Abstract
Purpose
Pathologic examination of prostate glands removed from patients with prostate cancer commonly reveals infiltrating CD4+ and CD8+ T cells. Little is known about the phenotype of these cells, despite accumulating evidence suggesting a potential role for chronic inflammation in the etiology of prostate cancer.
Experimental Design
We developed a technique that samples the majority of the peripheral prostate through serial needle aspirates. CD4+ prostate-infiltrating lymphocytes (PIL) were isolated using magnetic beads and analyzed for subset skewing using both flow cytometry and quantitative reverse transcription-PCR. The transcriptional profile of fluorescence-activated cell sorted prostate-infiltrating regulatory T cells (CD4+, CD25+, GITR+) was compared with naïve, peripheral blood T cells using microarray analysis.
Results
CD4+ PIL showed a paucity of TH2 (interleukin-4– secreting) cells, a surprising finding given the generally accepted association of these cells with chronic, smoldering inflammation. Instead, CD4+ PIL seemed to be skewed towards a regulatory Treg phenotype (FoxP3+) as well as towards the TH17 phenotype (interleukin-17+). We also found that a preponderance of TH17-mediated inflammation was associated with a lower pathologic Gleason score. These protein level data were reflected at the message level, as analyzed by quantitative reverse transcription-PCR. Microarray analysis of pooled prostate-infiltrating Treg revealed expected Treg-associated transcripts (FoxP3, CTLA-4, GITR, LAG-3) as well as a number of unique cell surface markers that may serve as additional Treg markers.
Conclusion
Taken together, these data suggest that TH17 and/or Treg CD4+ T cells (rather than TH2 T cells) may be involved in the development or progression of prostate cancer.
The presence of CD3+ tumor-infiltrating lymphocytes has been shown to correlate with favorable outcome in patients with several types of cancer including ovarian and colorectal carcinomas (1, 2). A similar scenario has been proposed for prostate cancer, yet studies have yielded disparate results (3, 4). Indeed, prostate-infiltrating lymphocytes (PIL) are frequently present both in and around the tumor-burdened areas of the gland. Few studies have investigated the phenotype of CD4+ T cells infiltrating the peripheral zone of the prostate, where most adenocarcinomas arise. In addition to the TH1 and TH2 subsets, which secrete IFN-γ and interleukin (IL)-4, respectively, a new subset of CD4+ (helper) T cells, termed TH17 cells, has been characterized by the production of IL-17. Whereas all three CD4+ T cell subtypes are known to play a role in immunomediated defense against intracellular or extracellular pathogens, TH17 cells are unique in that they are the key mediators in a number of autoimmune diseases, and may play a role in inflammation-associated cancer (reviewed in refs. 5, 6). Interestingly, increased expression of IL-17 at the mRNA level was shown in tissue from both prostate cancer and benign prostatic hyperplasia before the discovery that TH17 T cells represent a distinct lineage (7). Whether TH17 cells comprise a higher proportion of PIL than the peripheral blood lymphocytes of patients with prostate cancer has not been investigated.
Although CD4+ T cells are present in the human prostate, it is not yet clear whether these cells mediate an antitumor effector function, or whether they serve to dampen or regulate a CD8+ T-cell–mediated antitumor response. Important in this respect are regulatory T cells (Treg), a CD4+ T cell lineage involved in the suppression of autoreactive T cells, and thus prevention of autoimmunity. In light of this role in the suppression of self-reactive cells, Treg have recently been investigated as suppressors of antitumor immune responses (reviewed in ref. 8). Increased numbers of Treg have been reported in tumor-infiltrating lymphocytes of several human solid tumors including breast, pancreatic, hepatocellular, and prostate carcinomas (9–11). Furthermore, increased percentages of Treg in the peripheral blood of gastric and esophageal cancer patients and in the tumor tissue of ovarian cancer patients correlated with poor prognosis and decreased survival (12, 13). In earlier studies, CD25 was used as a cell surface marker for Treg (8). More recent data have suggested that the forkhead box transcription factor FoxP3 is a more specific marker for Treg (14). Here, we used intracellular staining for FoxP3 to determine the prevalence of Treg among PILs.
In addition to these protein level studies, we also used quantitative reverse transcription-PCR (qRT-PCR) analysis to conduct a comprehensive analysis of the presence of these subsets (TH1, TH2, TH17) in PIL and to determine the relative prevalence of CD4+ Treg that are potentially capable of regulating effective antitumor immune responses. Finally, we investigated correlations between T-cell subsets and tumor Gleason score in order to provide data regarding a potential tumor-promoting (or tumor-inhibiting) role for each subset.
Materials and Methods
Patient population and clinical samples
All specimens were acquired under a Johns Hopkins Medicine Institutional Review Board – approved protocol with written informed consent obtained from each patient. PIL samples were obtained from 20 patients (ages 37–66; mean, 56 years old) undergoing radical retropubic prostatectomy for localized adenocarcinoma of the prostate at the Johns Hopkins Hospital in Baltimore, MD (Supplementary Table S1). None of the patients were previously treated with immunosuppressive or radiation therapy. Within 1 h of resection, needle aspirates were taken from the posterior aspect (peripheral zone) of the prostate using a 20-gauge 1.5-inch needle and 5 mL syringe, and collected into RPMI 1640 supplemented with <1% FCS. Because prostate cancer typically presents multiple foci in the peripheral zone of the prostate (15), the aim was to sample T cells from the entire zone as opposed to a single, macroscopically identifiable tumor, as even macroscopically normal-appearing prostate tissue could contain multiple small tumor foci. Cells were resuspended in 1 mL of Dynal Buffer 1 and strained through a 100-µm strainer. CD4+ T cells were positively isolated using the Dynal CD4 Positive Isolation Kit (Invitrogen) and the manufacturer’s recommended protocol, including magnetic bead detachment. Positively isolated CD4+ T cells were then washed once with CTL medium (RPMI, 1% l-glutamate, 1% nonessential amino acids, 1% sodium pyruvate, 1% penicillin/streptomycin, 10% FCS, and 3.47 µL/L 14.4 mol/L B-mercaptoethanol).
Patient-matched peripheral blood samples were collected by venipuncture into 8.5 mL whole blood tubes with ACD solution A (BD Biosciences Vacutainer Systems) the evening prior to radical retropubic prostatectomy surgery and remained at room temperature with gentle shaking overnight. Multiple control experiments were done to verify that expression of the cytokines examined in this study did not vary over the short overnight incubation period (data not shown). CD4+ T cell isolation was done as described for PIL samples, except with 2 mL of whole blood as starting material. Using a directly conjugated monoclonal antibody (mAb) against CD4 (APC, Caltag), ~5,000 cells from each patient were stained to verify the purity of positively isolated CD4+ T cells. Samples were typically >95% CD4-positive.
CD4+ T cells used for qRT-PCR analyses were isolated from patient-matched peripheral blood and prostate samples as described above, but without bead detachment. These samples were snap-frozen and stored at −80°C until RNA extraction.
CD4 T-cell stimulation
Isolated CD4+ T cells from prostate and peripheral blood were resuspended in CTL medium with 0.05 µg/mL of phorbol 12-myristate 13-acetate, 0.5 µg/mL of ionomycin, and 1:1,000 GolgiStop (BD Biosciences) and plated at a density of <1 × 106 in 96-well U-bottomed plates. Cells were stimulated for 4 h at 37°C prior to intracellular staining.
Intracellular antibody staining and flow cytometry
Surface staining with a PE-Cy5 labeled mAb to CD45RO (BD Biosciences) was done prior to cell permeabilization. Intracellular staining with directly conjugated mAbs against IFN-γ (APC) and IL-17 (PE; eBioscience, clones 4S.B3 and eBio64Dec17, respectively) was done using the BD Cytofix/Cytoperm Fixation/Permeabilization Kit according to the manufacturer’s recommended protocol. Intracellular staining with directly conjugated mAbs against FoxP3 (APC, eBioscience, clone PCH101) and IL-4 (PE, BD Biosciences) was done using the eBioscience Human Regulatory T-cell Staining Kit and the manufacturer’s recommended protocol. Each of these mAbs (IFN-γ, IL-17, IL-4, and FoxP3) were titrated using naïve CD4+ T cells skewed towards the respective subset in vitro, and those skewing conditions served as positive controls for staining (Supplementary Materials and Methods). Flow cytometry for all in vitro controls as well as human peripheral blood and prostate samples was conducted using FACSCalibur (BD Biosciences), and data were analyzed using FlowJo software (Tree Star, Inc.).
qRT-PCR
These studies were done with the assistance of the Human Immunology Core Facility at the Johns Hopkins University School of Medicine (Baltimore, MD). RNA from patient-matched peripheral blood and prostate CD4+ T cells was extracted using the Trizol reagent (Invitrogen) with modifications for low cell numbers including the addition of glycogen as an RNA carrier. Synthesis of cDNA was done using random primers and Ready-To-Go beads (GE Life Sciences). The level of gene expression was determined by quantitative PCR done in triplicate with multiplexed target and control gene primer/probe sets using an ABI 7000 prism system (Applied Biosystems). Threshold cycle (CT) values were calculated as the average of three runs for each gene and were normalized to CD4 using the equation: Ratio = 2 ^ (CT CD4 − CT target gene). The target genes GAPDH, CD4, IFN-γ, IL-17, IL-4, FoxP3, IL-23R, IL-12, and IL-10 were analyzed using commercially available primer/probe pairs (Applied Biosystems).
Transcriptional analysis
Peripheral blood from four patients undergoing radical retropubic prostatectomy was collected for isolation of naïve CD4 T cells. From each patient, 2 mL of whole blood was lysed with ACK lysing buffer (Quality Biological, Inc.) and PBMC were stained with the following directly conjugated mAbs: CD4 (FITC, Caltag), CD45RA (PE-Cy5, BD Biosciences), and CD25 (PE, Miltenyi Biotec). The CD4+CD45RA+CD25− population from peripheral blood samples (representing naïve CD4+ T cells) was sorted to >95% purity using a FacsVantage instrument (BD Biosciences). Radical prostatectomy specimens from 11 patients were aspirated as described previously and stained with directly conjugated mAbs to CD4 (PC5, Beckman Coulter), CD25 (PE, Miltenyi Biotec), and GITR (FITC, R&D Systems). As above, the CD4+CD25highGITR+ (Treg) population from prostate samples was sorted to >90% purity using a FacsVantage instrument. Cells were frozen in 1 mL of Trizol and stored at −80°C prior to RNA extraction using the Trizol reagent. The integrity of extracted RNA from both peripheral blood and prostate T cells was analyzed using an Agilent 2100 Bioanalyzer and the RNA 6000 Pico and Nano Kits (Agilent Technologies) and concentrations were determined using a NanoDrop spectrophotometer (NanoDrop Technologies). Transcriptional analysis was done at the Johns Hopkins Microarray Core facility. Per standard protocol, RNA was amplified from 20 ng of starting total RNA with the Nugen Ovation RNA Amplification System V2, following the manufacturer’s protocol.4 cDNA was synthesized using the Nugen FL-Ovation cDNA Biotin Module V2 kit, following the manufacturer’s protocol.4 After standard labeling, each sample was hybridized to an Affymetrix U133Plus 2.0 Human Genome array, followed by examination with an Affymetrix GeneChip Scanner 3000.
Statistical analyses
Differences between peripheral blood and prostate T cell populations determined by flow cytometry were analyzed using a two-sided Student’s t test and the PRISM package (GraphPad Software, Inc.). Statistical analyses for the microarray experiment are described in the Supplementary Materials and Methods.
Results
TH17 skewing in PIL
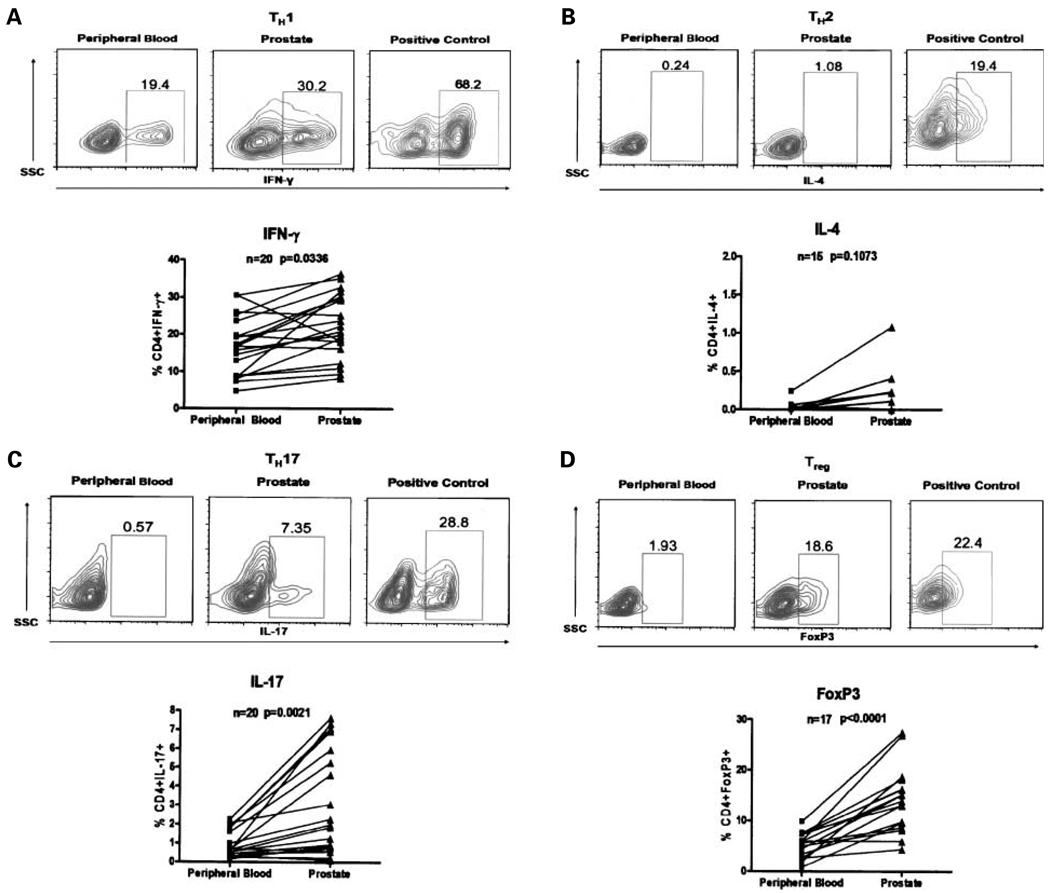
We first did a subset-specific phenotypic analysis of the CD4+ T cells in PIL from radical prostatectomy specimens. Numerically, the most abundant CD4+ T-cell subset observed in PIL were IFN-γ–secreting TH1 cells (Fig. 1A). Interestingly, we found almost no IL-4–secreting cells among these PIL, suggesting a skewing away from a TH2 phenotype when compared with a positive in vitro – derived TH2 control (Fig. 1B). These data are in contrast with suggestions that prostate cancer arises in the context of TH2-mediated inflammation (16). Remarkably, a significant skewing towards a TH17 phenotype was noted in the PIL (Fig. 1C). In some patients, up to 8% of the CD4+ PIL secreted IL-17 upon brief stimulation. A number of factors influence the differentiation of naïve CD4+ T cells towards a TH17 phenotype (5). Among these, transforming growth factor-β (TGF-β) and IL-6 have been associated with prostate cancer (17, 18), and might be present in the prostate microenvironment. It should be noted that although TGF-β and IL-6 have been shown to drive the initial lineage commitment of TH17 cells in mice (19–22), recent data using human naïve CD4+ T cells suggest that TH17 polarization is induced by IL-1β, possibly enhanced by IL-6, and may be suppressed by TGF-β (23, 24).
Fig. 1.
Frequency of Thelper subsets in peripheral blood and prostate tissue of patients with prostate cancer. Positively isolated CD4+ T cells were stimulated for 4 h in the presence of phorbol 12-myristate13-acetate and ionomycin and analyzed by flow cytometry. Representative fluorescence-activated cell sorting plots of CD4+IFN-γ+ (TH1), CD4+IL-4+ (TH2), CD4+IL-17+ (TH17), and CD4+FoxP3+ (Treg) T cells in peripheral blood and prostate tissue of prostate cancer patients with representative positive controls for each cellular marker from independent in vitro skewing experiments as well as a summary of the data. Percentage of CD4+ T cells positive for IFN-γ, IL-4, IL-17, and FoxP3. P values were calculated using two-sided Student’s t test.
A regulatory phenotype in PIL
Because TH17 cells were predominant among PIL, and these cells may differentiate under the influence of TGF-β, we queried whether Treg (which also differentiate in response to TGF-β) would be present in the prostate glands of men with cancer. In this regard, previous work had already identified CD4+CD25high T cells as a component of prostate tumor-infiltrating lymphocytes (11). To extend those studies, we quantified the Treg component of PIL based on intracellular FoxP3 staining, as FoxP3 represents a potentially more specific Treg marker (14). As shown in Fig. 1D, nearly all patients examined showed a relative enrichment of FoxP3+ Treg with respect to peripheral blood.
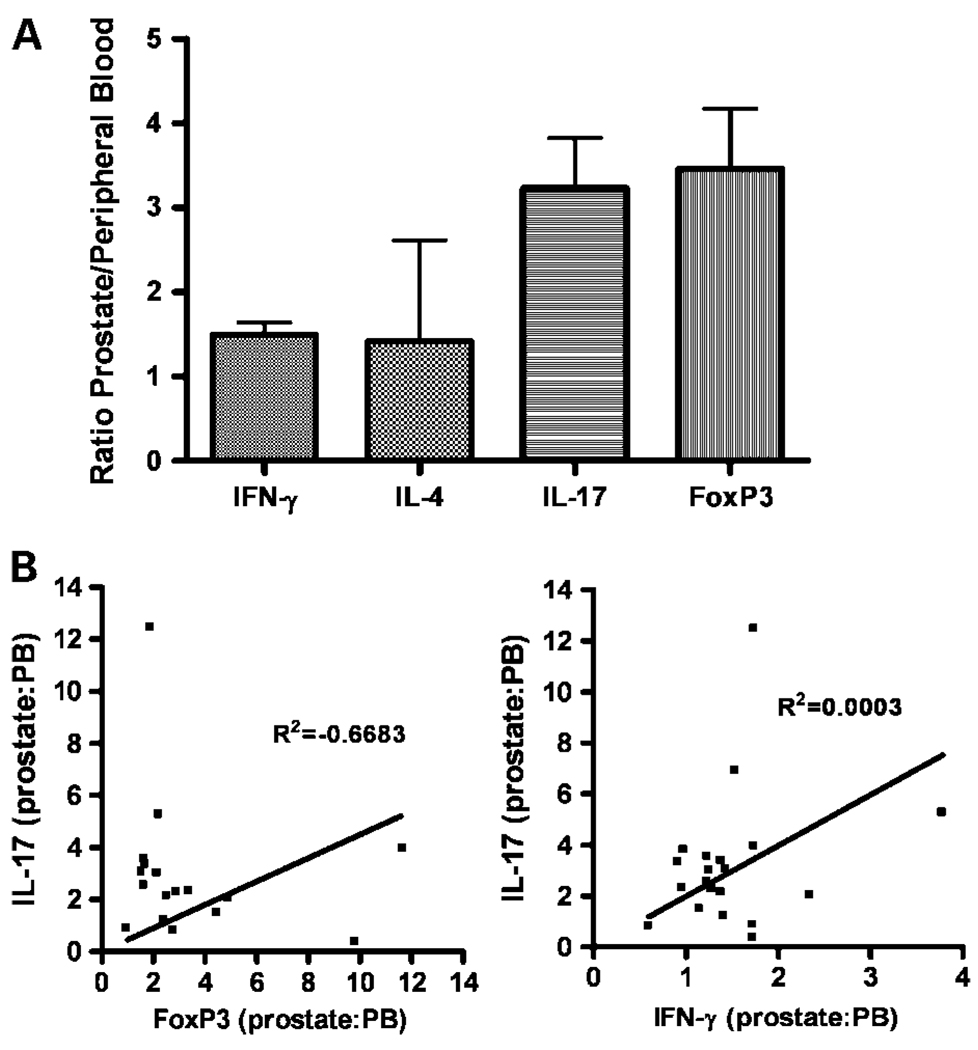
Taken together, this phenotypic analysis of PIL suggests that these cells are neither proinflammatory nor consistent with a “smoldering” TH2-mediated process. Rather, the CD4+ T cells in the prostate seem to be skewed towards either the TH17 phenotype associated with autoimmunity or the Treg phenotype, which down-regulates CD8+ T cell function (25). We next examined these data in summary, comparing the relative ratio of each T-cell subset in the PIL versus peripheral blood. These data (Fig. 2A) confirm the notion that PIL are generally not skewed towards a TH2 phenotype, but rather, seem to be biased towards TH17 and Treg. It should be noted that there is a weak skewing of PIL towards a TH1 phenotype as well. It is feasible that in some patients, these cells represent potential functional CD4+ effector cells, a point which will be discussed in further detail below.
Fig. 2.
Prostate-infiltrating T cells are not skewed toward a TH2 phenotype. A, ratio of prostate-infiltrating T cells positive for each of the markers shown/peripheral blood T cells for that marker. B, correlative data for FoxP3 vs. IL-17 and IFN-γ vs. IL-17 (patients were compared by ratio of prostate/peripheral blood for each marker). R2 value determined by linear least-squares regression.
Due to recent conflicting data regarding the potential influence of TGF-β in the development of Treg and TH17 cells, we sought to determine whether these two subsets cosegregated in patients, i.e., whether patients with a relative up-regulation of TH17 cells in the prostate also showed a relative upregulation of Treg. As shown in Fig. 2B, this was not the case, patients with a relative TH17 skewing seemed to show less of a Treg bias. Because one major difference between TH17 and Treg development is the presence of IL-6 during the differentiation process (5), it might be that patients with relative TH17 skewing had higher levels of IL-6. We tested this hypothesis by determining serum IL-6 levels in these patients using ELISA analysis (data not shown). Those results did not show a significant correlation between serum IL-6 levels and TH17 skewing; however, it is far more likely that local (i.e., intraprostatic) IL-6 levels would need to be examined to fully determine a role for IL-6 in this process. Recent data also indicate that IL-2 might play a role in regulating the TH17/Treg balance in the tumor microenvironment (26), but once again, those data would need to be determined at the tissue level rather than at the serum level. We also examined a potential relationship between TH17 and TH1 skewing (Fig. 2B). As expected, no such relationship was found (R2 = 0.0003).
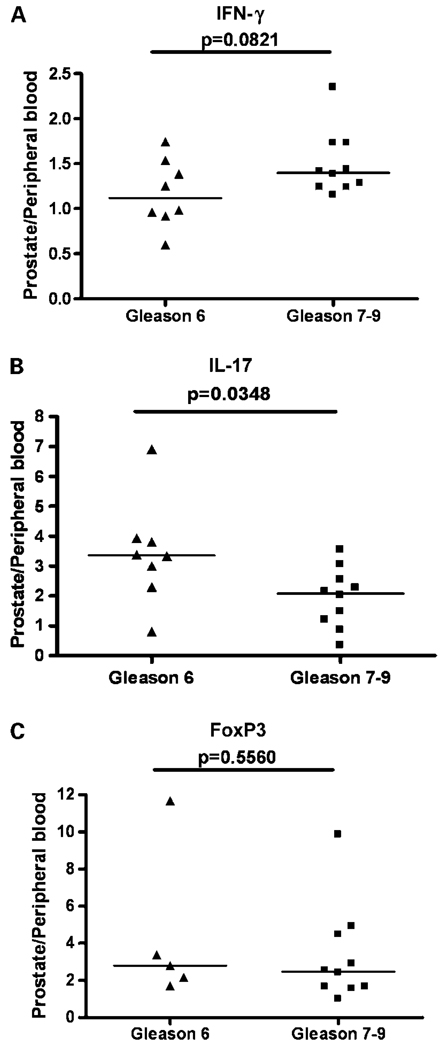
TH17 skewing of prostate-infiltrating CD4+ T cells inversely correlates with Gleason grade
As there is accumulating evidence for an association between increased numbers of tumor-infiltrating Treg and high-grade or high-risk disease for several types of cancer (12, 13, 27), we next sought to determine whether Treg skewing (as assayed at the protein level by intracellular FoxP3 staining) associated with disease grade in this group of men with prostate cancer. As shown in Fig. 3, such an association was not observed. We further hypothesized that an effector phenotype (TH1) might be associated with a relatively lower tumor grade, as in some systems, TH1-skewed T cells seemed to mediate an antitumor effect (28). This was also not observed; in these samples, there was a nonsignificant trend towards a more pronounced intraprostatic TH1 skewing in men with a higher Gleason grade. Finally, we examined the relationship between TH17 skewing and Gleason grade, testing the hypothesis that a more pronounced TH17 skewing would be associated with a higher tumor grade as suggested by the data of Langowski et al. (28). Interestingly, a statistically significant inverse correlation was found between TH17 skewing and tumor grade (Fig. 3). This finding is surprising and suggests the interesting possibility that TH17 T cells in the prostate might potentially mediate an antitumor effect. An alternative hypothesis is that TH17 skewing is an independent result of locally produced cytokines, i.e., as tumors progress, the local milieu progresses from one that favors TH17 skewing (TGF-β, IL-6, etc.) to one that favors TH1 skewing (IL-12 and IL-18). We are currently in the process of investigating a comprehensive series of tissue microarrays from patients with prostate cancer to confirm these results with a larger patient set.
Fig. 3.
TH17 skewing of prostate-infiltrating CD4+ T cells inversely correlates with Gleason grade. Y-axis, ratio of prostate-infiltrating CD4+ T cells positive for a given cytokine or marker/peripheral blood CD4+ T cells positive for that cytokine or marker. Bar, median; P values were calculated by two-sided Student’s t test.
qRT-PCR analysis of CD4+ PIL
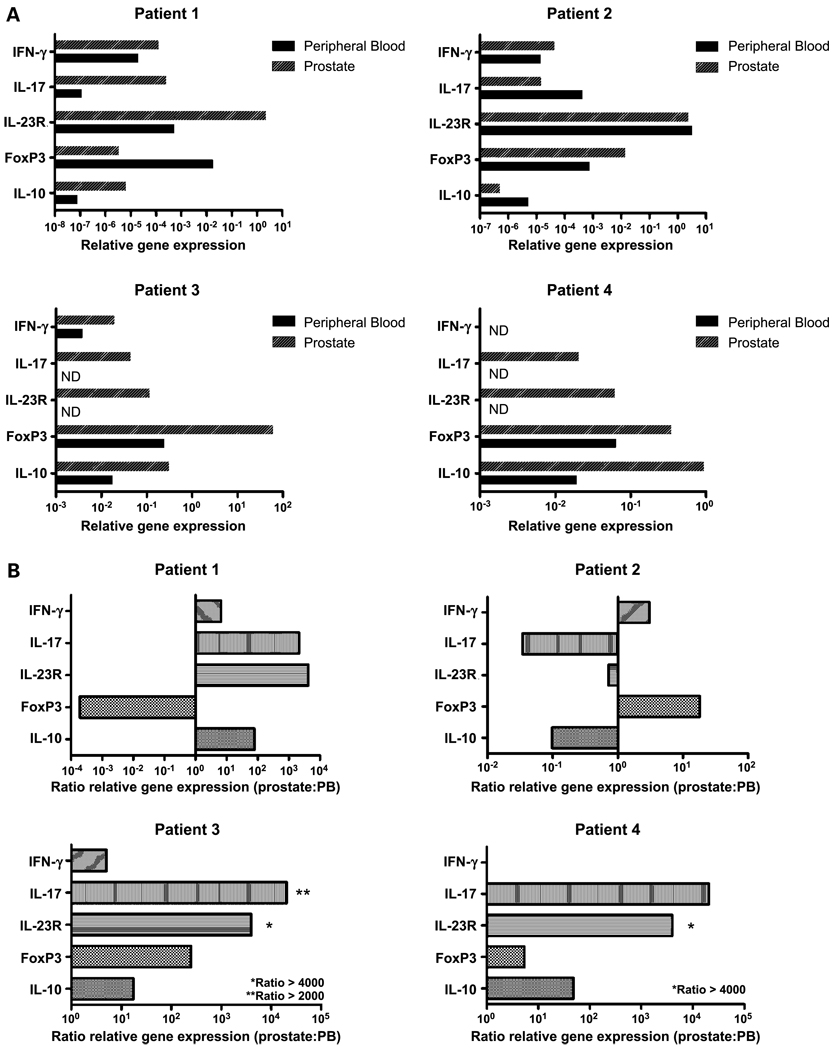
To determine whether the increased frequency of CD4+ T cells producing cytokines indicative of TH17 or Treg skewing in PIL by flow cytometry corresponds to increased transcription at the RNA level, we did qRT-PCR analyses. This technique also allows us to develop a more comprehensive picture of cytokine expression in the CD4+ T cells that infiltrate the prostate gland of an individual cancer patient, as multiple messages could be simultaneously analyzed from relatively small cell numbers. As shown in Fig. 4, the profiles suggested by fluorescence-activated cell sorting analysis were largely reflected at the message level. Three of four patients analyzed showed a relative up-regulation of IL-17 in the prostate. This was accompanied by an increase in IL-23 receptor message level. These data are interesting, as IL-23 signaling is required for TH17 maintenance (5). The FoxP3 message was also up-regulated in three of the four patients analyzed, again confirming the fluorescence-activated cell sorting data (Figs. 1 and 2). These data also address a potential correlation between Treg and TH17 infiltration (Fig. 2B); in two of the patients (patients 3 and 4) messages for FoxP3 and IL-17 seemed to correlate, but in the other two patients studied (patients 1 and 2), they seemed to digress. Overall, qRT-PCR analyses generally confirms the flow cytometry data, suggesting that the protein level cytokine changes are mediated at the message level; but these data also suggest that the situation is complex because, in some patients, the CD4+ T cell phenotype may be differentially skewed towards either a Treg or TH17 phenotype.
Fig. 4.
Prostate-infiltrating CD4+ T cells have heterogeneous cytokine gene expression profiles. A, qRT-PCR results normalized to CD4 message levels. B, ratio of message levels in prostate versus peripheral blood. IL-4 and IL-12 expression were not shown because message levels were undetectable in all patients. ND, not detectable. The Gleason scores for patients1 to 4 were as follows: 3 + 3 = 6; 3 + 4 = 7; 3 + 4 = 7; 4 + 3 = 7 (tertiary 5).
Microarray analysis of prostate-infiltrating Treg
Although several studies have explored the transcriptional phenotype of regulatory T cells, the profile of human tissue-resident Treg has yet to be reported (29–31). To address this question, we enriched Treg from the prostate gland by sorting CD4+CD25high− GITR+ cells. Sorted cells were >90% pure, but because the isolation procedure was stringent, we could not obtain sufficient cells for microarray analysis from a single patient. Therefore, sorted Treg were pooled from a number of patients. Although pooling Treg samples may introduce some “noise,” pooling also has the potential advantage of dampening low-level message level changes introduced by individual patient variability. As a reference CD4+ T cell population, we used naïve CD4+ T cells from the patients’ peripheral blood. Thus, differentially regulated transcripts will consist of Treg-specific messages, prostate homing markers as well as T cell activation markers associated with the CD25+ phenotype. The complete results of this analysis are presented in Supplementary Tables S2 and S3, and selected transcripts are highlighted in Table 1. As expected, we noted the up-regulation of Treg-associated transcripts such as FoxP3, providing a broad verification of our sorting and analysis procedures. The Treg in the prostate also seem to up-regulate a number of additional inhibitory markers, including CTLA-4 and LAG-3. In addition, we noted the increased expression of several TH17-associated transcripts, reflecting the notion that our sorting procedure also captures a prostate-infiltrating population of activated CD4+ T cells (CD25+), and providing further verification of both our flow cytometry data (Fig. 1) and our qRT-PCR data (Fig. 4). Interestingly, the CCR homing molecule most upregulated in these cells is CCR2, consistent with recent data describing CCR2 as a marker for human TH17 T cells (32). Finally, these prostate-infiltrating T cells also seem to display a relative up-regulation of TLR-3 and TLR-7. These receptors recognize double-stranded and single-stranded RNA, respectively; thus, these data are especially interesting in light of recent data demonstrating a novel retrovirus in some patients with prostate cancer (33).
Table 1.
Selected transcripts up-regulated in prostate-infiltrating Treg
| Probe ID | Fold increase | Gene definition | Gene symbol | Cellular component |
|---|---|---|---|---|
| Treg associated | ||||
| 211786_at | 122 | Tumor necrosis factor receptor superfamily, member 9 (4-1BB) | TNFRSF9 | Membrane |
| 234895_at | 95 | CTL-associated protein 4 | CTLA4 | Membrane |
| 206486_at | 86 | Lymphocyte-activation gene 3 | LAG3 | Membrane |
| 211269_s_at | 58 | IL-2 receptor-α (CD25) | IL2RA | Membrane |
| 223851_s_at | 31 | Tumor necrosis factor receptor superfamily, member 18 (GITR) | TNFRSF18 | Membrane |
| 224211_at | 17 | Forkhead box P3 | FOXP3 | Nucleus |
| Cytokines | ||||
| 216876_s_at | 262 | IL-17A | IL17A | Extracellular |
| 210354_at | 148 | IFNγ | IFNG | Extracellular |
| 207433_at | 116 | IL-10 | IL10 | Extracellular |
| 205207_at | 29 | IL-6 (IFNβ2) | IL6 | Extracellular |
| Cytokine receptors | ||||
| 202948_at | 254 | IL-1 receptor, type I | IL1R1 | Membrane |
| 1552912_a_at | 39 | IL-23 receptor | IL23R | Membrane |
| Chemokines | ||||
| 205476_at | 291 | Chemokine (C-C motif) ligand 20 | CCL20 | Extracellular |
| 204103_at | 189 | Chemokine (C-C motif) ligand 4 | CCL4 | Extracellular |
| 216598_s_at | 138 | Chemokine (C-C motif) ligand 2 | CCL2 | Extracellular |
| 205114_s_at | 52 | Chemokine (C-C motif) ligand 3 | CCL3 | Extracellular |
| Chemokine receptors | ||||
| 206978_at | 111 | Chemokine (C-C motif) receptor 2 | CCR2 | Membrane |
| 206991_s_at | 45 | Chemokine (C-C motif) receptor 5 | CCR5 | Membrane |
| 206983_at | 17 | Chemokine (C-C motif) receptor 6 | CCR6 | Membrane |
| Toll-like receptors | ||||
| 206271_at | 310 | Toll-like receptor 3 | TLR3 | Membrane |
| 220146_at | 26 | Toll-like receptor 7 | TLR7 | Membrane |
NOTE: See Supplementary Tables S2 and S3 for complete data set.
Discussion
Recent studies have suggested that chronic or recurrent inflammation may play a role in the development of many types of cancer—including prostate cancer (34). Although a TH2 phenotype has been associated with inflammation, the TH17 subset has been described as proinflammatory as well. In addition, TH17 cells have been determined to have a distinct lineage from Treg (5). Our data show that, in men with prostate cancer, a significant portion of the cells that infiltrate the gland are skewed towards a TH17 phenotype. This result is not in of itself surprising, as TH17 cells most likely develop under the influence of the cytokines TGF-β and IL-6, and both of these cytokines have been associated with advanced prostate cancer (17, 18). Indeed, the presence of TH17 cells in the prostate gland of men with benign prostatic hyperplasia has been previously reported (7), before these cells were known to represent a distinct lineage. What is surprising, however, is that a relative skewing of PIL with respect to peripheral blood seems to correlate inversely with Gleason grade. A seminal study by Langowski et al. (28) showed that TH17 cells seemed to be required for the development of cancer in an animal model, and a number of additional studies suggest that IL-23/IL-17 – mediated immunity thwarts active tumor surveillance (35). Our results may reflect a bystander role, wherein the tumor microenvironment shifts away from a TH17 skewing once prostate cancer progresses. They might also reflect a differential role for TH17 T cells in tumors of different grades. Most intriguingly, these data also raise the interesting possibility that TH17 T cells might, in some cancers, mediate an antitumor effect, a hypothesis best evaluated using animal models of tumor immunity. A major limitation of the present study is the lack of prostate tissue samples from patients with no evidence of cancer. The difficulties in obtaining such tissues are considerable. First, as radical retropubic prostatectomies are restricted to men with prostate cancer, this makes an identical isolation procedure difficult. Whereas in the present study, we were able to obtain lymphocyte samples within 1 hour of organ resection, autopsy samples would be challenging, if not impossible, to obtain in an identical time frame. Likewise, autopsy studies show that the incidence of occult prostate cancer is fairly high (~50% in men ages 50, and 70% in men >70 years of age; ref. 36). Thus, we would have to obtain tissue from an additional 40 to 50 age-matched men for such an analysis. We have explored the possibility of obtaining samples from men undergoing radical cystoprostatectomy for bladder cancer. Such studies are in the planning stages, however, even these are compromised by the possibility that cancer in an adjacent organ may alter the systemic cytokine profile and the PIL population.
Our data further confirm recent studies demonstrating that Treg infiltrate the prostate gland (11). As those earlier studies were done without FoxP3 staining, our data most likely represent a more specific evaluation of Treg in the prostate; however, it should be noted that FoxP3 is certainly not a perfect or exclusive Treg marker (37), and may mark a population of activated T cells in humans. These data were confirmed at both the protein and transcriptional level, using qRT-PCR as well as microarray analysis. The Treg-associated markers CTLA-4, 4-1BB, and LAG-3 seem to be highly up-regulated in prostate-infiltrating CD4+ T cells, suggesting potential targets for immunotherapeutic intervention. In addition, our data reveal a number of cell surface markers not previously associated with Treg. We are currently evaluating the specificity as well as the functional role of a number of these markers.
Several groups have suggested that cancers which arise in the context of inflammation arise out of a chronic, TH2-mediated inflammatory milieu (16), and such data are well supported by experimental studies. In the prostate, we were unable to find significant numbers of TH2-skewed CD4+ T cells either at the message or protein level. One possibility to explain these data would be that the cells are present, but nonfunctional—we are currently developing techniques to perform immunohistochemical staining for the major transcription factor of TH2 cells (GATA-3) in prostate tissue microarray collections. In contrast, TH1 cells were found to be the most predominantly enriched CD4+ T-cell subset in the prostate glands of these patients. This observation is particularly intriguing in light of recent evidence which supports the notion that at least a subset of tumors may exist in an “equilibrium state” specifically maintained by the adaptive immune system (38). However, the presence of these cells was not associated with a lower Gleason grade, as might be predicted. Indeed, our data suggested the opposing possibility, that TH1-mediated inflammation might be associated with a higher Gleason grade, again, a possibility best addressed through a comprehensive tissue microarray analysis.
In summary, these data provide the first comprehensive, protein level analyses of the phenotype of CD4+ T cells in the glands of men with prostate cancer. They document a relative skewing towards TH17, and raise important questions regarding a potential causal versus antitumor role for these cells in the development of human prostate cancer. We also document the presence of regulatory T cells, but the lack of correlation between Treg skewing and Gleason grade also raises interesting questions regarding the relative role of these cells in the etiology of prostate cancer versus other tumor types. Finally, we report the first microarray analyses of CD4+ Treg from the prostate glands of men with cancer—those data confirm several known cell surface markers which are targets for antitumor immunotherapy approaches and also suggest a number of additional cell surface markers and homing receptors potentially important in either the etiology or in the immunotherapeutic treatment of this common disease.
Supplementary Material
Acknowledgments
We thank the Johns Hopkins Microarray Core Facility and Dr. Chunfa Jie for microarray analyses, and Dr. Alan Hess of the Johns Hopkins immune monitoring core.
Grant support: C.G. Drake is a Damon Runyon Clinical Fellow. This work was also supported by NIH grants, a grant from the Patrick C. Walsh Prostate Cancer Research Fund (C.G. Drake), and an award from the Prostate Cancer Foundation (A.K. Meeker).
Footnotes
Reprints and Subscriptions To order reprints of this article or to subscribe to the journal, contact the AACR Publications Department at pubs@aacr.org.
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Zhang L, Conejo-Garcia JR, Katsaros D, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348:203–213. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 2.Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 3.Vesalainen S, Lipponen P, Talja M, Syrjanen K. Histological grade, perineural infiltration, tumour-infiltrating lymphocytes and apoptosis as determinants of long-term prognosis in prostatic adenocarcinoma. Eur J Cancer. 1994;30A:1797–1803. doi: 10.1016/0959-8049(94)e0159-2. [DOI] [PubMed] [Google Scholar]
- 4.McArdle PA, Canna K, McMillan DC, McNicol AM, Campbell R, Underwood MA. The relationship between T-lymphocyte subset infiltration and survival in patients with prostate cancer. Br J Cancer. 2004;91:541–543. doi: 10.1038/sj.bjc.6601943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM. Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity. 2006;24:677–688. doi: 10.1016/j.immuni.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Bettelli E, Oukka M, Kuchroo VK. T(H)-17 cells in the circle of immunity and autoimmunity. Nat Immunol. 2007;8:345–350. doi: 10.1038/ni0407-345. [DOI] [PubMed] [Google Scholar]
- 7.Steiner GE, Newman ME, Paikl D, et al. Expression and function of pro-inflammatory interleukin IL-17 and IL-17 receptor in normal, benign hyperplastic, and malignant prostate. Prostate. 2003;56:171–182. doi: 10.1002/pros.10238. [DOI] [PubMed] [Google Scholar]
- 8.Beyer M, Schultze JL. Regulatory T cells in cancer. Blood. 2006;108:804–804. doi: 10.1182/blood-2006-02-002774. [DOI] [PubMed] [Google Scholar]
- 9.Liyanage UK, Moore TT, Joo H-G, et al. Prevalence of regulatory T cells is increased in peripheral blood and tumor microenvironment of patients with pancreas or breast adenocarcinoma. J Immunol. 2002;169:2756–2761. doi: 10.4049/jimmunol.169.5.2756. [DOI] [PubMed] [Google Scholar]
- 10.Ormandy LA, Hillemann T, Wedemeyer H, Manns MP, Greten TF, Korangy F. Increased populations of regulatory T cells in peripheral blood of patients with hepatocellular carcinoma. Cancer Res. 2005;65:2457–2464. doi: 10.1158/0008-5472.CAN-04-3232. [DOI] [PubMed] [Google Scholar]
- 11.Miller AM, Lundberg K, Ozenci V, et al. CD4+CD25high T cells are enriched in the tumor and peripheral blood of prostate cancer patients. J Immunol. 2006;177:7398–7405. doi: 10.4049/jimmunol.177.10.7398. [DOI] [PubMed] [Google Scholar]
- 12.Curiel TJ, Coukos G, Zou L, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10:942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- 13.Kono K, Kawaida H, Takahashi A, et al. CD4(+)CD25high regulatory T cells increase with tumor stage in patients with gastric and esophageal cancers. Cancer Immunol Immunother. 2006;55:1064–1071. doi: 10.1007/s00262-005-0092-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- 15.Nelson WG, DeMarzo AM, Isaacs WB. Prostate cancer. N Engl J Med. 2003;349:366–381. doi: 10.1056/NEJMra021562. [DOI] [PubMed] [Google Scholar]
- 16.Balkwill F, Charles KA, Mantovani A. Smoldering and polarized inflammation in the initiation and promotion of malignant disease. Cancer Cell. 2005;7:211–217. doi: 10.1016/j.ccr.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Barrack ER. TGFβ in prostate cancer: a growth inhibitor that can enhance tumorigenicity. Prostate. 1997;31:61–70. doi: 10.1002/(sici)1097-0045(19970401)31:1<61::aid-pros10>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 18.Chung TD, Yu JJ, Spiotto MT, Bartkowski M, Simons JW. Characterization of the role of IL-6 in the progression of prostate cancer. Prostate. 1999;38:199–207. doi: 10.1002/(sici)1097-0045(19990215)38:3<199::aid-pros4>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 19.Bettelli E, Carrier Y, Gao W, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 20.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGF[β] in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Mangan PR, Harrington LE, O’Quinn DB, et al. Transforming growth factor-[β] induces development of the TH17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 22.McGeachy MJ, Bak-Jensen KS, Chen Y, et al. TGF-[β] and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain TH-17 cell-mediated pathology [advanced online publication] Nat Immunol. 2007;8:1390–1397. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- 23.Acosta-Rodriguez EV, Napolitani G, Lanzavecchia A, Sallusto F. Interleukins 1[β] and 6 but not transforming growth factor-[β] are essential for the differentiation of interleukin 17-producing human T helper cells. Nat Immunol. 2007;8:942–949. doi: 10.1038/ni1496. [DOI] [PubMed] [Google Scholar]
- 24.Wilson NJ, Boniface K, Chan JR, et al. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol. 2007;8:950–957. doi: 10.1038/ni1497. [DOI] [PubMed] [Google Scholar]
- 25.Chen M-L, Pittet MJ, Gorelik L, et al. Regulatory T cells suppress tumor-specific CD8 T cell cytotoxicity through TGF-{β} signals in vivo. Proc Natl Acad Sci U S A. 2005;102:419–424. doi: 10.1073/pnas.0408197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kryczek I, Wei S, Zou L, et al. Cutting edge: Th17 and regulatory T cell dynamics and the regulation by IL-2 in the tumor microenvironment. J Immunol. 2007;178:6730–6733. doi: 10.4049/jimmunol.178.11.6730. [DOI] [PubMed] [Google Scholar]
- 27.Sasada T, Kimura M, Yoshida Y, Kanai M, Takabayashi A. CD4+CD25+ regulatory T cells in patients with gastrointestinal malignancies: possible involvement of regulatory T cells in disease progression. Cancer. 2003;98:1089–1099. doi: 10.1002/cncr.11618. [DOI] [PubMed] [Google Scholar]
- 28.Langowski JL, Zhang X, Wu L, et al. IL-23 promotes tumour incidence and growth. Nature. 2006;442:461–465. doi: 10.1038/nature04808. [DOI] [PubMed] [Google Scholar]
- 29.Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 30.Marson A, Kretschmer K, Frampton GM, et al. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445:931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng Y, Rudensky AY. Foxp3 in control of the regulatory T cell lineage. Nat Immunol. 2007;8:457–462. doi: 10.1038/ni1455. [DOI] [PubMed] [Google Scholar]
- 32.Sato W, Aranami T, Yamamura T. Cutting edge: human Th17 cells are identified as bearing CCR2+CCR5− phenotype. J Immunol. 2007;178:7525–7529. doi: 10.4049/jimmunol.178.12.7525. [DOI] [PubMed] [Google Scholar]
- 33.Urisman A, Molinaro RJ, Fischer N, et al. Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006;2:e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.DeMarzo AM, Platz EA, Sutcliffe S, et al. Inflammation in prostate carcinogenesis. Nat Rev Cancer. 2007;7:256–269. doi: 10.1038/nrc2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langowski JL, Kastelein RA, Oft M. Swords into plowshares: IL-23 repurposes tumor immune surveillance. Trends Immunol. 2007;28:207–212. doi: 10.1016/j.it.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 36.Sakr WA, Grignon DJ, Crissman JD, et al. High grade prostatic intraepithelial neoplasia (HGPIN) and prostatic adenocarcinoma between the ages of 20–69: an autopsy study of 249 cases. In Vivo. 1994;8:439–443. [PubMed] [Google Scholar]
- 37.Shevach EM. From vanilla to 28 flavors: multiple varieties of T regulatory cells. Immunity. 2006;25:195–201. doi: 10.1016/j.immuni.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 38.Koebel CM, Vermi W, Swann JB, et al. Adaptive immunity maintains occult cancer in an equilibrium state. Nature. 2007;450:903–907. doi: 10.1038/nature06309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.