Abstract
This chapter describes how to use the Brainbow strategy to label neurons in many different hues. The Brainbow system uses a random Cre/lox recombination to create varied combinations of red, blue, and green fluorescent proteins in each cell. The differences in color allow users to follow multiple cells, regardless of how closely they are positioned. We use the zebrafish trigeminal sensory ganglion as an example and discuss potential modifications for the general use of this technique.
INTRODUCTION
Brainbow Imaging in Zebrafish
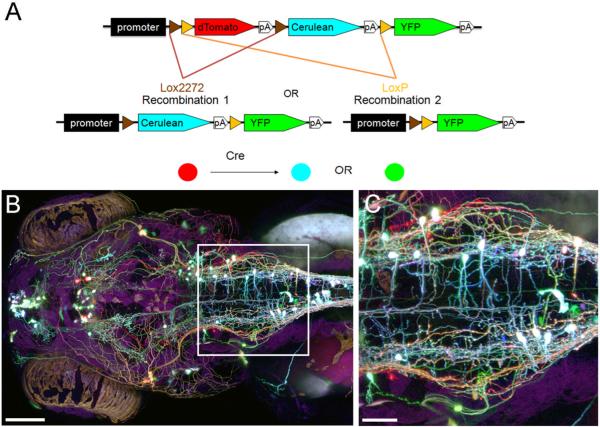
Brainbow is an imaging strategy that provides multicolor labeling of cells using a single transgenic cassette (Livet et al. 2007, Lichtman et al. 2008). More details on Brainbow can be found in Weissman et al. (2011). Briefly, red, blue, and green fluorescent proteins are arranged in a tandem array, with two pairs of lox sites (recognition sequences for the Cre recombinase) flanking the first two fluorescent proteins (Fig 1A). Without Cre-induced recombination, the first protein (red) in the array will be expressed. Cre expression results in one of three outcomes: red (no recombination), blue (recombination 1), or green (recombination 2). When additional copies of the Brainbow cassette coexist inside a cell, these three primary tones can be mixed, rapidly increasing possible color combinations. This diversity of color using a single promoter provides a powerful platform for studying neuronal morphology and cell movements. Brainbow can thus be considered a multicolor version of the Golgi stain that can be used to analyze brain circuitry or mutant phenotypes (Fig 1B, C).
FIGURE 1. Brainbow labeling in zebrafish.
(A) Schematic of a Brainbow transgenic construct. A promoter is followed by three fluorescent proteins, interspersed with two pairs of lox sites (lox2272 and loxP). lox2272 and loxP are only recognized by Cre as identical pairs; thus, only two possible recombinations can occur (1 and 2). Recombinations 1 and 2 will allow expression of Cerulean and yellow fluorescent protein (YFP), respectively. (B) A 120-hpf (hours post fertilization) larval zebrafish labeled with Islet1-Brainbow, viewed dorsally. (C) High-magnification image of the boxed region in B. Scale bars: 50 μm (B), 20 μm (C).
There are two unique advantages of using Brainbow in zebrafish compared to many other model organisms. First, zebrafish is transparent during early development. Live fluorescent cells of embryos, larvae, and juveniles can be visualized directly using optical microscopy. Secondly, generating Brainbow transgenic fish by DNA injection is time-efficient, since injected embryos immediately express Brainbow in a mosaic pattern. Thus, mutants can be analyzed using Brainbow without the need for several generations of breeding.
PROTOCOL
Brainbow Imaging in Zebrafish
This protocol describes how to use Brainbow imaging in zebrafish and provides examples of how to use color as a guide to trace axonal processes.
IMAGING SETUP
We use an Olympus laser-scanning confocal imaging system (FV1000) on an upright microscope. Required laser lines are 440 nm, 515 nm, and 561 nm. The recommended objective is the Olympus 20x/0.95-NA XLUMPlanFl. Although we will only describe Brainbow imaging by confocal microscopy, it is also possible to use wide-field fluorescence or two-photon microscopy. See Microscope Setup, below, to test if your imaging setup can adequately separate the three different fluorophores used in Brainbow. See Step 12 for specific excitation wavelength and emission filter settings.
MATERIALS
CAUTION: See Appendix 7 for proper handling of materials marked with <!>.
See the end of the chapter for recipes for reagents marked with <R>.
Reagents
Agarose, 1.5% low melting point in water
Blue water (20 mL of methylene blue <!> [0.1% weight-to-volume ratio stock in water], 6 g of Instant Ocean sea salt [Instant Ocean], and 20 L distilled deionized water)
Brainbow cassette cytomegalovirus (CMV)-Brainbow-1.0 L (see Selection of Brainbow Versions, below)
MESAB <R>, 30×
1-phenyl-2-thiourea (PTU) water <R>
Zebrafish, 1–5-d-old larvae
Equipment
Conical tube, 50-mL
Culture dishes, glass-bottomed (P35G-0-14-C, MatTek)
Dry block heater (40°C; VWR International)
Glass slides
Incubator (28°C)
Petri dishes
Tape, double-sided
METHOD
General Zebrafish Methods
General methods for zebrafish use can be found online in “The zebrafish book” (Westerfield, 2000; http://zfin.org/zf_info/zfbook/zfbk.html). Methods for generating transgenic fish by DNA injection can be found in Chapter 27.
Selection of Brainbow Plasmids
Brainbow plasmid constructs described here can be obtained from Addgene.com, a nonprofit plasmid sharing resource. Search with keyword “Brainbow,” and select those most suitable for your needs. The CMV versions can be used for ubiquitous expression in zebrafish; the Thy1 promoter used the Thy1-Brainbow plasmids, which do not work in zebrafish. In this protocol, we use the CMVBrainbow-1.0 L plasmid, which encodes three cytoplasmic fluorescent proteins—dTomato (red), monomeric-Cerulean (blue), and monomeric-YFP (green). Because these three fluorescent proteins have nonoverlapping excitation/emission profiles, their colors can be separated with optical filters. Four-color versions of the Brainbow cassette (CMV-Brainbow-1.1 M and CMV-Brainbow-2.1 R) generate more color diversity, but the overlapping spectra of the four fluorescent proteins require computational processing (linear unmixing or spectral analysis) for color channel separation (see Livet et al. 2007). We have also noticed that the membrane-tagged version of Brainbow (R.W. Draft and J.W. Lichtman, unpubl.) confers more neuronal toxicity than the cytoplasmic version, rendering the cytoplasmic version more suitable when a high expression level is necessary.
Microscope Setup
Before starting, it is crucial to test if your imaging set up can adequately separate the three different fluorophores used in Brainbow. A good tool to use is the spectra viewer on the Invitrogen website (http://probes.invitrogen.com/servlets/spectraviewer). Minimize bleed-through between channels by acquiring each channel independently (i.e. sequential scan), as bleed-through reduces color diversity.
Generation of Brainbow Expression Vectors
In terms of cloning, the Brainbow cassette behaves no differently than a single fluorescent protein. We have placed various neural-specific promoters upstream of the Brainbow cassette.
Introducing Cre Recombinase
Brainbow can work both when it is stably integrated into the genome (e.g. stable Brainbow transgenic lines) or as an extra-chromosomal element (e.g. embryos injected with Brainbow DNA). To induce Cre expression, we generated transgenic fish that express Cre under the control of a heat-shock promoter (Hsp:Cre; B. Ciruna and A. F. Schier, unpubl.), thereby inducing Cre expression and hence recombination after 37°C incubation in a water bath. This heat shock should take place before the Brainbow promoter initiates transcription. Alternatively, in vitro synthesized Cre mRNA can be injected along with Brainbow plasmid.
One quick way to test for Cre expression is to directly inject the ubiquitously-expressed CMV-Brainbow-1.0 L plasmid at the one cell stage. Cerulean and YFP fluorescence should appear in a variety of tissues, including muscle, brain, skin, and notochord. Sufficient Cre expression will yield cells in a variety of colors.
Cross Hsp:Cre+/+ transgenic fish.
Inject Brainbow plasmid DNA into embryos at the one-cell stage. Transfer the embryos to a Petri dish with blue water, and incubate at 28°C.
At 12 hpf, transfer injected embryos into a 50-mL conical tube. The total volume of embryos plus blue water should be 35 mL.
Place the conical tube into a 37°C water bath, and incubate for 1 h to induce Cre expression.
Transfer the tube to a 28°C incubator for 1 h, and then transfer the embryos back to original Petri dish.
At 24 hpf, move the fish to PTU water to prevent pigment formation. Alternatively, pigment mutants such as nacre (available from http://zebrafish.org) can be used.
At the desired time point of development, anesthetize the fish with 1× MESAB in PTU water (see Recipes). Gently poke the fish with forceps to make sure they are immobile.
Transfer the fish to a glass-bottomed dish. Remove as much water as possible.
Add 1.5% low-melt agarose (kept on a dry heat block set at 40°C) to the dish. Arrange the fish so that the surface to be imaged is facing the glass bottom.
When the agarose has solidified, add 1× MESAB to prevent the agarose from drying.
Place the dish directly onto the slide mount of an inverted microscope. If an upright microscope is being used, pour out the MESAB solution in the dish, and invert the dish so that the glass bottom is facing up. Attach two pieces of double-sided tape to a glass slide, and adhere the top of the dish to the slide (glass slide up).
- The excitation wavelength and emission filter settings for our confocal microscope are as follows:
- dTomato, excitation 561 nm; emission filter, band pass 585–685 nm
- YFP, excitation 515 nm; emission filter, band pass 535–565 nm
- Cerulean, excitation 440 nm; emission filter, band pass 465–495 nm
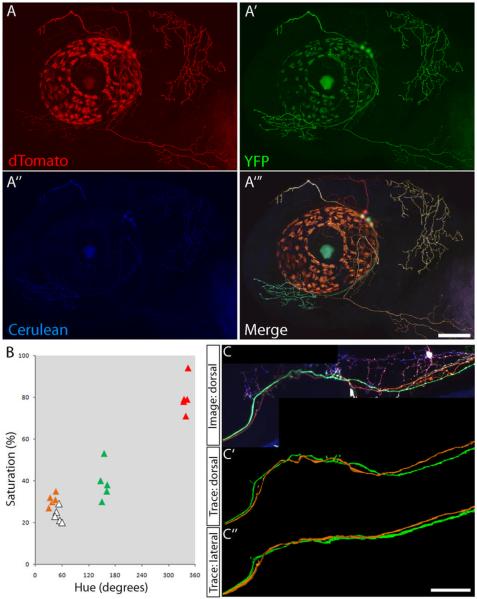
Gray scale images acquired from all three channels are pseudocolored and merged to obtain a red/green/blue (RGB) color image (Fig. 2A–A'”). This can be done by image processing software such as ImageJ or Adobe Photoshop. In the examples shown in Figures 1 and 2, dTomato is pseudocolored red, YFP is green, and Cerulean is blue. Differences in color can then be determined visually.
Color difference can also be measured using quantitative color models (Livet et al. 2007). We use the HSB (hue, saturation, and brightness) color model in Adobe Photoshop to measure the hue and saturation of different neurons (Fig. 2B). HSB values will be displayed in the color picker window upon selection of the “color picker” tool in Adobe Photoshop and clicking on a point of interest. Brightness is not taken into consideration as it varies greatly between different regions of the same cell (e.g., between axon and cell body).
Figure 2. Brainbow assisted tracing of trigeminal sensory neuron central axons.
Trigeminal sensory neurons were labeled by transient expression of Islet1-Brainbow. (A–A‴) A 2-d-old larva with trigeminal sensory neurons labeled in different colors. Confocal images were acquired in the dTomato (A), YFP (A′), and Cerulean (A″) channels. These channels are combined to generate a color image (A‴). The distinct peripheral axon territories can be discerned based on their color (white, red, green, and orange). (B) To ensure that color separation is consistent throughout the length of the axon, the color of each neuron is defined quantitatively. The hue and saturation of randomly selected points along different axons are measured in Adobe Photoshop and plotted. Points from different neurons, colored to match the image in A, form distinct clusters on the graph. This indicates that the color separation in axons correctly reflects separate cellular origins. (C) Trigeminal central axon of the same larva at 5 d of development, imaged dorsally. (C′–C″) The green and orange axons are traced digitally and viewed dorsally and laterally. Scale bars, 50 μm.
Troubleshooting
Problem
Seeing the blue fluorescence is difficult.
Solution
The blue fluorescent protein, Cerulean, is the least bright of the three fluorophores. If overall expression is weak, it may be difficult to see blue fluorescence. It is important to use the optimal excitation wavelength for Cerulean; that is, 405-nm and 458-nm laser lines are less capable of exciting Cerulean compared to the 440-nm laser line. Alternatively, the more recently developed monomeric teal fluorescent protein 1 (mTFP1) is brighter and more photostable than Cerulean and is a promising candidate to replace Cerulean in Brainbow (Ai et al. 2006).
Problem
Poor color diversity is exhibited.
Solution
Diversity of colors depends on the copy number of Brainbow cassettes. If there is only one copy of Brainbow in a transgenic line, there will be at most three colors (red, green, and blue). Additional copies of Brainbow are necessary for more colors. Copy number is usually high in transient expression by DNA injection. In stable transgenic, however, a low number of inserts may lead to less color diversity. It is necessary to screen for lines with multiple Brainbow copies or intercross multiple Brainbow lines. The efficiency of Cre recombination also affects color diversity. If most Brainbow cassettes have recombined and excised dTomato, the overall expression of dTomato will be low. If most Brainbow cassettes are not recombined, very little Cerulean and YFP will be expressed. Finally, the stochastic nature of Cre/lox recombination will lead to different color combination in individual animals. It is often necessary to screen many larvae to find ones where all labeled cells are differently colored.
EXAMPLE APPLICATION
We used the sensory neuron–specific element of the Islet1 gene to drive Brainbow expression in trigeminal sensory neurons (Higashijima et al. 2000; Sagasti et al. 2005). After Cre recombination, trigeminal neurons were imaged in three different channels (red, green, and blue), which were then combined to form a single color image (Fig. 2). Because of the stochastic expression of fluorescent proteins, different trigeminal neurons had different fluorescent profiles. The resulting image showed trigeminal neuron cell bodies and peripheral axons that were white, red, green, and orange. The color of each neuron was visually and quantitatively distinct, which allowed for identification of neuronal processes and their cell bodies of origin for different trigeminal sensory neurons (Fig. 2A,B). The central axons of the green and orange trigeminal neuron were digitally traced using the 3D-reconstruction software Reconstruct (Fig. 2C) (Lu et al. 2009). Axons could be followed easily even after they crossed paths.
COCLUSION
Rapid advances in the fields of optical microscopy and fluorescent dyes provide strong incentives to use an optically accessible model organism such as the zebrafish. Although Brainbow was originally developed for mice, the technique can easily be adapted for zebrafish. Because fish have fewer neurons than mice, it may be possible to determine the complete connectivity between different brain regions—for instance, between the retina and the optic tectum or between the trigeminal ganglion and the hindbrain. Moreover, live Brainbow fish can be imaged over days, allowing the analysis of highly dynamic processes such as axon growth and pruning. Brainbow also holds great potential for the study of other organs and tissues, using color diversity for easier cell tracing in lineage experiments.
RECIPES
CAUTION: See Appendix 7 for proper handling of materials marked with <!>.
Recipes for reagents marked with <R> are included in this list.
MESAB (30×)
| Reagent (for 1200 mL) | Quantity |
| Ethyl 3-aminobenzoate methane-sulfonate salt (0.5 mM) (Sigma-Aldrich) <!> | 5 g |
| Distilled deionized water | 1174.8 mL |
| Tris base <!> (1 M, pH 9.0) | 25.2 mL |
Make 10-mL aliquots, and store in freezer at −80°C. Dilute to 1x with blue water.
PTU water
Add 150 mg of PTU <!> (Sigma-Aldrich) to 100-mL post fertilization blue water to make a 50× (10-mM) stock. PTU may precipitate at room temperature. Heat to 60°C to re-dissolve PTU. Dilute to 1× (0.2 mM) by adding blue water.
ACKNOLEDGEMENTS
We thank Shu-Hsien Sheu, Cindy Wang, and Ian Woods for critical reading of the manuscript. Our research is supported by a NRSA postdoctoral fellowship (Y.A.P) and research grants from the National Institutes of Health (A.F.S.).
REFERENCES
- Ai HW, Henderson JN, Remington SJ, Campbell RE. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: Structural characterization and applications in fluorescence imaging. Biochem J. 2006;400:531–540. doi: 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima S, Hotta Y, Okamoto H. Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J Neurosci. 2000;20:206–218. doi: 10.1523/JNEUROSCI.20-01-00206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtman JW, Livet J, Sanes JR. A technicolour approach to the connectome. Nat Rev Neurosci. 2008;9:417–422. doi: 10.1038/nrn2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Lu J, Tapia JC, White OL, Lichtman JW. The interscutularis muscle connectome. PLoS Biol. 2009;7:e32. doi: 10.1371/journal.pbio.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagasti A, Guido MR, Raible DW, Schier AF. Repulsive interactions shape the morphologies and functional arrangement of zebrafish peripheral sensory arbors. Curr Biol. 2005;15:804–814. doi: 10.1016/j.cub.2005.03.048. [DOI] [PubMed] [Google Scholar]
- Weissman TA, Sanes JR, Lichtman JW, Livet J. Generating and imaging multicolor Brainbow mice. In: Helmchen F, Konnerth A, editors. In Imgaing in neuroscience: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2011. (in press) [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book: A guide for the laboratory use of zebrafish Danio (Brachydanio) rerio. 4th ed. University of Oregon Press; Eugene, OR: 2000. [Google Scholar]
WWW RESOURCES
- Addgene.com Addgene, a nonprofit plasmid sharing resource.
- http://probes.invitrogen.com/servlets/spectraviewer Invitrogen Spectra Viewer.
- http://zfin.org/zf_info/zfbook/zfbk.html Westerfield M. 2000. The zebrafish book: A guide for the laboratory use of zebrafish Danio (Brachydanio) rerio, 4th ed. University of Oregon Press, Eugene, OR.