Abstract
Tailed-bacteriophage virions contain a single linear dsDNA chromosome which can range in size from about 18 to 500 kbp across the known tailed-phage types. These linear chromosomes can have one of several known types of termini as follows: cohesive ends (5′- or 3′-single-strand extensions), circularly permuted direct terminal repeats, short or long exact direct terminal repeats, terminal host DNA sequences, or covalently bound terminal proteins. These different types of ends reflect differing DNA replication strategies and especially differing terminase actions during DNA packaging. In general, complete genome sequence determination does not by itself elucidate the nature of these ends, so directed experimental analysis is usually required to understand the nature of the virion chromosome ends. This chapter discusses these methods.
Keywords: Terminase, tailed-phages, cohesive ends, terminal redundancy, DNA packaging
1 Introduction
1.1 General Background
The tailed-bacteriophage virions all contain single, linear dsDNA molecules that are packaged into a procapsid by similar DNA translocase molecular motors; however, their DNA replication strategies and the resulting nature of ends of the packaged DNAs are not all the same. Their virion DNAs have six well-studied types of termini which are characterized by the presence of (i) single-stranded cohesive ends, (ii) circularly permuted direct terminal repeats, (iii) short, several hundred base pairs exact (non-permuted) direct terminal repeats, (iv) long, several thousand base pairs exact (non-permuted) direct terminal repeats, (v) terminal host DNA sequences, and (vi) covalently bound terminal proteins (Table 7.1). Five of the above six types of virion chromosome ends are generated by nucleolytic cleavage from replicating concatemeric or circular DNA molecules; only the phage genomes with terminal proteins replicate as monomeric linear molecules. These cleavages are in all cases tightly coupled to the DNA packaging process and are performed by a phage-encoded enzyme called “terminase,” so named because this cleavage creates the termini of the virion DNA (1,2).
Table 7.1.
Termini of tailed-phage virion DNAs.
| Terminus type | Prototype phage | Replication strategy |
|---|---|---|
| Cohesive ends | ||
| 5′-single-strand extension | λ P2 | Rolling circle→concatemer |
| Circle→circle | ||
| 3′-single-strand extension | HK97 | Rolling circle→ concatemer* |
| Circularly permuted direct terminal repeats† | ||
| T4 | Complex→concatemer | |
| P22 | Rolling circle→concatemer | |
| P1 | Rolling circle→concatemer | |
| Host DNA at termini | ||
| Mu | Duplicative transposition into host DNA | |
| Exact direct terminal repeats | ||
| Short (few hundred base pairs) | T7 | Linear→concatemer |
| Long (thousands of base pairs) | SPO1 | Complex→concatemer |
| T5 | Complex→concatemer | |
| Covalent terminal protein | ||
| ϕ29 | Protein-primed linear→linear | |
Individual virions of these phages have chromosomes that terminate at many different places on the genome sequence, and the length of the terminal repeat varies among individual virions (see text).
Genomic analysis predicts this mode of replication, but it has not been studied experimentally.
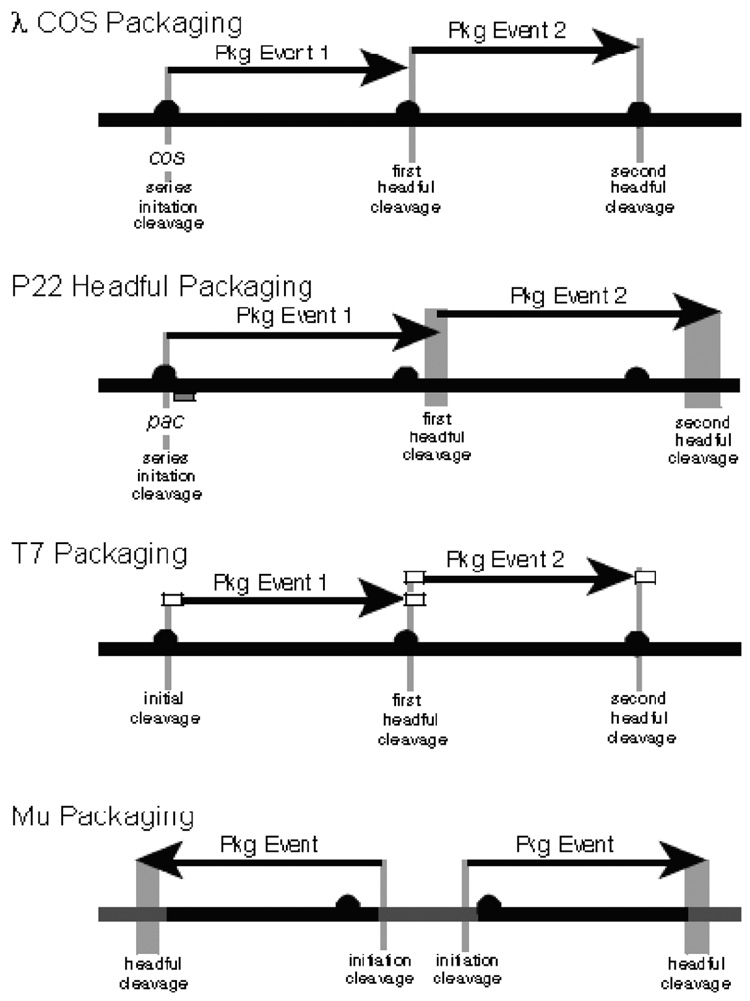
Most tailed-phage package DNA from concatemeric substrates that result from rolling-circle or more complex replication mechanisms. Among those studied, only the P2-like phages (subgroup of phage whose chromosomes have 5′-cohesive “COS” ends), the ϕ29-like phages whose chromosomes have covalently bound terminal proteins, and the Mu-like phages whose DNA is integrated into host DNA, have packaging substrates that are monomeric; note that the first and last of these require that cleavage by terminase linearize the molecule for packaging and release the integrated phage DNA, respectively. The phages that package from concatemers typically engage in unidirectional “packaging series” on the DNA concatemers, where non-series-initiating packaging events begin at a DNA end generated by the previous event. This has been studied in most detail in mid-sized phages, such as P22 and λ, and the details of concatemer handling during packaging in the larger, more complex phages like T4 and SPO1 remain more poorly understood. Figure 7.1 shows packaging series for four well-studied phages λ (COS ends), T7 (short direct terminal repeat or short DTR), P22 (terminally redundant and circularly permuted), and Mu (terminal host DNA).
Fig. 7.1.
Four tailed-phage DNA packaging strategies. Packaging strategies of phages λ, P22, T7, and Mu are shown diagrammatically. Thick black horizontal lines represent phage concatemeric DNAs, or, in the case of Mu, phage genomes that are integrated into the host bacteria’s chromosome (the latter represented by thick gray line). Black circles mark the packaging recognition sites and horizontal black arrows represent individual packaging events. Vertical black lines indicate precise terminase cleavages and vertical gray lines indicate imprecise cleavages (see text). In each case except Mu, sequential series of packaging events occur, in which subsequent events (event 2 in figure) on the same concatemer molecule begin at the concatemer end created by the previous event (event 1); although only two successive events are shown, packaging series can in some cases be up to 10 or more events long. In phages λ and T7, each event begins and ends at a packaging recognition site, and in phage T7 the white rectangles show the region (the direct terminal repeat) that is duplicated in concert with packaging. In phage P22, the increasing width of the vertical gray boxes to the right, denotes the increased range of cleavage site locations as events proceed rightward. The small gray horizontal rectangle below the first P22 event is the optimal location of the Southern probe used to analyze pac fragments (see text).
The first event in such packaging series is recognition of the DNA by terminase, and then a double-strand cleavage is made at or near the packaging recognition site (typically called pac in headful packaging phages (3) and cos in cohesive end phages (4)). Only one of the two DNA ends created by this first (packaging series initiation) cleavage is threaded into a viral procapsid so that the packaging motor inserts DNA into the procapsid in only one direction from the cleavage point (Fig. 7.1). When DNA has filled the procapsid, a second nucleolytic cut (the “headful” cleavage) is made by terminase, which releases the packaged DNA from the concatemer, thus terminating the first packaging event. A second packaging event on that concatemer is then initiated by insertion of the unpackaged concatemer end created by the previous headful cleavage into a new procapsid. The second event is terminated like the first, with a headful cleavage (the second headful cleavage of the series). Subsequent packaging events then follow sequentially in the same manner as the second. Such unidirectional packaging series are usually two to five packaging events long, but can be upto 10 or more events long, depending on infection conditions (5).
Below we discuss briefly how the different kinds of virion DNA ends result from different replication/terminase cleavage/ packaging mechanisms. Demonstration of the presence of each of these types of ends requires specialized, directed analysis. Successful analysis of these ends requires more understanding of the various possible DNA end styles and how they are generated than it does technically difficult experimental analysis. Particular attention is paid here to phages whose genomes are completely sequenced, since at present this is very often the case for phages whose end structures are of interest. When phage genomes are sequenced by random shotgun sequencing (or even by primer walking on a phage chromosome template) apparently circular sequences are generated for circularly permuted and terminal exact direct repeat genomes, and even for cohesive end phages if the plasmid-cloned phage DNA inserts include cohesive ends that have been ligated together. Of course these are all artificially circular sequences, since all known tailed-phage virion chromosomes are linear. Our current knowledge of the mechanism of DNA packaging and injection suggests that covalently-circular virion DNA molecules will never be found in a tailed-phage, since the dsDNA must be threaded into the virion during packaging and out of the virion during injection through a narrow “portal” passage that will not accommodate two parallel dsDNAs simultaneously (which would be required if the chromosome were circular) (6). Thus, even when the complete genome sequence has been determined, additional experiments are usually required to understand the true nature of the linear virion DNA.
1.2 Cohesive Ends
1.2.1 Best Studied Phages—λ, HK97, and P2
The two ends of cohesive end-containing phage chromosomes have protruding single-strands of identical length that are complementary to one another in sequence; upon injection these two ends anneal to each other, and each strand is closed by DNA ligase (of the host in those cases studied) to generate the covalently-closed circular molecule that serves as a template for DNA replication. Such cohesive ends can have either 5′- or 3′- protruding strands (7–9) and have been reported to be between 7 and 19 nucleotides in length in various phages (e.g., P2 has 19 nucleotide 5′-protruding strands (10) and HP1 has 7 nucleotide 5′-protruding strands (11)). Such ends are generated when the terminase makes staggered, sequence-specific cuts in the two DNA strands as it is being packaged. On a concatemer, a pair of staggered cuts (separated by the length of the eventual single-strand extension) generates the right end of one chromosome and the left end of the next chromosome to be packaged (except for the first and last cuts in a packaging series, where DNA one only one side of the cleavage is packaged; Fig. 7.1). Thus, the cohesive end termini of all individuals of a given COS phage are present at identical locations on the genome sequence.
DNAs with cohesive ends can be recognized by the ability of the opposite ends to anneal in the test tube, and the simplest way to detect such annealing is by the joining or “coherence” (hence the name (7)) of the two terminal restriction fragments. This annealing is most easily observed in agarose electrophoresis gels. Thus, if restricted DNA that contains cohesive ends is heated to a temperature that separates the cohesive ends but does not separate the strands of the remainder of the DNA (e.g., 75–80 °C) and cooled either slowly or rapidly. The cohesive-ended fragments will, under slow cooling conditions, anneal to one another and be visible as a larger band in such gels, but under fast cooling conditions, the two terminal bands do not have time to anneal with one another (Fig. 7.2A). We note that restriction enzyme-generated ends typically have ≤ 5 bp single-strand protrusions, which are insufficient to keep two fragments together in such a gel. In some cases, for example Bacillus subtilis phage ϕ 105, the seven nucleotide 3′-cohesive ends have been reported to associate unusually rapidly and addition of formamide and/or treatment with single-strand specific nucleases were required to separate the joined end fragments (9).
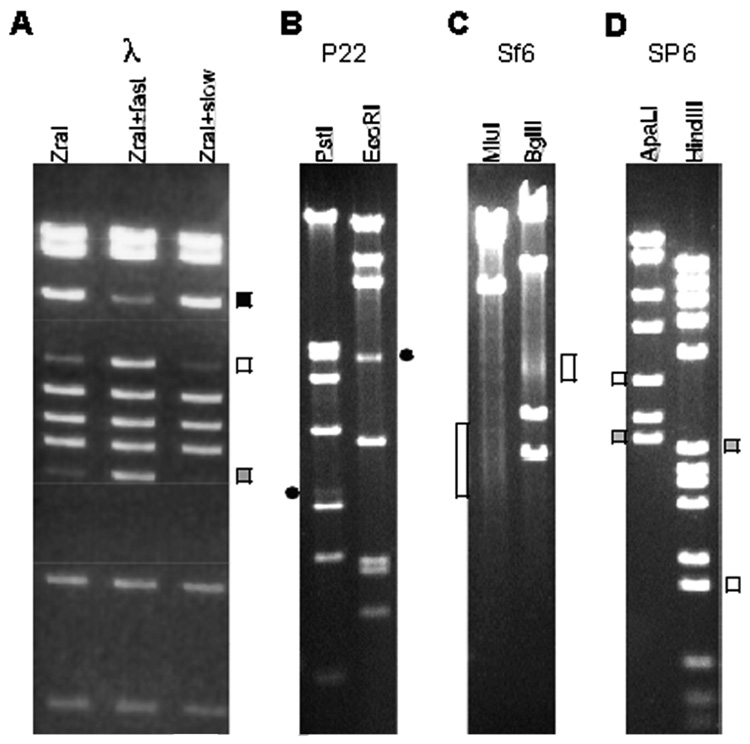
Fig. 7.2.
Restriction enzyme generated fragments from tailed-phage virion DNA. DNA fragments were separated by 0.8% agarose gel electrophoresis and visualized by staining with ethidium bromide. The phage virion source of the DNA is indicated above each panel, and the restriction enzymes used are indicated above each lane. A. Phage λ DNA after normal isolation and storage. DNA in the second and third lanes was heated to 75 °C for 15 min and then fast or slow cooled to room temperature, respectively, as described in the Methods. Terminal DNA fragments are indicated as follows: white square, left end fragment; gray square, right end fragment; black square, left and right end fragments annealed together by their cohesive ends. B. Phage P22 DNA. The pac fragments generated by PstI and EcoRI are indicated by black circles on the left and right, respectively. C. Phage Sf6 DNA. The locations of the diffuse pac fragment bands generated by imprecise packaging series initiation are indicated by white rectangles (see also ref. (31)). D. Phage SF6 DNA. Terminal DNA fragments are indicated as follows: white square, left end fragment; gray square, right end fragment.
This simple analysis can indicate whether cohesive ends are present, but does not determine the single-strand extension length or which strand protrudes. Typically, the exact nature of such ends is determined by running dideoxynucleotide sequencing reactions of both ends of a native chromosome template DNA to determine exactly where the template ends at each terminus. The locations of the 5′-end of each strand can then be deduced by comparison to sequences across closed, ligated ends (12), and being aware that Taq DNA polymerase tends to add at least one non-templated A as it runs off the end of a template. The 3′-ends of each strand are more difficult to determine directly, and their positions are typically deduced by assuming, since the strand breaks at annealed cohesive ends are ligatable, that the two ends were generated by single-strand nicks as described above. The above determination is straightforward if one knows about where the ends are on the genome. The approximate locations of the terminase-generated ends (cos site) can be determined from the sizes of the termini-containing restriction fragments (in the above fast vs. slow cooling experiment) and a restriction map or the location of the restriction sites in the genome sequence. In addition, the location of the cos site is quite highly conserved, and in a large majority of the cases studied to date, it is within about one Kbp and transcriptionally upstream of the gene that encodes the small terminase subunit. Although the small subunit genes are very variable in sequence, the more highly conserved terminase large subunit and portal proteins and the (nearly always) highly stereotyped order of the phage head assembly genes can often allow an informed guess for the location of the small terminase subunit gene and hence the cos site (13). We hasten to mention that exceptions to this generality do exist, which include the P2-like phages, Mycobacterium tuberculosis phages L5 and D29 (14, 15) and Lactococcus lactis phages r1t and c2 (16, 17); in each of these cases, the head genes appear have an atypical order and in the latter four cases the small terminase gene is not yet recognizable. In such cases, the cohesive ends must be located by restriction mapping before they can be characterized in detail.
1.3 Headful Packaging
1.3.1 Best Studied Phages—P22, P1, SPP1 and T4
Phages that contain chromosomes that are terminally redundant and circularly permuted are called “headful packaging” phages (18). Phage P22 is the best characterized headful packaging phage. In this case, a specific site is recognized on the replicated concatemer to initiate a packaging series (19), but it is the available volume inside of the head, not DNA sequence, that determines the location of subsequent cleavages in the packaging series (Fig. 7.1) (20–22). This packaging series initiation site is called pac, and this name is typically reserved for the site that terminase recognizes to begin a headful packaging series. The terminase makes a sequence-specific cleavage (see below) to initiate series, but has little sequence specificity at the following headful cleavages, that are made only when the procapsid (phage head precursor) is “full” of DNA (23). The packaged DNA length in the headful packaging phages is typically between 102% and 110% the length of the genome sequence, so these chromosomes have direct terminal repeats that vary from 2% to 10% of the genome length in different phages. Upon infection, homologous recombination between these direct, terminal repeats generates the circular genome that is the template for DNA replication. One consequence of this packaging strategy is that the ends of each successive packaging event in a series “move” along the genome sequence (rightward in Fig. 7.1) from those generated by the previous event by the length of the terminal repeat. The result of such packaging series is that the virion chromosome is circularly permuted and terminally redundant. The DNA is often only “partially permuted” in that the ends are not completely randomly distributed across the whole genome sequence, but are found distributed over only a portion of the genome. This is the expected result if packaging series all start at the same place (a pac site), and terminal redundancy size and series lengths are limited; i.e., ends are all located in a region adjacent to the pac site for a distance that depends on the size of the terminal redundancy and on the number of events in packaging series (Fig. 7.1). Headful packaging phages do not have cohesive ends; their DNA ends are usually thought to be blunt due to their ability to be ligated to other blunt DNA ends (24), but in fact the precise nature of their ends has been difficult to determine unambiguously, because of the many different end positions present in any DNA preparation.
A further complication to the analysis of headful packaging phage chromosomes is that during packaging the determination of when the capsid is full of DNA is imprecise, so somewhat different lengths of DNA are packaged in different individual virions (Fig. 7.1). This variation is about ±2% the genome length or ±700–1, 000 bp in the few cases that have been studied (23, 25, 26). Yet another complication is that the site of the initiation cleavage for headful packaging series is not precise in the headful phages analyzed to date, and alternative initiation cuts are scattered over regions that range from 9 bp in phage SPP1 to about 2,000 bp in phage Sf6 (27–32). The overall results of such packaging events, where both packaging initiation and chromosome length are imprecise, are DNA molecules from different individual virions whose end locations can lie at many if not all of the possible positions within a substantial region of the genome. We discuss below the diagnostic experimental features of headful packaging.
Since their virion chromosomes are not all the same length, one way of determining whether a phage utilizes a heedful-type packaging strategy is the greater width of the whole chromosome band relative to a similar-sized DNA molecule of precise length (e.g., phage λ virion DNA) after pulsed-field electrophoresis. This is not described in detail here, but see. for example. Fig. 7.2c in ref. (31) and Fig. 7.2b in ref. (32). We note that phage Mu uses a heedful-type packaging mechanism in spite of its very different packaging substrate (see below), so like the other headful packaging phages discussed in this section, it also generates chromosomes of somewhat variable length (33). Another experimental indicator of headful packaging is generalized transduction. Because of the lack of terminase sequence specificity in the headful cleavages, if packaging mistakenly initiates on host DNA, it is efficiently packaged into functional virion-like transducing particles (unlike in the COS phages, where sequence specificity is required at both the initiation and the headful cleavages and special experimental strategies are required to show generalized transduction (34)).
A more informative analysis of headful packaging can be obtained from analysis of the virion DNA’s restriction fragment pattern. When terminally redundant, circularly permuted, headful packaged DNA is restricted, all of the true restriction fragments (fragments with restriction enzyme cleavages at both ends) that would be generated from a circular version of the genome sequence are typically present in at least some virion DNA molecules. Thus, the electrophoresis band pattern is at minimum that would have been generated by circular DNA. If the series initiation cleavage near the pac site is relatively precise (all cleavages occur within a few hundred base pair regions), restriction of DNA from the first packaging event in any series will generate a discrete DNA fragment with a packaging initiation cleavage at its left end and a restriction cleavage at its right end (orientation as in Fig. 7.1). This series initiation-end fragment is called the “pac fragment” (3), and it is present in fewer copies than the true restriction fragments, since it is only created from the first DNA packaging event of any packaging series (the molar ratio of the pac fragment to the true restriction fragments that are present in essentially all molecules correspond to the average packaging series length) (3, 27, 35). Because of the substantial imprecision in the headful measurement of packaged DNA length (above), the packaging termination-generated end is imprecise. Thus, the right end restriction fragment from the first event in a packaging series and all terminal fragments (from both ends) generated by subsequent events in that series are variable in size and are so spread out in the gel background that they are nearly “invisible” in ethidium bromide-stained electrophoresis gels. Thus, if series initiation is relatively precise (as it is in phages P22, SPP1, and P1, for example), the restriction pattern of headful packaged DNA will consist of all the fragments expected from a circular genome plus a submolar pac fragment. The phage P22 restriction pattern is shown in Fig. 7.2B; see also Fig. 7.1A in (3). When present, this type of restriction pattern is considered to be diagnostic of headful packaging. We note that the position of the terminase-generated end of the pac fragment is very near the pac site in the cases studied (summarized in ref. 19).
There are potential complications to such an analysis that must be understood. Of course, for any given restriction enzyme, the pac fragment can be small and run off the gel or be obscured by a true restriction fragment, so the absence of an apparent pac fragment does not prove the absence of headful packaging, especially if no genome sequence is available (see also below); it may be necessary to try a number of different restriction enzymes to find ones that display the pac fragment unambiguously. This type of analysis can also be complicated somewhat if the circular permutation is sufficiently limited that all ends fall in a relatively small fraction of the genome; under these conditions, the true restriction fragments that extend across this region can also be present in submolar amounts or even missing altogether. This type of analysis is much more robust if a restriction map of the genome is available. Thus, several restriction patterns should agree on the location of the terminase-generated end of the pac fragment. Finally, certain special combinations of fragment size, terminal redundancy length, and headful precision can give rise to visible diffuse gel bands from the non-series initiation ends (see Fig. 7.2a in ref. 23).
Some headful terminases appear to be able to move substantial distances along the DNA between pac site recognition and cleavage of the DNA to initiate a packaging series. If it moves only a short distance, a discrete pac fragment is generated as discussed above. However, if it can move long distances, the series initiation cleavage by terminase becomes too imprecise to allow the generation of an easily visualized pac fragment in electrophoresis gels. This is the case for phages Sf6 and ES18 (31, 32) and perhaps T4 (36). Then it too is an “invisible” diffuse band in a stained electrophoresis gel of restricted virion DNA, and the pattern of restriction fragments will simply be that expected from a circular genome; i.e., all the terminal fragments are so variable in length that they are lost in the background. Note in Fig. 7.2 that these terminal fragments in headful phages P22 and Sf6 show as DNA staining in the background (between bands), and phages λ and SP6 which have unique ends have much less background straining material. In some fortuitous situations, a diffuse pac fragment can be seen as a faint, fuzzy-stained bands as in the MluI and BglII digested phage Sf6 DNAs shown in Fig. 7.2C. However, with this type of phage, Southern analysis using a probe that hybridizes to all of the variably sized pac fragments will specifically visualize the diffuse pac fragments band (and the true restriction fragment that includes the pac fragment) (31, 32). Nucleotide sequence information is required in choosing the DNA probe. Where they have been studied, headful packaging phage pac sites usually lie within or near the small terminase subunit gene and packaging proceeds in the direction in which that gene is transcribed. As in the COS phages (above) in nearly all characterized headful packaging phages the large terminase subunit gene and portal protein genes are close and transcriptionally downstream from the small subunit gene. Thus, a Southern probe from within the large terminase or portal gene will usually hybridize to any (diffuse or not) pac fragments that might initiate near a pac recognition site near the small terminase gene. One known exception is Streptococcus phage MM1, where the pac site is about 2 Kbp transcriptionally downstream of the large terminase gene, however, it is possible that this is a case of the small terminase gene, which is not recognizable in the MM1 sequence, being in a non-canonical location (37). Identifying these diffuse bands generated by several different restriction enzymes effectively locates the variable end of the pac fragment and thus the region where packaging series begins.
Any method that can locate the chromosome ends can in the-ory elucidate the same things that the above restriction analysis does, and electron microscopic heteroduplex and partial denaturation mapping as well as cloning of terminal fragments have been used in this context. In fact, Tye et al. (20) used this type of electron microscopic analysis to be the first to deduce the “pac site— packaging series” strategy utilized by phage P22. More recently, Loessner et al. (38) have applied this technology to the analysis of Listeria phage A118. Plunkett et al. (39) deduced tentative chromosome end locations by analysis of the locations of DNA clones in a random library of phage 933W chromosomal DNA. Both methods are labor-intensive because of the need to manually examine many independent DNA molecules to gain the necessary statistical power. We will not describe these methods in detail here.
A final note of caution is warranted regarding interpretation of apparent, completely “random permutation” of phage chromosomes. Phages have not been studied to the point that we have cataloged all of their molecular lifestyles in detail, so packaging strategies other than those discussed here are possible. For example, the existence of headful packaging phages that initiate packaging, without a pac site, at genuinely random locations on the phage DNA remains possible. The observation of apparently completely randomly distributed virion DNA ends could be the result of the presence multiple pac sites, long packaging series, long terminal redundancy and/or terminase movements between recognition and DNA cleavage. On the other hand, it could be due to a “new” packaging strategy that somehow starts packaging randomly on the phage DNA without a pac site. The former explanations seem more likely since all phages that have been analyzed preferentially package their own DNA, and no truly random situation has been documented. The most relevant experimentally studied case is phage T4, which destroys all the non-T4 DNA in the infected cell and so might not need a nucleotide sequence target to recognize its own DNA. Its packaging strategy is not yet understood in every detail, but the current best working model is that it too initiates packaging imprecisely at a recognition site in or near its small terminase gene (36).
1.4 Short Exact Direct Repeat Ends
1.4.1 Best Studied Phages—T3 and T7
Phages of this type have direct double-stranded repeats at their termini which are a few hundred base pairs long and are exactly the same in every virion chromosome (i.e., they are not permuted). These chromosomes, when characterized, are thought to have blunt ends, again by the criterion of ligatability to other blunt ends. The terminal repeats are generated by a duplication of the direct repeat DNA in concert with packaging (40, 41) (Fig. 7.1). This type of DNA end structure could be overlooked when a phage genome sequence is determined by shotgun methods, since sequence assembly can merge the two ends to give a circular sequence. Analysis of appropriate restriction digests of this type of virion DNA will give rise to a gel pattern of fragments that has all equimolar fragments (and heating and cooling will not alter the pattern as with cos site phages) and terminal fragments which are not correctly predicted by such an artificially circularized sequence (except in the unlikely event of a fortuitous cleavage site in the short duplicated region; thus, multiple restriction digests should be analyzed in this way); Fig. 7.2D shows two restriction digests of T7-like phage SP6 DNA. In this type of phage, the approximate location of the ends can be determined by restriction mapping since the restriction maps of such molecules are linear, or if nucleotide sequence is available, approximate repeat locations can sometimes be predicted from the end locations in related phages. When the approximate locations of the chromosome ends are known, sequencing reactions initiated by primers that anneal to unique whole genome template sites internal to the terminal repeat and program synthesis across the repeats to the two ends can determine the exact end of the template strands by the position that synthesis stops and thus the length of the terminal repeats (42–44).
1.5 Long Exact Direct Repeat Ends
1.5.1 Best Studied Phages—T5 and SPO1
Most “exact terminal repeat” phages that have been characterized have terminal repeats that have lengths in the one to a few hundred base pairs range as described in the previous section; however, several large, complex phages with genomes in the 130 kbp range, the best studied of which are E. coli phage T5 and Bacillus subtilis phage SPO1, have chromosomes with very long exact terminal direct repeats that are 10139 and 13185 bp, respectively ((45); R. Hendrix, W. Huang, S. Casjens, G. Hatfull, M. Padulla and C. Stewart, unpublished results). The mechanism by which these long repeats are generated is not known, but as with the short exact repeat phages, there appears to be only one copy of the terminal repeat between genomes in replication-generated concatemers. This suggests that the repeat region is duplicated prior to or during packaging. In these cases, shotgun sequencing methods will determine an apparently circular sequence. The long terminal redundancy can be noticed genetically (46, 47) or by electron microscopic analysis of heteroduplexes of nuclease-trimmed DNA (48), but it is best discovered and studied by restriction mapping, which can locate the approximate position of the ends of such molecules on the sequence (49, 50). Determination of the precise end sequences is more difficult since no uniquely templated primer can program a sequencing run all the way across the long terminal redundancy to the molecular end of a virion chromosome template. The best strategy is to isolate or clone terminal restriction fragments and use them to template sequencing reactions over the ends (51).
1.6 Host DNA at Ends
1.6.1 Best Studied Phage—Mu
Phages like Mu, which replicate their genomes by duplicative transposition into host DNA, must package viral genomes that are randomly integrated into host DNA. The Mu terminase has not been studied in detail but it appears to recognize a pac site near one end of the genome and reach out from that point to initiate a packaging event by cutting in the adjacent host DNA (52, 53). A headful type packaging event then extends from this initial cleavage, across the integrated phage DNA and beyond, to include about 1,800 bp of host DNA at the other side (Fig. 7.1) (54). Again the ends are thought to be blunt by their ability to be ligated to other blunt ends (e.g., EcoRV ends in ref. (55)). Thus, each Mu DNA molecule has unique host DNA attached at both ends that is different in different virions. This terminal host DNA can be recognized “automatically” during a genome sequence determination project by the presence of these joined host sequences (55, 56), but can be more difficult to recognize in the absence of this information. One straightforward way to detect is the presence of “frayed single-stranded ends” upon electron microscopic examination of heteroduplex molecules after strand separation and reannealing of virion DNA, since the host DNA at the ends has huge variety and strands with matching terminal host sequences almost never find one another during the reannealing. Details of this technique are not given here, but see ref. (54).
1.7 Covalently Bound Terminal Proteins
1.7.1 Best Studied Phage—ϕ29
Bacillus subtilis phage ϕ29 and its relatives that infect other Gram-positive bacteria are the only phages currently known that have covalently bound proteins at the ends of their virion and replicating chromosomes. Such DNAs are typically recognized by the abnormally slow migration of their terminal DNA fragments in electrophoresis gels, and restoration to “normal” mobility by protease treatment. This is not discussed in detail here, but see refs. (57, 58).
1.8 Prediction of Packaging Strategy and DNA End Structure from Terminase Amino Acid Sequence
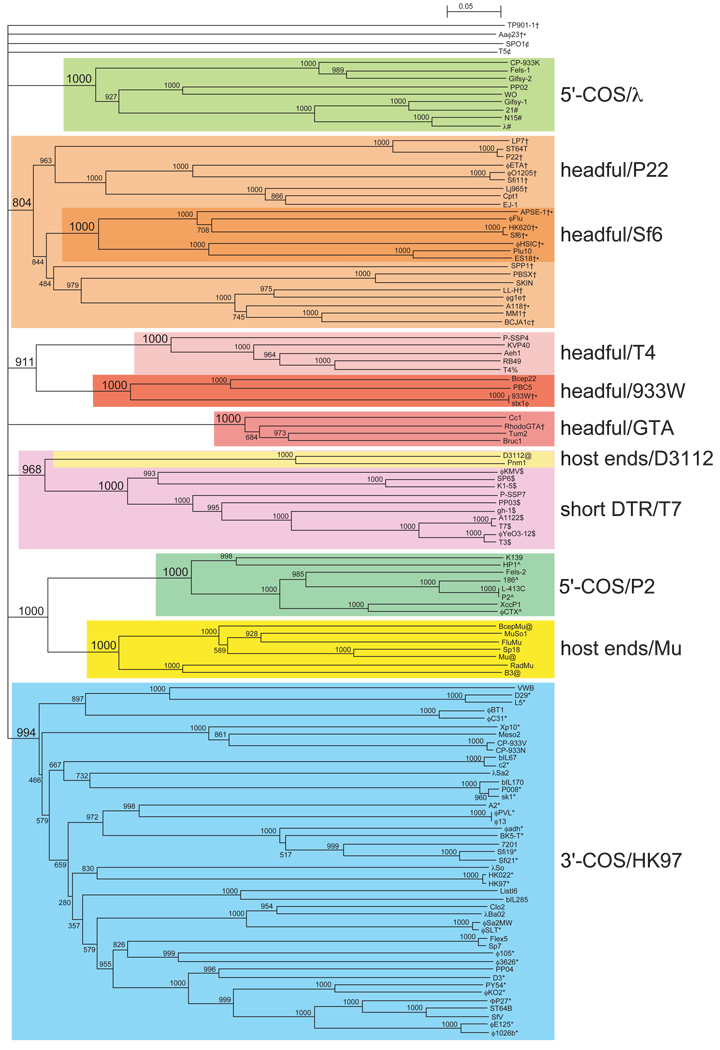
If the amino acid sequence of a phage’s large terminase subunit is known, the packaging strategy (and hence type of DNA ends) that a phage utilizes can often be successfully predicted from it. The terminase enzymes that create the virion DNA ends are quite varied, but still are among the most conserved tailed-phage proteins (13). Comparative analysis among them has shown that they cluster according to the type of DNA end that they create (32). Thus, if a terminase amino acid sequence falls robustly into one of the characterized clusters it very likely forms DNA ends that are similar to the other members of the group. Fig. 7.3 shows such a tree where 3′-COS phages, two groups of 5′-COS phages, short exact direct terminal repeat phages, two apparent types of terminal host sequence phages, and at least five separable groups of headful packaging phages can be distinguished by their terminase amino acid sequences. It is also important to realize that the current picture of terminase diversity is not complete, and if a terminase sequence does not fall convincingly within a characterized group (currently including, for example, the long direct terminal repeat terminases of SPO1 and T5, headful terminases of P1, TP901-1, and Aaϕ23; COS phage VP16C, and short terminal repeat phage VpV260), the predictive value of such a comparison falls precipitously. In addition, as was pointed out by Casjens et al. (32), the inclusion of some terminases such as those of 3′-COS phages TM4, MS1, and r1t in the tree shown in Fig. 7.3 can lower some of the groups’ bootstrap values. This type of prediction should of course not replace experimental analysis, but can be helpful in pointing the best way to proceed experimentally in a determination of tailed-phage chromosome end structure.
Fig. 7.3.
Neighbor-joining tree of large terminase subunit amino acid sequences. A neighbor-joining tree of 123 tailed-phage terminase amino acid sequences was generated by CLUSTAL X (59). The numbers near bifurcations are bootstrap values for 1,000 trials. Short bifurcating branches linking the major groups shown here were manually merged, since all had low bootstrap values, and the major deep branches are all shown as radiating from a single source. The names of the phages or prophages are shown at the right of each terminal branch (some prophage names are those proposed by Casjens (13)). Major robust, related groups of terminases are highlighted with gray boxes, and the packaging strategy and a prototype phage for each group is given at the far right. Those phages whose virion DNA termini structures have been experimentally determined are indicated as follows: #, 5′-cohesive ends in lambdoid phages;^, 5′-cohesive ends in P2-like phages; *, 3′-cohesive ends; $, T7-like phages with direct terminal repeats and no circular permutation; , phages with long direct terminal repeats and no circular permutation; @, phages with host DNA at termini; †, headful packaging in non-T4-like phages, including P22 and the gene transfer agents (GTA); %, headful packaging in T4-like phages; ●, headful packaging phages for which no obvious pac fragment band has been identified in ethidium stained electrophoresis gels of restricted DNA (see text; the darker orange boxed “headful/Sf6” subgroup within the “headful/P22” group appear to be one branch of this type of terminase). This figure is modified from Fig. 7.6 of Casjens et al. (32).
2 Materials
No unusual or special materials or techniques are required for the analysis of tailed-phage virion chromosomal restriction patterns in electrophoresis gels, and protocols for such analyses can be found in any molecular biology laboratory manual (60).
3 Methods and Practical Considerations
3.1 DNA Release and Isolation from Virions
A solution of CsCl equilibrium density or step gradient-purified virions is made in 0.25% sodium lauryl sulfate (SDS), 50mM Tris–Cl (pH 8.0), and 25 mM ethylenediamine-tetraacetic acid (EDTA) by addition of appropriate amounts from 20% SDS, 1 M Tris–Cl, 0.5 M EDTA stock solutions.
Incubate at 75–80 °C for 15 min.
Add potassium acetate to a final concentration of 0.625 M from a 2 M stock and mix well.
Chill on ice for 60 min.
Remove the potassium-SDS precipitate by centrifugation at 10,000 rpm in a microfuge for 15 min at 4 °C.
Add two volumes of ethanol and wind the released DNA out of solution on the tip of a sterile Pasteur pipet.
Rinse the DNA on the pipet tip by dipping it into room temperature 70% ethanol, air dry for a few minutes until no obvious liquid remains, and dissolve by dipping the tip into 200 µl of 10 mM Tris–Cl (pH 8.0), 1 mM EDTA. The partially hydrated DNA will release from the pipet tip within a few minutes; then let the DNA dissolve at 4 °C for at least 12 h before use. Over drying can make resuspension more difficult.
DNA prepared from purified phage in this way should be suitable for subsequent manipulations. If it appears to be degraded (as assayed by electrophoretic analysis) by contaminating nucleases upon incubation in Mg++ containing buffer, shake it gently with an equal volume of equilibrated phenol at room temperature for 10 min, ethanol precipitate, and resuspend in 10 mM Tris–Cl (pH 8.0), 1 mM EDTA.
3.2 Cohesive End DNA Analysis
Cleave 1 µg of purified virion DNA with a restriction enzyme of choice (one that results in the display the two end fragments, joined and separated, at uncrowded gel positions, as determined by trial-and-error or from analysis of a genome sequence).
Heat the reaction mix from step 1 to 75–80 °C for 15 min, and divide into two equal portions. Chill one rapidly by placing it quickly in wet ice, and cool the other to room temperature slowly. Useful slow cooling can be achieved by programming a PCR cycler to cool from 75 to 24 °C over 40 min or by simply placing the tube in a 75 °C or 80 °C metal heating block and letting the block cool to room temperature on the bench top.
Separate and display the resulting DNA bands in an agarose electrophoresis gel and visualize bands by ethidium bromide staining (60).
If the phage has COS ends, two fragments will be visible in the quick chilled sample that are missing (or nearly so) in the slow cooled sample. In the slow cooled sample, these two fragments should be joined as a larger fragment whose molecular weight should be their sum (see Fig. 7.2A).
3.3 Headful DNA Analysis
3.3.1 P22 Headful Type—Discrete Pac Fragment
Cleave 1 µg of purified virion DNA with a restriction enzyme that results in the display the DNA pac fragment at an uncrowded gel position. Enzyme choice can be determined by trial-and-error by searching for enzymes that yield a single sub-molar DNA fragment or from analysis of a genome sequence and assuming that the pac site is within the small terminase gene, just upstream of the large terminase gene (the latter is not foolproof since there are known exceptions to this location, see above).
Separate and display the resulting DNA bands in an agarose electrophoresis gel and visualize bands by ethidium bromide staining (60).
Since submolar DNA bands can be present due to partial digestion, excess restriction enzyme should be used, and several restriction digests that each display a single submolar band should be visualized before this type of headful packaging is considered to be convincing.
In order for this to be a robust conclusion, a restriction site map or genome sequence should be available to show that the putative pac cleavage-generated ends of the putative pac fragments lie at the same location in the genome for each of the different enzymes used.
3.3.2 Sf6 Type—Diffuse Pac Fragment that Is not Obviously Visible by Staining
Cleave 1 µg of purified virion DNA with a restriction enzyme that results in the display, the single submolar diffuse DNA pac fragment at an uncrowded gel position. Such an analysis in phages with Sf6-type type of packaging strategy is not recommended without some DNA sequence and other information on the phage under study; for example, one might know that the phage DNA is somewhat variable in length (above) or the terminase is highly related to known headful terminases, but efforts to visualize a pac fragment by staining have proven unsuccessful. Sequence information is useful because the choice of probe for the Southern analysis (below) requires at least a good estimate of where the pac fragments will lie on the genome and so how they will be displayed in the electrophoresis gel relative to the true restriction fragments. As with the P22-type phages (sharp pac fragment band) above, this “best guess” is that the pac site is within the small terminase gene and that packaging proceeds from there in the direction of the portal protein gene (again, the latter is not foolproof, since there are known exceptions, and the large virulent phages are not yet “well-studied” in this regard, see above).
Separate and display the resulting DNA bands in an agarose electrophoresis gel (60).
Transfer the DNA to a membrane and perform Southern analysis (31, 60, 61) with a suitable DNA probe. The DNA probe can be either a cloned or a PCR amplified fragment of the phage DNA that is chosen to hybridize to the pac fragments; typically the probe will be within the large terminase or portal gene. This probe will also hybridize to the true restriction fragment that covers the pac fragment and restriction enzymes and gel conditions should be chosen that results in good separation of these two gel bands.
As with the P22 “sharp-pac fragment band” type analysis above, for this to be a robust conclusion one should show by restriction site mapping that the putative pac cleavagegenerated ends of the pac fragments lie at the same location in the genome for each of the different enzymes used.
Notes
As was mentioned above, the actual laboratory techniques used are standard ones that molecular biology laboratories will be familiar with. Thus, success or failure in the analysis of tailed-phage virion DNA end structure and packaging strategy is much more dependent upon the specific analysis strategy chosen than on technical process or protocol details. We have therefore emphasized strategic issues in our discussion. Perhaps the most important technical aspect in such an analysis is the use of DNA that is prepared from highly purified virions. Virions should whenever possible be purified by a method that takes advantage of their unusual buoyant density (between those of nucleic acids and protein), and cesium chloride gradient centrifugation has historically been the method of choice for purifying phage particles according to particle density. In general, true equilibrium sedimentation is not essential and considerable time can be saved by the use of CsCl “step gradients” as described in Earnshaw et al. (62). There are a few exceptional tailed-phage virions (e.g., phage ES18 (32)) which are apparently impermeable to Cs+ ions and so band in such gradients a position that does not separate them from the bulk of the proteinaceous cellular materials; then one is largely relegated to methods that separate on the basis of size such as differential and sucrose gradient centrifugation.
Acknowledgements
We thank Roger Hendrix for phage SF6 and Miriam Susskind for the essence of the protocol for DNA isolation from virions. The authors’ research was supported by NSF grant MCB-990526 to SRC.
References
- 1.Mousset S, Thomas R. Ter, a function which generates the ends of the mature lambda chromosome. Nature. 1969;221:242–244. doi: 10.1038/221242a0. [DOI] [PubMed] [Google Scholar]
- 2.Feiss M, Campbell A. Duplication of the bacteriophage lambda cohesive end site: genetic studies. J. Mol. Biol. 1974;83:527–540. doi: 10.1016/0022-2836(74)90512-9. [DOI] [PubMed] [Google Scholar]
- 3.Jackson EN, Jackson DA, Deans RJ. EcoRI analysis of bacteriophage P22 DNA packaging. J. Mol. Biol. 1978;118:365–388. doi: 10.1016/0022-2836(78)90234-6. [DOI] [PubMed] [Google Scholar]
- 4.Emmons SW. Bacteriophage lambda derivatives carrying two copies of the cohesive end site. J. Mol. Biol. 1974;83:511–525. doi: 10.1016/0022-2836(74)90511-7. [DOI] [PubMed] [Google Scholar]
- 5.Adams MB, Hayden M, Casjens S. On the sequential packaging of bacteriophage P22 DNA. J. Virol. 1983;46:673–677. doi: 10.1128/jvi.46.2.673-677.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage f29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hershey AD, Burgi E. Complementary structure of interacting sites at the ends of lambda DNA molecules. Proc. Natl. Acad. Sci. USA. 1965;53:325–330. doi: 10.1073/pnas.53.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu R, Taylor E. Nucleotide sequence analysis of DNA. II. Complete nucleotide sequence of the cohesive ends of bacteriophage lambda DNA. J. Mol. Biol. 1971;57:491–511. doi: 10.1016/0022-2836(71)90105-7. [DOI] [PubMed] [Google Scholar]
- 9.Ellis DM, Dean DH. Nucleotide sequence of the cohesive single-stranded ends of Bacillus subtilis temperate bacteriophage f105. J. Virol. 1985;55:513–515. doi: 10.1128/jvi.55.2.513-515.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Padmanabhan R, Wu R, Calendar R. Complete nucleotide sequence of the cohesive ends of bacteriophage P2 deoxyribonucleic acid. J. Biol. Chem. 1974;249:6197–6207. [PubMed] [Google Scholar]
- 11.Fitzmaurice WP, Waldman AS, Benjamin RC, Huang PC, Scocca JJ. Nucleotide sequence and properties of the cohesive DNA termini from bacteriophage HP1c1 of Haemophilus influenzae Rd. Gene. 1984;31:197–203. doi: 10.1016/0378-1119(84)90210-5. [DOI] [PubMed] [Google Scholar]
- 12.Juhala RJ, Ford ME, Duda RL, Youlton A, Hatfull GF, Hendrix RW. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 2000;299:27–51. doi: 10.1006/jmbi.2000.3729. [DOI] [PubMed] [Google Scholar]
- 13.Casjens S. Prophages in bacterial genomics: What have we learned so far? Molec. Microbiol. 2003;249:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- 14.Hatfull GF, Sarkis GJ. DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Molec. Microbiol. 1993;7:395–405. doi: 10.1111/j.1365-2958.1993.tb01131.x. [DOI] [PubMed] [Google Scholar]
- 15.Ford ME, Sarkis GJ, Belanger AE, Hendrix RW, Hatfull GF. Genome structure of mycobacteriophage D29: implications for phage evolution. J. Mol. Biol. 1998;279:143–164. doi: 10.1006/jmbi.1997.1610. [DOI] [PubMed] [Google Scholar]
- 16.Lubbers M, Ward L, Beresford T, Jarvis B, Jarvis A. Sequencing and analysis of the cos region of the lactococcal bacteriophage c2. Mol. Gen. Genet. 1994;245:160–166. doi: 10.1007/BF00283263. [DOI] [PubMed] [Google Scholar]
- 17.van Sinderen D, Karsens H, Kok J, Terpstra P, Ruiters MH, Venema G, Nauta A. Sequence analysis and molecular characterization of the temperate lactococcal bacteriophage r1t. Molec. Microbiol. 1996;19:1343–1355. doi: 10.1111/j.1365-2958.1996.tb02478.x. [DOI] [PubMed] [Google Scholar]
- 18.Streisinger G, Enrich J, Stahl M. Chromosome structure in T4. III. Terminal redundancy and length determination. Proc. Natl’l. Acad. Sci., U.S.A. 1967;57:292–295. doi: 10.1073/pnas.57.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu H, Sampson L, Parr R, Casjens S. The DNA site utilized by bacteriophage P22 for initiation of DNA packaging. Molec. Microbiol. 2002;45:1631–1646. doi: 10.1046/j.1365-2958.2002.03114.x. [DOI] [PubMed] [Google Scholar]
- 20.Tye BK, Huberman JA, Botstein D. Non-random circular permutation of phage P22 DNA. J. Mol. Biol. 1974;85:501–528. doi: 10.1016/0022-2836(74)90312-x. [DOI] [PubMed] [Google Scholar]
- 21.Moore SD, Prevelige PE., Jr Bacteriophage P22 portal vertex formation in vivo. J. Mol. Biol. 2002;315:975–994. doi: 10.1006/jmbi.2001.5275. [DOI] [PubMed] [Google Scholar]
- 22.Weigele PR, Sampson L, Winn-Stapley D, Casjens SR. Molecular genetics of bacteriophage P22 scaffolding protein’s functional domains. J. Mol. Biol. 2005;348:831–844. doi: 10.1016/j.jmb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 23.Casjens S, Hayden M. Analysis in vivo of the bacteriophage P22 headful nuclease. J. Mol. Biol. 1988;199:467–474. doi: 10.1016/0022-2836(88)90618-3. [DOI] [PubMed] [Google Scholar]
- 24.Schmieger H, Taleghani KM, Meierl A, Weiss L. A molecular analysis of terminase cuts in headful packaging of Salmonella phage P22. Mol. Gen. Genet. 1990;221:199–202. doi: 10.1007/BF00261721. [DOI] [PubMed] [Google Scholar]
- 25.Chow LT, Bukhari AI. Heteroduplex electron microscopy of phage Mu mutants containing IS1 insertions and chloramphenicol resistance transposons. Gene. 1978;3:333–346. doi: 10.1016/0378-1119(78)90042-2. [DOI] [PubMed] [Google Scholar]
- 26.Humphreys GO, Trautner TA. Maturation of bacteriophage SPPI DNA: limited precision in the sizing of mature bacteriophage genomes. J. Virol. 1981;37:832–835. doi: 10.1128/jvi.37.2.832-835.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casjens S, Huang WM. Initiation of sequential packaging of bacteriophage P22 DNA. J. Mol. Biol. 1982;157:287–298. doi: 10.1016/0022-2836(82)90235-2. [DOI] [PubMed] [Google Scholar]
- 28.Deichelbohrer I, Alonso JC, Luder G, Trautner TA. Plasmid transduction by Bacillus subtilis bacteriophage SPP1: effects of DNA homology between plasmid and bacteriophage. J. Bacteriol. 1985;162:1238–1243. doi: 10.1128/jb.162.3.1238-1243.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sternberg N, Coulby J. Recognition and cleavage of the bacteriophage P1 packaging site (pac). II. Functional limits of pac and location of pac cleavage termini. J. Mol. Biol. 1987;194:469–479. doi: 10.1016/0022-2836(87)90675-9. [DOI] [PubMed] [Google Scholar]
- 30.Casjens S, Sampson L, Randall S, Eppler K, Wu H, Petri JB, Schmieger H. Molecular genetic analysis of bacterio-phage P22 gene 3 product, a protein involved in the initiation of headful DNA packaging. J. Mol. Biol. 1992;227:1086–1099. doi: 10.1016/0022-2836(92)90523-m. [DOI] [PubMed] [Google Scholar]
- 31.Casjens S, Winn-Stapley D, Gilcrease E, Moreno R, Kühlewein C, Chua JE, Manning PA, Inwood W, Clark AJ. The chromosome of Shigella flexneri bacteriophage Sf6: complete nucleotide sequence, genetic mosaicism, and DNA packaging. J. Mol. Biol. 2004;339:379–394. doi: 10.1016/j.jmb.2004.03.068. [DOI] [PubMed] [Google Scholar]
- 32.Casjens SR, Gilcrease EB, Winn-Stapley DA, Schicklmaier P, Schmieger H, Pedulla ML, Ford ME, Houtz JM, Hatfull GF, Hendrix RW. The generalized transducing Salmonella bacteriophage ES18: complete genome sequence and DNA packaging strategy. J. Bacteriol. 2005;187:1091–1104. doi: 10.1128/JB.187.3.1091-1104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chow LT, Bukhari AI. Bacteriophage Mu genome: structural studies on Mu DNA and Mu mutants carrying insertions. In: Bukhari AI, Shapiro JA, Adhya SL, editors. DNA insertion elements, plasmids, and episomes. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1977. pp. 295–306. [Google Scholar]
- 34.Sternberg N. The production of generalized transducing phage by bacteriophage lambda. Gene. 1986;50:69–85. doi: 10.1016/0378-1119(86)90311-2. [DOI] [PubMed] [Google Scholar]
- 35.Bachi B, Arber W. Physical mapping of BglII, BamHI, EcoRI, HindIII and PstI restriction fragments of bacteriophage P1 DNA. Mol. Gen. Genet. 1977;153:311–324. doi: 10.1007/BF00431596. [DOI] [PubMed] [Google Scholar]
- 36.Lin H, Black LW. DNA requirements in vivo for phage T4 packaging. Virology. 1998;242:118–127. doi: 10.1006/viro.1997.9019. [DOI] [PubMed] [Google Scholar]
- 37.Obregon V, Garcia JL, Garcia E, Lopez R, Garcia P. Peculiarities of the DNA of MM1, a temperate phage of Streptococcus pneumoniae. Int. Microbiol. 2004;7:133–137. [PubMed] [Google Scholar]
- 38.Loessner MJ, Inman RB, Lauer P, Calendar R. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Molec. Microbiol. 2000;35:324–340. doi: 10.1046/j.1365-2958.2000.01720.x. [DOI] [PubMed] [Google Scholar]
- 39.Plunkett G, 3rd, Rose DJ, Durfee TJ, Blattner FR. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 1999;181:1767–1778. doi: 10.1128/jb.181.6.1767-1778.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung YB, Nardone C, Hinkle DC. Bacteriophage T7 DNA packaging. III. A “hairpin” end formed on T7 concatemers may be an intermediate in the processing reaction. J. Mol. Biol. 1990;216:939–948. doi: 10.1016/S0022-2836(99)80012-6. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Studier FW. Multiple roles of T7 RNA polymerase and T7 lysozyme during bacteriophage T7 infection. J. Mol. Biol. 2004;340:707–730. doi: 10.1016/j.jmb.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 42.Dunn J, Studier W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J. Mol. Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 43.Dobbins AT, George M, Jr, Basham DA, Ford ME, Houtz JM, Pedulla ML, Lawrence JG, Hatfull GF, Hendrix RW. Complete genomic sequence of the virulent Salmonella bacteriophage SP6. J. Bacteriol. 2004;186:1933–1944. doi: 10.1128/JB.186.7.1933-1944.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scholl D, Kieleczawa J, Kemp P, Rush J, Richardson CC, Merril C, Adhya S, Molineux IJ. Genomic analysis of bacteriophages SP6 and K1-5, an estranged subgroup of the T7 supergroup. J. Mol. Biol. 2004;335:1151–1171. doi: 10.1016/j.jmb.2003.11.035. [DOI] [PubMed] [Google Scholar]
- 45.Wang J, Jiang Y, Vincent M, Sun Y, Yu H, Wang J, Bao Q, Kong H, Hu S. Complete genome sequence of bacteriophage T5. Virology. 2005;332:45–65. doi: 10.1016/j.virol.2004.10.049. [DOI] [PubMed] [Google Scholar]
- 46.Fischhoff D, MacNeil D, Kleckner N. Terminal redundancy heterozygotes involving the first-step-transfer region of the bacteriophage T5 chromosome. Genetics. 1976;82:145–159. doi: 10.1093/genetics/82.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cregg JM, Stewart CR. Terminal redundancy of “high frequency of recombination” markers of Bacillus subtilis phage SPO1. Virology. 1978;86:530–541. doi: 10.1016/0042-6822(78)90091-0. [DOI] [PubMed] [Google Scholar]
- 48.Rhoades M, Rhoades EA. Terminal repetition in the DNA of bacteriophage T5. J. Mol. Biol. 1972;69:187–200. doi: 10.1016/0022-2836(72)90224-0. [DOI] [PubMed] [Google Scholar]
- 49.Perkus ME, Shub DA. Mapping the genes in the terminal redundancy of bacteriophage SPO1 with restriction endonucleases. J. Virol. 1985;56:40–48. doi: 10.1128/jvi.56.1.40-48.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiest JS, McCorquodale DJ. Characterization of pre-early genes in the terminal repetition of bacteriophage BF23 DNA by nucleotide sequencing and restriction mapping. Virology. 1990;177:745–754. doi: 10.1016/0042-6822(90)90541-x. [DOI] [PubMed] [Google Scholar]
- 51.Panganiban AT, Whiteley HR. Bacillus subtilis RNAase III cleavage sites in phage SP82 early mRNA. Cell. 1983;33:907–913. doi: 10.1016/0092-8674(83)90033-8. [DOI] [PubMed] [Google Scholar]
- 52.George M, Bukhari AI. Heterogeneous host DNA attached to the left end of mature bacteriophage Mu DNA. Nature. 1981;292:175–176. doi: 10.1038/292175a0. [DOI] [PubMed] [Google Scholar]
- 53.Groenen MA, van de Putte P. Mapping of a site for packaging of bacteriophage Mu DNA. Virology. 1985;144:520–522. doi: 10.1016/0042-6822(85)90292-2. [DOI] [PubMed] [Google Scholar]
- 54.Bukhari AI, Taylor AL. Influence of insertions on packaging of host sequences covalently linked to bacteriophage Mu DNA. Proc. Natl. Acad. Sci., U S A. 1975;72:4399–4403. doi: 10.1073/pnas.72.11.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morgan G, Hatfull G, Casjens S, Hendrix R. Bacteriophage Mu genome sequence: analysis and comparison with Mulike prophages in Haemophilus, Neisseria and Deinococcus. J. Mol. Biol. 2002;317:337–359. doi: 10.1006/jmbi.2002.5437. [DOI] [PubMed] [Google Scholar]
- 56.Summer EJ, Gonzalez CF, Carlisle T, Mebane LM, Cass AM, Savva CG, LiPuma J, Young R. Burkholderia cenocepacia phage BcepMu and a family of Mu-like phages encoding potential pathogenesis factors. J. Mol. Biol. 2004;340:49–65. doi: 10.1016/j.jmb.2004.04.053. [DOI] [PubMed] [Google Scholar]
- 57.Ito J. Bacteriophage f29 terminal protein: its association with the 5′ termini of the f29 genome. J. Virol. 1978;28:895–904. doi: 10.1128/jvi.28.3.895-904.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salas M, Mellado RP, Vinuela E. Characterization of a protein covalently linked to the 5′ termini of the DNA of Bacillus subtilis phage f29. J. Mol. Biol. 1978;119:269–291. doi: 10.1016/0022-2836(78)90438-2. [DOI] [PubMed] [Google Scholar]
- 59.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 60.Maniatis T, Fritsch E, Sambrook J. Molecular cloning A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Labortory; 1982. pp. 150–163. [Google Scholar]
- 61.Southern E. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 62.Earnshaw W, Casjens S, Harrison S. Assembly of the head of bacteriophage P22, X-ray diffraction from heads, proheads and related structures. J. Mol. Biol. 1976;104:387–410. doi: 10.1016/0022-2836(76)90278-3. [DOI] [PubMed] [Google Scholar]