Abstract
The synthesis and enzymatic characterization of DUPAAA, a novel fluorogenic substrate for RNases of the pancreatic type is described. It consists of the dinucleotide uridylyl-3',5'-deoxyadenosine to which a fluorophore, o-aminobenzoic acid, and a quencher, 2,4-dinitroaniline, have been attached by means of phosphodiester linkages. Due to intramolecular quenching the intact substrate displayed very little fluorescence. Cleavage of the phosphodiester bond at the 3'-side of the uridylyl residue by RNase caused a 60-fold increase in fluorescence. This allowed the continuous and highly sensitive monitoring of enzyme activity. The substrate was turned over efficiently by RNases of the pancreatic type, but no cleavage was observed with the microbial RNase T1. Compared to the dinucleotide substrate UpA, the specificity constant with RNase A, RNase PL3 and RNase U(s) increased 6-, 18-, and 29-fold, respectively. These differences in increased catalytic efficiency most likely reflect differences in the importance of subsites on the enzyme in the binding of elongated substrates. Studies on the interactions of RNase inhibitor with RNase A using DUPAAA as a reporter substrate showed that it was well suited for monitoring this very tight protein-protein interaction using pre-steady-state kinetic methods.
Full text
PDF


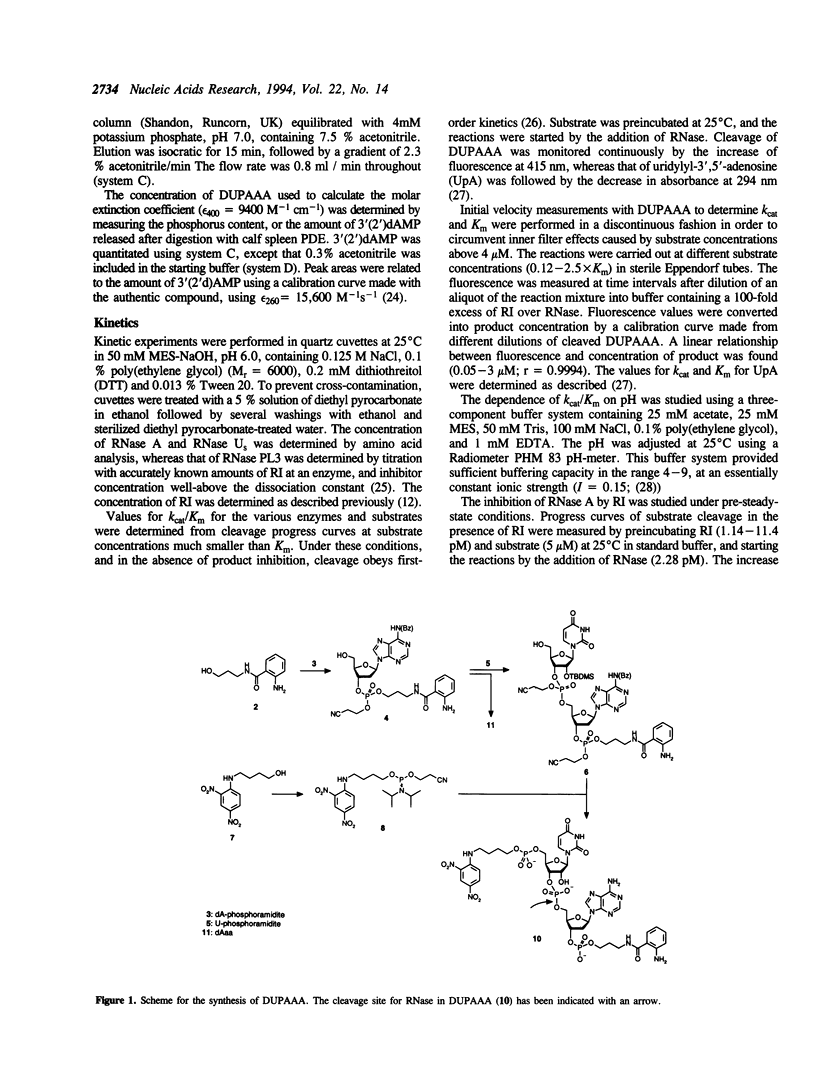
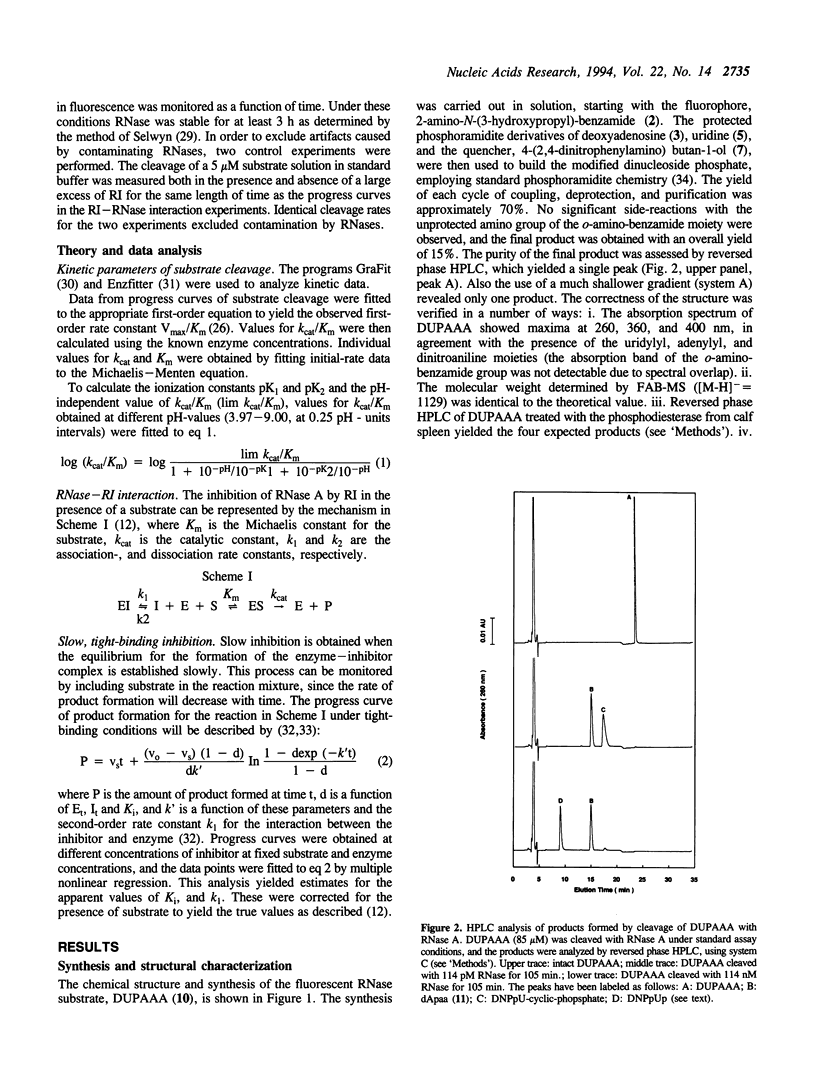
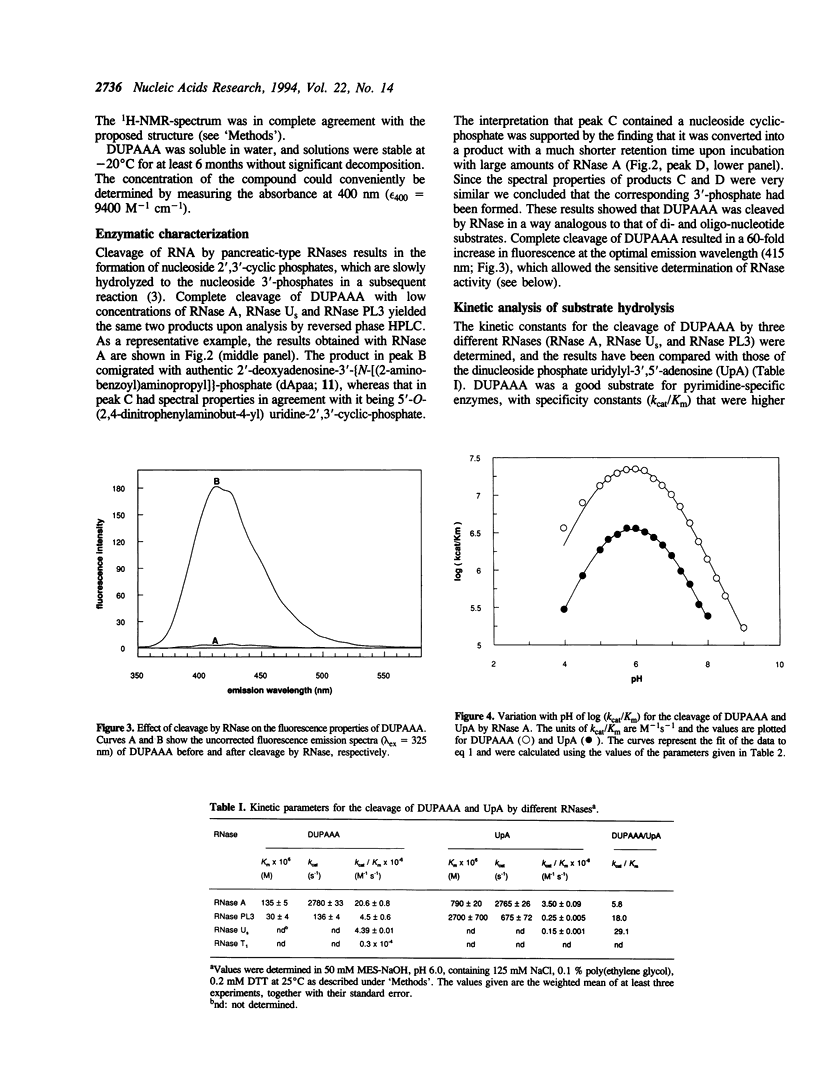
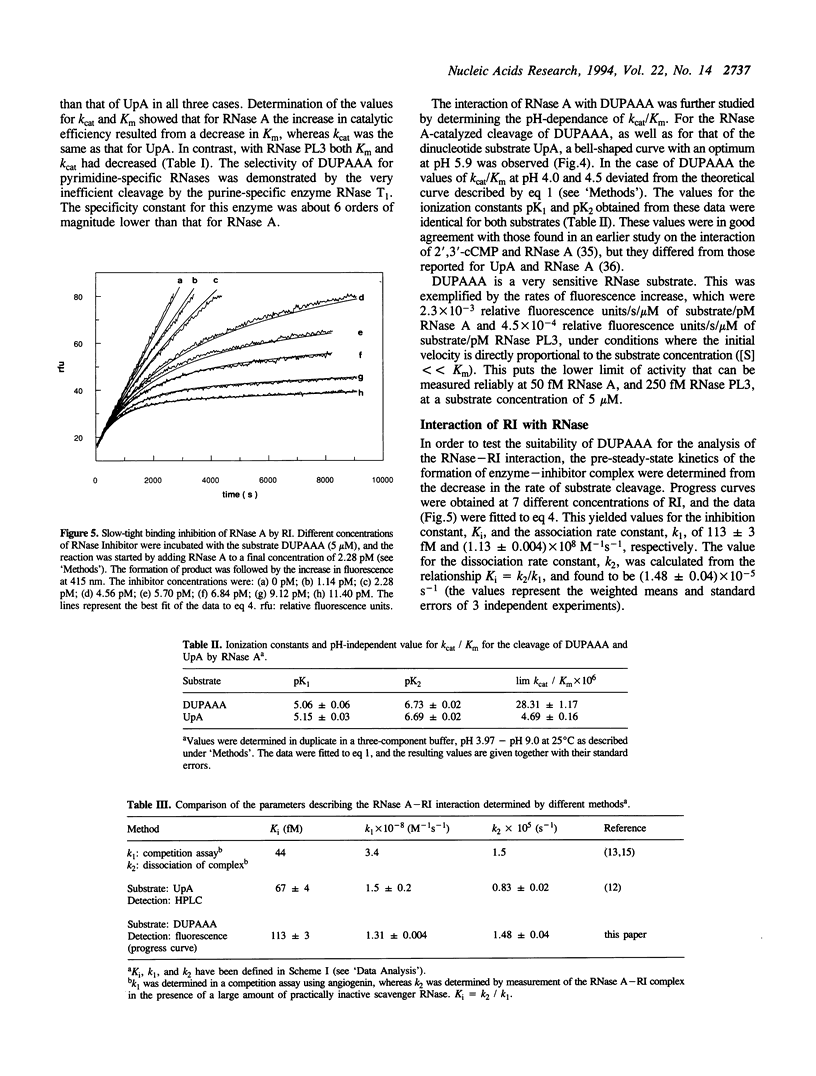


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cha S. Tight-binding inhibitors-I. Kinetic behavior. Biochem Pharmacol. 1975 Dec 1;24(23):2177–2185. doi: 10.1016/0006-2952(75)90050-7. [DOI] [PubMed] [Google Scholar]
- Durack D. T., Ackerman S. J., Loegering D. A., Gleich G. J. Purification of human eosinophil-derived neurotoxin. Proc Natl Acad Sci U S A. 1981 Aug;78(8):5165–5169. doi: 10.1073/pnas.78.8.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis K. J., Morrison J. F. Buffers of constant ionic strength for studying pH-dependent processes. Methods Enzymol. 1982;87:405–426. doi: 10.1016/s0076-6879(82)87025-0. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Renard M. pH dependence of chymotrypsin catalysis. Appendix: substrate binding to dimeric alpha-chymotrypsin studied by x-ray diffraction and the equilibrium method. Biochemistry. 1974 Mar 26;13(7):1416–1426. doi: 10.1021/bi00704a016. [DOI] [PubMed] [Google Scholar]
- Fett J. W., Strydom D. J., Lobb R. R., Alderman E. M., Bethune J. L., Riordan J. F., Vallee B. L. Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma cells. Biochemistry. 1985 Sep 24;24(20):5480–5486. doi: 10.1021/bi00341a030. [DOI] [PubMed] [Google Scholar]
- Fitzgerald P. C., Hartley R. W. Polyethenoadenosine phosphate as a fluorogenic substrate for barnase. Anal Biochem. 1993 Nov 1;214(2):544–547. doi: 10.1006/abio.1993.1536. [DOI] [PubMed] [Google Scholar]
- Gleich G. J., Loegering D. A., Bell M. P., Checkel J. L., Ackerman S. J., McKean D. J. Biochemical and functional similarities between human eosinophil-derived neurotoxin and eosinophil cationic protein: homology with ribonuclease. Proc Natl Acad Sci U S A. 1986 May;83(10):3146–3150. doi: 10.1073/pnas.83.10.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HERRIES D. G., MATHIAS A. P., RABIN B. R. The active site and mechanism of action of bovine pancreatic ribonuclease. 3. The pH-dependence of the kinetic parameters for the hydrolysis of cytidine 2',3'-phosphate. Biochem J. 1962 Oct;85:127–134. doi: 10.1042/bj0850127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofsteenge J., Matthies R., Stone S. R. Primary structure of a ribonuclease from porcine liver, a new member of the ribonuclease superfamily. Biochemistry. 1989 Dec 12;28(25):9806–9813. doi: 10.1021/bi00451a040. [DOI] [PubMed] [Google Scholar]
- Hosoya K., Nagareda Y., Hasemi S., Sanda A., Takizawa Y., Watanabe H., Ohgi K., Irie M. Primary structure of an alkaline ribonuclease from bovine liver. J Biochem. 1990 Apr;107(4):613–618. doi: 10.1093/oxfordjournals.jbchem.a123095. [DOI] [PubMed] [Google Scholar]
- Irie M., Ohgi K., Yoshinaga M., Yanagida T., Okada Y., Teno N. Roles of lysine1 and lysine7 residues of bovine pancreatic ribonuclease in the enzymatic activity. J Biochem. 1986 Oct;100(4):1057–1063. doi: 10.1093/oxfordjournals.jbchem.a121785. [DOI] [PubMed] [Google Scholar]
- Iwama M., Kunihiro M., Ohgi K., Irie M. Purification and properties of human urine ribonucleases. J Biochem. 1981 Apr;89(4):1005–1016. [PubMed] [Google Scholar]
- Lee F. S., Shapiro R., Vallee B. L. Tight-binding inhibition of angiogenin and ribonuclease A by placental ribonuclease inhibitor. Biochemistry. 1989 Jan 10;28(1):225–230. doi: 10.1021/bi00427a031. [DOI] [PubMed] [Google Scholar]
- Lee F. S., Vallee B. L. Structure and action of mammalian ribonuclease (angiogenin) inhibitor. Prog Nucleic Acid Res Mol Biol. 1993;44:1–30. doi: 10.1016/s0079-6603(08)60215-9. [DOI] [PubMed] [Google Scholar]
- Neumann U., Hofsteenge J. Interaction of semisynthetic variants of RNase A with ribonuclease inhibitor. Protein Sci. 1994 Feb;3(2):248–256. doi: 10.1002/pro.5560030209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira M. C., Hirata I. Y., Chagas J. R., Boschcov P., Gomes R. A., Figueiredo A. F., Juliano L. Intramolecularly quenched fluorogenic peptide substrates for human renin. Anal Biochem. 1992 May 15;203(1):39–46. doi: 10.1016/0003-2697(92)90040-e. [DOI] [PubMed] [Google Scholar]
- Parés X., Nogués M. V., de Llorens R., Cuchillo C. M. Structure and function of ribonuclease A binding subsites. Essays Biochem. 1991;26:89–103. [PubMed] [Google Scholar]
- Postek K. M., LaDue T., Nelson C., Sandwick R. K. Spectrophotometric ribonuclease assays using dinucleoside monophosphate substrates. Anal Biochem. 1992 May 15;203(1):47–52. doi: 10.1016/0003-2697(92)90041-5. [DOI] [PubMed] [Google Scholar]
- Reimert C. M., Minuva U., Kharazmi A., Bendtzen K. Eosinophil protein X/eosinophil derived neurotoxin (EPX/EDN). Detection by enzyme-linked immunosorbent assay and purification from normal human urine. J Immunol Methods. 1991 Jul 26;141(1):97–104. doi: 10.1016/0022-1759(91)90214-z. [DOI] [PubMed] [Google Scholar]
- Sato E., Itoh S., Kanaoka Y. New fluorogenic substrates containing bimane system for ribonuclease T1. J Pharmacobiodyn. 1991 Jan;14(1):43–46. doi: 10.1248/bpb1978.14.43. [DOI] [PubMed] [Google Scholar]
- Selwyn M. J. A simple test for inactivation of an enzyme during assay. Biochim Biophys Acta. 1965 Jul 29;105(1):193–195. doi: 10.1016/s0926-6593(65)80190-4. [DOI] [PubMed] [Google Scholar]
- Shapiro R., Riordan J. F., Vallee B. L. Characteristic ribonucleolytic activity of human angiogenin. Biochemistry. 1986 Jun 17;25(12):3527–3532. doi: 10.1021/bi00360a008. [DOI] [PubMed] [Google Scholar]
- Shapiro R., Vallee B. L. Interaction of human placental ribonuclease with placental ribonuclease inhibitor. Biochemistry. 1991 Feb 26;30(8):2246–2255. doi: 10.1021/bi00222a030. [DOI] [PubMed] [Google Scholar]
- Vicentini A. M., Hemmings B. A., Hofsteenge J. Residues 36-42 of liver RNase PL3 contribute to its uridine-preferring substrate specificity. Cloning of the cDNA and site-directed mutagenesis studies. Protein Sci. 1994 Mar;3(3):459–466. doi: 10.1002/pro.5560030311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicentini A. M., Kieffer B., Matthies R., Meyhack B., Hemmings B. A., Stone S. R., Hofsteenge J. Protein chemical and kinetic characterization of recombinant porcine ribonuclease inhibitor expressed in Saccharomyces cerevisiae. Biochemistry. 1990 Sep 18;29(37):8827–8834. doi: 10.1021/bi00489a046. [DOI] [PubMed] [Google Scholar]
- Ward D. C., Reich E., Stryer L. Fluorescence studies of nucleotides and polynucleotides. I. Formycin, 2-aminopurine riboside, 2,6-diaminopurine riboside, and their derivatives. J Biol Chem. 1969 Mar 10;244(5):1228–1237. [PubMed] [Google Scholar]
- Wierenga R. K., Huizinga J. D., Gaastra W., Welling G. W., Beintema J. J. Affinity chromatography of porcine pancreatic ribonuclease reinvestigation of the N-terminal amino acid sequence. FEBS Lett. 1973 Apr 15;31(2):181–185. doi: 10.1016/0014-5793(73)80098-5. [DOI] [PubMed] [Google Scholar]
- Williams J. W., Morrison J. F., Duggleby R. G. Methotrexate, a high-affinity pseudosubstrate of dihydrofolate reductase. Biochemistry. 1979 Jun 12;18(12):2567–2573. doi: 10.1021/bi00579a021. [DOI] [PubMed] [Google Scholar]
- Witmer M. R., Falcomer C. M., Weiner M. P., Kay M. S., Begley T. P., Ganem B., Scheraga H. A. U-3'-BCIP: a chromogenic substrate for the detection of RNase A in recombinant DNA expression systems. Nucleic Acids Res. 1991 Jan 11;19(1):1–4. doi: 10.1093/nar/19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron A., Carmel A., Katchalski-Katzir E. Intramolecularly quenched fluorogenic substrates for hydrolytic enzymes. Anal Biochem. 1979 May;95(1):228–235. doi: 10.1016/0003-2697(79)90210-0. [DOI] [PubMed] [Google Scholar]
- Youle R. J., Newton D., Wu Y. N., Gadina M., Rybak S. M. Cytotoxic ribonucleases and chimeras in cancer therapy. Crit Rev Ther Drug Carrier Syst. 1993;10(1):1–28. [PubMed] [Google Scholar]
- Zhou H. M., Strydom D. J. The amino acid sequence of human ribonuclease 4, a highly conserved ribonuclease that cleaves specifically on the 3' side of uridine. Eur J Biochem. 1993 Oct 1;217(1):401–410. doi: 10.1111/j.1432-1033.1993.tb18259.x. [DOI] [PubMed] [Google Scholar]
- del Rosario E. J., Hammes G. G. Kinetic and equilibrium studies of the ribonuclease-catalyzed hydrolysis of uridine 2',3'-cyclic phosphate. Biochemistry. 1969 May;8(5):1884–1889. doi: 10.1021/bi00833a017. [DOI] [PubMed] [Google Scholar]



