Abstract
Tumors express a wide variety of both mutated and nonmutated Ags. Whether these tumor Ags are broadly recognized as self or foreign by the immune system is currently unclear. Using an autochthonous prostate cancer model in which hemagglutinin (HA) is specifically expressed in the tumor (ProHA × TRAMP mice), as well as an analogous model wherein HA is expressed in normal tissues as a model self-Ag (C3HAhigh), we examined the transcriptional profile of CD4 T cells undergoing Ag-specific division. Consistent with our previous data, transfer of Ag-specific CD4 T cells into C3HAhigh resulted in a functionally inactivated CD4 T cell profile. Conversely, adoptive transfer of an identical CD4 T cell population into ProHA × TRAMP mice resulted in the induction of a regulatory phenotype of the T cell (Treg) both at the transcriptional and functional level. Interestingly, this Treg skewing was a property of even early-stage tumors, suggesting Treg induction as an important tolerance mechanism during tumor development.
The development of tumors in immunocompetent hosts is generally thought to be accompanied by the subversion of an effective antitumor immune response (1–4). This process, known as immune evasion, is multifactorial and includes alterations in Ag processing, Ag presentation, expression of immune-inhibitory molecules such as TGF-β and IDO, as well as the expression of cell surface molecules such as B7-H1 that inhibit immune function (2–8). As a consequence of these and other mechanisms, the T cell response to tumors is correspondingly blunted or absent, as evidenced by a lack of lytic function in CD8 T cells and a lack of effector cytokine production by specific CD4 T cells (9–17).
To better understand the nature of this functional CD4 T cell tolerance, we examined the transcriptional profile of CD4 T cells that recognize nonmutated tumor Ag, using both self-Ag and viral Ag recognition for comparison. For these studies, we used a well-described adoptive transfer system in which TCR-transgenic T cells specific for hemagglutinin (HA)4 are introduced into mice expressing their cognate Ag as a self-Ag or as tumor-restricted Ag (14, 18–21). In the self-Ag system, the C3 promoter drives HA expression in the lung, as well as in other normal tissues (19). In the tumor system, prostate-specific expression of HA is driven by the minimal rat probasin promoter (14, 22). These mice are termed ProHA (Probasin HA). For a tumor model, we utilized the transgenic adenocarcinoma of the mouse prostate (TRAMP) model, which develops autochthonous prostate tumors in a progressive manner (22, 23). By intercrossing ProHA with TRAMP mice, we created a murine system in which a well-defined tumor Ag is expressed in a tumor- and tissue-specific manner (14). In both ProHA × TRAMP and C3HAhigh mice, CD4 T cell recognition of cognate Ag is accompanied by division as well as by functional tolerance as evidenced by a lack of effector cytokine production (20, 24). Our data, involving transcriptional profiling of CD4 T cells isolated from these distinct models, supports the notion that CD4 T cell recognition in the context of an evolving prostate tumor results in a phenotype different from that induced by self-Ag recognition, one characterized by a relative up-regulation of the transcription factor FoxP3 and the development of a regulatory T cell phenotype.
These data are consistent with previously published studies suggesting that tumors may specifically expand or induce regulatory T cells and that multiple tumor types are infiltrated with regulatory T cells (Treg) in humans with cancer (25–32). In addition, our data suggest that these cells represent induced as opposed to natural Tregs, which arise spontaneously in the thymus (33–38). Because tumors in TRAMP mice evolve slowly over time, we were also able to examine the relative effect of tumor stage on Treg induction/expansion by evaluating the results of adoptive transfer to younger vs older animals (39). Surprisingly, Treg induction/expansion was a characteristic of even very early precancerous lesions (prostatic intraepithelial neoplasia, or PIN), suggesting that Treg induction by evolving tumors might be a relatively early contributor to T cell tolerance (40).
Materials and Methods
Mice
All mice were on the B10.D2 (H-2d) background. C3-HA-transgenic mice express influenza HA under the control of the C3 promoter and have been previously described (19, 20). The strain of 6.5 mice are CD4 TCR-transgenic animals that recognize an I-Ed-restricted HA epitope (110SFERFEIF-PKE120; Ref. 18). These mice were backcrossed to a Thy1.1-congenic B10.D2 background for >12 generations. TRAMP mice on the C57BL/6J background were backcrossed to the B10.D2 background for >14 generations (22). ProHA mice express HA in a prostate-restricted manner under the control of the same minimal rat probasin promoter used to generate TRAMP mice and have been previously described (14). ProHA × TRAMP mice were generated by backcrossing ProHA to TRAMP mice for >16 generations (14). Animal care and experimental procedures were performed under pathogen-free conditions in accordance with established institutional protocols from Institutional Animal Care and Use Committees of Johns Hopkins University.
Adoptive transfer
Donor TCR-transgenic (6.5) mice were sacrificed via CO2 asphyxiation. Spleens and lymph nodes were collected and homogenized, and RBCs were lysed. CD4 T cells were purified using Miltenyi magnetically labeled beads according to the manufacturer’s protocol. For some experiments, purified cells were labeled with for 8 min with CFSE (Invitrogen) by adding 0.5 μl of 5 mM stock per 1 ml of cells. After labeling, cells were washed twice and resuspended in HBSS for i.v. injections. Purified cells (5 × 106) were injected per mouse in 0.2 ml total volume by tail vein injection. For CD4 T cell activation controls, nontransgenic B10.D2 mice were infected with recombinant Vaccinia virus expressing wild-type HA protein as previously described (14).
Flow cytometry/intracellular staining
Lymph nodes were harvested 3–10 days post-adoptive transfer, and single-cell suspensions were prepared. RBCs were lysed with ammonium chloride-potassium lysis buffer. All staining reagents were purchased from Pharmingen, with the exception of FoxP3, which was analyzed using a prepared kit according to the manufacturer’s instructions (Ebiosciences). After 10 min of incubation, samples were washed once in PBS plus 1% FBS solution and analyzed using a FACSCalibur instrument (BD Biosciences). Intracellular cytokine analysis was performed as previously described (41). Data were analyzed using the FloJo software package (Treestar).
Direct ex vivo Ag detection
A direct ex vivo Ag detection assay was performed as previously described (42, 43). Briefly, HA-specific memory cells were generated by adoptive transfer of cells into nontransgenic B10.D2 recipients that received primary vaccination of Vaccinia-hemagglutinin (VaccHA). Two weeks after initial transfer; B10.D2 recipient received a secondary boost of Listeria monocytogenes-HA (LMHA). Two weeks after LMHA boost, all lymph nodes and spleena were harvested from host mice, and responder CD4 T cells were enriched using anti-Thy1.1-biotin Ab and streptavidin microbeads (Miltenyi Biotech). class IIhighCD11chigh dendritic cells (DC) were enriched from the iliac lymph nodes from ProHA × TRAMP, C3HAhigh, and VaccHA-vaccinated B10.D2 animals as stimulators. Responder CD4 T cells (2 × 104) were incubated with 1 × 103 stimulators, resulting in a 20:1 ratio of responder to stimulator. Reactions were set up in triplicates. Forty-eight hours later, cultures were pulsed with 1 μCi of [3H]TdR and incubated for an additional 24 h before harvest with a Packard Micromate cell harvester. Determination of the amount of incorporated radioactive counts was performed with a Packard Matrix 96 direct beta counter (Packard Biosciences).
Donor CD4 T cell isolation
From 3 to 7 days post-adoptive transfer of HA-specific CD4 T cells, recipient animals were euthanized, and lymph nodes were harvested. CD4+ T cells were enriched by depleting CD8 and B cells using biotinylated anti-CD8, anti-B220, and MACS LS separation columns (Miltenyi Biotech) as previously described (21). Ag-specific T cells that had undergone specific division in vivo were sorted using a FACSVantage SE cell sorter (BD Biosciences), gating on CFSE-diluted, CD4+Thy1.1+ cells. As all recipient animals are of the Thy1.2 phenotype, this technique results in >95% pure donor cells and avoids the use of TCR-specific or CD4 core-ceptor-specific Abs that could potentially alter TCR- or CD4-dependent gene expression patterns. Control naive T cells were isolated in a similar manner, using nontransgenic B10.D2 animals as recipients, but instead of gating on CFSElow cells, the undivided, CFSEhigh cells were isolated.
Transcriptional analysis
Sorted cells were pelleted, frozen under 1 ml of Trizol, and stored at −80°C before RNA extraction using the Trizol reagent. Our microarray experiment was controlled using paired biological replicates. For these experiments, an identical population of CD4 T cells was adoptively transferred to the two groups of mice at a time. Then, cells were flow-sorted from each group of >10 mice separately, and further processing was performed in parallel; i.e., sorting, RNA extraction, template preparation, and analysis were done in parallel, using two separate Affymetrix chips. The integrity of extracted RNA from T cells was analyzed using an Agilent 2100 bioanalyzer and the RNA 6000 Pico and Nano Kits (Agilent Technologies), and concentrations were determined using a NanoDrop spectrophotometer (NanoDrop Technologies). Transcriptional analysis was performed at the Johns Hopkins Microarray Core facility. Per the standard protocol, RNA was amplified from 20 ng of starting total RNA with the Nugen Ovation RNA Amplification System V2, following the manufacturer’s protocol (http://www.nugeninc.com/pdfs/ov-v2_userguide.pdf). cDNA was synthesized using the Nugen FL-Ovation cDNA Biotin Module V2 kit, following the manufacturer’s protocol (http://www.nugeninc.com/pdfs/flbv2_userguide.pdf). After standard labeling, each sample was hybridized to an Affymetrix Mouse 430 Plus2 expression array, followed by interrogation with an Affymetrix GeneChip Scanner 3000. RMA analysis of paired replicates revealed a striking concordance, with fewer than 300 of 36,000 transcripts different at the p < 0.05 level.
Statistical analysis
To estimate the gene expression signals, data analysis was conducted on the CEL file probe signal values of the chip at the Affymetrix probe pair (perfect match probe and mismatch probe) level, using the statistical algorithm robust multiarray average expression measure (44) with Affy. This probe level data processing includes a normalization procedure using a quantile normalization method to reduce the obscuring variation between microarrays, which might be introduced during the processes of sample preparation, manufacture, fluorescence labeling, hybridization, and/or scanning (45). Using the signal intensities estimated above, an empirical bias method with the γ- γ modeling, as implemented in the bioconductor package EBarrays, was used to estimate the posterior probabilities of the differential expression of genes between the sample conditions. The criterion of the posterior probability >0.5, which means the posterior odds favoring change, was used to produce a differentially expressed gene list. Heatmaps were created using TreeView package version 1.60 (46).
Quantitative PCR
Quantitative PCR was performed as previously described (21). Briefly, RNA was immediately extracted from sorted 6.5 CD4+ T cells using the Trizol reagent (Invitrogen). Reverse transcription was performed with the Superscript First Strand Synthesis System (Invitrogen). cDNA levels were analyzed by real-time quantitative PCR with the TaqMan system (Applied Biosystems). Each sample was assayed in triplicates for the target gene together with 18S rRNA as the internal reference in a 25-μl final reaction volume, using the TaqMan Universal PCR Master Mix and the ABI Prism 7700 Sequence Detection system. The relative mRNA frequency was determined by normalization to the internal control 18S RNA. Relative mRNA frequencies were calculated as 2ΔΔCt, where ΔΔCt = (ΔCt calibration − ΔCt sample).
Suppression assay
In vitro suppression assays were performed as previously described (34). Briefly, 1 × 104 purified T cells (responders) were mixed with 1 × 103 6.5 CD4+ T cells sorted from various recipients (suppressors), giving a 1:10 suppressor-to-responder ratio. Cells were then incubated in flat-bottom 96-well tissue culture plates precoated with 5 μg/ml anti-CD3e in 200 μl of CTL media. After 72 h, cultures were pulsed with 1 μCi of [3H]TdR and incubated for an additional 16 h before harvest with a Packard Micromate cell harvester. Determination of the amount of incorporated radioactive counts was performed with a Packard Matrix 96 direct beta counter (Packard Biosciences).
Immunohistochemistry
For immunohistochemistry, dorsal lobes of the prostate were harvested from a 6-, 10-, and 24-wk-old ProHA × TRAMP animals and frozen in Sakura Tissue-tek OCT compound (Accurate Chemical and Scientific) as described by the manufacturer. Serial unstained sections from the dorsal prostate lobes were then cut. Unstained slides were fixed in 75% acetone, 25% ethanol for 5 min. Dry slides were washed three times in PBS, followed by a 30-min incubation with Image-iT FX signal enhancer (Molecular Probes). Slides were washed in PBS, blocked with serum, washed, and incubated with polyclonal rabbit Ki67 (Abcam) for 45 min. Slides were washed and incubated with anti-rabbit IgG HRP for 30 min. Diaminobenzidine was used as the chromogen. Slides were then imaged on a microscope.
In vivo Ab blocking
For in vivo TGF-β blocking experiments, ProHA × TRAMP and control B10.d2 mice were injected with 0.2 mg of anti-TGF-β (2G7 or 1D11) Ab (R&D Systems) or the control IgG (Jackson ImmunoResearch Laboratories) at the time of adoptive transfer of naive transgenic T cells with a second dose administered 3 days later. As above, mice were harvested 5–10 days post-adoptive T cell transfer, and T cells were analyzed as described above.
Results
Specific CD4 T cell proliferation to self, viral, and tumor Ag
Previously, we demonstrated that adoptive transfer of HA-specific CD4 clonotypic cells into C3HAhigh-transgenic mice, in which there is widespread expression of the HA Ag (self Ag model), generates functionally tolerized T cells that are prone to deletion (19, 20). These CD4 T cells are characterized by a lack of IFN-γ or IL-2 secretion upon restimulation ex vivo. Similarly, adoptive transfer of identical HA-specific CD4 T cells to animals in which HA is expressed as a prostate/prostate cancer-restricted Ag also results in functional tolerance (14). To assess whether the proliferative response to a self Ag or tumor Ag was broadly comparable in these two models, naive HA-specific CD4 T cells were CFSE labeled and transferred into the following recipients: 1) no Ag (B10.D2); 2) viral Ag (VaccHA); 3) self Ag (C3HAhigh); and 4) prostate tissue/tumor Ag (ProHA × TRAMP; Fig. 1A). Adoptively transferred HA-specific CD4 T cells proliferated robustly in response to recognition of self or viral Ag. However, proliferation in response to the prostate/prostate cancer-restricted Ag was less robust and was primarily restricted to the prostate-draining (iliac) lymph nodes (14). Additionally, proliferation in single-transgenic ProHA mice is barely detectable under these conditions (data not shown), consistent with the notion that tumorigenesis is required for Ag recognition in this model (14).
FIGURE 1.
Ag-specific proliferation of HA-specific CD4 T cells. A, CFSE-labeled HA-specific Thy1.1+CD4+ T cells were adoptively transferred into recipients presenting HA Ag in varying contexts; nontransgenic, no Ag; VaccHA, viral Ag; C3HAhigh, self Ag; and ProHA × TRAMP, tumor Ag. Flow cytometric analysis of Ag-driven CFSE dilution gated on donor (Thy1.1+) CD4+ T cells. B, Ag presentation assayed using a direct ex vivo ag detection assay. HA-specific memory CD4 T cells generated from VaccHA-vaccinated recipient mice were incubated with CD11c+-enriched DCs from the iliac lymph nodes of the indicated groups. *, p < 0.05 by two-sided Student’s t test; **, p < 0.01.
We next sought to determine whether DCs were capable of driving HA-specific CD4 proliferation by performing a direct ex vivo Ag detection (DEAD) assay (42, 43). Memory HA-specific CD4 T cells were generated by adoptively transferring cells into B10.D2 (nontransgenic) hosts with a primary immunization of VaccHA and boosted with LMHA. Four weeks after initial transfer, memory CD4 T cells were enriched from vaccinated hosts using Thy1.1 microbeads and incubated with CD11chigh DCs isolated from the lymph nodes of various experimental animals. As a positive control, DCs were isolated from the lymph nodes of VaccHA-infected nontransgenic mice. DCs isolated from single-transgenic ProHA and TRAMP (without HA) mice were used as negative controls given that the proliferation of HA-specific clonotypic cells is minimal following adoptive transfer. DCs from the lymph nodes of both tumor- and self Ag-bearing animals were capable of driving significant (p < 0.05) proliferation of memory HA-specific CD4+ T cells, confirming similar reports that HA is cross-presented in the C3HAhigh self Ag model and demonstrating DC as a likely APC in the ProHA × TRAMP model (Refs. 19 and 47 and Fig. 1B).
Transcriptional analysis of CD4 T cells specific for tumor or self Ag
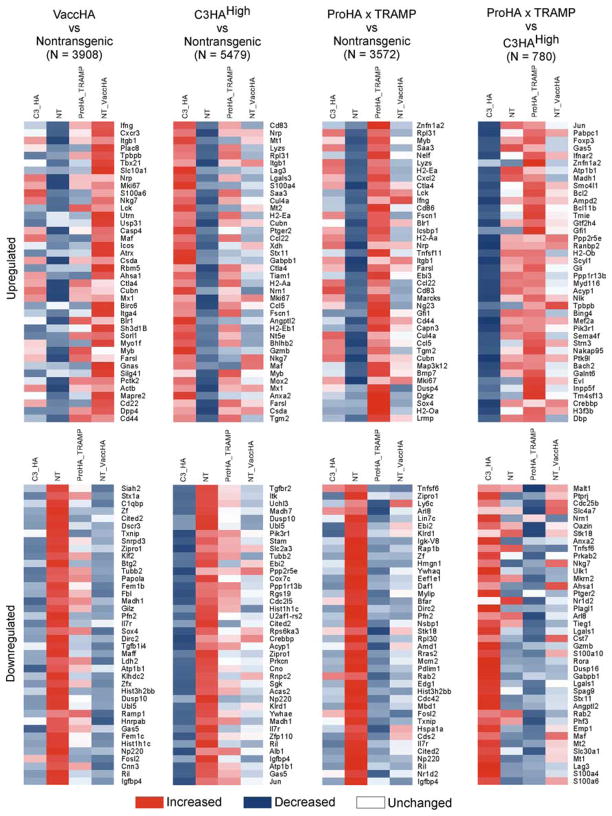
We next sought to determine whether these various Ag-specific proliferative stimuli would result in distinct transcriptional profiles in vivo. Naive, CFSE-labeled, Thy1.1+ HA-specific CD4 T cells were adoptively transferred into nontransgenic (no Ag), VaccHA-vaccinated (viral Ag), C3HAhigh (self-Ag), and ProHA × TRAMP (tumor/tissue Ag) recipients. As shown in Fig. 1A, Thy1.1+ CFSE diluted clonotypic cells were FACS sorted from VaccHA, C3HAhigh, and ProHA × TRAMP recipients, whereas CFSE undiluted cells from nontrangenic recipients were sorted as a negative control for the adoptive transfer and isolation procedures. Transcriptional profiles were determined using the Affymetrix 430A GeneChip microarray representing 39,700 transcripts and analyzed using the statistical criteria previously described (48). Compared with naive cells isolated from nontransgenic controls, microarray analysis revealed 3908 transcripts differentially expressed during viral Ag recognition, 5479 transcripts in self Ag recognition, and 3572 differentially expressed transcripts in tumor Ag recognition (Fig. 2 and supplemental Tables I–III).5 Only 780 unique transcripts were significantly differentially expressed between CFSE diluted tumor vs self Ag recognizing CD4 T cells. A curated list of the top 40 up-regulated (Fig. 2A) and down-regulated (Fig. 2B) genes involved in tumor vs self Ag recognition is presented in Tables I and II. These genes represent a broad array of molecules, including transcription factors such as FoxP3, Madh1, and Znfn1a2; anti-apoptosis molecules such as Bcl2 and Bcl-11b; signaling molecules; and cell surface markers. A similar comparison revealed 1930 transcripts that were differentially expressed in tumor Ag-recognizing CD4 T cells as compared with viral-associated Ag-recognizing T cells (Table III and IV). Overall, these data appear to be consistent with previously published data from our group and others (21, 37, 49, 50). Among many others, we noted the expected up-regulation of IFN-γ in CD4 T cells recognizing HA in the context of viral infection, and a relative up-regulation of the T cell surface protein LAG-3 in the self-tolerance (C3HAhigh) model (21).
FIGURE 2.
Gene expression profiling of Ag-specific CD4 T cells. Gene expression profiling was performed on pooled FACS-sorted, CFSE-diluted, HA-specific Thy1.1+CD4+ T cells from the lymph nodes of no Ag (nontransgenic; NT), viral Ag (VaccHA), self Ag (C3HAhigh), or tumor (ProHA × TRAMP) Ag recognition models using Affymetrix Mouse 430 Plus2. Differentially expressed genes were identified by comparing the expression profile of the indicated group to the control group in a pairwise analysis. The top 40 up-regulated (fold change >4) or down-regulated (fold change <–2) genes were selected from each comparison pair to generate the heatmap. N, Number of genes statistically different between indicated groups.
Table I.
Genes differentially up-regulated by CD4 T cells experiencing Ag derived from tumor (ProHA × TRAMP) or self (C3HAhigh)
| Probe Set IDa | Gene Symbol | Gene Title | ProHA × TRAMP | C3-HA | Fold Change |
|---|---|---|---|---|---|
| 1417409_at | Jun | Jun oncogene | 809 | 38 | 21.5 |
| 1453840_at | Pabpc1 | Poly(A)b-binding protein, cytoplasmic 1 | 1075 | 83 | 13.0 |
| 1455805_x_at | FoxP3 | Forkhead box P3 | 340 | 32 | 10.5 |
| 1436222_at | Gas5 | Growth arrest-specific 5 | 357 | 37 | 9.8 |
| 1440169_x_at | Ifnar2 | IFN (α and β) receptor 2 | 497 | 55 | 9.0 |
| 1456956_at | Zfn1a2 | Zinc finger protein, subfamily 1A, 2 | 859 | 103 | 8.4 |
| 1439036_a_at | Atp1b1 | ATPase, Na +/K +-transporting, β1 polypeptide | 125 | 15 | 8.1 |
| 1448208_at | Madh1 | MAD homolog 1 | 206 | 26 | 7.8 |
| 1427275_at | Smc4I1 | SMC4 structural maintenance of chromosomes 4-like 1 | 1290 | 186 | 7.0 |
| 1440770_at | Bcl2 | B cell leukemia/lymphoma 2 | 686 | 100 | 6.9 |
| 1438941_x_at | Ampd2 | AMP deaminase 2 | 722 | 105 | 6.9 |
| 1450339_a_at | Bcl11b | B cell leukemia/lymphoma 11B | 2529 | 369 | 6.9 |
| 1441926_x_at | Tmie | Transmembrane inner ear | 705 | 107 | 6.6 |
| 1437163_x_at | Gtf2h4 | General transcription factor II H, polypeptide 4 | 312 | 48 | 6.5 |
| 1417679_at | Gfi1 | Growth factor-independent 1 | 855 | 135 | 6.3 |
| 1428463_a_at | Ppp2r5e | Protein phosphatase 2, regulatory subunit B (B56), ε isoform | 128 | 20 | 6.3 |
| 1440104_at | Ranbp2 | RAN-binding protein 2 | 674 | 107 | 6.3 |
| 1440837_at | H2-Ob | Histocompatibility 2, O region β locus | 219 | 35 | 6.3 |
| 1436804_s_at | Scyl1 | SCY1-like 1 | 250 | 40 | 6.3 |
| 1419810_x_at | Gli | GLI-Kruppel family member GLI | 1392 | 227 | 6.1 |
| 1434895_s_at | Ppp1r13b | Protein phosphatase 1, regulatory (inhibitor) subunit 13B | 720 | 118 | 6.1 |
| 1448325_at | Myd116 | Myeloid differentiation primary response gene 116 | 2204 | 363 | 6.1 |
| 1450095_a_at | Acyp1 | Acylphosphatase 1, erythrocyte (common) type | 133 | 22 | 6.0 |
| 1456467_s_at | Nik | Nemo-like kinase | 314 | 53 | 5.9 |
| 1448471_a_at | Tpbpb | Trophoblast-specific protein β | 737 | 125 | 5.9 |
| 1437828_s_at | Bing4 | BING4 protein | 415 | 71 | 5.9 |
| 1427186_a_at | Mef2a | Myocyte enhancer factor 2A | 187 | 32 | 5.9 |
| 1425514_at | Pik3r1 | PI3K, regulatory subunit, polypeptide 1 | 156 | 27 | 5.7 |
| 1439768_x_at | Sema4f | Sema domain, Ig domain, TM domain, and short cytoplasmic domain | 419 | 74 | 5.7 |
| 1438259_at | Strn3 | Striatin, calmodulin-binding protein 3 | 328 | 58 | 5.6 |
| 1417734_at | Nakap95 | Neighbor of A kinase-anchoring protein 95 | 891 | 160 | 5.6 |
| 1439440_x_at | Ptk 9I | Protein tyrosine kinase 9-like | 1344 | 246 | 5.5 |
| 1437667_a_at | Bach2 | BTB and CNC homology 2 | 1291 | 237 | 5.4 |
| 1434399_at | Galnt6 | UDP-N-acetyl-α-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 | 1516 | 278 | 5.4 |
| 1440885_at | Evl | Ena-vasodilator-stimulated phosphoprotein | 909 | 168 | 5.4 |
| 1447757_x_at | Inpp5f | Inositol polyphosphate-5-phosphatase F | 180 | 34 | 5.3 |
| 1418643_at | Tm4sf13 | Transmembrane 4 superfamily member 13 | 5332 | 1004 | 5.3 |
| 1436983_at | Crebbp | CREB-binding protein | 673 | 127 | 5.3 |
| 1430357_at | H3f3b | H3 histone, family 3B | 527 | 100 | 5.3 |
| 1438211_s_at | Dbp | D site albumin promoter-binding protein | 382 | 73 | 5.2 |
ID, identification number; poly(A), polyadenylate; RAN, Ras-related nuclear protein; GLI, gliotactin; sema, semaphorin; TM, transmembrane; BTB, bric-a-brac, tramtrack, and broad complex; CNC, canary complex; Ena, enabled.
Table II.
Genes differentially down-regulated by CD4 T cells experiencing Ag derived from tumor (ProHA × TRAMP) or self (C3HAhigh)
| Probe Set IDa | Gene Symbol | Gene Title | ProHA × TRAMP | C3-HA | Fold Change |
|---|---|---|---|---|---|
| 1421375_a_at | S100a6 | S100 calcium-binding protein A6 | 73.7 | 683.6 | −9.3 |
| 1424542_at | S100a4 | S100 calcium-binding protein A4 | 31.0 | 267.8 | −8.6 |
| 1449911_at | Lag3 | Lymphocyte activation gene 3 | 48.6 | 391.3 | −8.1 |
| 1422557_s_at | Mt1 | Metallothionein 1 | 91.8 | 739.2 | −8.1 |
| 1436164_at | Slc30a1 | Solute carrier family 30 (zinc transporter), member 1 | 137.8 | 1055.9 | −7.7 |
| 1428942_at | Mt2 | Metallothionein 2 | 209.4 | 1175.5 | −5.6 |
| 1447849_s_at | Maf | Avian musculoaponeurotic fibrosarcoma (v-maf) AS42 oncogene homolog | 19.4 | 108.4 | −5.6 |
| 1416529_at | Emp1 | Epithelial membrane protein 1 | 12.6 | 59.5 | −4.7 |
| 1420119_s_at | Phf3 | PHD finger protein 3 | 19.0 | 87.8 | −4.6 |
| 1418622_at | Rab2 | RAB2, member RAS oncogene family | 24.0 | 105.0 | −4.4 |
| 1455090_at | Angptl2 | Angiopoietin-like 2 | 156.6 | 672.9 | −4.3 |
| 1453228_at | Stx11 | Syntaxin 11 | 114.6 | 483.7 | −4.2 |
| 1433648_at | Spag9 | Sperm-associated Ag 9 | 127.0 | 501.0 | −3.9 |
| 1455439_a_at | Lgals1 | Lectin, galactose binding, soluble 1 | 743.7 | 2915.4 | −3.9 |
| 1419761_a_at | Gabpb1 | GA repeat binding protein, β1 | 47.8 | 183.4 | −3.8 |
| 1418401_a_at | Dusp16 | Dual-specificity phosphatase 16 | 30.2 | 115.6 | −3.8 |
| 1436325_at | Rora | RAR-related orphan receptor α | 21.2 | 79.6 | −3.8 |
| 1439348_at | S100a10 | S100 calcium-binding protein A10 | 65.2 | 244.2 | −3.7 |
| 1419060_at | Gzmb | Granzyme B | 79.2 | 295.2 | −3.7 |
| 1419202_at | Cst7 | Cystatin F | 152.3 | 554.8 | −3.6 |
| 1419573_a_at | Lgals1 | Lectin, galactose binding, soluble 1 | 520.8 | 1853.3 | −3.6 |
| 1416029_at | Tieg1 | TGF- β-inducible early growth response 1 | 92.8 | 322.3 | −3.5 |
| 1455166_at | Arl8 | ADP ribosylation factor-like 8 | 140.0 | 484.2 | −3.5 |
| 1426208_x_at | Plagl1 | Pleiomorphic adenoma gene-like 1 | 94.0 | 318.2 | −3.4 |
| 1416958_at | Nr1d2 | Nuclear receptor subfamily 1, group D, member 2 | 39.5 | 130.9 | −3.3 |
| 1449310_at | Ptger2 | PGER2 | 42.4 | 138.4 | −3.3 |
| 1444500_at | Ahsa1 | AHA1, activator of heat shock 90-kDa protein ATPase homolog 1 | 50.9 | 162.9 | −3.2 |
| 1417935_at | Mkrn2 | Makorin, ring finger protein, 2 | 243.9 | 775.2 | −3.2 |
| 1448370_at | Ulk1 | Unc-51-like kinase 1 | 260.0 | 814.9 | −3.1 |
| 1450753_at | Nkg7 | Natural killer cell group 7 sequence | 111.4 | 346.7 | −3.1 |
| 1435874_at | Prkab2 | Protein kinase, AMP-activated, β2 noncatalytic subunit | 104.8 | 323.6 | −3.1 |
| 1449235_at | Tnfsf6 | TNF (ligand) superfamily, member 6 | 79.8 | 230.8 | −2.9 |
| 1419091_a_at | Anxa2 | Annexin A2 | 345.1 | 965.4 | −2.8 |
| 1419838_s_at | Stk18 | Serine/threonine kinase 18 | 33.6 | 93.6 | −2.8 |
| 1450714_at | Oazin | Ornithine decarboxylase antizyme inhibitor | 96.7 | 266.8 | −2.8 |
| 1428393_at | Nrn1 | Neuritin 1 | 393.1 | 1073.4 | −2.7 |
| 1457528_at | Slc4a7 | Solute carrier family 4, sodium bicarbonate cotransporter, member 7 | 48.7 | 131.0 | −2.7 |
| 1421963_a_at | Cdc25b | Cell division cycle 25 homolog B | 97.8 | 262.8 | −2.7 |
| 1427629_at | Ptprj | Protein tyrosine phosphatase, receptor type, J | 17.5 | 46.8 | −2.7 |
| 1456126_at | Malt1 | Mucosa-associated lymphoid tissue lymphoma translocation gene 1 | 75.2 | 200.9 | −2.7 |
ID, identification number; PHD, plant homeodomain.
Table III.
Genes differentially up-regulated by CD4 T cells experiencing Ag derived from tumor (ProHA × TRAMP) vs virus (VaccHA)
| Probe Set IDa | Gene Symbol | Gene Title | ProHA × TRAMP | VaccHA | Fold Change |
|---|---|---|---|---|---|
| 1456956_at | Zfpn1a2 | Zinc finger protein, subfamily 1A, 2 | 859 | 105 | 8.2 |
| 1417481_at | Ramp 1 | Receptor (calcitonin) activity-modifying protein 1 | 470 | 63 | 7.5 |
| 1455265_a_at | Rgs16 | Regulator of G-protein signaling 16 | 337 | 47 | 7.2 |
| 1428834_at | Dusp4 | Dual-specificity phosphatase 4 | 1139 | 173 | 6.6 |
| 1436759_x_at | Cnn3 | Calponin 3, acidic | 232 | 44 | 5.2 |
| 1419156_at | Sox4 | SRY-box containing gene 4 | 261 | 54 | 4.8 |
| 1450826_a_at | Saa3 | Serum amyloid A 3 | 1370 | 286 | 4.8 |
| 1441760_at | Rps25 | Ribosomal protein S25 | 866 | 187 | 4.6 |
| 1416514_a_at | Fscn1 | Fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) | 611 | 136 | 4.5 |
| 1449984_at | Cxcl2 | Chemokine (C-X-C motif) ligand 2 | 208 | 47 | 4.4 |
| 1418449_at | Lad1 | Ladinin | 542 | 125 | 4.3 |
| 1438148_at | Gm1 960 | Gene model 1 960 | 52 | 12 | 4.3 |
| 1433575_at | Sox4 | SRY box-containing gene 4 | 64 | 15 | 4.2 |
| 1422892_s_at | H2-Ea | Histocompatibility 2, class II Ag Eα | 1108 | 268 | 4.1 |
| 1434499_a_at | Ldh2 | Lactate dehydrogenase 2, B chain | 186 | 46 | 4.1 |
| 1426738_at | Dgkz | Diacylglycerol kinase ζ | 926 | 232 | 4.0 |
| 1428329_a_at | Wdr5 6 | WD repeat domain 56 | 370 | 95 | 3.9 |
| 1423547_at | Lyzs | Lysozyme | 867 | 223 | 3.9 |
| 1438370_x_at | Dos | Downstream of Stk11 | 430 | 111 | 3.9 |
| 1437807_x_at | Ctnna1 | Catenin (cadherin-associated protein), α1 | 350 | 92 | 3.8 |
| 1426112_a_at | Cd72 | CD72 Ag | 267 | 71 | 3.8 |
| 1452543_a_at | Scgb1a1 | Secretoglobin, family 1A, member 1 | 101 | 27 | 3.7 |
| 1417925_at | Ccl2 2 | Chemokine (C-C motif) ligand 22 | 1254 | 338 | 3.7 |
| 1416714_at | Icsbp 1 | IFN consensus sequence-binding protein 1 | 617 | 166 | 3.7 |
| 1416111_at | Cd83 | CD83 Ag | 449 | 121 | 3.7 |
| 1419297_at | H2-Oa | Histocompatibility 2, O region alpha locus | 541 | 146 | 3.7 |
| 1421038_a_at | Kcnn4 | Potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 | 539 | 146 | 3.7 |
| 1456700_x_at | Marcks | Myristoylated alanine-rich protein kinase C substrate | 220 | 60 | 3.7 |
| 1421073_a_at | Ptger4 | PGER 4 | 54 | 15 | 3.7 |
| 1436959_x_at | Nelf | Nasal embryonic LHRH factor | 1288 | 351 | 3.7 |
| 1456941_at | Tert | Telomerase reverse transcriptase | 76 | 21 | 3.6 |
| 1437270_a_at | Bsf3 | Cardiotrophin-like cytokine factor 1 | 278 | 78 | 3.6 |
| 1420404_at | Cd86 | CD86 Ag | 474 | 135 | 3.5 |
| 1428643_at | Mgat5 | Mannoside acetylglucosaminyltransferase 5 | 263 | 75 | 3.5 |
| 1419549_at | Arg1 | Arginase 1, liver | 68 | 20 | 3.5 |
| 1451285_at | Fus | Fusion, derived from t(12;16) malignant liposarcoma (human) | 183 | 53 | 3.5 |
| 1432466_a_at | Apoe | Apolipoprotein E | 73 | 21 | 3.4 |
| 1438274_at | Zfpn1a4 | Zinc finger protein, subfamily 1A, 4 | 341 | 99 | 3.4 |
| 1419083_at | Tnfsf1 1 | TNF (ligand) superfamily, member 11 | 321 | 94 | 3.4 |
| 1435290_x_at | H2-Aa | Histocompatibility 2, class II Ag A, α | 1307 | 384 | 3.4 |
ID, identification number; PHD, plant homeodomain; RAB, Ras-related; RAR, retinoic acid receptor; Ras, rat sarcoma.
Table IV.
Genes differentially down-regulated by CD4 T cells experiencing Ag derived from tumor (ProHA × TRAMP) vs virus (VaccHA)
| Probe Set IDa | Gene Symbol | Gene Title | ProHA × TRAMP | VaccHA | Fold Change |
|---|---|---|---|---|---|
| 1458947_at | Fancc | Fanconi anemia, complementation group C | 420 | 787 | −1.9 |
| 1444676_at | Ctcf | CCCTC-binding factor | 60 | 112 | −1.9 |
| 1441669_at | Centb2 | Centaurin, β2 | 125 | 233 | −1.9 |
| 1447903_x_at | Ap1s2 | Adaptor-related protein complex 1, σ2 subunit | 263 | 493 | −1.9 |
| 1459736_at | Stk 10 | Serine/threonine kinase 10 | 134 | 250 | −1.9 |
| 1438683_at | Wasf2 | WAS protein family, member 2 | 125 | 233 | −1.9 |
| 1445337_at | Dnajc13 | DnaJ (Hsp40) homolog, subfamily C, member 13 | 137 | 255 | −1.9 |
| 1423727_at | Cnih | Cornichon homolog | 260 | 483 | −1.9 |
| 1445928_at | March6 | Membrane-associated ring finger (C3HC 4) 6 | 140 | 261 | −1.9 |
| 1455886_at | Cbl | Casitas B-lineage lymphoma | 448 | 833 | −1.9 |
| 1440729_at | Eps15 | Epidermal growth factor receptor pathway substrate 15 | 19 | 36 | −1.9 |
| 1443480_at | Rassf3 | Ras association (RalGDS/AF-6) domain family 3 | 159 | 295 | −1.9 |
| 1426559_at | Sbno1 | Sno, strawberry notch homolog 1 | 635 | 1177 | −1.9 |
| 1448885_at | Rap 2b | RAP2B, member of RAS oncogene family | 230 | 426 | −1.9 |
| 1423423_at | Pdia3 | Protein disulfide isomerase-associated 3 | 63 | 116 | −1.9 |
| 1417270_at | Wdr12 | WD repeat domain 12 | 39 | 72 | −1.9 |
| 1422533_at | Cyp51 | Cytochrome P450, family 51 | 26 | 49 | −1.9 |
| 1415838_at | Tde2 | Tumor differentially expressed 2 | 124 | 230 | −1.8 |
| 1419758_at | Abcb 1a | ATP-binding cassette, subfamily B (MDR/TAP), member 1A | 22 | 40 | −1.8 |
| 1420021_s_at | Suz12 | Suppressor of zeste 12 homolog | 328 | 604 | −1.8 |
| 1420497_a_at | Cebpz | CCAAT/enhancer binding protein zeta | 89 | 164 | −1.8 |
| 1419497_at | Cdkn1 b | Cyclin-dependent kinase inhibitor 1B | 36 | 65 | −1.8 |
| 1423819_s_at | Arl6ip1 | ADP ribosylation factor-like 6-interacting protein 1 | 805 | 1480 | −1.8 |
| 1450093_s_at | Zbtb7a | Zinc finger and BTB domain containing 7a | 30 | 56 | −1.8 |
| 1459635_at | Dlgh1 | Discs, large homolog 1 | 55 | 101 | −1.8 |
| 1453361_at | Hells | Helicase, lymphoid specific | 18 | 33 | −1.8 |
| 1417371_at | Peli1 | Pellino 1 | 528 | 968 | −1.8 |
| 1446205_at | Nfyc | Nuclear transcription factor-Yγ | 89 | 162 | −1.8 |
| 1429525_s_at | Myo1f | Myosin IF | 33 | 61 | −1.8 |
| 1448339_at | Tmem30 a | Transmembrane protein 30A | 66 | 121 | −1.8 |
| 1441460_at | Fgfr1op 2 | FGFR1 oncogene partner 2 | 71 | 130 | −1.8 |
| 1459457_at | Camk2d | Calcium/calmodulin-dependent protein kinase II, delta | 170 | 310 | −1.8 |
| 1431197_at | Arl6ip2 | ADP ribosylation factor-like 6-interacting protein 2 | 82 | 149 | −1.8 |
| 1422769_at | Syncrip | Synaptotagmin binding, cytoplasmic RNA interacting protein | 15 | 27 | −1.8 |
| 1434039_at | Appbp2 | Amyloid β precursor protein (cytoplasmic tail)-binding protein 2 | 22 | 40 | −1.8 |
| 1452115_a_at | Plk4 | Polo-like kinase 4 | 17 | 31 | −1.8 |
| 1449480_at | Sap 18 | Sin 3-associated polypeptide 18 | 46 | 84 | −1.8 |
| 1424443_at | Tm6sf1 | Transmembrane 6 superfamily member 1 | 12 | 22 | −1.8 |
| 1441319_at | Rbm5 | RNA-binding motif protein 5 | 52 | 95 | −1.8 |
| 1438064_at | Nsep 1 | Nuclease-sensitive element binding protein 1 | 262 | 478 | −1.8 |
| 1437884_at | Arl8 | ADP ribosylation factor-like 8 | 71 | 129 | −1.8 |
ID, identification number; MDR, multidrug resistant; FGFR, fibroblast growth factor receptor; WAS, Wiskott-Aldrich syndrome; Ras, rat sarcoma; BTB, bric-a-brac, tramtrack, and broad complex.
CD4 T cells that recognized tumor-associated Ag have a Treg phenotype
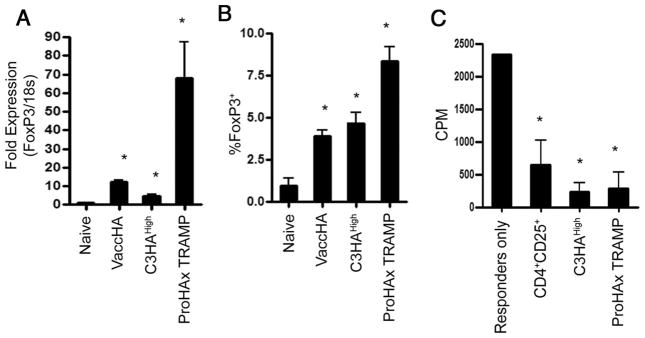
In the set of transcripts up-regulated in response to tumor-recognition, we noted the transcription factor FoxP3, a relatively well-established marker of regulatory T cell (Treg) (33, 34, 51–53). To verify these data, we performed quantitative PCR analysis on CFSE-diluted HA-specific CD4+ T cells isolated after adoptive transfer. As shown in Fig. 3A, CD4 T cells isolated from ProHA × TRAMP tumor-bearing animals expressed significantly higher FoxP3 mRNA levels than cells harvested from C3HAhigh or Vacc-HA-infected nontransgenic mice (p < 0.05). We next tested whether these transcriptional changes were reflected at the protein level by intracellular staining for FoxP3 (Fig. 3B), and found the expected up-regulation. Tregs may be functionally distinguished from other T cell subsets by their capacity to suppress the proliferative response of activated T cells (54). Thus, we next sought to determine whether these tumor-induced Tregs were capable of suppressing proliferation of responder cells using an in vitro suppression assay. For these studies, anti-CD3-activated T cells from nontransgenic mice were used as responders. Naturally occurring CD4+ CD25+ T cells were FACS sorted as control suppressors from the same mice that were the source for responders. FACS-sorted HA-specific CD4 T cells from ProHA × TRAMP recipients were able to suppress the proliferation of responders in a manner similar to that of natural Tregs (Fig. 3C). In accordance with our previously published data, Ag-experienced CD4 T cells isolated from self Ag-expressing animals were also able to suppress to some degree in this assay, despite a lack of relative overexpression of FoxP3 (21). These data are in agreement with multiple reports suggesting that FoxP3 expression is not an absolute requirement for suppressive activity in vitro (55).
FIGURE 3.
FoxP3 expression in clonotypic CD4 T cells that recognize tumor-restricted Ag. CFSE-labeled HA-specific Thy1.1+ CD4+ T cells were adoptively transferred into the indicated Ag recognition models. CSFE-diluted Thy1.1+ CD4+ T cell were FACS sorted on day 3 from recipient mice. A, FoxP3 mRNA expression as determined by real-time PCR. B, Intracellular staining for FoxP3. C, CSFE-diluted CD4 T cells from a tumor Ag recognition model were able to suppress proliferation of activated responders. An in vitro suppression assay was performed by FACS sorting CFSE-diluted Thy1.1+ CD4+ T cells from C3HAhigh and ProHA × TRAMP recipients. Sorted suppressors were coincubated at a 1:10 ratio with 105 T cell responders activated by anti-CD3-coated plate for 72 h. Proliferation was measured by [3H]TdR incorporation during the final 18 h of incubation. A p value of <0.05 is denoted by a single asterisk (*) for statistical significance.
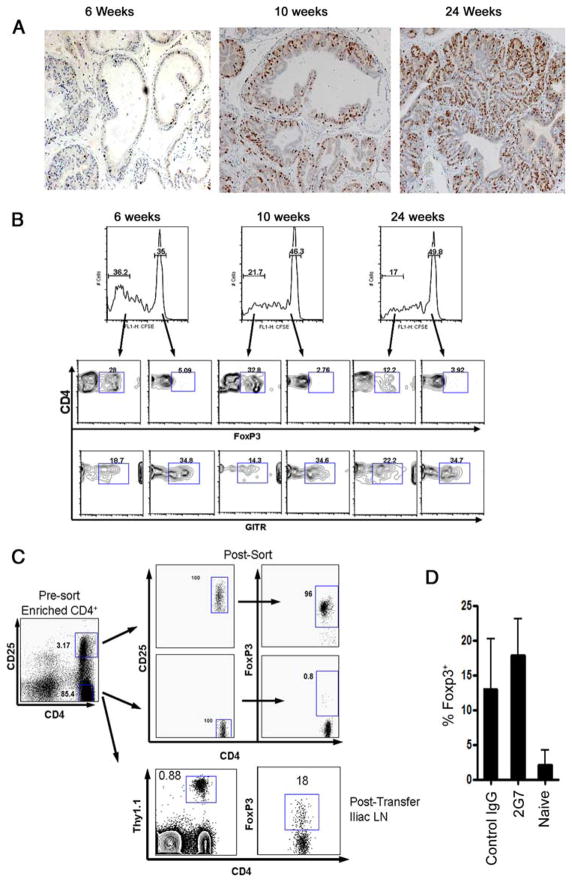
Treg development is an early event in ProHA × TRAMP animals
In the same manner as TRAMP mice, ProHA × TRAMP mice reliably develop autochthonous prostate tumors in an age-dependent manner (39). In young mice, typically <6 wk, the prostate glands appear grossly normal (Fig. 4A). Between 6 and 10 wk, hyperplasia and cribriform structures appear, a lesion that has been termed murine PIN (39). Finally, older animals develop overtly neoplastic lesions, with metastatic spread to the draining lymph nodes and liver. We took advantage of the progressive nature of this model to query whether Treg development was a function of tumor stage. Naive CFSE-labeled, HA-specific CD4+ T cells were adoptively transferred to ProHA × TRAMP mice of various ages, harvested from the prostate-draining lymph nodes, and evaluated for FoxP3 expression by intracellular staining. Divided cells appeared to up-regulate FoxP3 even in mice in which tumor development was not obvious on gross pathological examination (Fig. 4B). These data suggest that induction of Treg might be a relatively early event in tumor development. Glucocorticoid-induced TNFR family-related gene staining did not correlate with the FoxP3-stained population in our system.
FIGURE 4.
Treg phenotype in tumor Ag-experienced CD4 T cells. A, Ki67 immunohistochemistry staining on prostate dorsal lob sections of different age group ProHA × TRAMP animals indicating pathological progression prostate cancer. B, FoxP3 intracellular staining on CFSE-labeled HA-specific Thy1.1+CD4+ T cells that were adoptively transferred into the tumor Ag recognition model (ProHA × TRAMP recipients) at various stages of disease; 6 wk with no apparent disease pathology, 10 wk with low-disease-grade pathology, and 24 wk with high-disease-grade pathology. C, Apparent de novo FoxP3 expression in HA-specific Thy1.1+CD4+CD25− T cells adoptively transferred into ProHA × TRAMP mice. D, In vivo TGF-β blocking with 2G7 in ProHA × TRAMP and B10.d2 hosts did not affect the frequency FoxP3+ T cells in the iliac lymph node (LN).
Although CFSE labeling experiments (Fig. 4B) suggested that the Tregs observed in this model represent induced Tregs, rather than the expansion of natural CD4+ CD25+ Tregs, we attempted to address this issue more completely by depleting donor T cells of CD25+ T cells before adoptive transfer using FACS sorting. As shown (Fig. 4C), this process was reasonably efficient, the CD25low CD4 fraction was <1% FoxP3+, as opposed to the CD25high CD4 fraction, which was 95% FoxP3+. These CD4+CD25low-sorted Thy1.1+ HA-specific T cells were adoptively transferred to ProHA × TRAMP mice and recovered from the tumor-draining iliac lymph nodes 10 days posttransfer. By this time point, >15% of these HA-specific cells were FoxP3+ by intracellular staining, supporting the hypothesis that tumor recognition may induce specific Tregs in vivo. Finally, we attempted to investigate a potential role for TGF-β in the development of Tregs in this system, by administering TGF-β blocking mAb (2G7) during and after adoptive transfer of HA-specific CD4 T cells into ProHA × TRAMP mice and naive B10.d2 hosts. As shown in Fig. 4D, blocking TGF-β in vivo did not significantly affect the percentage of FoxP3+ Thy1.1 cells compared with control IgG group. Similar results were obtained using a second TGF-β blocking mAb 1D11 (data not shown).
Discussion
In these studies, we used well-established tolerance models to study the transcriptional profiles of CD4 T cells responding to either tumor Ag or self Ag recognition in vivo. For a tumor model, we used TRAMP-transgenic mice (22). These mice develop PIN during puberty followed by a progressive invasive carcinoma and metastasis of epithelial origin in the adult male animals (56). For our studies, TRAMP mice were crossed with our ProHA mice that express influenza HA under the control of the identical probasin promoter used in TRAMP mice (14). Thus, in ProHA × TRAMP mice, expression of the nonmutated tumor Ag HA is a function of tumor progression. C3HAhigh-transgenic mice were developed to study T cell tolerance to self-Ag; in these animals HA is widely expressed under the control of the rat C3 promoter (19, 20). We found that both self and tumor recognition resulted in relative up-regulation and down-regulation of large numbers of transcripts as compared with naive cells, but that a number of genes appeared to be differentially expressed between the two conditions. These data generally suggest that tumor and self-recognition result in different CD4 T cell profiles, but this interpretation must be tempered by several other differences between the two models, including a more robust division in the self Ag model, and the far more localized nature of Ag in the ProHA × TRAMP model.
Our transcriptional data also suggested a significant up-regulation of FoxP3 in tumor vs self-tolerance. This was somewhat surprising because our transcriptional profiling studies were designed in an open-ended manner, evaluating tens of thousands of transcripts in parallel without a specific hypothesis regarding the gene sets that would be found. Up-regulation of FoxP3 was confirmed at both the transcriptional level and at the protein level, and correlated with a suppressive phenotype in the in vitro suppression assay (Fig. 3C). These data are consistent with multiple reports of tumor-induced Tregs, by our group and others (27, 57, 58). Tumors may not be unique in their ability to induce or expand Tregs from a naive CD4 T cell population, as the Von Boehmer group has reported generation of regulatory T cells to both ubiquitously expressed Ag and low-level Ag encountered chronically (59–61).
Several other groups have reported a transcriptional profile for Tregs using in vivo models (37, 49, 50). Our data are somewhat unique in this respect, given that we did not initially set out with the goal of profiling Tregs. However, our data appear to be broadly consistent with prior work. For example, both Sakaguchi’s group (49)and Rudensky’s group (37) found up-regulation of ZFN1a2 in natural Tregs, and here we report a similar up-regulation. Similarly, all three studies report up-regulation of Nrp, ZFN1a4, CD86, Myb, CTL4, and BCL2 family members. However, our dataset also includes a number of unique transcripts that were previously not associated with Tregs such as CD83, Madh1, Infra2, and Scy1. Our complete transcriptional dataset is deposited in the Geo database at the National Center for Biotechnology Information (accession number GSE14662; www.ncbi.nlm.nih.gov/geo/); in-depth examination of these profiles using pathway or comparative assays could conceivably provide additional insight into the biochemical pathways involved in Treg induction and function.
Previous studies in TRAMP mice and their derivatives demonstrated T cell tolerance to prostate-associated Ag(s), with several of these reports suggesting an age dependence to this mechanism (9, 16, 62– 65). However, the relationship between Tregs and T cell tolerance in the TRAMP model is less clear, as at least one study reported that Treg depletion in these animals did not affect the vaccine response in vivo (47, 62). Our data provide additional insight into this issue, suggesting that the ability to induce and/or expand Tregs might occur when tumors are barely pathologically recognizable. These data are also consistent with recent data presented by the Blankenstein group, who used a spontaneous transgenic tumor model to show that early recognition of evolving tumors was a critical event in later tumor tolerance (40, 66). Our findings in this regard have potential clinical relevance as well, confirming other work highlighting a potential role for Treg depletion in tumor immunotherapy (67–70).
Several aspects of these data suggest that the Tregs we observe here represent induced Tregs rather than the expansion of naturally occurring CD4+CD25+ Tregs. First, we observe FoxP3 expression at the protein level in Ag-specific CD4 T cells that have divided in response to tumor recognition, but not in undivided (CF-SEhigh) cells (Fig. 4B). These data are not consistent with those of Valzasina et al. (58), who showed that up-regulation of FoxP3 is independent of division of tumor Ag-experienced CD4 T cells. These differences may reflect differences in TCR affinity or Ag expression in the two systems. We attempted to address this question more fully by transferring naive CD4 T cells that were sorted to be CD25low; only a very small fraction (<1%) of such cells are FoxP3+ at the time of transfer. Posttransfer, these cells showed FoxP3 expression in tumor-draining lymph nodes, consistent with possible Treg induction. We are cognizant, however, that such studies are not definitive and that it is certainly possible that these FoxP3 cells represent the result of a substantial expansion of a small population of natural Tregs in the adoptively transferred population. To investigate this more fully will require crossing FoxP3-GFP mice into the 6.5 TCR B10.D2-transgenic background animals, such that only cells that are truly FoxP3− can be definitively sorted out before adoptive transfer.
In summary, we report here the comparative expression profile of Ag-specific CD4 T cells that have divided in response to tumor Ag, self Ag, and viral Ag. Although these profiles are interesting in and of themselves, our data demonstrate the induction and/or expansion of Tregs by tumor recognition, as evidenced by FoxP3 up-regulation at both the transcriptional and protein levels, as well as the acquisition of a regulatory phenotype in vitro. Ag-experienced CD4 T cells both from the self or tumor Ag models suppressed proliferating responder cells, suggesting that the Treg skewing of CD4 T cells is a tumor recognition phenomenon, whereas the ability to suppress is a shared phenomenon between the two models. The observation of a relative skewing of Tregs at a very early stage of disease further supports the notion that the expansion and/or induction of the Treg is a tumor-related event, a result with clear clinical implications. The Tregs in this system appear to represent an induced population, although far more extensive studies will be required to address that particular point more directly. Nevertheless, the transcriptional profile of these cells appears to be unique, and evaluation of particular transcripts or pathways could conceivably provide additional therapeutic targets to potentiate an antitumor immune response. Finally, this study provides evidence that tumor Ag recognition results in a distinct transcriptional profile in CD4 T cells that is unique from that induced by self Ag recognition.
Supplementary Material
Footnotes
This work was supported by National Institutes of Health Grants R01 CA127153 (to C.G.D.) and K08 CA096948 (to C.G.D.) and by the Patrick C. Walsh Fund. C.G.D. is a Damon Runyon-Lilly Clinical Investigator. D.M.P. is a Januey Scholar, holds the Seraph Chair for Cancer Research, and is supported in part by gifts from William and Betty Toperer, Dorothy Needle, and the Commonwealth Foundation.
Abbreviations used in this paper: HA, hemagglutinin; TRAMP, transgenic adenocarcinoma of the mouse prostate; PIN, prostatic intraepithelial neoplasia; Treg, regulatory T cell; VaccHA, Vaccinia-hemagglutinin; LMHA, Listeria monocytogenes-HA; DC, dendritic cell.
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Drake CG, Jaffee E, Pardoll DM. Mechanisms of immune evasion by tumors. Adv Immunol. 2006;90:51–81. doi: 10.1016/S0065-2776(06)90002-9. [DOI] [PubMed] [Google Scholar]
- 2.Pardoll D. Does the immune system see tumors as foreign or self? Annu Rev Immunol. 2003;21:807–839. doi: 10.1146/annurev.immunol.21.120601.141135. [DOI] [PubMed] [Google Scholar]
- 3.Dunn GP, Old LJ, Schreiber RD. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 2004;21:137–148. doi: 10.1016/j.immuni.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 4.Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol. 2004;22:329–360. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- 5.Marincola FM, Jaffee EM, Hicklin DJ, Ferrone S. Escape of human solid tumors from T-cell recognition: molecular mechanisms and functional significance. Adv Immunol. 2000;74:181–273. doi: 10.1016/s0065-2776(08)60911-6. [DOI] [PubMed] [Google Scholar]
- 6.Munn DH, Mellor AL. Indoleamine 2,3-dioxygenase and tumor-induced tolerance. J Clin Invest. 2007;117:1147–1154. doi: 10.1172/JCI31178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massague J. TGFβ in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenwald RJ, Freeman GJ, Sharpe AH. The B7 family revisited. Annu Rev Immunol. 2005;23:515–548. doi: 10.1146/annurev.immunol.23.021704.115611. [DOI] [PubMed] [Google Scholar]
- 9.Shafer-Weaver K, Anderson M, Malyguine A, Hurwitz AA. T cell tolerance to tumors and cancer immunotherapy. Adv Exp Med Biol. 2007;601:357–368. doi: 10.1007/978-0-387-72005-0_38. [DOI] [PubMed] [Google Scholar]
- 10.Willimsky G, Blankenstein T. Sporadic immunogenic tumours avoid destruction by inducing T-cell tolerance. Nature. 2005;437:141–146. doi: 10.1038/nature03954. [DOI] [PubMed] [Google Scholar]
- 11.Degl’Innocenti E, Grioni M, Boni A, Camporeale A, Bertilaccio MT, Freschi M, Monno A, Arcelloni C, Greenberg NM, Bellone M. Peripheral T cell tolerance occurs early during spontaneous prostate cancer development and can be rescued by dendritic cell immunization. Eur J Immunol. 2005;35:66–75. doi: 10.1002/eji.200425531. [DOI] [PubMed] [Google Scholar]
- 12.Anderson MJ, Shafer-Weaver K, Greenberg NM, Hurwitz AA. Tolerization of tumor-specific T cells despite efficient initial priming in a primary murine model of prostate cancer. J Immunol. 2007;178:1268–1276. doi: 10.4049/jimmunol.178.3.1268. [DOI] [PubMed] [Google Scholar]
- 13.Morgan DJ, Kreuwel HT, Sherman LA. Antigen concentration and precursor frequency determine the rate of CD8+ T cell tolerance to peripherally expressed antigens. J Immunol. 1999;163:723–727. [PubMed] [Google Scholar]
- 14.Drake CG, Doody AD, Mihalyo MA, Huang CT, Kelleher E, Ravi S, Hipkiss EL, Flies DB, Kennedy EP, Long M, et al. Androgen ablation mitigates tolerance to a prostate/prostate cancer-restricted antigen. Cancer Cell. 2005;7:239–249. doi: 10.1016/j.ccr.2005.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohashi PS, Oehen S, Buerki K, Pircher H, Ohashi CT, Odermatt B, Malissen B, Zinkernagel RM, Hengartner H. Ablation of “tolerance” and induction of diabetes by virus infection in viral antigen transgenic mice. Cell. 1991;65:305–317. doi: 10.1016/0092-8674(91)90164-t. [DOI] [PubMed] [Google Scholar]
- 16.Bai A, Higham E, Eisen HN, Wittrup KD, Chen J. Rapid tolerization of virus-activated tumor-specific CD8+ T cells in prostate tumors of TRAMP mice. Proc Natl Acad Sci USA. 2008;105:13003–13008. doi: 10.1073/pnas.0805599105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poggi A, Zocchi MR. Mechanisms of tumor escape: role of tumor microenvironment in inducing apoptosis of cytolytic effector cells. Arch Immunol Ther Exp (Warsz) 2006;54:323–333. doi: 10.1007/s00005-006-0038-7. [DOI] [PubMed] [Google Scholar]
- 18.Kirberg J, Baron A, Jakob S, Rolink A, Karjalainen K, von Boehmer H. Thymic selection of CD8+ single positive cells with a class II major histocompatibility complex-restricted receptor. J Exp Med. 1994;180:25–34. doi: 10.1084/jem.180.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adler AJ, Marsh DW, Yochum GS, Guzzo JL, Nigam A, Nelson WG, Pardoll DM. CD4+ T cell tolerance to parenchymal self-antigens requires presentation by bone marrow-derived antigen-presenting cells. J Exp Med. 1998;187:1555–1564. doi: 10.1084/jem.187.10.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adler AJ, Huang CT, Yochum GS, Marsh DW, Pardoll DM. In vivo CD4+ T cell tolerance induction versus priming is independent of the rate and number of cell divisions. J Immunol. 2000;164:649–655. doi: 10.4049/jimmunol.164.2.649. [DOI] [PubMed] [Google Scholar]
- 21.Huang CT, Workman CJ, Flies D, Pan X, Marson AL, Zhou G, Hipkiss EL, Ravi S, Kowalski J, Levitsky HI, et al. Role of LAG-3 in regulatory T cells. Immunity. 2004;21:503–513. doi: 10.1016/j.immuni.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Greenberg NM, DeMayo F, Finegold MJ, Medina D, Tilley WD, Aspinall JO, Cunha GR, Donjacour AA, Matusik RJ, Rosen JM. Prostate cancer in a transgenic mouse. Proc Natl Acad Sci USA. 1995;92:3439–3443. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gingrich JR, Greenberg NM. A transgenic mouse prostate cancer model. Toxicol Pathol. 1996;24:502–504. doi: 10.1177/019262339602400414. [DOI] [PubMed] [Google Scholar]
- 24.Huang CT, Huso DL, Lu Z, Wang T, Zhou G, Kennedy EP, Drake CG, Morgan DJ, Sherman LA, Higgins AD, Pardoll DM, Adler AJ. CD4+ T cells pass through an effector phase during the process of in vivo tolerance induction. J Immunol. 2003;170:3945–3953. doi: 10.4049/jimmunol.170.8.3945. [DOI] [PubMed] [Google Scholar]
- 25.Miller AM, Lundberg K, Ozenci V, Banham AH, Hellström M, Egevad L, Pisa P. CD4+CD25high T cells are enriched in the tumor and peripheral blood of prostate cancer patients. J Immunol. 2006;177:7398–7405. doi: 10.4049/jimmunol.177.10.7398. [DOI] [PubMed] [Google Scholar]
- 26.Ochando JC, Homma C, Yang Y, Hidalgo A, Garin A, Tacke F, Angeli V, Li Y, Boros P, Ding Y, et al. Alloantigen-presenting plasmacytoid dendritic cells mediate tolerance to vascularized grafts. Nat Immunol. 2006;7:652–662. doi: 10.1038/ni1333. [DOI] [PubMed] [Google Scholar]
- 27.Zhou G, Levitsky HI. Natural regulatory T cells and de novo-induced regulatory T cells contribute independently to tumor-specific tolerance. J Immunol. 2007;178:2155–2162. doi: 10.4049/jimmunol.178.4.2155. [DOI] [PubMed] [Google Scholar]
- 28.Bluestone JA, Abbas AK. Natural versus adaptive regulatory T cells. Nat Rev Immunol. 2003;3:253–257. doi: 10.1038/nri1032. [DOI] [PubMed] [Google Scholar]
- 29.Barnett B, Kryczek I, Cheng P, Zou W, Curiel TJ. Regulatory T cells in ovarian cancer: biology and therapeutic potential. Am J Reprod Immunol. 2005;54:369–377. doi: 10.1111/j.1600-0897.2005.00330.x. [DOI] [PubMed] [Google Scholar]
- 30.Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, Evdemon-Hogan M, Conejo-Garcia JR, Zhang L, Burow M, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10:942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- 31.Ormandy LA, Hillemann T, Wedemeyer H, Manns MP, Greten TF, Korangy F. Increased populations of regulatory T cells in peripheral blood of patients with hepatocellular carcinoma. Cancer Res. 2005;65:2457–2464. doi: 10.1158/0008-5472.CAN-04-3232. [DOI] [PubMed] [Google Scholar]
- 32.Kono K, Kawaida H, Takahashi A, Sugai H, Mimura K, Miyagawa N, Omata H, Fujii H. CD4+CD25high regulatory T cells increase with tumor stage in patients with gastric and esophageal cancers. Cancer Immunol Immunother. 2006;55:1064–1071. doi: 10.1007/s00262-005-0092-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 34.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 35.Shevach EM. From vanilla to 28 flavors: multiple varieties of T regulatory cells. Immunity. 2006;25:195–201. doi: 10.1016/j.immuni.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 36.Vignali DA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol. 2008;8:523–532. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 38.von Boehmer H. Mechanisms of suppression by suppressor T cells. Nat Immunol. 2005;6:338–344. doi: 10.1038/ni1180. [DOI] [PubMed] [Google Scholar]
- 39.Gingrich JR, Barrios RJ, Foster BA, Greenberg NM. Pathologic progression of autochthonous prostate cancer in the TRAMP model. Prostate Cancer Prostatic Dis. 1999;2:70–75. doi: 10.1038/sj.pcan.4500296. [DOI] [PubMed] [Google Scholar]
- 40.Willimsky G, Czeh M, Loddenkemper C, Gellermann J, Schmidt K, Wust P, Stein H, Blankenstein T. Immunogenicity of premalignant lesions is the primary cause of general cytotoxic T lymphocyte unresponsiveness. J Exp Med. 2008;205:1687–1700. doi: 10.1084/jem.20072016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grosso JF, Kelleher CC, Harris TJ, Maris CH, Hipkiss EL, De Marzo A, Anders R, Netto G, Getnet D, Bruno TC, et al. LAG-3 regulates CD8+ T cell accumulation and effector function in murine self- and tumor-tolerance systems. J Clin Invest. 2007;117:3383–3392. doi: 10.1172/JCI31184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Badovinac VP, Porter BB, Harty JT. Programmed contraction of CD8+ T cells after infection. Nat Immunol. 2002;3:619–626. doi: 10.1038/ni804. [DOI] [PubMed] [Google Scholar]
- 43.Badovinac VP, Porter BB, Harty JT. CD8+ T cell contraction is controlled by early inflammation. Nat Immunol. 2004;5:809–817. doi: 10.1038/ni1098. [DOI] [PubMed] [Google Scholar]
- 44.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 45.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 46.Saldanha AJ. Java Treeview: extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 47.Hagymasi AT, Slaiby AM, Mihalyo MA, Qui HZ, Zammit DJ, Lefrancois L, Adler AJ. Steady state dendritic cells present parenchymal self-antigen and contribute to, but are not essential for, tolerization of naive and Th1 effector CD4 cells. J Immunol. 2007;179:1524–1531. doi: 10.4049/jimmunol.179.3.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sfanos KS, Bruno TC, Maris CH, Xu L, Thoburn CJ, DeMarzo AM, Meeker AK, Isaacs WB, Drake CG. Phenotypic analysis of prostate-infiltrating lymphocytes reveals Th17 and Treg skewing. Clin Cancer Res. 2008;14:3254–3261. doi: 10.1158/1078-0432.CCR-07-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sugimoto N, Oida T, Hirota K, Nakamura K, Nomura T, Uchiyama T, Sakaguchi S. Foxp3-dependent and -independent molecules specific for CD25+CD4+ natural regulatory T cells revealed by DNA microarray analysis. Int Immunol. 2006;18:1197–1209. doi: 10.1093/intimm/dxl060. [DOI] [PubMed] [Google Scholar]
- 50.Hill JA, Feuerer M, Tash K, Haxhinasto S, Perez J, Melamed R, Mathis D, Benoist C. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27:786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 51.Zheng Y, Rudensky AY. Foxp3 in control of the regulatory T cell lineage. Nat Immunol. 2007;8:457–462. doi: 10.1038/ni1455. [DOI] [PubMed] [Google Scholar]
- 52.Kim JM, Rudensky A. The role of the transcription factor Foxp3 in the development of regulatory T cells. Immunol Rev. 2006;212:86–98. doi: 10.1111/j.0105-2896.2006.00426.x. [DOI] [PubMed] [Google Scholar]
- 53.Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat Immunol. 2005;6:1142–1151. doi: 10.1038/ni1263. [DOI] [PubMed] [Google Scholar]
- 54.Thornton AM, Shevach EM. CD4+CD25+ immunoregulatory T cells suppress polyclonal T cell activation in vitro by inhibiting interleukin 2 production. J Exp Med. 1998;188:287–296. doi: 10.1084/jem.188.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maynard CL, Harrington LE, Janowski KM, Oliver JR, Zindl CL, Rudensky AY, Weaver CT. Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3− precursor cells in the absence of interleukin 10. Nat Immunol. 2007;8:931–941. doi: 10.1038/ni1504. [DOI] [PubMed] [Google Scholar]
- 56.Huss WJ, Maddison LA, Greenberg NM. Autochthonous mouse models for prostate cancer: past, present and future. Semin Cancer Biol. 2001;11:245–260. doi: 10.1006/scbi.2001.0373. [DOI] [PubMed] [Google Scholar]
- 57.Zhou G, Drake CG, Levitsky HI. Amplification of tumor-specific regulatory T cells following therapeutic cancer vaccines. Blood. 2006;107:628–636. doi: 10.1182/blood-2005-07-2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Valzasina B, Piconese S, Guiducci C, Colombo MP. Tumor-induced expansion of regulatory T cells by conversion of CD4+CD25− lymphocytes is thymus and proliferation independent. Cancer Res. 2006;66:4488–4495. doi: 10.1158/0008-5472.CAN-05-4217. [DOI] [PubMed] [Google Scholar]
- 59.Verginis P, McLaughlin KA, Wucherpfennig KW, von Boehmer H, Apostolou I. Induction of antigen-specific regulatory T cells in wild-type mice: visualization and targets of suppression. Proc Natl Acad Sci USA. 2008;105:3479–3484. doi: 10.1073/pnas.0800149105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Apostolou I, Sarukhan A, Klein L, von Boehmer H. Origin of regulatory T cells with known specificity for antigen. Nat Immunol. 2002;3:756–763. doi: 10.1038/ni816. [DOI] [PubMed] [Google Scholar]
- 61.von Boehmer H. Peptide-based instruction of suppressor commitment in naive T cells and dynamics of immunosuppression in vivo. Scand J Immunol. 2005;62(Suppl 1):49–54. doi: 10.1111/j.1365-3083.2005.01609.x. [DOI] [PubMed] [Google Scholar]
- 62.Degl’Innocenti E, Grioni M, Capuano G, Jachetti E, Freschi M, Bertilaccio MT, Hess-Michelini R, Doglioni C, Bellone M. Peripheral T-cell tolerance associated with prostate cancer is independent from CD4+CD25+ regulatory T cells. Cancer Res. 2008;68:292–300. doi: 10.1158/0008-5472.CAN-07-2429. [DOI] [PubMed] [Google Scholar]
- 63.Neeley YC, Arredouani MS, Hollenbeck B, Eng MH, Rubin MA, Sanda MG. Partially circumventing peripheral tolerance for oncogene-specific prostate cancer immunotherapy. Prostate. 2008;68:715–727. doi: 10.1002/pros.20689. [DOI] [PubMed] [Google Scholar]
- 64.Mihalyo MA, Hagymasi AT, Slaiby AM, Nevius EE, Adler AJ. Dendritic cells program non-immunogenic prostate-specific T cell responses beginning at early stages of prostate tumorigenesis. Prostate. 2007;67:536–546. doi: 10.1002/pros.20549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lees JR, Charbonneau B, Swanson AK, Jensen R, Zhang J, Matusik R, Ratliff TL. Deletion is neither sufficient nor necessary for the induction of peripheral tolerance in mature CD8+ T cells. Immunology. 2006;117:248–261. doi: 10.1111/j.1365-2567.2005.02293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Willimsky G, Blankenstein T. The adaptive immune response to sporadic cancer. Immunol Rev. 2007;220:102–112. doi: 10.1111/j.1600-065X.2007.00578.x. [DOI] [PubMed] [Google Scholar]
- 67.Morse MA, Hobeika AC, Osada T, Serra D, Niedzwiecki D, Lyerly HK, Clay TM. Depletion of human regulatory T cells specifically enhances antigen-specific immune responses to cancer vaccines. Blood. 2008;112:610–618. doi: 10.1182/blood-2008-01-135319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dannull J, Su Z, Rizzieri D, Yang BK, Coleman D, Yancey D, Zhang A, Dahm P, Chao N, Gilboa E, Vieweg J. Enhancement of vaccine-mediated antitumor immunity in cancer patients after depletion of regulatory T cells. J Clin Invest. 2005;115:3623–3633. doi: 10.1172/JCI25947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mahnke K, Schonfeld K, Fondel S, Ring S, Karakhanova S, Wiedemeyer K, Bedke T, Johnson TS, Storn V, Schallenberg S, Enk AH. Depletion of CD4+CD25+ human regulatory T cells in vivo: kinetics of Treg depletion and alterations in immune functions in vivo and in vitro. Int J Cancer. 2007;120:2723–2733. doi: 10.1002/ijc.22617. [DOI] [PubMed] [Google Scholar]
- 70.Vieweg JW. Prostate cancer: targeting complexity. Curr Opin Urol. 2008;18:261–262. doi: 10.1097/MOU.0b013e3282f9b413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.