Abstract
Background
The scientific community has proven the value of circulating tumor cells (CTCs) as a prognostic factor in the development of cancer and progress to metastases [1, 2, 3, 4]. Simultaneously, a new type of cancer stem cell-like (CSC-like) cells has also been established as a progenitor of metastases and relapses in cancer patients [5, 6]. The present research attempts to support the hypothesis that CTCs have all the cellular hallmarks of CSC-like cells which play a crucial role in cancer spreading.
Materials and Methods
Two methods have been chosen: a cellular-based and a molecular-based method. The first method is based on the fact that CSCs form microspheres in culture. In the second method, microspheres develop in the presence of specific markers that define the CSC phenotype [6].
Results
In cellular-based assays, it has been shown that microspheres form in semi-suspension in a culture flask. In the second panel of the test, Nanog was chosen as a marker and the tested sample was positive when grown under specific conditions.
Conclusion
Our analysis has demonstrated that in this particular case, CSCs-like cells are included in the vast majority of CTCs.
Key Words: Breast cancer, Cancer stem cell-like cells, Circulating tumor cells
Introduction
It is well known that circulating tumor cells (CTCs) are a distinct population of cancer cells that have detached from the primary tumor and flow into the blood circulation, creating a secondary tumor. Their role in the metastatic pathway has proven to be essential [4, 9, 10, 11, 12].
Initiation of metastasis involves CTCs creation which includes cell-to-cell adhesion mechanisms and cell mobility. Several growth factors act in order to stimulate the epithelial-to-mesenchymal transition (EMT). The primary epithelial cancer cells interact with the basement membrane via multiple biochemical changes and acquire a mesenchymal cell phenotype. Through the blood vessels, they migrate into distant healthy tissues. Their survival depends on the interaction between them and the host. As this biological process progresses, during the cancer cells’ extravasation many angiogenic factors and signaling agents contribute to the colonization. Accordingly, through mesenchymal-to-epithelial transition (MET) – the opposite of EMT – a micrometastasis occurs followed by a macrometastasis [10, 11, 13, 14, 15].
Thus, CTCs can be considered as the progenitors of relapses. This indicates that they may have all the hallmarks of cancer stem cell-like (CSC-like) cells, as CSCs have the ability to give rise to a new tumor [5, 16]. This assumption needs to be confirmed in a case report or extended analysis.
Materials and Methods
To provide more accurate evidence for the existence of CSC-like cells within CTCs in the primary tumor, it was necessary to use more than one approach: the first method was cellular-based and the second one was molecular-based.
CTC Isolation
To isolate CTCs, whole blood cells were centrifuged with polysucrose solution [Histopaque 1077 (10771; Sigma) layered on Histopaque 1119 (11191; Sigma)]. Mononuclear cells, lymphocytes, platelets, and granulocytes were collected after centrifugation and washed twice with PBS. Cells were then incubated at 4°C for 30 min with EpCAM magnetic beads (39-EPC-50; Gentaur). EpCAM is an epithelial cell adhesion molecule-associated antigen that is expressed in the vast majority of carcinomas. As breast cancer is of epithelial origin, EpCAM beads were used for breast cancer CTC isolation. Following incubation, the sample was placed in a magnetic field, selected and washed with PBS. Isolated breast cancer cells were then divided and cultured in a 25-cm2 flask (5520100; Orange Scientific) with RPMI-1640 medium (R6504; Sigma). Since the cancer cells have an infinite division potential, the cells that remained in the flask after 1 week of culture were the cells of preference [4, 17].
Blood Sample Collection
Cancer cells were obtained from a 55-year-old female patient with breast cancer stage II (lobular invasive ductal carcinoma, grade 2). Thereafter, 20 ml of peripheral blood was collected from the patient and placed in a tube with EDTA as anticoagulant (Vacutainer K3E; BD) and rotated for 30 min to prevent coagulation.
Flow Cytometry
To determine the presence of breast cancer cells in our sample, flow cytometry was performed as method of choice. Briefly, 10 μl of CD45-PC5 (MCA1719C; AbD Serotec) and 10 μl of CD31-PE (MCA1738PE; AbD Serotec) antibodies were added to 100 μl of whole blood. In a second tube, 10 μl of CD45-PC5 and 10 μl of c-Met (FAB3582P; RnD Systems) antibodies were added to 100 μl of whole blood. Both of the above samples were prepared twice, followed by incubation for 25 min at room temperature. After incubation, 100 μl of medium A (Leucoperm, BUF09; AbD Serotec), containing formaldehyde (0.1% v/v), was added to each of the 4 sample tubes. The samples were then washed with 3 ml of PBS (P3813; Sigma) containing 5% w/v FBS (10106-169; Invitrogen) and 0.1% v/v NaN3 (S8032; Sigma) and centrifuged for 5 min at 1,600 rpm. Supernatant was removed and 100 μl of medium B (Leucoperm) was added to each tube. To the first couple of tubes (CD31 and c-Met), containing intracellular pan-cytokeratin (ABIN144814; GeneTex), antibody was added. To the second couple of tubes, CD227 (MCA1742F; AbD Serotec) was added. Samples were incubated for 25 min at room temperature. After a second wash, the samples were analyzed by Accuri C6 cytometer (Accuri C6; Accuri).
Cell Culture
As positive control, we used Human Breast Cancer Stem Cell Culture (36102-29-T25; Celprogen) which has been provided by Celprogen. Both CTCs and Human Breast Cancer Stem Cell Culture were incubated in 25-cm2 flasks (5520100; Orange Scientific) at 37°C in 5% CO2 atmosphere. As culture medium for the Human Breast Cancer Stem cell culture, the STEMPRO hESC SFM kit was used (A10007-01; Invitrogen) which stimulates the growth of human mammary epithelial cells and provides the highest quality for microsphere culture. The STEMPRO product (complete medium) is composed of a DMEM/F-12 + GlutaMAX (1X) cultivation medium and by the following additional ingredients: 10 μg/ml of FGF-basic factor (F029; Sigma), STEMPRO hESC SFM Growth Supplement (50X), 25% of BSA, and 55 mmol/l of 2-mercaptoethanol (M3148; Sigma). CTCs were cultivated both in RPMI-1640 (R0883; Sigma), with the appropriate amount of heat-inactivated fetal bovine serum (10106-169, FBS; Invitrogen), and 2 mmol/l glutamine (G5792; Sigma), and in the STEMPRO kit in order to make a comparison under different conditions from the starting population. All the experiments were done during the exponential phase and after a culture confluence of 80–90%.
Cellular-Based Evaluation Method
This method is based on the ability of CSCs to form microspheres. To analyze the microsphere-positive population, after 48 h of cultivation, we compared the capacity of RPMI-1640-cultivated cells and STEMPRO-cultivated cells based on their ability to form spherical colonies in semi-suspension using the Human Breast Cancer Stem Cell Culture as positive control. Light microscope was used for assessment and evaluation.
Molecular-Based Evaluation Method
This scientific approach is based on the fact that the CSC phenotype is defined by multiple molecular markers such as Nanog, oct4, nestin, E-cadherin, AML1, ALDH, etc. For gene expression analysis, endpoint and quantitative RT-PCR protocols have been used (Maxima Sybr Green K0221; Fermentas), and flow cytometry analysis has been used for protein expression (Accuri C6; Accuri) [7, 8, 18, 19, 20].
Gene Expression Analysis
The primers used in the endpoint RT-PCR protocol were designed by the Genamics expression program (Genamics Expression, version 1.100© 2000). The endpoint RT-PCR protocol used to test the gene expression of interest includes a 10-min template initial denaturing step at 95°C, a 45-second main denaturing step at 94°C, a 45-second primer annealing step at 59°C, and a 2-min primer extension step at 72°C. Usually, a 10-min final extension step is performed and the reaction runs for 45 cycles to obtain gene patterns (Nanog gene: forward: 5′-TGAGATGCCTCACACGGAGACTG-3′, reverse: 5′-GGGTTGTTTGCCTTTGGGACTG-3′).
Protein Expression Analysis
The quantitative evaluation of Nanog protein-expressing cells has been performed using a murine antihuman antibody conjugated with fluorescein isothiocyanate, a major fluorescein derivative (ABIN278021; Antibodies-online GmbH), and by the FCS express software (FCS Express V3, version 3.00.0504, Professional Standalone, 2001–2007; DeNovo Software).
Results
The outcome of the cellular-based assays has pointed out the formation of microspheres in semi-suspension inside a culture flask [9]. As mentioned before, CSC-like cells have the ability to form spherical colonies under enabling conditions. To analyze this type of formation in our cultures, we compared RPMI-1640-cultivated cells and STEMPRO-cultivated cells with the formation of human breast CSCs (control cell line) (fig. 1).
Fig. 1.
Human breast CSCs.
In the second panel of the test, the representative marker (Nanog gene) of choice [5, 21, 22] was found to be positive in the human breast CSC sample, as expected. STEMPRO-cultivated cells were also found to be positive, in contrast to RPMI-cultivated cells, where an expression of the Nanog gene was missing (fig. 2).
Fig. 2.
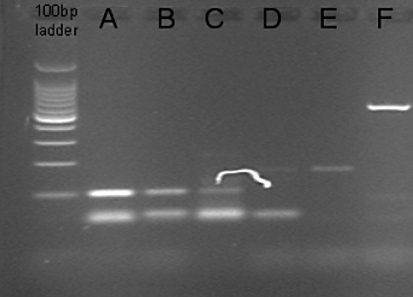
The CSC-like cell phenotype is associated with the expression of the Nanog gene [7]. Here, its expression was determined by RT-endpoint PCR. The predicted size of the PCR product was 104 bp. The Nanog gene was highly expressed in both CSC-like cells (A, B) and in one of the two samples of STEMPRO-cultivated cells (C, D). In samples E and F (RPMI-1640-cultivated cells) the Nanog gene was not expressed.
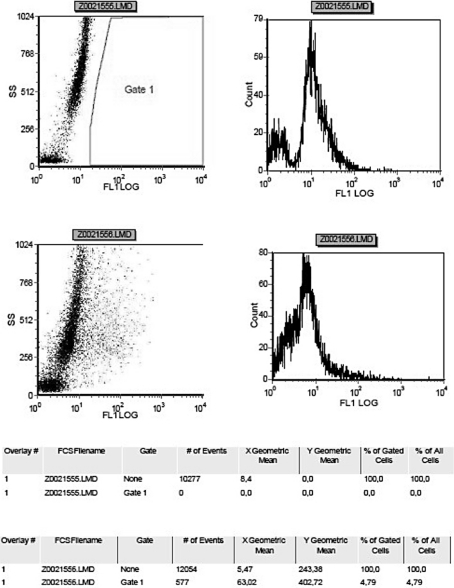
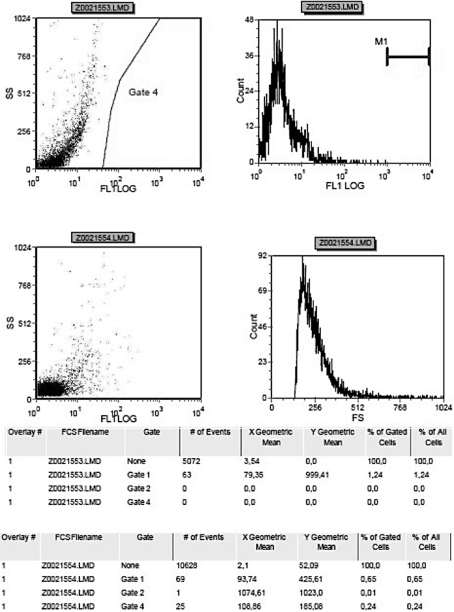
In order to check the protein expression of Nanog, flow cytometry analysis has been used. The cells shown in fig. 3 and in fig. 4 represent the percentage of RPMI-1640-cultivated cells, which expressed 1.24% of Nanog protein, and the percentage of STEMPRO-cultivated cells, which expressed 4.79% of Nanog protein of the total amount. Consequently, Nanog protein has been observed to be highly expressed in STEMPRO-cultivated cells.
Fig. 3.
Expression of Nanog protein in RPMI-cultivated cells.
Fig. 4.
Expression of Nanog protein in STEMPRO-cultivated cells.
Using all the evaluating methods described above, we tested the expression of Nanog from transcription to translation in order to draw the final conclusions.
Discussion
The most striking or crucial characteristic of cancer disease is its ability to spread and produce new metastatic sites. In alignment with the scientific indications cited above, CTCs, which are part of the primary tumor, play an important role for the development of the disease, and later, of metastasis [15, 16, 23, 24, 25].
Shortly, a tumor is formed when cancer cells grow as colonies. Many cellular and biochemical changes in the phenotype are responsible for the loss of epithelial hallmarks by the tumor cells.
Loss of epithelial polarity (interaction between tumor cells and stromal cells) drives CTCs to alter themselves and form spheres. This shape facilitates their immune escape, and finally reacts with the target. Ras/MAPK pathway is activated and EMT takes place. Expression levels of epithelial markers like E-cadherin and cytokeratin decrease and, as a result, the linkage between CTCs and actin cytoskeleton is weakened. In contrast, many mesenchymal markers, such as vimentin and fibronectin, are expressed in high levels and so cancer cells (CTCs) acquire a fibroblast-like shape and their mobility and invasiveness increases. As a result, circulation of tumor cells via proliferation through the blood circulation (intravasation) is taking place. CFS-1 and EGF are the major stimulators of this process, as well as a great range of metalloproteinases (MMPs), especially MMP-9 and G proteins (e.g., Ras, Rho). MMP-9 expression correlates with the tumorigenesis/metastasis cascade. It is well known that metalloproteinase plays a crucial role in normal tissues, as it is initially synthesized as soluble inactive pro-enzyme and acts like a caspase. Other caspases act then in order to stimulate the metastatic attitude of MMPs. On the contrary, G proteins like Ras and Rho are members of a signal transduction pathway, and their activation causes cell growth, differentiation and survival. Thus, using blood vessels, CTCs flow and reach the targeted place, where they are released via extravasation and produce new tumors (micrometastases). Since the environment lacks the previously owned signals and growth factors, CTCs need time in order to proliferate and find all necessary growth supplements to finally produce the detectable mass (macrometastasis) [10, 11, 14, 17].
Many efforts have been done by others in order to prove that cells which have detached from the primary tumor are CTCs [17, 24, 25, 26]. However, these methods present advantages and disadvantages. The first one is a PCR-based method which does not sustain cells as entity for analysis, meaning that there are limitations concerning further molecular analysis of CTCs. Furthermore, even if we have the ability to enumerate CTCs [4], we have identified only few molecular characteristics. The second widely used method is a magnetic bead-based method which uses a small range of markers in order to enrich the CTC population; hence, it is based on their positive selection [17].
In this research attempt, more than one method has been used to render the results more accurate and complete. CTCs isolation from the patient's blood sample was made using EpCAM magnetic beads, which is a pan-epithelial carcinoma-associated antigen that is expressed in almost all carcinomas, as mentioned before. Following isolation, CD45-negative selection was performed in order to isolate cells of epithelial origin from the remaining blood cells. Moreover, CD31-positive cells, endothelial cells or pericytes were excluded because they were considered normal.
The MET gene encodes protein that triggers tumor proliferation, neovascularization and metastases to distinct organs. Therefore, c-Met-positive cells are cells of interest. Expression of cytokeratins is organ and tissue specific. Moreover, the cytokeratin profile tends to remain intact when an epithelial cell undergoes a malignant transformation. Therefore, a cytokeratin profile study is helpful for tumor characterization [4, 24, 25].
The CD227 (Mucin-1 or Ca 15.3) antigen is highly expressed by virtually all mucosal epithelial tissues. It is aberrantly expressed in most human breast cancers, and in this study it was used as a breast cancer marker. CTCs were quantified in the sample [3, 27, 28].
The hypothesis that a population of CTCs has many CSC-like hallmarks is reinforced by the expression of related molecular markers. NANOG is a transcription factor which plays an important role in the self-renewal of undifferentiated embryonic stem cells. It is a nuclear protein and its function is correlated with other factors such as OCT4 [POU5F1 (POU class 5 homeobox 1)]. It has been shown that these genes play a key role in forming the identity of embryonic stem cells. Oct4 is initially active in the oocyte, and it remains active even later in the preimplantation period. It forms a dimmer with the Sox2 gene and so binds to the DNA. Studies claim that this dimmer activates the Nanog protein, and so this molecular cascade is responsible for the capability of tumor cells to differentiate into any of the 3 germ layers (pluripotency). Another protein that expresses early during cell development is Nestin. It is a marker that is not expressed in adults because it is replaced by tissue-specific intermediate filament proteins. Due to the above-mentioned reasons, these markers were chosen in order to prove the initial hypothesis. Since CSCs must hold the hallmark of self-renewal and asymmetric cell division, which is driven by the above factors, the present attempt shows that this hallmark also exists in CTCs [7, 19, 29, 30].
Conclusion
Our analysis has well pointed out that in this particular case, CSC-like cells are included in the vast majority of CTCs. It is also hypothesized that this model could be applied in general for all cancer metastasis processes. Furthermore, it is necessary to perform further extended studies in different types of malignancies and in a larger scale of sample types in order to prove the relevance between CTCs and CSCs.
Footnotes
This is an Open Access article licensed under the terms of the Creative Commons Attribution-NonCommercial-NoDerivs 3.0 License (www.karger.com/OA-license), applicable to the online version of the article only. Distribution for non-commercial purposes only.
References
- 1.Cristofanilli M, Hayes DF, Budd GT, Ellis MJ, Stopeck A, Reuben JM, Doyle GV, Matera J, Allard WJ, Miller MC, Fritsche HA, Hortobagyi GN, Terstappen LW. Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J Clin Oncol. 2005;23:1420–1430. doi: 10.1200/JCO.2005.08.140. [DOI] [PubMed] [Google Scholar]
- 2.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller M C, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 3.Hayes DF, Smerage J. Is there a role for circulating tumor cells in the management of breast cancer? Clin Cancer Res. 2008;14:3646–3650. doi: 10.1158/1078-0432.CCR-07-4481. [DOI] [PubMed] [Google Scholar]
- 4.Tibbe AG, Miller MC, Terstappen LW. Statistical considerations for enumeration of circulating tumor cells. Cytometry A. 2007;71:154–162. doi: 10.1002/cyto.a.20369. [DOI] [PubMed] [Google Scholar]
- 5.Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, Hur MH, Diebel ME, Monville F, Dutcher J, Brown M, Viens P, Xerri L, Bertucci F, Stassi G, Dontu G, Birnbaum D, Wicha MS. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, Visvader J, Weissman IL, Wahl GM. Cancer stem cells – perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 7.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 8.Zieglschmid V, Hollmann C, Mannel J, Albert W, Jaeschke-Melli S, Eckstein B, Hillemann T, Greten TF, Gross E, Bocher O. Tumor-associated gene expression in disseminated tumor cells correlates with disease progression and tumor stage in colorectal cancer. Anticancer Res. 2007;27:1823–1832. [PubMed] [Google Scholar]
- 9.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanchez-Garcia I, Vicente-Duenas C, Cobaleda C. The theoretical basis of cancer-stem-cell-based therapeutics of cancer: can it be put into practice? Bioessays. 2007;29:1269–1280. doi: 10.1002/bies.20679. [DOI] [PubMed] [Google Scholar]
- 13.Figueira RC, Gomes LR, Neto JS, Silva FC, Silva ID, Sogayar MC. Correlation between MMPs and their inhibitors in breast cancer tumor tissue specimens and in cell lines with different metastatic potential. BMC Cancer. 2009;9:20. doi: 10.1186/1471-2407-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tien YW, Jeng YM, Hu RH, Chang KJ, Hsu SM, Lee PH. Intravasation-related metastatic factors in colorectal cancer. Tumour Biol. 2004;25:48–55. doi: 10.1159/000077723. [DOI] [PubMed] [Google Scholar]
- 15.Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, Kasimir-Bauer S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009;11:R46. doi: 10.1186/bcr2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill RP. Identifying cancer stem cells in solid tumors: case not proven. Cancer Res. 2006;66:1891–1895. doi: 10.1158/0008-5472.CAN-05-3450. discussion 1890. [DOI] [PubMed] [Google Scholar]
- 17.Riethdorf S, Fritsche H, Muller V, Rau T, Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Janicke F, Jackson S, Gornet T, Cristofanilli M, Pantel K. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin Cancer Res. 2007;13:920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 18.Fehm T, Hoffmann O, Aktas B, Becker S, Solomayer EF, Wallwiener D, Kimmig R, Kasimir-Bauer S. Detection and characterization of circulating tumor cells in blood of primary breast cancer patients by RT-PCR and comparison to status of bone marrow disseminated cells. Breast Cancer Res. 2009;11:R59. doi: 10.1186/bcr2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiou SH, Yu CC, Huang CY, Lin SC, Liu CJ, Tsai TH, Chou SH, Chien CS, Ku HH, Lo J F. Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin Cancer Res. 2008;14:4085–4095. doi: 10.1158/1078-0432.CCR-07-4404. [DOI] [PubMed] [Google Scholar]
- 20.Hattori N, Imao Y, Nishino K, Ohgane J, Yagi S, Tanaka S, Shiota K. Epigenetic regulation of Nanog gene in embryonic stem and trophoblast stem cells. Genes Cells. 2007;12:387–396. doi: 10.1111/j.1365-2443.2007.01058.x. [DOI] [PubMed] [Google Scholar]
- 21.Li C, Lee CJ, Simeone DM. Identification of human pancreatic cancer stem cells. Methods Mol Biol. 2009;568:161–173. doi: 10.1007/978-1-59745-280-9_10. [DOI] [PubMed] [Google Scholar]
- 22.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 23.Tewes M, Aktas B, Welt A, Mueller S, Hauch S, Kimmig R, Kasimir-Bauer S. Molecular profiling and predictive value of circulating tumor cells in patients with metastatic breast cancer: an option for monitoring response to breast cancer related therapies. Breast Cancer Res Treat. 2009;115:581–590. doi: 10.1007/s10549-008-0143-x. [DOI] [PubMed] [Google Scholar]
- 24.Marrinucci D, Bethel K, Lazar D, Fisher J, Huynh E, Clark P, Bruce R, Nieva J, Kuhn P. Cytomorphology of circulating colorectal tumor cells: a small case series. J Oncol. 2010;2010:861341. doi: 10.1155/2010/861341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng S, Tripathy D, Frenkel EP, Shete S, Naftalis EZ, Huth JF, Beitsch PD, Leitch M, Hoover S, Euhus D, Haley B, Morrison L, Fleming TP, Herlyn D, Terstappen LW, Fehm T, Tucker TF, Lane N, Wang J, Uhr JW. Circulating tumor cells in patients with breast cancer dormancy. Clin Cancer Res. 2004;10:8152–8162. doi: 10.1158/1078-0432.CCR-04-1110. [DOI] [PubMed] [Google Scholar]
- 26.Lankiewicz S, Zimmermann S, Hollmann C, Hillemann T, Greten TF. Circulating tumour cells as a predictive factor for response to systemic chemotherapy in patients with advanced colorectal cancer. Mol Oncol. 2008;2:349–355. doi: 10.1016/j.molonc.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dick JE. Breast cancer stem cells revealed. Proc Natl Acad Sci USA. 2003;100:3547–3549. doi: 10.1073/pnas.0830967100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hauch S, Zimmermann S, Lankiewicz S, Zieglschmid V, Bocher O, Albert WH. The clinical significance of circulating tumour cells in breast cancer and colorectal cancer patients. Anticancer Res. 2007;27:1337–1341. [PubMed] [Google Scholar]
- 29.Jung M, Peterson H, Chavez L, Kahlem P, Lehrach H, Vilo J, Adjaye J. A data integration approach to mapping OCT4 gene regulatory networks operative in embryonic stem cells and embryonal carcinoma cells. PLoS One. 2010;5:e10709. doi: 10.1371/journal.pone.0010709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P. Transcriptional regulation of Nanog by OCT4 and SOX2. J Biol Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]