profiles represented by fluorescent characterization of FITC-dextran (8 KDa): linear shape (top), hill shape (middle), and cliff shape (bottom). Reproduced and adapted with permission from N. L. Jeon et al., Nat. Biotechnol. 20, 826 (2002). Copyright © 2002 by Nature Publishing group.
Flow-based gradient generators have great control over the gradient’s spatiotemporal resolution. Gradients can be changed very rapidly (a few seconds) by adjusting the relative flow rates of the constituent solutions. However, flow-based gradient generators suffer from a number of shortcomings. Control of the gradient profile requires maintaining constant flow rates, which consumes a great amount of reagent. The constant flow will remove autocrine, paracrine, and other secreted molecules from cells, which could be critical in cancer cell migration regulation.45 Moreover, due to the flow, the cells are subject to undesired shear stress, which is proven to affect cell migration.46
Diffusion-based
A number of emerging microfluidic technologies have been developed to generate soluble gradients based on passive diffusion. They can achieve reduction or elimination of the fluid flow within the cell surroundings using hydrogels or high resistance microchannels. In this way, diffusion becomes the predominant transport activity for molecular exchange. Typically, two reservoirs are used to maintain constant molecular exchange, with a high chemical concentration in one (source) and low chemical concentration in the other (sink). As flow is eliminated or reduced, diffusion occurs to buffer the concentration difference between source and sink. The diffusion rate can be maintained by either initially supplying a large amount in the source or frequently replenishing the source and sink. In such devices, steady-state gradients can form with little reagent consumption compared to flow-based devices. The bridging channels would also provide mechanical confinement for chemotaxis in amoeboid cells [Fig. 5a].47 In such a device, migrating cells crawl up to the gradient through the bridging channels and completely occlude the microchannels that perturb spatially heterogeneous intracellular signaling. While gradients require longer time to develop, there is minimal shear stress, and potential autocrine, paracrine, and other secreted molecules from cells are preserved. Furthermore, the absence of flow makes it possible to evaluate nonadhesive migration, which can be disrupted by shear stress under flow. Amoeboid movement by cancer cells is of particular interest to biologists because this type of movement may be responsible for the highly invasive behavior of cancers in vivo.
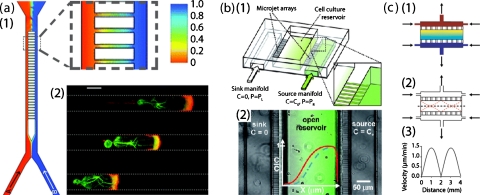
Figure 5.
Diffusion-based gradient generator using microchannels. (a) Gradient generator is composed of two parallel perfusion chamber connected with an array of microchannels. Two streams are mixed in bridging microchannels through diffusion predominantly. As illustrated in simulation (a1), the linear gradient of content B is created in microchannels. (a2) Cytoskeleton polarity is observed with cells that migrating along this attractant gradient. Actin (in red) is mainly located at the leading edge and on the side in contract with the microchannel wall. Microtubules (in green) are localized behind the nucleus but extend from the leading edge to the uropod. No thin lamellipodium is seen at the leading edge. Scale bar: 5 μm. Reproduced and adapted with permission from D. Irimia et al., Lab Chip 7, 1783 (2007). Copyright © 2007 by Royal Society of Chemistry. (b) Microjets gradient generator. (b1) A 3D schematic of the device shows the parallel source and sink chambers that are connected with microchannel and an open structure of the cell culture/gradient chamber. (b2) Top view of the device with a fluorescence image overlay of an Alexa 488 gradient that is generated through diffusion (see Ref. 75). Reproduced and adapted with permission from T. M. Keenan et al., Lab Chip 10, 116 (2010). Copyright © 2010 by Royal Society of Chemistry. (c) Additional manipulation of perfusion chamber can introduce further control over convection in cell culture/gradient chamber. (c1) Simulation of gradient. (c2) Fluid flow streamlines showing the cross-current flow pattern produced due to the pressure drop between inlets/outlets. (c3) The resulting fluid velocity along the dashed line is shown in (C2) (see Ref. 76). Reproduced and adapted with permission from A. Shamloo et al., Lab Chip 8, 1292 (2008). Copyright © 2008 by Royal Society of Chemistry.
Planer microfluidic chambers are patterned with underlying microchannels and 1 μm apertures to generate a locally intense and consistent gradient of EGF.48 Unlike conventional methods, the fluid and channels in this microfluidic device are delivered from beneath, providing convenience for imaging and further manipulation.
Microscopic imaging and antibody-labeling enable the localization of cell organelles, such as cytoskeleton and actin cortex, which can then be correlated with the gradient. Furthermore, occlusion of the channels by migrating cells made it easy to deliver in situ agonist∕antagonist to the front or back, providing an excellent platform for drug screening.
Some microchannel-based devices have three parallel large channels [Figs. 5b, 5c], with the middle channel used as a gradient chamber. As flow is reduced due to the microchannel confinement, diffusion becomes the predominant mode of molecule transport within the microchannels and the center gradient chamber. Thus, a steady-state gradient is formed in microchannels and gradient chamber that is stable for hours. The source and sink are maintained by a constant or intermittent flow in the outer channels. It has been shown that migratory cells experiencing the concentration gradients exhibit higher activity in their filopodia dynamics, while forming more filopodia protrusions toward high EGF concentration along the gradients.49
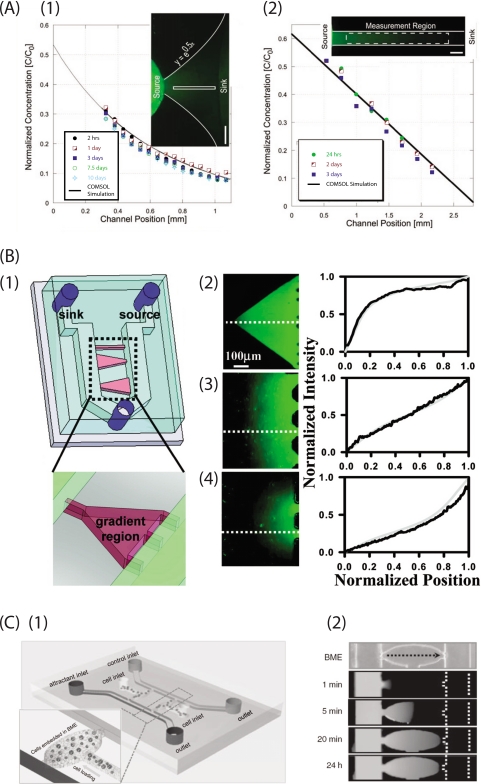
In contrast, a hydrogel-filled channel completely blocks the flow caused by hydraulic pressure and local convective flows, which can disturb and even completely destroy the established gradients. Meanwhile, hydrogels provide a space for the diffusion of biomolecules that are water-soluble and smaller than the gel pores. All these features are similar to the physiological function of ECM under in vivo conditions, providing an excellent scaffold for 3D culturing. Thus, hydrogels (e.g., collagen) would accurately mimic the microenvironment of tumors, especially for cell invasion associated with proteolytic activity.50 Beebe and colleagues51 developed such a gradient generator in a hydrogel-filled channel [Fig. 6a]. In this device, gradient profiles can be maintained for up to 10 days, consuming as little as 34.5 μl of solute with appropriate source replenishment. In order to establish a gradient more quickly with minimal reagent consumption, the timing and concentration of the source replenishment are critical. Furthermore, the steady-state gradient profile is determined by the geometry profile of the channel and can be predicted with computer simulation, allowing more complex gradient profiles to be formed. An EGF linear gradient in type-I collagen was used in such a device to study the invasive metastatic behavior of invasive rat mammary adenocarcinoma (MTLn3) cells. Compared to an unstimulated control, locomotion of the cancer cells in 24–48 h is found to be statistically faster, suggesting a strong metastatic activity in that time range.
Figure 6.
Diffusion-based gradient generator using hydrogel. (a) Submillimeter-sized channel that connects between source and sink reservoirs can generate gradient profile in exponential shape (a1) and linear shape (a2), visualized with Alexa 488 solution. Reproduced and adapted with permission from V. V. Abhyanka et al., Lab Chip 8, 1507 (2008). Copyright © 2008 by Royal Society of Chemistry Publishing group. (b) Hydrogel-confined microchannels can generate gradients profile in a variety shapes. (b1) Hydrogel is confined in microchannels as indicated in red, while source and sink fluid flows toward the same outlet (in green), keeping constant molecule exchange with 3D hydrogel. Geometry of microchannel, therefore the hydrogel compartment determines the gradient profile in a concave down nonlinear shape (b2), linear shape (b3), and convex up nonlinear shape (b4). Reproduced and adapted with permission from B. Mosadegh et al., Langmuir 23, 10910 (2007). Copyright © 2007 by American Chemistry Society Publications. (c) A microfluidic gradient generator designed for the study of cancer cell directed invasion in a 3D environment. (c1) Two parallel perfusion channels were introduced with attractant-containing medium and control medium that served as source and sink reservoirs, respectively. Cell-BME mixture is loaded from cell inlets into cell culture chambers (shown in zoom view). (c2) The perfusion channels are introduced with FAM-DNA (MW 6000 D) and PBS, respectively. The dashed line indicates the channel location. In 20 min, a linear gradient is established and is maintained for over 24 h (see Ref. 77). Reproduced and adapted with permission from T. Liu et al., Electrophoresis 30, 4285 (2009). Copyright © 2009 by WILEY-VCH Verlag GmbH&Co.
However, hydrogel-based gradient generators suffer from several limitations. Gradients usually take hours to form. Rapid adjustment of the gradient profiles and molecule species are too difficult to be practical. An alternative approach can significantly reduce the gradient formation time by combining hydrogel and microchannels in the same device.52 In this device, a hydrogel is confined in microchannels that are similar to the ones we described earlier in this section. The source and sink reservoirs are filled with biomolecule-contained fluid, which can be rapidly changed and make mass exchange with the hydrogel in the microchannels. Compared to bulk hydrogel, biomolecules diffuse into the hydrogel in the microscale long microchannel much more quickly and establish a steady-state gradient in less than 1h. Reshaping the microchannel geometry also leads to a more complex gradient profile, including nonlinear gradients [Fig. 6b].53
Micropatterning
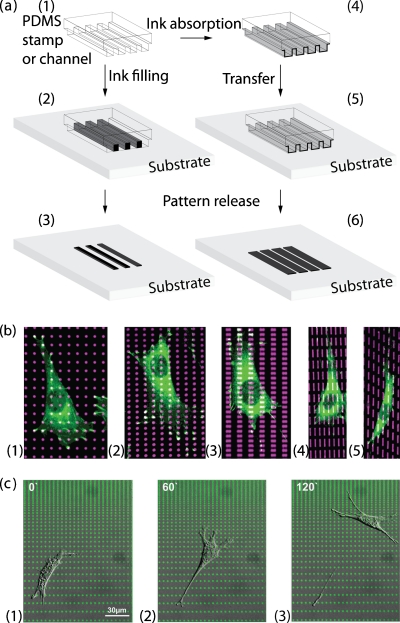
In addition to diffusive cues, cell migration is also guided by surface attachment between the cell and the surrounding tissues. Surface attachment also plays an essential role in cell signaling and cellular haptotaxis (haptotaxis is the directional motility or outgrowth of cells in response to surface stimuli). In this context, the interplay of integrin and surface-bound signal has been one of the essential targets in the study of cancer cell migration.54 It is believed that the surface-bound signal mediates morphological polarization and migration behaviors of cell through pattern shape and concentration differences,55, 56 which are difficult to track in conventional dish culturing conditions.56 Numerous micropatterning tools have been developed for generating such micro-∕nanometer surface patterns with adhesion-promoting biomolecules such as polylysine and fibronectin.57 The most widely used tool for making surface-bound gradients is microcontact printing, as illustrated in Fig. 7. Although microcontact is not technically microfluidics, its fabrication processes use similar technologies as microfluidic device fabrication. Micropatterning fabrication begins with a PDMS stamp containing the desired microfeatures, which is made via soft lithography and then employed through two methods. In one way [Fig. 7a, 4→6], the PDMS stamp is adsorbed with biomolecule-contained ink by immersing in the solution and then contacts with the substrate. This allows for biomolecules transferring onto the substrate. In another way [Fig. 7a, 1→3], the microfabricated PDMS stamp is temporarily attached on the substrate to form microsized channels or wells, which are then filled with ink. The ink will then be coated to the substrate surface, according to these microsized features of PDMS stamp, by self-assembly or surface adsorption. Either technique is able to achieve micropatterns of biomolecules in a resolution up to 10 nm. With such a high resolution, cells restrict their morphology and cytoskeleton distribution almost exclusively to the ECM pattern.58 On the other hand, the imposed asymmetric microgeometries bias the cytoskeleton distribution such that an array of ratchetlike patterns is able to polarize cells [Fig. 7b],59 promote their protrusion, and guide their migration [Fig. 7c].60, 61 Interestingly, cancer cells, during this type of adhesion-mediated migration, display a larger lamellipodia protrusion as compared to noncancerous cells [Fig. 8a],62 implying a strong regulation capability of cancer cells in cytosol volume and cytoskeleton organization.
Figure 7.
Microcontacting printing of surface cues. (a) A PDMS stamp that contains the desired microfeatures is made via soft lithography and is used for patterning through two techniques. Either by adsorbing on PDMS features (1→4→5→6) or filling in the PDMS channel (1→2→3), the biomolecule-contained ink makes contact with substrate and is geometrically patterned. The ink will then be coated on the substrate surface, according to these microsized features of the PDMS stamp, by self-assembly or surface adsorption. (b) The shape and distribution of vinculin-containing focal adhesions correspond to those of the micropatterned fibronectin islands. Immunofluorescence microscopic overlay images of cells cultured for 8 h on different micropatterned distributions of rhodamine-FN-coated islands (magenta) and stained with antivinculin antibodies (green). Reproduced and adapted with permission from N. Xia et al., FASEB J. 22, 1649 (2008). Copyright © 2008 by Federation of American Societies for Experimental Biology. (c) Time lapse of a fibroblast cell migrating along the surface gradient of fibronectin. Round-shaped fibronectin islands are patterned in gradient by varying interisland spacing. Cell is found to generate protrusion and polarity in response to the variation of interisland spacing and then migrate toward denser pattern. Personal communication/unpublished data with permission from Tatjana Autenrieth and Martin Bastmeyer [Universitaet Karlsruhe (TH), Germany].
Figure 8.
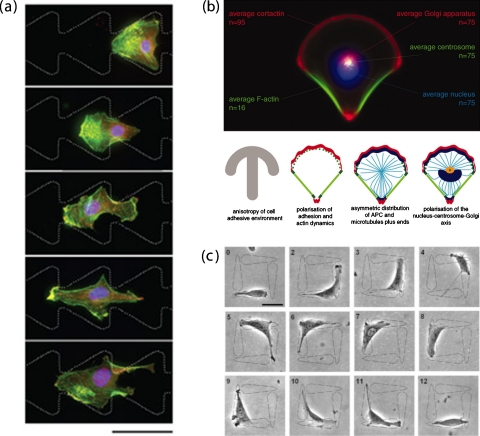
Single cell migration in response to geometry confinement of surface pattern. (a) B16F1 cell on a pattern of two reservoirs (not shown) connected by ratchets. The cell migrates through the bridging ratchets (from right to left) by repeating a cycle of protrusion (at the cell’s leading edge, in the funneling direction) and retraction (at its rear). The polarity markers (Arp2/3 complex, seen as yellow on the overlay images with actin) and organization of actin cytoskeleton (fluorescent phalloidin, green) is stained to reveal the migration cycles. The scale bar represents 50 microns. Reproduced and adapted with permission from G. Mahmud et al., Nat. Phys. 5, 606 (2009). Copyright © 2009 by Nature Publishing group. (b) Polarized single cell in response to geometry constraint by surface pattern. (b) Top: cell membrane polarity propagates to cell internal polarity, as shown in the average of 75 cell fluorescent images. (b) Bottom, from left to right: carton illustration of this propagation process. In response to the anisotropic distribution of fibronectin in micropattern (gray), the distribution of adhesions (green) and actin network (red) becomes polarized with polymerizing meshwork on adhesive edges and stress fibers on nonadhesive edges. This in turn triggers the reorganization of actin-MT connectors and the location of Golgi apparatus (blue). Reproduced and adapted with permission from M. Théry et al., Proc. Natl. Acad. Sci. U.S.A. 103, 19771 (2006). Copyright © 2006 by National Academy of Sciences. (c) One-way microarrays direct the cell migration. Time-lapse phase-contrast images (numbered in hours upon cell seeding) show the continuous directional migration of individual NIH 3T3 fibroblast from the blunt end of the teardrop island to the tip of an adjacent island. Scale bar: 50 μm. Reproduced and adapted with permission from G. Kumar et al., Adv. Mater. (Weinheim, Ger.) 19, 1084 (2007). Copyright © 2007 by WILEY-VCH Verlag GmbH&Co.
Microcontact printing combined with soft lithography is able to create patterned islands of chemoattractants. These islands seem to be able to precisely position focal adhesions and control the direction of cell movement by patterning their shape. In fibroblasts, arrays of fibronectin islands were patterned to study the influence of surface adhesion to cell motility and Rac activity.63 The fibroblast cells seem to spread and align preferentially along the main axis of the linear islands even when they were isotropically distributed [Fig. 8b]. For the round islands, cells orient along the direction with smaller interisland separation. Such orientation during the initial cell attachment affects the cell polarization and governs the cell moving direction through Rac GTPase activation [Fig. 8b].63 In this context, ligand density that mediates cell attachment certainly plays an important role. Therefore, chemical surface gradients are also expected to guide cell haptotaxis. One method of generating surface gradients is to vary the size and the spacing of the above patterned islands such that the surface cover with them ramps up from one end to the other.64 In this way, adhesion promotion activity of cells due to surface ligands is biased and so are the lamellipodia protrusion and cell migration direction. Adhesion response and cell distribution are found to linearly correlate to such surface-bound gradients.65 Recent progress of Autenrieth and Bastmeyer66 demonstrates that 80% of haptotactic fibroblasts do migrate toward high fibronectin density in such gradient, with Golgi complex located behind nucleus [Fig. 7c]. This haptotaxis activity is highly repressed in the presence of taxol.66
NEW FRONTIER-MIGRATION OF BRAIN TUMOR CELLS
In this section, we would like to highlight some recent progress regarding migration of glioblastoma multiforme (GBM) tumor and tumor stem cells (BTSC).
GBM tumor and tumor stem cells
Cancer is the number one killer in the world today, causing up to 13% of all human deaths. Over the past 40 years, the United States has invested over 0.2 trillion dollars on cancer research looking for effective therapies, but the effort has resulted in only a 5% decrease in the death rate between the years 1950 and 2005. To a great extent, the lack of therapeutic advances arises from incomplete understanding of some important cancer mechanisms, especially oncogenesis and invasion.67 The inability to prevent tumor spreading and recurrence is especially true for GBM.
GBM is the most prevalent adult primary malignant brain tumor68 with over 14 000 cases diagnosed annually in the United States. Despite the aggressive therapies, the median survival of patients is only 12–14 months.68 Standard of care for patients with GBM includes maximal safe surgical resection, radiotherapy, and systemic chemotherapy. However, the prognosis is poor, largely due to the near universal recurrence of tumors after initial treatment. Recurrence in GBM can, in part, be attributed to the migratory capacity of glioma cells that allows spreading into the surrounding healthy tissue and precluding eradication with current therapies. Interestingly, extraneural metastasis of GBM is particularly rare.69 Recent work suggests that intermingled within the GBM tumor bulk is a small subpopulation of BTSC, which may be the perpetuating cell of GBM.67, 70 Compared to tumor cells, BTSCs are more efficient in driving tumor growth in animal models and display a resistance to conventional therapies including radiation, similar to the actual disease.71 BTSC mirrors characteristics of neural stem cells including self-renewal, extensive brain migration, and divergent differentiation, and thus harbor characteristics consistent with the tumor stem cell theory.72
Migratory signature in microfluidic approach
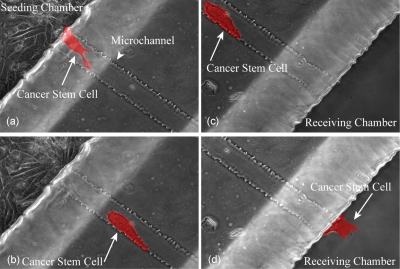
Preliminary work done in our laboratories demonstrates the feasibility of culturing BTSC in microfluidic devices. Isolation and characterization of BTSC have been reported in our previous study.73 BTSC had the ability to migrate from a seeding chamber to a parallel receiving chamber through the microchannels of a microfluidic device [Fig. 9a, arrow].74 Cells in the seeding chamber migrated in mesenchymal mode, forming cell polarization and protrusion [Fig. 9b, arrow]. The cells that were approximate to the microchannels exhibited transient long protrusions and compete with each other such that only one cell at a time was able to initiate spontaneous migration through the microchannel. However, once the cell entered the microsized space (40 μm wide, 5 μm tall, and 400 μm long) of the microchannel, it lost the polarity and transformed into amoeboid mode by generating blebs [Fig. 9d, arrow]. Furthermore, we tailored the device such that the seeding and receiving chambers can hold a sufficient amount of media to allow BTSC survival for 4–5 days without media exchange. Using Accutase to enzymatically detach cells from the microfluidic device, we were able to isolate two populations of BTSC, migratory and nonmigratory subsets, which can be subject to gene analysis to determine a migratory signature in the future. This type of approach illustrates the potential for using microfluidic devices in the BTSC study.
Figure 9.
BTSC migration through the microfluidic device. Real time microscopy demonstrating a BTSC (highlighted in red) migrating from seeding chamber to the receiving chamber through a bridging microchannel
SUMMARY
In recent years, there has been a great progress in the application of microfluidics in cell migration research, particularly for cancer cell migration. The microfluidic devices provide both mechanical and 3D environments, as well as complicated chemical environment where chemotaxis is guided by concentration gradients of growth factors, chemokines, and surface ligands. These studies have enhanced our understanding of cell deformation and focal adhesion dynamics as well as their responses to subtle variations in their microenvironment. We hope that this review will serve as a guide for both biologists and engineers as they further develop the microfluidic toolbox toward applications in cancer research, especially in cancer stem cell research.
ACKNOWLEDGMENTS
This work was supported, in part, by NIH Grant No. R01NS38118 (D. Sun) and NIH Grant No. P30 HD03352 (Waisman Center). J.S.K. was partially supported by the HEADRUSH Brain Tumor Research Professorship, Roger Loff Memorial GBM Research Fund, and the UW Foundation∕Neurosurgery Brain Tumor Research and Education Fund. J.C.W. and Y.H. are partially supported by NIH Grant No. NIBIB 1R01EB009103-01.
References
- Pearce T. M. and Williams J. C., Lab Chip 7, 30 (2007). 10.1039/b612856b [DOI] [PubMed] [Google Scholar]
- Whitesides G. M., Nature (London) 442, 368 (2006). 10.1038/nature05058 [DOI] [PubMed] [Google Scholar]
- Forster E. M., Integrative Biology 1, 13 (2009). 10.1039/b819961m [DOI] [Google Scholar]
- Chao T. C. and Ros A., J. R. Soc., Interface 5, S139 (2008). 10.1098/rsif.2008.0233.focus [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Thomas Parsons J., and Rick Horwitz A., Science 302, 1704 (2003). 10.1126/science.1092053 [DOI] [PubMed] [Google Scholar]
- Sahai E., Curr. Opin. Genet. Dev. 15, 87 (2005). 10.1016/j.gde.2004.12.002 [DOI] [PubMed] [Google Scholar]
- Tournaviti S., Hannemann S., Terjung S., Kitzing T. M., Stegmayer C., Ritzerfeld J., Walther P., Grosse R., Nickel W., and Fackler O. T., J. Cell Sci. 120, 3820 (2007) 10.1242/jcs.011130 [DOI] [PubMed] [Google Scholar]; Sahai E. and Marshall C. J., Nat. Cell Biol. 5, 711 (2003). 10.1038/ncb1019 [DOI] [PubMed] [Google Scholar]
- Gutjahr M. C., Rossy J., and Niggli V., Exp. Cell Res. 308, 422 (2005). 10.1016/j.yexcr.2005.05.001 [DOI] [PubMed] [Google Scholar]
- Lämmermann T. and Sixt M., Curr. Opin. Cell Biol. 21, 636 (2009). 10.1016/j.ceb.2009.05.003 [DOI] [PubMed] [Google Scholar]
- Paluch E., Piel M., Prost J., Bornens M., and Sykes C., Biophys. J. 89, 724 (2005). 10.1529/biophysj.105.060590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham C. C., J. Cell Biol. 129, 1589 (1995). 10.1083/jcb.129.6.1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova T. and Segall J. E., Cell Adhesion & Migration 1, 165 (2007). 10.4161/cam.1.4.5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Mazo I., Leung H., Engelke K., von Andrian U. H., Deryugina E. I., Strongin A. Y., Bröcker E. -B., and Friedl P., J. Cell Biol. 160, 267 (2003). 10.1083/jcb.200209006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen J. J. and Rajasekaran A. K., Cancer Res. 66, 8319 (2006). 10.1158/0008-5472.CAN-06-0410 [DOI] [PubMed] [Google Scholar]
- Rørth P., Annu. Rev. Cell Dev. Biol. 25, 407 (2009). 10.1146/annurev.cellbio.042308.113231 [DOI] [PubMed] [Google Scholar]
- Radisky D. C., J. Cell Sci. 118, 4325 (2005). 10.1242/jcs.02552 [DOI] [PubMed] [Google Scholar]
- Condeelis J. and Segall J. E., Nat. Rev. Cancer 3, 921 (2003). 10.1038/nrc1231 [DOI] [PubMed] [Google Scholar]
- Karnoub A. E., Dash A. B., Vo A. P., Sullivan A., Brooks M. W., Bell G. W., Richardson A. L., Polyak K., Tubo R., and Weinberg R. A., Nature (London) 449, 557 (2007). 10.1038/nature06188 [DOI] [PubMed] [Google Scholar]
- Soon L., Mouneimne G., Segall J., Wyckoff J., and Condeelis J., Cell Motil. Cytoskeleton 62, 27 (2005) 10.1002/cm.20082 [DOI] [PubMed] [Google Scholar]; Bryan H. and Paul K., Sci. STKE 2003, l5. [Google Scholar]
- Xia Y. N. and Whitesides G. M., Angew. Chem., Int. Ed. 37, 550 (1998). [DOI] [PubMed] [Google Scholar]
- Qin D., Xia Y. N., and Whitesides G. M., Nat. Protoc. 5, 491 (2010). 10.1038/nprot.2009.234 [DOI] [PubMed] [Google Scholar]
- Taylor A. M., Rhee S. W., Tu C. H., Cribbs D. H., Cotman C. W., and Jeon N. L., Langmuir 19, 1551 (2003). 10.1021/la026417v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guck J., Lautenschläger F., Paschke S., and Beil M., Integrative Biology (Critical review) 2, 575 (2010). 10.1039/c0ib00050g [DOI] [PubMed] [Google Scholar]
- Irimia D. and Toner M., Integrative Biology 1, 506 (2009). 10.1039/b908595e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Wu Y. I., Liu Y., Geiger J., Tam E., Overall C., Sharon Stack M., and Friedl P., Nat. Cell Biol. 9, 893 (2007). 10.1038/ncb1616 [DOI] [PubMed] [Google Scholar]
- Beil M., Micoulet A., von Wichert G., Paschke S., Walther P., Bishr Omary M., Van Veldhoven P. P., Gern U., Wolff-Hieber E., Eggermann J., Waltenberger J., Adler G., Spatz J., and Seufferlein T., Nat. Cell Biol. 5, 803 (2003). 10.1038/ncb1037 [DOI] [PubMed] [Google Scholar]
- Rolli C. G., Seufferlein T., Kemkemer R., and Spatz J. P., PLoS ONE 5, e8726 (2010). 10.1371/journal.pone.0008726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lämmermann T., Bader B. L., Monkley S. J., Worbs T., Wedlich-Söldner R., Hirsch K., Keller M., Förster R., Critchley D. R., Fässler R., and Sixt M., Nature (London) 453, 51 (2008). 10.1038/nature06887 [DOI] [PubMed] [Google Scholar]
- Hawkins R. J., Piel M., Faure-Andre G., Lennon-Dumenil A. M., Joanny J. F., Prost J., and Voituriez R., Phys. Rev. Lett. 102, 058103 (2009). 10.1103/PhysRevLett.102.058103 [DOI] [PubMed] [Google Scholar]
- Niggemann B., T. L.DrellIV, Joseph J., Weidt C., Lang K., Zaenker K. S., and Entschladen F., Exp. Cell Res. 298, 178 (2004). 10.1016/j.yexcr.2004.04.001 [DOI] [PubMed] [Google Scholar]
- Provenzano P. P., Inman D. R., Eliceiri K. W., Trier S. M., and Keely P. J., Biophys. J. 95, 5374 (2008). 10.1529/biophysj.108.133116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung K. E., Su G., Pehlke C., Trier S. M., Eliceiri K. W., Keely P. J., Friedl A., and Beebe D. J., Biomaterials 30, 4833 (2009). 10.1016/j.biomaterials.2009.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J. L., Tien J., Pirone D. M., Gray D. S., Bhadriraju K., and Chen C. S., Proc. Natl. Acad. Sci. U.S.A. 100, 1484 (2003). 10.1073/pnas.0235407100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoen I., Hu W., Klotzsch E., and Vogel V., Nano Lett. 10, 1823 (2010). 10.1021/nl100533c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A., Ghibaudo M., Buguin A., Silberzan P., and Ladoux B., Proc. Natl. Acad. Sci. U.S.A. 104, 8281 (2007) 10.1073/pnas.0702259104 [DOI] [PMC free article] [PubMed] [Google Scholar]; Rabodzey A., Alcaide P., Luscinskas F. W., and Ladoux B., Biophys. J. 95, 1428 (2008) 10.1529/biophysj.107.119156 [DOI] [PMC free article] [PubMed] [Google Scholar]; Moon M. W., Cha T. G., Lee K. R., Vaziri A., and Kim H. -Y., Soft Matter 6, 3924 (2010) 10.1039/c0sm00126k [DOI] [Google Scholar]; Lemmon C. A., Chen C. S., and Romer L. H., Biophys. J. 96, 729 (2009) 10.1016/j.bpj.2008.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]; Greiner C., del Campo A., and Arzt E., Langmuir 23, 3495 (2007) 10.1021/la0633987 [DOI] [PubMed] [Google Scholar]; Cesa C. M., Kirchgeßner N., Mayer D., Schwarz U. S., Hoffmann B., and Merkel R., Rev. Sci. Instrum. 78, 034301 (2007). 10.1063/1.2712870 [DOI] [PubMed] [Google Scholar]
- Li Z., Song J. H., Mantini G., Lu M. -Y., Fang H., Falconi C., Chen L. -J., and Wang Z. L., Nano Lett. 9, 3575 (2009). 10.1021/nl901774m [DOI] [PubMed] [Google Scholar]
- Sniadecki N. J., Anguelouch A., Yang M. T., Lamb C. M., Liu Z., Kirschner S. B., Liu Y., Reich D. H., and Chen C. S., Proc. Natl. Acad. Sci. U.S.A. 104, 14553 (2007). 10.1073/pnas.0611613104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan T. M. and Folch A., Lab Chip 8, 34 (2008). 10.1039/b711887b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. S., Lee S. H., Kwon T. H., and Ahn C. H., Lab Chip 5, 739 (2005) 10.1039/b418314b [DOI] [PubMed] [Google Scholar]; Bhagat A. A. S., Peterson E. T. K., and Papautsky I., J. Micromech. Microeng. 17, 1017 (2007). 10.1088/0960-1317/17/5/023 [DOI] [Google Scholar]
- Jeon N. L., Baskaran H., Dertinger S. K. W., Whitesides G. M., van de Water L., and Toner M., Nat. Biotechnol. 20, 826 (2002). [DOI] [PubMed] [Google Scholar]
- Kricka L. J., Nozaki O., Heyner S., Garside W. T., and Wilding P., Clin. Chem. 39, 1944 (1993). [PubMed] [Google Scholar]
- Lin F., Saadi W., Rhee S. W., Wang S. -J., Mittal S., and Jeon N. L., Lab Chip 4, 164 (2004). 10.1039/b313600k [DOI] [PubMed] [Google Scholar]
- Chung B. G., Park J. W., Hu J. S., Huang C., Monuki E. S., and Jeon N. L., BMC Biotechnol. 7, 60 (2007). 10.1186/1472-6750-7-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. J., Saadi W., Lin F., Minh-Canh Nguyen C., and Jeon N. L., Exp. Cell Res. 300, 180 (2004). 10.1016/j.yexcr.2004.06.030 [DOI] [PubMed] [Google Scholar]
- Mosadegh B., Saadi W., Wang S. J., and Jeon N. L., Biotechnol. Bioeng. 100, 1205 (2008). 10.1002/bit.21851 [DOI] [PubMed] [Google Scholar]
- Lawler K., O’Sullivan G., Long A., and Kenny D., Cancer Sci. 100, 1082 (2009). 10.1111/j.1349-7006.2009.01160.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia D., Charras G., Agrawal N., Mitchison T., and Toner M., Lab Chip 7, 1783 (2007). 10.1039/b710524j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fok S., Domachuk P., Rosengarten G., Krause N., Braet F., Eggleton B. J., and Soon L. L., Biophys. J. 95, 1523 (2008). 10.1529/biophysj.107.115246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T. H., Yen M. H., Liao W. Y., Cheng J. -Y., and Lee C. -H., Lab Chip 9, 884 (2009). 10.1039/b814405b [DOI] [PubMed] [Google Scholar]
- Haessler U., Kalinin Y., Swartz M. A., and Wu M., Biomed. Microdevices 11, 827 (2009). 10.1007/s10544-009-9299-3 [DOI] [PubMed] [Google Scholar]
- Abhyankar V. V., Toepke M. W., Cortesio C. L., Lokuta M. A., Huttenlocher A., and Beebe D. J., Lab Chip 8, 1507 (2008). 10.1039/b803533d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadi W., Rhee S. W., Lin F., Vahidi B., Chung B. G., and Jeon N. L., Biomed. Microdevices 9, 627 (2007). 10.1007/s10544-007-9051-9 [DOI] [PubMed] [Google Scholar]
- Mosadegh B., Huang C., Park J. W., Shin H. S., Chung B. G., Hwang S. -K., Lee K. -H., Kim H. J., Brody J., and Jeon N. L., Langmuir 23, 10910 (2007). 10.1021/la7026835 [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Choi C. K., and Horwitz A. R., J. Cell Sci. 122, 1473 (2009) 10.1242/jcs.052894 [DOI] [PMC free article] [PubMed] [Google Scholar]; Hood J. D. and Cheresh D. A., Nat. Rev. Cancer 2, 91 (2002). 10.1038/nrc727 [DOI] [PubMed] [Google Scholar]
- Chicurel M., Science 295, 606 (2002) 10.1126/science.295.5555.606 [DOI] [PubMed] [Google Scholar]; Smith J. T., Elkin J. T., and Reichert W. M., Exp. Cell Res. 312, 2424 (2006). 10.1016/j.yexcr.2006.04.005 [DOI] [PubMed] [Google Scholar]
- Brandley B. K. and Schnaar R. L., Dev. Biol. 135, 74 (1989). 10.1016/0012-1606(89)90159-0 [DOI] [PubMed] [Google Scholar]
- Pulsipher A. and Yousaf M. N., ChemBioChem 11, 745 (2010). 10.1002/cbic.200900787 [DOI] [PubMed] [Google Scholar]
- Théry M., Pépin A., Dressaire E., Chen Y., and Bornens M., Cell Motil. Cytoskeleton 63, 341 (2006) 10.1002/cm.20126 [DOI] [PubMed] [Google Scholar]; Brock A., Chang E., Ho C. C., LeDuc P., Jiang X., Whitesides G. M., and Ingber D. E., Langmuir 19, 1611 (2003). 10.1021/la026394k [DOI] [PubMed] [Google Scholar]
- Théry M., Racine V., Piel M., Pépin A., Dimitrov A., Chen Y., Sibarita J. -B., and Bornens M., Proc. Natl. Acad. Sci. U.S.A. 103, 19771 (2006). 10.1073/pnas.0609267103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar G., Ho C. C., and Co C. C., Adv. Mater. (Weinheim, Ger.) 19, 1084 (2007). 10.1002/adma.200601629 [DOI] [Google Scholar]
- Jiang X. Y., Bruzewicz D. A., Wong A. P., Piel M., and Whitesides G. M., Proc. Natl. Acad. Sci. U.S.A. 102, 975 (2005). 10.1073/pnas.0408954102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmud G., Campbell C. J., Bishop K. J. M., Komarova Y. A., Chaga O., Soh S., Huda S., Kandere-Grzybowska K., and Grzybowski B. A., Nat. Phys. 5, 606 (2009). 10.1038/nphys1306 [DOI] [Google Scholar]
- Xia N., Thodeti C. K., Hunt T. P., Xu Q., Ho M., Whitesides G. M., Westervelt R., and Ingber D. E., FASEB J. 22, 1649 (2008). 10.1096/fj.07-090571 [DOI] [PubMed] [Google Scholar]
- von Philipsborn A. C., Lang S., Bernard A., Loeschinger J., David C., Lehnert D., Bastmeyer M., and Bonhoeffer F., Nat. Protoc. 1, 1322 (2006). 10.1038/nprot.2006.251 [DOI] [PubMed] [Google Scholar]
- Hjortø G. M., Hansen M., Larsen N. B., and Kledal T. N., Biomaterials 30, 5305 (2009). 10.1016/j.biomaterials.2009.06.018 [DOI] [PubMed] [Google Scholar]
- Autenrieth T. and Bastmeyer M., presented at the Proceedings of the 33rd Annual Meeting of the German-Society-for-Cell-Biology/German Melanoma Research Network, University of Regensburg, Regensburg, Germany, 2010.
- Clarke M. F., Dick J. E., Dirks P. B., Eaves C. J., Jamieson C. H. M., Jones D. Leanne, Visvader J., Weissman I. L., and Wahl G. M., Cancer Res. 66, 9339 (2006). 10.1158/0008-5472.CAN-06-3126 [DOI] [PubMed] [Google Scholar]
- Stupp R., Mason W. P., van den Bent M. J., Weller M., Fisher B., Taphoorn M. J. B., Belanger K., Brandes A. A., Marosi C., Bogdahn U., Curschmann J., Janzer R. C., Ludwin S. K., Gorlia T., Allgeier A., Lacombe D., Cairncross J. G., Eisenhauer E., Mirimanoff R. O. et al. ,N. Engl. J. Med. 352, 987 (2005). 10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- Robert M. C. and Wastie M. L., Biomed Imaging Interv J 4, e3 (2008) 10.2349/biij.4.1.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]; Newton H. B., Rosenblum M. K., and Walker R. W., Cancer 69, 2149 (1992). [DOI] [PubMed] [Google Scholar]
- Shackleton M., Quintana E., Fearon E. R., and Morrison S. J., Cell 138, 822 (2009). 10.1016/j.cell.2009.08.017 [DOI] [PubMed] [Google Scholar]
- Bao S. D., Wu Q. L., McLendon R. E., Hao Y., Shi Q., Hjelmeland A. B., Dewhirst M. W., Bigner D. D., and Rich J. N., Nature (London) 444, 756 (2006) 10.1038/nature05236 [DOI] [PubMed] [Google Scholar]; Piccirillo S. G. M., Reynolds B. A., Zanetti N., Lamorte G., Binda E., Broggi G., Brem H., Olivi A., Dimeco F., and Vescovi A. L., Nature (London) 444, 761 (2006). 10.1038/nature05349 [DOI] [PubMed] [Google Scholar]
- Lapidot T., Sirard C., Vormoor J., Murdoch B., Hoang T., Caceres-Cortes J., Minden M., Paterson B., Caligiuri M. A., and Dick J. E., Nature (London) 367, 645 (1994) 10.1038/367645a0 [DOI] [PubMed] [Google Scholar]; Lee J., Kotliarova S., Kotliarov Y., Li A., Su Q., Donin N. M., Pastorino S., Purow B. W., Christopher N., Zhang W., Park J. K., and Fine H. A., Cancer Cells 9, 391 (2006). 10.1016/j.ccr.2006.03.030 [DOI] [PubMed] [Google Scholar]
- Clark P. A., Treisman D. M., Ebben J., and Kuo J. S., Dev. Dyn. 236, 3297 (2007). 10.1002/dvdy.21381 [DOI] [PubMed] [Google Scholar]
- Agrawal B., Huang Y., Williams J., and Kuo J., presented at the Neurosurgical Forum, San Francisco, CA, 2010.
- Keenan T. M., Frevert C. W., Wu A., Wong V., and Folch A., Lab Chip 10, 116 (2010). 10.1039/b913494h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamloo A., Ma N., Poo M. -m., Sohn L. L., and Heilshorn S. C., Lab Chip 8, 1292 (2008). 10.1039/b719788h [DOI] [PubMed] [Google Scholar]
- Liu T., Li C., Li H., Zeng S., Qin J., and Lin B., Electrophoresis 30, 4285 (2009). 10.1002/elps.200900289 [DOI] [PubMed] [Google Scholar]