Abstract
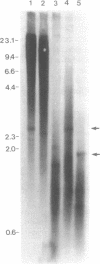
A repetitive element of approximately 200 bp was cloned from harbour seal (Phoca vitulina concolour) genomic DNA. The sequence of the element revealed putative RNA polymerase III control boxes, a poly A tail and direct terminal repeats characteristic of SINEs. Sequence and secondary structural similarities suggest that the SINE is derived from a tRNA, possibly tRNA-alanine. Southern blot analysis indicated that the element is predominately dispersed in unique regions of the seal genome, but may also be present in other repetitive sequences, such as tandemly arrayed satellite DNA. Based on slot-blot hybridization analysis, we estimate that 1.3 x 10(6) copies of the SINE are present in the harbour seal genome; SINE copy number based on the number of clones isolated from a size-selected library, however, is an order of magnitude lower (1-3 x 10(5) copies), an estimate consistent with the abundance of SINEs in other mammalian genomes. Database searches found similar sequences have been isolated from dog (Canis familiaris) and mink (Mustela vison). These, and the seal SINE sequences are characterized by an internal CT dinucleotide microsatellite in the tRNA-unrelated region. Hybridization of genomic DNA from representative species of a wide range of mammalian orders to an oligonucleotide (30mer) probe complementary to a conserved region of the SINE confirmed that the element is unique to carnivores of the superfamily Canoidea.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Brutlag D. L. Molecular arrangement and evolution of heterochromatic DNA. Annu Rev Genet. 1980;14:121–144. doi: 10.1146/annurev.ge.14.120180.001005. [DOI] [PubMed] [Google Scholar]
- Deininger P. L., Batzer M. A., Hutchison C. A., 3rd, Edgell M. H. Master genes in mammalian repetitive DNA amplification. Trends Genet. 1992 Sep;8(9):307–311. doi: 10.1016/0168-9525(92)90262-3. [DOI] [PubMed] [Google Scholar]
- Goode B. L., Feinstein S. C. "Speedprep" purification of template for double-stranded DNA sequencing. Biotechniques. 1992 Mar;12(3):374–375. [PubMed] [Google Scholar]
- Jelinek W. R., Schmid C. W. Repetitive sequences in eukaryotic DNA and their expression. Annu Rev Biochem. 1982;51:813–844. doi: 10.1146/annurev.bi.51.070182.004121. [DOI] [PubMed] [Google Scholar]
- Kido Y., Aono M., Yamaki T., Matsumoto K., Murata S., Saneyoshi M., Okada N. Shaping and reshaping of salmonid genomes by amplification of tRNA-derived retroposons during evolution. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2326–2330. doi: 10.1073/pnas.88.6.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrent'eva M. V., Rivkin M. I., Shilov A. G., Kobets M. L., Rogozin I. B., Serov O. L. B2-podobnaia povtoriaiushchaiasia posledovatel'nost'' v genome amerikanskoi norki. Dokl Akad Nauk SSSR. 1989 Jul-Aug;307(1):226–228. [PubMed] [Google Scholar]
- Minnick M. F., Stillwell L. C., Heineman J. M., Stiegler G. L. A highly repetitive DNA sequence possibly unique to canids. Gene. 1992 Jan 15;110(2):235–238. doi: 10.1016/0378-1119(92)90654-8. [DOI] [PubMed] [Google Scholar]
- Ohshima K., Koishi R., Matsuo M., Okada N. Several short interspersed repetitive elements (SINEs) in distant species may have originated from a common ancestral retrovirus: characterization of a squid SINE and a possible mechanism for generation of tRNA-derived retroposons. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6260–6264. doi: 10.1073/pnas.90.13.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada N., Ohshima K. A model for the mechanism of initial generation of short interspersed elements (SINEs). J Mol Evol. 1993 Aug;37(2):167–170. doi: 10.1007/BF02407352. [DOI] [PubMed] [Google Scholar]
- Rogers J. H. The origin and evolution of retroposons. Int Rev Cytol. 1985;93:187–279. doi: 10.1016/s0074-7696(08)61375-3. [DOI] [PubMed] [Google Scholar]
- Russo T., Duilio A., Ammendola R., Costanzo F., Cimino F. Nucleotide sequence of a mouse tRNA gene cluster. Nucleic Acids Res. 1987 Oct 26;15(20):8562–8562. doi: 10.1093/nar/15.20.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J. Mutation at VNTRs: Are minisatellites the evolutionary progeny of microsatellites? Genome. 1994 Apr;37(2):345–347. doi: 10.1139/g94-047. [DOI] [PubMed] [Google Scholar]
- Yoshioka Y., Matsumoto S., Kojima S., Ohshima K., Okada N., Machida Y. Molecular characterization of a short interspersed repetitive element from tobacco that exhibits sequence homology to specific tRNAs. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6562–6566. doi: 10.1073/pnas.90.14.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]