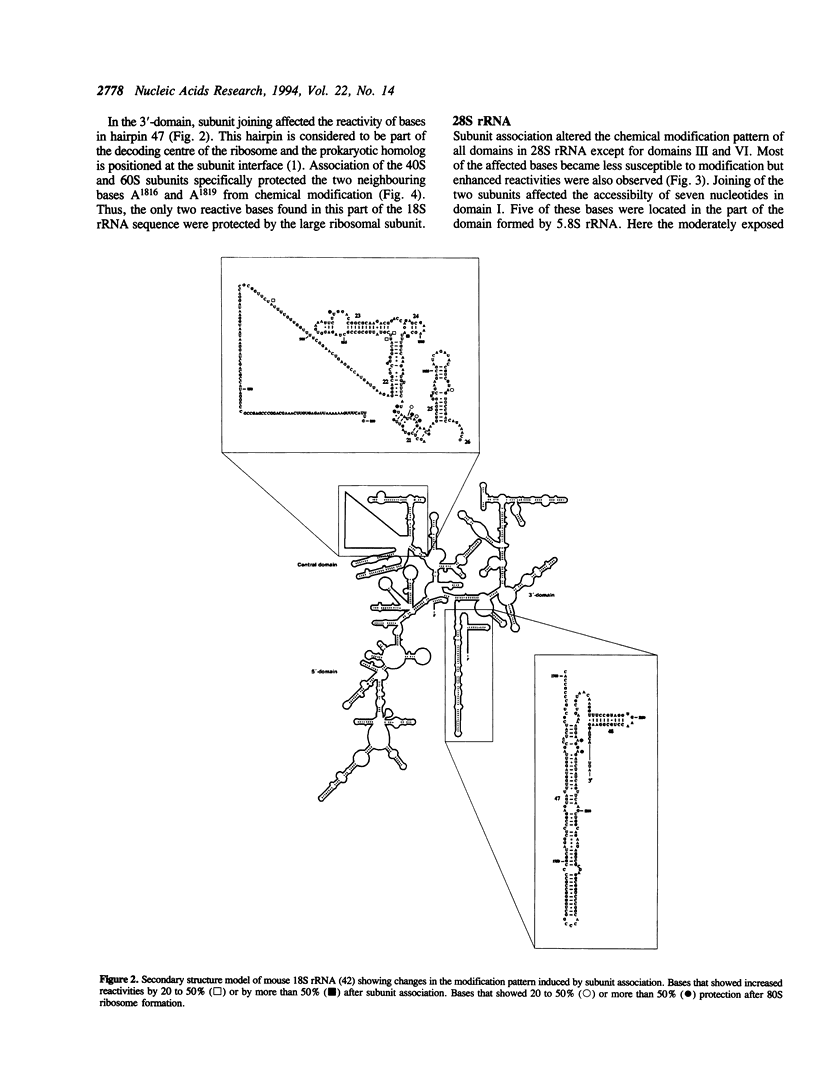
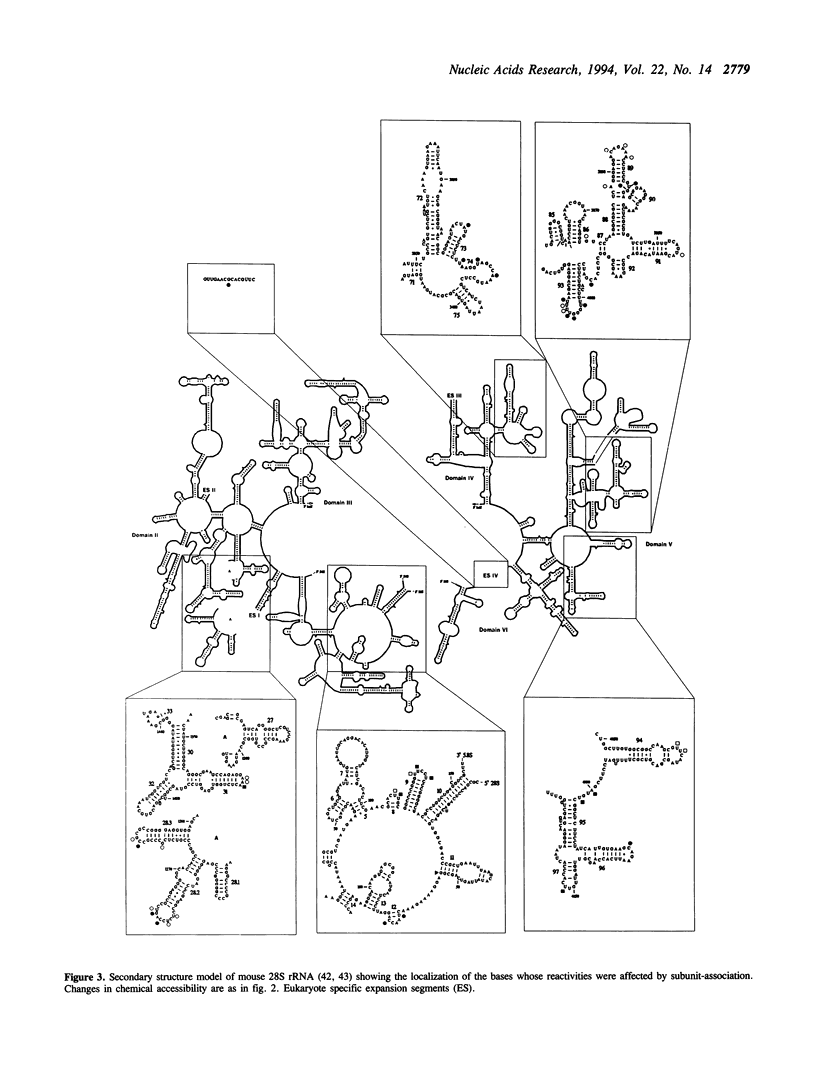
Abstract
The participation of 18S, 5.8S and 28S ribosomal RNA in subunit association was investigated by chemical modification and primer extension. Derived 40S and 60S ribosomal subunits isolated from mouse Ehrlich ascites cells were reassociated into 80S particles. These ribosomes were treated with dimethyl sulphate and 1-cyclohexyl-3-(morpholinoethyl) carbodiimide metho-p-toluene sulfonate to allow specific modification of single strand bases in the rRNAs. The modification pattern in the 80S ribosome was compared to that of the derived ribosomal subunits. Formation of complete 80S ribosomes altered the extent of modification of a limited number of bases in the rRNAs. The majority of these nucleotides were located to phylogenetically conserved regions in the rRNA but the reactivity of some bases in eukaryote specific sequences was also changed. The nucleotides affected by subunit association were clustered in the central and 3'-minor domains of 18S rRNA as well as in domains I, II, IV and V of 5.8/28S rRNA. Most of the bases became less accessible to modification in the 80S ribosome, suggesting that these bases were involved in subunit interaction. Three regions of the rRNAs, the central domain of 18S rRNA, 5.8S rRNA and domain V in 28S rRNA, contained bases that showed increased accessibility for modification after subunit association. The increased reactivity indicates that these regions undergo structural changes upon subunit association.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baudin F., Ehresmann C., Romby P., Mougel M., Colin J., Lempereur L., Bachellerie J. P., Ebel J. P., Ehresmann B. Higher-order structure of domain III in Escherichia coli 16S ribosomal RNA, 30S subunit and 70S ribosome. Biochimie. 1987 Oct;69(10):1081–1096. doi: 10.1016/0300-9084(87)90008-3. [DOI] [PubMed] [Google Scholar]
- Baudin F., Mougel M., Romby P., Eyermann F., Ebel J. P., Ehresmann B., Ehresmann C. Probing the phosphates of the Escherichia coli ribosomal 16S RNA in its naked form, in the 30S subunit, and in the 70S ribosome. Biochemistry. 1989 Jul 11;28(14):5847–5855. doi: 10.1021/bi00440a022. [DOI] [PubMed] [Google Scholar]
- Brawerman G., Mendecki J., Lee S. Y. A procedure for the isolation of mammalian messenger ribonucleic acid. Biochemistry. 1972 Feb 15;11(4):637–641. doi: 10.1021/bi00754a027. [DOI] [PubMed] [Google Scholar]
- Caruthers M. H., Beaton G., Wu J. V., Wiesler W. Chemical synthesis of deoxyoligonucleotides and deoxyoligonucleotide analogs. Methods Enzymol. 1992;211:3–20. doi: 10.1016/0076-6879(92)11003-2. [DOI] [PubMed] [Google Scholar]
- Chapman N. M., Noller H. F. Protection of specific sites in 16 S RNA from chemical modification by association of 30 S and 50 S ribosomes. J Mol Biol. 1977 Jan 5;109(1):131–149. doi: 10.1016/s0022-2836(77)80049-1. [DOI] [PubMed] [Google Scholar]
- Döring T., Greuer B., Brimacombe R. The three-dimensional folding of ribosomal RNA; localization of a series of intra-RNA cross-links in 23S RNA induced by treatment of Escherichia coli 50S ribosomal subunits with bis-(2-chloroethyl)-methylamine. Nucleic Acids Res. 1991 Jul 11;19(13):3517–3524. doi: 10.1093/nar/19.13.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring T., Greuer B., Brimacombe R. The topography of the 3'-terminal region of Escherichia coli 16S ribosomal RNA; an intra-RNA cross-linking study. Nucleic Acids Res. 1992 Apr 11;20(7):1593–1597. doi: 10.1093/nar/20.7.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerman D. J., Symons R. H. Sequence at the site of attachment of an affinity-label derivative of puromycin on 23-S ribosomal RNA of Escherichia coli ribosomes. Eur J Biochem. 1978 Jan 2;82(1):225–234. doi: 10.1111/j.1432-1033.1978.tb12015.x. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber N. M., Cantor C. R. Accessibility and structure of ribosomal RNA monitored by slow tritium exchange of ribosomes. J Mol Biol. 1981 Feb 25;146(2):241–257. doi: 10.1016/0022-2836(81)90434-4. [DOI] [PubMed] [Google Scholar]
- Gornicki P., Baudin F., Romby P., Wiewiorowski M., Kryzosiak W., Ebel J. P., Ehresmann C., Ehresmann B. Use of lead(II) to probe the structure of large RNA's. Conformation of the 3' terminal domain of E. coli 16S rRNA and its involvement in building the tRNA binding sites. J Biomol Struct Dyn. 1989 Apr;6(5):971–984. doi: 10.1080/07391102.1989.10506525. [DOI] [PubMed] [Google Scholar]
- Gutell R. R., Schnare M. N., Gray M. W. A compilation of large subunit (23S- and 23S-like) ribosomal RNA structures. Nucleic Acids Res. 1992 May 11;20 (Suppl):2095–2109. doi: 10.1093/nar/20.suppl.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Weiser B., Woese C. R., Noller H. F. Comparative anatomy of 16-S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- Herr W., Chapman N. M., Noller H. F. Mechanism of ribosomal subunit association: discrimination of specific sites in 16 S RNA essential for association activity. J Mol Biol. 1979 Jun 5;130(4):433–449. doi: 10.1016/0022-2836(79)90433-9. [DOI] [PubMed] [Google Scholar]
- Herr W., Noller H. F. Protection of specific sites in 23 S and 5 S RNA from chemical modification by association of 30 S and 50 S ribosomes. J Mol Biol. 1979 Jun 5;130(4):421–432. doi: 10.1016/0022-2836(79)90432-7. [DOI] [PubMed] [Google Scholar]
- Holmberg L., Melander Y., Nygård O. Probing the structure of mouse Ehrlich ascites cell 5.8S, 18S and 28S ribosomal RNA in situ. Nucleic Acids Res. 1994 Apr 25;22(8):1374–1382. doi: 10.1093/nar/22.8.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg L., Melander Y., Nygård O. Ribosome-bound eukaryotic elongation factor 2 protects 5 S rRNA from modification. J Biol Chem. 1992 Oct 25;267(30):21906–21910. [PubMed] [Google Scholar]
- Kühlbrandt W., Unwin P. N. Distribution of RNA and protein in crystalline eukaryotic ribosomes. J Mol Biol. 1982 Apr 15;156(3):431–448. doi: 10.1016/0022-2836(82)90259-5. [DOI] [PubMed] [Google Scholar]
- Liu W., Lo A. C., Nazar R. N. Structure of the ribosome-associated 5.8 S ribosomal RNA. J Mol Biol. 1983 Dec 5;171(2):217–224. doi: 10.1016/s0022-2836(83)80354-4. [DOI] [PubMed] [Google Scholar]
- Marconi R. T., Hill W. E. Evidence for a tRNA/rRNA interaction site within the peptidyltransferase center of the Escherichia coli ribosome. Biochemistry. 1989 Jan 24;28(2):893–899. doi: 10.1021/bi00428a073. [DOI] [PubMed] [Google Scholar]
- Meier N., Göringer H. U., Kleuvers B., Scheibe U., Eberle J., Szymkowiak C., Zacharias M., Wagner R. The importance of individual nucleotides for the structure and function of rRNA molecules in E. coli. A mutagenesis study. FEBS Lett. 1986 Aug 11;204(1):89–95. doi: 10.1016/0014-5793(86)81392-8. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Osswald M., Brimacombe R. Identification of intermolecular RNA cross-links at the subunit interface of the Escherichia coli ribosome. Biochemistry. 1992 Mar 24;31(11):3004–3011. doi: 10.1021/bi00126a023. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Binding of tRNA to the ribosomal A and P sites protects two distinct sets of nucleotides in 16 S rRNA. J Mol Biol. 1990 Jan 5;211(1):135–145. doi: 10.1016/0022-2836(90)90016-F. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989 May 19;57(4):585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989 Nov 9;342(6246):142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- Noller H. F. Ribosomal RNA and translation. Annu Rev Biochem. 1991;60:191–227. doi: 10.1146/annurev.bi.60.070191.001203. [DOI] [PubMed] [Google Scholar]
- Nygård O., Nika H. Identification by RNA-protein cross-linking of ribosomal proteins located at the interface between the small and the large subunits of mammalian ribosomes. EMBO J. 1982;1(3):357–362. doi: 10.1002/j.1460-2075.1982.tb01174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygård O., Nilsson L. Characterization of the ribosomal properties required for formation of a GTPase active complex with the eukaryotic elongation factor 2. Eur J Biochem. 1989 Feb 15;179(3):603–608. doi: 10.1111/j.1432-1033.1989.tb14589.x. [DOI] [PubMed] [Google Scholar]
- Nygård O., Westermann P. Purification of protein synthesis initiation factor eIF-3 from rat liver microsomes by affinity chromatography on rRNA-cellulose. Biochim Biophys Acta. 1982 Jun 30;697(3):263–269. doi: 10.1016/0167-4781(82)90088-4. [DOI] [PubMed] [Google Scholar]
- Rottmann N., Kleuvers B., Atmadja J., Wagner R. Mutants with base changes at the 3'-end of the 16S RNA from Escherichia coli. Construction, expression and functional analysis. Eur J Biochem. 1988 Oct 15;177(1):81–90. doi: 10.1111/j.1432-1033.1988.tb14347.x. [DOI] [PubMed] [Google Scholar]
- Santer M., Bennett-Guerrero E., Byahatti S., Czarnecki S., O'Connell D., Meyer M., Khoury J., Cheng X., Schwartz I., McLaughlin J. Base changes at position 792 of Escherichia coli 16S rRNA affect assembly of 70S ribosomes. Proc Natl Acad Sci U S A. 1990 May;87(10):3700–3704. doi: 10.1073/pnas.87.10.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santer M., Shane S. Area of 16S ribonucleic acid at or near the interface between 30S and 50S ribosomes of Escherichia coli. J Bacteriol. 1977 May;130(2):900–910. doi: 10.1128/jb.130.2.900-910.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge K. D., Maxwell E. S. Intermolecular hybridization of 5S rRNA with 18S rRNA: identification of a 5'-terminally-located nucleotide sequence in mouse 5S rRNA which base-pairs with two specific complementary sequences in 18S rRNA. Biochim Biophys Acta. 1991 Jan 17;1088(1):57–70. doi: 10.1016/0167-4781(91)90153-d. [DOI] [PubMed] [Google Scholar]
- Stebbins-Boaz B., Gerbi S. A. Structural analysis of the peptidyl transferase region in ribosomal RNA of the eukaryote Xenopus laevis. J Mol Biol. 1991 Jan 5;217(1):93–112. doi: 10.1016/0022-2836(91)90614-c. [DOI] [PubMed] [Google Scholar]
- Steiner G., Kuechler E., Barta A. Photo-affinity labelling at the peptidyl transferase centre reveals two different positions for the A- and P-sites in domain V of 23S rRNA. EMBO J. 1988 Dec 1;7(12):3949–3955. doi: 10.1002/j.1460-2075.1988.tb03281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapprich W. E., Goss D. J., Dahlberg A. E. Mutation at position 791 in Escherichia coli 16S ribosomal RNA affects processes involved in the initiation of protein synthesis. Proc Natl Acad Sci U S A. 1989 Jul;86(13):4927–4931. doi: 10.1073/pnas.86.13.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapprich W. E., Hill W. E. Involvement of bases 787-795 of Escherichia coli 16S ribosomal RNA in ribosomal subunit association. Proc Natl Acad Sci U S A. 1986 Feb;83(3):556–560. doi: 10.1073/pnas.83.3.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilenko S. K., Carbon P., Ebel J. P., Ehresmann C. Topography of 16 S RNA in 30 S subunits and 70 S ribosomes accessibility to cobra venom ribonuclease. J Mol Biol. 1981 Nov 15;152(4):699–721. doi: 10.1016/0022-2836(81)90123-6. [DOI] [PubMed] [Google Scholar]
- Vester B., Garrett R. A. The importance of highly conserved nucleotides in the binding region of chloramphenicol at the peptidyl transfer centre of Escherichia coli 23S ribosomal RNA. EMBO J. 1988 Nov;7(11):3577–3587. doi: 10.1002/j.1460-2075.1988.tb03235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann P., Nygård O., Bielka H. Cross-linking of Met-tRNAf to eIF-2 beta and to the ribosomal proteins S3a and S6 within the eukaryotic inhibition complex, eIF-2 .GMPPCP.Met-tRNAf.small ribosomal subunit. Nucleic Acids Res. 1981 May 25;9(10):2387–2396. doi: 10.1093/nar/9.10.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wower J., Hixson S. S., Zimmermann R. A. Labeling the peptidyltransferase center of the Escherichia coli ribosome with photoreactive tRNA(Phe) derivatives containing azidoadenosine at the 3' end of the acceptor arm: a model of the tRNA-ribosome complex. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5232–5236. doi: 10.1073/pnas.86.14.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C., Jemiolo D. K., Jacob W. F., Wagner R., Dahlberg A. E. Characterization of a collection of deletion mutants at the 3'-end of 16S ribosomal RNA of Escherichia coli. Mol Gen Genet. 1986 May;203(2):256–264. doi: 10.1007/BF00333963. [DOI] [PubMed] [Google Scholar]