Abstract
Background
While considerable genomic and transcriptomic data are available for Schistosoma mansoni, many of its genes lack significant annotation. A transcriptomic study of individual tissues and organs of schistosomes could play an important role in functional annotation of the unknown genes, particularly by providing rapid localisation data and thus giving insight into the potential roles of these molecules in parasite development, reproduction and homeostasis, and in the complex host-parasite interaction.
Methodology/Principal Findings
Quantification of gene expression in tissues of S. mansoni was achieved by a combination of laser microdissection microscopy (LMM) and oligonucleotide microarray analysis. We compared the gene expression profile of the adult female gastrodermis and male and female reproductive tissues with whole worm controls. The results revealed a total of 393 genes (contigs) that were up-regulated two-fold or more in the gastrodermis, 4,450 in the ovary, 384 in the vitelline tissues of female parasites, and 2,171 in the testes. We have also supplemented these data with the identification of highly expressed genes in different regions of manually dissected male and female S. mansoni. Though relatively crude, this dissection strategy provides low resolution localisation data for critical regions of the adult parasites that are not amenable to LMM isolation.
Conclusions
This is the first detailed transcriptomic study of the reproductive tissues and gastrodermis of S. mansoni. The results obtained will help direct future research on the functional aspects of these tissues, expediting the characterisation of currently unannotated gene products of S. mansoni and the discovery of new drug and vaccine targets.
Author Summary
There is currently only one drug available for treatment of schistosomiasis mansoni and no vaccine. The searches for possible new drug and vaccine candidates remain two major areas of current research in schistosomiasis. There are considerable amounts of data available on the genomics, transcriptomics and proteomics of Schistosoma mansoni from which useful candidates for future drug and vaccine development can be identified. Arranging these data into a biologically relevant context through the characterisation of gene expression profiles of the different tissues of this complex metazoan parasite, is an essential step in identifying molecules with potential therapeutic value. We have used laser microdissection microscopy and microarray analysis to show that many tissue-specific genes are up-regulated in the digestive and reproductive tissues of S. mansoni. This new knowledge provides an avenue to investigate the molecular components associated with fundamental aspects of schistosome biology.
Introduction
The last decade has seen an exponential growth in the availability of genomic and proteomic data for Schistosoma mansoni and S. japonicum through genomic sequencing projects [1], [2] and numerous transcriptomic and proteomic studies [3], [4], [5], [6], [7], [8], [9]. However, the functions of many schistosome gene products remain unsolved. As argued by Jones et al [10], a major step towards elucidating function of these uncharacterised gene products lies in defining their sites of expression. Large-scale investigation of the expression repertoire of specific tissues is difficult to achieve because schistosome tissues generally cannot be isolated, due to their small tubiform bodies and the absence of a body cavity [10], [11], [12]. Laser micro-dissection microscopy (LMM), a tool that enables selection and sampling of small cell populations from histological sections, has been used to overcome the difficulty in separating the tissues for cell-enriched studies of these parasites [10], [13]. The approach has been used successfully by our team to study tissue specific transcriptomics of gastrodermal, ovary and vitelline tissues of adult females of S. japonicum [11].
In the current study, we describe the application of LMM, in conjunction with microarray gene expression analysis, to map the transcriptome of biologically important tissues of S. mansoni, augmenting our study on S. japonicum tissue specific transcriptomics [11]. The gastrodermis (absorptive gut lining) of females and the reproductive tissues of male (testes) and female (ovary, vitelline glands) worms were targeted with this gene atlasing strategy to determine the expression repertoire of these tissues.
Materials and Methods
Research involving animals in this study was approved by the Animal Ethics Committee of the Queensland Institute of Medical Research. The study was conducted according to guidelines of the National Health and Medical Research Council of Australia, as published in the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes, 7th edition, 2004 (www.nhmrc.gov.au).
Sample preparation
The Puerto Rican strain of Schistosoma mansoni is maintained in ARC Swiss mice and Biomphalaria glabrata snails at QIMR from stocks originating from the National Institute of Allergy and Infectious Diseases Schistosomiasis Resource Centre, Biomedical Research institute (Rockville, Maryland, USA).
Worm pairs were perfused from mice six weeks post-infection. Males and females were separated and single-sex samples, consisting of 30–40 worms, were transferred immediately into Tissue-Tek Optimal Cutting Temperature compound (OCT) (ProSciTech, Thuringowa, Australia) and frozen in dry ice. Four OCT blocks, each containing 30–40 worms, were used for the microdissection of each sex of S. mansoni. Worm sections were prepared for microdissection as described [11]. Cryosections were cut onto PALM 0.17 mm polyethylene naphthalene membrane covered slides (Carl Zeiss Microimaging GmbH, Bernried, Germany).
Laser microdissection of schistosome tissues
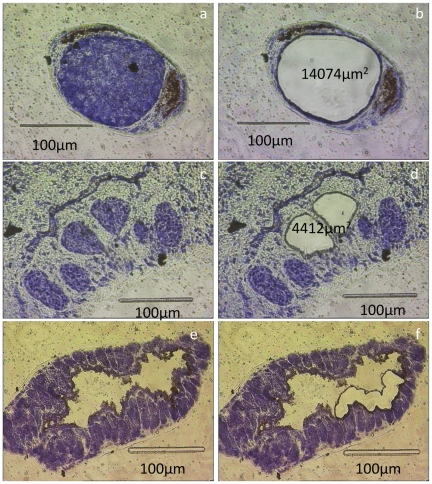
Thawed cryosections of worms were stained with 1% (w/v) toluidine blue (CHROMA, Stuttgart, Germany), washed in distilled water and air dried in a sterile biohazard hood immediately prior to microdissection [11]. A PALM MicroBeam Laser Catapult Microscope (Carl Zeiss Microimaging) was used to microdissect the gut, ovary, vitelline glands of females and testes from males from the stained sections using a ×40 microdissection objective lens. The RoboLPC mode in PALMRobo 2.2.2 software (Carl Zeiss Microimaging) was used to cut and catapult the selected tissues from sections of adult schistosomes. Approximately 4×106 µm2 of each tissue was catapulted into 200 µl opaque adhesive Teflon-coated caps (Carl Zeiss Microimaging) located in the laser path, a few millimetres above the specimen stage (Figure 1).
Figure 1. Laser Microdissection of adult S. mansoni.
Sections of male and female S. mansoni, before and after microdissection, stained with 1% (w/v) toluidine blue. The left column shows the ovary (a), testes (c) and gastrodermis (e) before microdissection, while the right column shows the same sections of the ovary (b), testes (d) and gastrodermis (f) after microdissection.
The caps containing the dissected material were stored at −80°C until used. Sex-matched controls were obtained by randomly microdissecting sections of female or male worms to obtain microdissected samples, representing the entire expression repertoire of the adult worms. For female controls, an area approximating those of the three target tissues was dissected randomly from sections of females. For males, it was necessary to dissect an area of 20×106 µm2 in order to obtain sufficient sample material as control tissue.
Macro-dissection
As a complement to the microdissection studies, a set of macro-dissected samples was also obtained from S. mansoni male and female worms using a dissecting microscope and sterile razor blade. Females were cut transversely into three parts: namely, the head region, extending from the anterior extremity of the schistosome to the ventral sucker; the middle region, extending from the posterior extremity of the ventral sucker to the posterior margin of the ovary; and the hind region, extending from the posterior extremity of the ovary to the posterior extremity of the parasite. The tissue types included in each of these regions are shown (Table S1). Because of its extremely small size, the head region of 100 females was dissected for RNA extraction, whereas the middle and hind regions were obtained from only 15 females. Males were dissected into two regions; namely, the head region, which was that extending from the anterior extremity of the schistosome to the ventral sucker, and the hind region, representing the remainder of the worm. Head region samples were dissected from 100 worms whereas the hind region was dissected from only 3 parasites. A total of 5 female and 3 male sex-matched controls were used. In each case, the worm number was determined by the maximum amount of tissue amenable for efficient RNA extraction using an RNAqueous-Micro kit (Ambion, Austin, TX, USA) (see below).
Total RNA isolation and hybridisation
Total RNA was isolated from LMM samples and manually dissected samples using RNAqueous-Micro kits using the manufacturer's protocol for LMM samples and whole tissue samples, respectively. The quality of RNA was assessed using the Bioanalyser RNA Pico Lab Chip (Agilent Technologies, Santa Clara, CA, USA) as described [11].
A 44k oligonucleotide microarray platform (Agilent Technologies) designed specifically for S. mansoni was used for microarray analyses [14]. Access to the design was kindly provided by Dr Sergio Verjovski-Almeida (Universidade de São Paulo) [14]. The microarray platform has 39,342 probes representing 19,907 putative genes [14]. The array was designed using publicly available EST data for S. mansoni and S. japonicum to include orthologous S. japonicum genes not cloned and sequenced by the time of microarray construction [14]. The coverage and similarity of microarray used in this current study was compared to the microarray used in our previous reported study of S. japonicum [11], by performing a local BLASTn using BioEdit with default settings [15]. The BLASTn was performed with the ESTs used in the design of the original microarray used in the S. japonicum transcriptomic study, queried against the nucleotide database consisting of ESTs from the microarray used in the current study (Table S2).
Gene expression analysis was performed for all the samples and biological replicates. Two and three biological replicates were used for testes and vitelline tissue samples, respectively, and single samples were used for the rest of the tissues due to limited availability of material for analysis. Two technical replicates ( = two hybridisations) were performed for each sample.
Microarray hybridisation used 200 ng of RNA from each sample according to the manufacturer's instructions (One-colour Microarray-Based Gene Expression Analysis Protocol; Version 5.7, March 2008 Agilent) [7]. GENESPRING software version 7.3.1 (Agilent Technologies/Silicon Genetics) was used for the in silico analysis of microarray data [16]. Microarray signal intensities from laser microdissected female tissues were normalised against those of laser microdissected whole female controls, while signal intensities from testis samples were normalised against those of laser microdissected whole male controls. Macro-dissected male and female samples were normalised against whole male and female control samples, respectively.
Hybridized slides were scanned using an Agilent Microarray Scanner (B version) as Tiff files and processed with the Feature Extraction 9.5.3.1 Image Analysis program (Agilent) to produce standardised data for statistical analysis. All microarray slides were assessed for background evenness by viewing the Tiff image by Feature Extraction. Feature extracted data were analysed using GENESPRING (version 7.3.1; Agilent Technologies/Silicon Genetics, Redwood City, CA). Microarray data were normalised using a normalisation scenario for “Agilent FE one-color” which including “Data Transformation: Set measurements less than 5.0 to 5.0”, “Per Chip: Normalize to 50th percentile” and “Per Gene: Normalize to median”. Data sets were further analysed using published procedures based on one-colour experiments [17]. The gProcessedSignal values were determined in GENESPRING using Agilent's Feature Extraction software including aspects of signal/noise ratio, spot morphology and homogeneity. Thus, gProcessedSignal represents signal after localised background subtraction and includes corrections for surface trends. Features were deemed Absent when the processed signal intensity was less than two fold the value of the processed signal error value. Features were deemed Marginal when the measured intensity was at a saturated value or if there was a substantial amount of variation in the signal intensity within the pixels of a particular feature. Features that were not Absent or Marginal were deemed Present. Data points were included only if Present or Present/Marginal and probes or contigs retained if all data points were Present or Present/Marginal.
Microarray data have been submitted to the Gene Expression Omnibus public database, under accession numbers GPL10875 and GSE23942.
Gene ontology analysis
Updating of annotation was performed on 20 October 2010 on all of the differentially expressed genes (6,295) detected in the four tissue types, using the Blast2GO online annotation pipeline [18]. BlastX was performed using the NCBI blast server URL (http://www.ncbi.nlm.nih.gov/Blast.cgi), ExpectedValue 1.0.E-3, and QBlast mode. The Gene Ontologies were assigned based on the hits received from the Blast using default settings. InterPro annotations were identified and retrieved domain/motif information used to transfer GO terms with already existent GO terms. Genes with GO annotation were submitted for Enzyme code and KEGG map assignment. The sequence description based is on BlastX analysis, sequence length and the number of hits (Table S3).
Real-time PCR validation of microarray data
Gene expression patterns determined by microarray analysis were validated using real-time PCR. A subset of genes was selected for each tissue for validation. The selections were made based on the high level of expression and the importance of function in each tissue. Forward and reverse primers (Sigma-Aldrich, Castle Hill, Australia) were designed for 14 selected contigs (Table S4). All total RNA samples were DNase-treated (Promega, Annandale, Australia) before complementary DNA (cDNA) synthesis [11]. Real-time PCR was performed and analysed using previously described protocols [19]. DNA segregation ATPase [16] (TC15682- Smp176580, Contig809759.1) was used as housekeeping gene for the real time PCR reactions. Real-time PCR and microarray data were compared using GraphPad Prism Version 5.00 (GraphPad Software, San Diego, USA) [16], [20]. Data from microarray and real time PCR analyses were examined to ascertain if they fitted normal distributions, using the D'Agostino and Pearson omnibus and the Shapiro-Wilk normality tests.
Results
Laser microdissected tissues
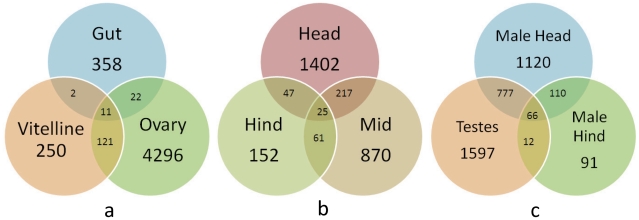
Signal from female microdissected tissues (Figure 1), normalised against the whole female microdissected sample and subsequent filtering for flags revealed 15,109 contigs of which 5,060 were expressed 2-fold or higher than the female control in at least one of the three target tissues. The number of genes identified as being enriched in each tissue is shown (Figure 2a, Table 1, Table 2, Table 3, Table 4, Table S5). The majority of enriched genes were detected in the ovarian sample. A total of 121 genes were enriched for both vitellaria and ovary, which was as expected, considering the ontogenetic and functional similarities of the two tissues. Fewer genes were enriched in both the gastrodermis and either of the two female germinal tissues. Three hundred and fifty eight (358) genes were enriched for the gastrodermis only; of these 112 were significantly up-regulated (P<0.05). A representative list of genes highly expressed in the gastrodermis is shown (Table 1). Among those highly expressed were digestive proteases with diverse mechanistic activity, transmembrane transporters, lysosomal proteins and many unknown proteins. A large proportion (>40%) of the contigs enriched in the gut encoded hypothetical proteins with or without conserved domains, or unknown proteins with no sequence similarity to any known proteins from other organisms.
Figure 2. Genes enriched in specific tissues of adult S. mansoni.
Venn diagram showing the numbers of S. mansoni genes up-regulated at least 2-fold in (a) microdissected female tissues, (b) macrodissected female tissues, (c) macro-dissected male tissue and testes.
Table 1. A selection of differentially up-regulated genes of the female gastrodermis normalised to the whole female control.
| *Systematic name | Gene name | Fold change | Contig | Protein description |
| Q2_P17138 | Smp_127380 | 147.3 | C918435.1 | huntingtin associated protein-1 domain, putative |
| Q2_P37910 | Smp_014610 | 8.83 | C919425.1 | conserved hypothetical protein |
| Q2_P27373 | Smp_166170 | 7.98 | C900002.1 | hypothetical protein |
| Q2_P35922 | Smp_128430 | 6.58 | C915671.1 | cd36 antigen, putative |
| Q2_P17781 | Smp_095980 | 4.32 | C920836.1 | superoxide dismutase precursor (EC 1.15.1.1), putative |
| Q2_P04863 | Smp_151910 | 4.1 | C808892.1 | multidrug resistance protein 1, 2, 3 (p glycoprotein 1, 2, 3) |
| Q2_P17708 | Smp_138570 | 4.04 | C920388.1 | spore germination protein, putative |
| Q2_P35898 | Smp_014570 | 3.91 | C915640.1 | saposin- saposin -containing protein |
| Q2_P37714 | Smp_085010 | 3.41 | C919093.1 | putative cathepsin B, |
| Q2_P07155 | Smp_136730 | 3.4 | C812496.1 | cathepsin D2-like protein |
| Q2_P35878 | Smp_194850 | 3.23 | C915617.1 | Niemann Pick type C2 protein homolog |
| Q2_P40679 | Smp_006760 | 3.09 | JAP06514.C | fasting-inducible integral membrane protein tm6p1-related |
| Q2_P23552 | Smp_152050 | 3.08 | C807737.1 | tetraspanin 18, isoform 1, putative |
| Q2_P30192 | Smp_004190 | 2.76 | C904653.1 | cationic amino acid transporter, putative |
| Q2_P19424 | Smp_138380 | 2.57 | C802143.1 | leucine aminopeptidase- related |
| Q2_P19417 | Smp_139160 | 2.33 | C802136.1 | cysteine protease family C1-related |
| Q2_P03089 | Smp_162770 | 2.7 | C806088.1 | lysosome-associated membrane glycoprotein |
| Q2_P03953 | Smp_173160 | 2.3 | C807425.1 | zinc metalloprotease, putative |
| Q2_P04728 | Smp_137170 | 2.25 | C808691.1 | plasma membrane calcium-transporting atpase, putative |
| Q2_P25963 | Smp_159350 | 2.22 | C811300.1 | platelet binding protein-related |
*The systematic naming of probes follows [14].
Table 2. A selection of differentially up-regulated genes of the vitelline tissue normalised to the whole female control.
| *Systematic name | Gene name | Fold change | Contig | Protein description |
| Q2_P25336 | Smp_164320 | 12.67 | C810082.1 | hypothetical protein |
| Q2_P41174 | Smp_041540 | 4.02 | JAP07007.C | hormone receptor 4 (dHR4), putative |
| Q2_P39884 | Smp_056350 | 3.33 | JAP08870.C | mitotic spindle assembly checkpoint protein mad2- |
| Q2_P40473 | Smp_170990 | 3.05 | JAP07382.S | vinculin, putative |
| Q2_P39851 | Smp_174820 | 2.85 | JAP08581.C | serine/threonine kinase, putative |
| Q2_P36026 | Smp_173320 | 2.58 | C915812.1 | tubulin tyrosine ligase-related |
| Q2_P37617 | Smp_169380 | 2.53 | C918667.1 | protein-tyrosine phosphatase, putative |
| Q2_P36808 | Smp_162900 | 2.37 | C917244.1 | tyrosine-protein kinase pr2, putative |
| Q2_P38755 | Smp_148680 | 2.28 | JAP01788.S | DNA double-strand break repair rad50 ATPase, |
| Q2_P27568 | Smp_076950.2 | 2.24 | C900263.1 | solute carrier family 33 (acetyl-CoA transporter) |
| Q2_P26890 | Smp_008580 | 2.23 | C812627.1 | sugar transporter, putative |
| Q2_P31891 | Smp_124110 | 2.13 | C907823.1 | smad nuclear interacting protein, putative |
| Q2_P35951 | Smp_137080 | 2.11 | C915708.1 | multidrug resistance protein 1, 2, 3 |
| Q2_P33700 | Smp_158400 | 2.04 | C911301.1 | receptor protein tyrosine phosphatase r (pcptp1), |
*The systematic naming of probes follows [14].
Table 3. A selection of differentially up-regulated genes of the ovary normalised to the whole female control.
| *Systematic name | Gene name | Fold change | Contig | Protein description |
| Q2_P35767 | Smp_141900 | 70.62 | C915481.1 | expressed protein |
| Q2_P17568 | Smp_144080 | 69.56 | C919966.1 | proprotein convertase subtilisin/kexin-related |
| Q2_P35553 | Smp_150350 | 57.65 | C915220.1 | synaptotagmin, putative |
| Q2_P22372 | Smp_080360 | 52.15 | C806137.1 | hypothetical protein |
| Q2_P06908 | Smp_011870 | 50.87 | C812082.1 | hypothetical protein |
| Q2_P32715 | Smp_026400 | 38.6 | C909489.1 | thyrotroph embryonic factor related |
| Q2_P34493 | Smp_027920 | 24.25 | C912754.1 | tubulin alpha chain, putative |
| Q2_P32366 | Smp_082490 | 17.99 | C908755.1 | cyclin B, putative |
| Q2_P02801 | Smp_153660 | 15.49 | C805565.1 | cyclin d, putative |
| Q2_P39175 | Smp_067260 | 14.77 | JAP01374.C | transforming growth factor-beta receptor type I, putative |
| Q2_P41215 | Smp_145580 | 14.09 | JAP08203.C | progesterone-induced-blocking factor, putative |
| Q2_P14033 | Smp_160630 | 13.32 | C911654.1 | DNA repair protein rad51 homolog 3, r51h3, putative |
| Q2_P24711 | Smp_004060 | 11.98 | C809241.1 | cell division cycle 20 (cdc20) (fizzy), putative |
| Q2_P29040 | Smp_162710 | 11.46 | C903052.1 | aurora kinase-related |
| Q2_P40215 | Smp_135900 | 9.43 | JAP01762.S | DNA double-strand break repair rad50 ATPase, putative |
| Q2_P00792 | Smp_160650.2 | 5.43 | C801902.1 | smad1, 5, 8, and, putative |
| Q2_P28420 | Smp_145900 | 4.69 | C902171.1 | adam, putative |
| Q2_P14621 | Smp_089700 | 4.16 | C912832.1 | integrin beta subunit, putative |
| Q2_P36767 | Smp_144390 | 3.21 | C917178.1 | transforming growth factor-beta receptor type I and II, putative |
| Q2_P05400 | Smp_165220 | 3.15 | C809991.1 | embryonic ectoderm development protein, putative |
| Q2_P11728 | Smp_033950 | 2.66 | C907151.1 | Smad4, putative |
| Q2_P18126 | Smp_157540 | 2.54 | C800274.1 | smad, putative |
| Q2_P22966 | Smp_072370 | 2.31 | C806933.1 | major egg antigen (p40) |
*The systematic name of probes follows [14].
Table 4. A selection of differentially up-regulated genes of testis normalised to the whole male control.
| *Systematic name | Gene name | Fold change | Contig | Protein dscription |
| Q2_P28420 | Smp_145900 | 32.89 | C902171.1 | ADAM, putative |
| Q2_P39027 | Smp_165970 | 31.26 | JAP00180.C | acidic fibroblast growth factor intracellular binding protein, putative |
| Q2_P38111 | Smp_143970 | 26.69 | C920217.1 | kelch-like protein |
| Q2_P35735 | Smp_157820 | 26.09 | C915438.1 | ataxia telangiectasia mutated (atm), putative |
| Q2_P30827 | Smp_074830 | 20.54 | C905692.1 | tubulin tyrosine ligase-related |
| Q2_P40105 | Smp_067260 | 19.78 | JAP11452.C | transforming growth factor-beta receptor type I, putative |
| Q2_P02282 | Smp_032490 | 19.72 | C804679.1 | tropomyosin, putative |
| Q2_P14033 | Smp_160630 | 15.73 | C911654.1 | DNA repair protein rad51 homolog 3, r51h3, putative |
| Q2_P38558 | Smp_169970 | 14.78 | JAP06742.C | Paramyosin, putative |
| Q2_P39506 | Smp_136210 | 10.12 | JAP03797.C | testis development protein nyd-sp29, putative |
| Q2_P34493 | Smp_027920 | 9.08 | C912754.1 | tubulin alpha chain, putative |
| Q2_P25694 | Smp_167000 | 8.73 | C810809.1 | testis specific protein, putative |
| Q2_P32904 | Smp_042740 | 8.47 | C909800.1 | sperm associated antigen 6 |
| Q2_P06063 | Smp_167610 | 7.02 | C810843.1 | multidrug resistance-associated protein 4 (mrp/cmoat-related abc transporter |
| Q2_P12548 | Smp_139730 | 5.04 | C908741.1 | intraflagellar transport 81 |
| Q2_P21892 | Smp_075140 | 3.96 | C805511.1 | spermatogenesis associated 18 |
| Q2_P10649 | Smp_058320 | 3.68 | C904995.1 | nuclear autoantigenic sperm protein (nasp), putative |
| Q2_P20054 | Smp_078040 | 3.45 | C802934.1 | tubulin beta chain, putative |
| Q2_P39599 | Smp_152680 | 3.41 | JAP05136.C | epidermal growth factor receptor, putative |
| Q2_P22159 | Smp_104730 | 3.17 | C805862.1 | DNAj homolog subfamily B member 4, putative |
*The systematic naming of probes follows [14].
For the testes, after filtering the microarray data for flagged signal and normalising against the whole male control, a total of 16,334 contigs were revealed, of which 2,171 were up-regulated 2 fold or higher than the whole male control (Table S5). We also compared gene transcription between the male and female reproductive tissues. The ovary had more genes in common with testes than with the vitelline tissue, while all reproductive tissues expressed 35 enriched contigs in common (Table S6, Figure S1).
GO distribution for each tissue type is shown in Figure S2 using level 2 annotations. A pie chart representation and the number of contigs at level 2 annotation are shown for either “Biological Process” or “Molecular Function”. The top 10 GO annotations, based on overall contig numbers for each ontology, are shown (Figure S3). Results are shown for either “Biological Process” or “Molecular Function”. Annotations for the 6,295 contigs that were differentially expressed in any 1 of the 4 tissue types isolated by LMM, as determined by microarray analysis, are shown (Table S5).
Macro-dissected female tissues
Of the 41,799 probes on the microarray, 18,359 were present in all the macro- dissected female tissue samples when normalised against the whole female control and filtered for significant signal. Three thousand one hundred and nine genes were up-regulated two-fold or more in at least one region of the parasite, compared with the control sample, and their presentation in each region is shown (Figure 2b). The head and middle regions had fewer genes in common with the hind region (Table S7, Table S8, Table S9) and this may be due to the dominance of vitelline tissue in the hind region of adult female schistosomes. The microdissected ovarian sample and the female middle region showed 574 genes enriched in common, which is probably due to the anatomical location of the ovary within this region (Table S1), but the vitelline tissue had only 4 common genes with the hind region. This latter result is interpreted as being due to the dominance of vitelline gland transcriptional activity in females overall.
There were 3,773 genes up-regulated in manually dissected male tissues and the testes when compared with the whole male worm. Genes that were up-regulated two fold or higher in each male tissue sample are shown (Figure 2c and Table S10, Table S11). The testes were included in the male hind region during manual dissection, but the analysis showed that the testis had more genes in common with the head than the hind region. This may reflect similar gene expression in tissues bordering or surrounding the testes, or may reflect the gross manual dissection procedure. Either way, the accuracy of LMM provides the best isolation method for this type of tissue, and this type of experimental approach.
Real-time PCR validation of the microarray data
The D'Agostino & Pearson omnibus and the Shapiro-Wilk normality tests showed that the data were not normally distributed. Therefore, Spearman correlation was used [20] to compare the real-time and microarray data. Comparison of 31 data pairs using the Spearman correlation showed a statistically significant correlation (P<0.0001, alpha = 0.05) between the real-time and microarray data (Figure S4).
Comparison of expression profiles of S. japonicum and S. mansoni adult tissues
In our previous study, we investigated expression profiles of three female tissues of S. japonicum using a different microarray platform [11]. To compare more directly the expression profiles of these tissues in S. japonicum and those of S. mansoni presented here, BLASTn searches of the two datasets were performed to identify potential homologues enriched in tissue for both species (Table S2).
Approximately 72% of the contigs on the microarray used in the previous investigation of S. japonicum tissue-specific expression corresponded to putative orthologues of genes represented on the current microarray, with homology greater than 75% and covering more than 40% of the sequence length (e-value<1E-10). When the previous reported S. japonicum/S. mansoni microarray [11] was compared to that used here on the basis of design, approximately 95% of contigs derived from S. mansoni (TC) had homologues on the current microarray, whereas only approximately 33% of contigs derived from S. japonicum ESTs on that platform had strong homology to sequences on the current microarray. Overall there are strong similarities between the combined (S. japonicum and S. mansoni) microarray [11] and the microarray used in the current study [14] which was based on a more recent EST assembly.
The considerable overlap between the two microarrays allowed us to make direct comparisons of expression profiles of tissues of the two schistosome species. Of the 393 up-regulated genes in S. mansoni gastrodermal tissue, 180 had homologues in the microarray used in the previous S. japonicum study [11], and 31 of these were transcriptionally enriched in the S. japonicum gastrodermis. Similarly, of the 384 enriched genes in S. mansoni vitelline samples, 188 had homologues in the combined microarray, of which 33 were enriched also in the S. japonicum vitelline tissue. Four thousand four hundred and fifty (4,450) upregulated genes in S. mansoni ovary had 2675 homologues, and 917 of these were enriched in the S. japonicum ovary (Table S5).
Discussion
In this paper we present transcriptomic data for four tissues in adult worms of Schistosoma mansoni. This study complements our previous description of tissue-specific transcriptomics for the related species S. japonicum [11], but, additionally, provides the first expression analysis of the testes of a schistosome. As a means to defining the transcriptome of other less accessible tissues, we have also incorporated data from crude dissection of adults. Although this latter dataset cannot replace detailed localisation investigations of specific tissues, it can be used to give a crude overview of sites of expression of genes.
For this study, we pooled multiple schistosome parasites, from separate infections, so as to allow the microdissection of sufficient volumes of tissues necessary to obtain sufficient total RNA for downstream analysis. This approach limits any impact that variation between individual parasites may have on gene expression. Due to extreme technical constraints in the LMM method, the numbers of strict biological replicates was restricted, but we are confident that the use of pooled individual parasites limits any drawbacks normally overcome with biological replicates. Our data fulfils the criterion of biological relevance, that is, genes, for which we know function and sites of expression, were largely expressed in the tissue in which we expected them to be. Thus, while there is a limitation brought about by the sheer immensity of the undertaking, we argue that the results are robust. Further surveys will augment our dataset.
Gene ontology analysis performed on the differentially expressed genes of all four tissue types gave insight into which common biological processes and molecular functions are present in the individual tissues. For all tissues examined here, most differentially expressed genes were involved in catalytic activity and binding functions. For the gastrodermis, probes for genes encoding cystine-type endopeptidase activity followed by RNA binding were well represented. With regard to biological processes and molecular function (Figures S2, S3), genes related to proteolytic process were, as expected, the most abundant in the gastrodermis (Figure 3), followed by genes for RNA dependent DNA replication and genes related to auxin biosynthesis process, such as those encoding protein kinases, RNA and DNA helicases, and ATP-dependent transporters. The expression profile of the gastrodermis thus reflects the digestive and absorptive nature of the tissue as well as its high synthetic activity.
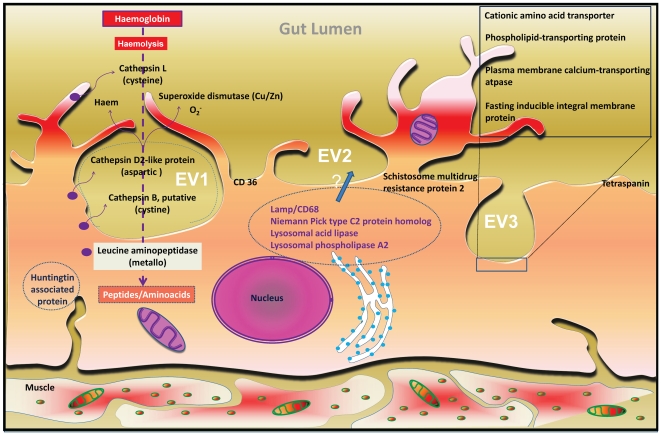
Figure 3. Diagrammatic representation of the gastrodermis of female S. mansoni.
The diagram is based on an illustration originally published by Morris [52]. The illustration shows some highly enriched genes identified in this transcriptomic survey (Table 1, Table S3). Many of the genes products are putatively released into, or onto the membrane of epicellular vacuoles (EV) present at the luminal surface of the syncytial epithelium. Three EVs are depicted (EV1–3), each showing different physiological activities proposed to occur in the vacuoles. EV1 depicts haemoglobinolysis pathways. Many proteases are released into the EV, resulting in catabolism of haemoglobin into amino acids or small peptides for uptake. Haem, a byproduct of catabolism, is sequestered into haematin (see illustration in [53]) for subsequent egestion. EV2 depicts a potential series of molecules associated with lysosomal functions, which has been transferred to these vacuolar compartments. EV3 shows a series of membrane proteins, involved in a variety of functions, notably transmembrane transport of metabolites, which would be preferentially associated with the epicellular vacuoles. All three physiological activities may occur in the same vacuole, probably concurrently.
The gastrodermis transcribes genes encoding a range of endo- and exo-peptidases [21]–[24], saposins, structural genes, and numerous genes associated with intracellular compartments (Table 1, Table S5, Figure 3). Full knowledge of the proteolytic repertoire of the schistosome gut, however, is limited because we are largely ignorant about the expression repertoire of the proximal region of the gut, the oesophageal gland and oesophagus. This lack of data arises from the minute size of this gland in female schistosomes [12]. The oesophageal gland, a syncytial mass surrounding the tegument-like foregut, is thought to be responsible, among other activities, for lysis of red cells after their ingestion by the parasite [12], [25]. Our crude analysis of the expression profiles of the anterior regions of females suggest the expression of candidate genes that might contribute to host cell lysis, including a number of saposins and a diversity of apical peptidases (Table S7). Full characterisation of the haemoglobinolytic cascade in the entire schistosome gut requires detailed transcriptional analysis of the oesophageal gland. To address this, we have begun to analyse the expression activity of the male oesophageal gland to partially address this information deficit.
Our two transcriptomic surveys have shown strong overlap in the genes enriched for the gastrodermis of S. japonicum and S. mansoni [11] (Table S5). We detected a range of molecules encoding proteins involved in lipid binding, uptake or metabolism, such as saposins (for S. mansoni, these include Smp_130100 Smp_105420.1 Smp_014570 Smp_028870). As schistosomes are unable to synthesise sterols and lipids [1], the abundance of molecules for fatty acid transport emphasises their nutritional dependency on the host. Niemann Pick type C2 protein homologue (NPC2) genes (Smp_194850) (Figure 3, Table S5) are enriched for the gastrodermis for both schistosome species, as well as for the ovary of S. mansoni (Table S5), an observation consistent with the detection of abundant NPC2 in the secretome of S. mansoni eggs [26]. NPC2 is a lysosomal cholesterol-binding protein which, in conjunction with NPC1, helps in the trafficking of cholesterol and other sterols and glycolipids in eukaryotic cells [27], [28]. While NPC2 was found to be enriched in the gastrodermis of S. mansoni, its putative binding partner, NPC1, was enriched for the head region of females (Table S7), a region that contains the secretory oesophageal gland.
The gastrodermis of schistosomes bears numerous epicellular vacuolar compartments [22] that are suggested to function in a lysosomal capacity. We detected numerous genes that in mammals are associated with membrane trafficking through endocytosis, endocytic compartments and lysosomes in the gastrodermis of S. mansoni. Among these enriched genes were a homologue of human Huntingtin associated protein 1 (HAP 1) (Smp_127380) [29], and numerous lysosomal molecules including 1-o-acylceramide synthase (Smp_166530.1) (lysosomal phospholipase A2 or lcat-like lysophospholipase) and lysosomal acid lipase-related protein (Smp_146180). In addition, a homolog of a lysosome-associated membrane glycoprotein (LAMP)/CD68 (Table 1, Figure 3) (Smp_162770) was enriched for the gastrodermis of both schistosome species [11] and is also abundant in the ovary of S. mansoni. LAMP family members sit in the membranes of lysosomes, where they cover the majority of the lysosomal luminal surface [30], [31]. Other LAMPs occur in the plasma membrane of cells such that much of the LAMP protein is extracellular [32], [33]. If the epicellular vacuoles of the schistosome gastrodermis function as part of the endocytic or lysosomal system, then such molecules as schistosome NPC2 and LAMP may function in these luminal components of the gastrodermis, and may represent accessible drug or vaccine targets.
Gene ontology analysis of reproductive tissues, including the vitellaria, of S. mansoni showed that the greatest number of enriched genes had molecular functions associated with ATP and protein binding, while biological processes associated with auxin metabolism, signal transduction and transmembrane transport were also prominent. Expression profiling of the three reproductive tissues indicated that in all some 35 genes were enriched for all three tissues. Many of these genes encode products with no annotation, and thus no assigned function. Of genes that did have annotations, some encoded proteins involved in cell cycle and DNA repair, as would be expected for high turnover tissues. The ovary had more genes in common with the testes, than with the vitellaria (Table S6).
Egg shell precursor proteins were highly up-regulated in the vitellaria relative to the ovary, with one (Smp_000430.1) being enriched some 78-fold in the former tissue. Hormone receptor 4 (dHR4) (Smp_041540), which was up-regulated 4-fold in S. mansoni vitelline tissue (Table 2), is a nuclear receptor which is a transcription regulator [34]. King-Jones and colleagues [35] have suggested that dHR4 could be one of the factors that coordinate hormone-mediated growth and maturation in Drosophila.
In the ovary, we found a number of genes encoding proteins that in mammals are thought to dock sperm during fertilisation, such as the alpha and beta integrins [36], [37] and an ADAM (A Disintegrin And Metalloproteinase) (Smp_145900) [36] (discussed below). Tyrotrophic embryonic factor related protein (Smp_026400) was one of the highest transcripts up-regulated (30-fold) in the ovary and is known to regulate transcription [38]. Several other proteins involved in DNA repair and cell cycle were enriched in the ovary. Cyclins and their protein kinases play a key regulatory function in the cell cycle. Cyclins, mainly Cyclin B (Smp_082490, Smp_047620), which is associated with the mitotic phase [39], were highly enriched in both ovary (17.99 fold) and testis (19.3 fold) (Table 3, Table S5).
An ADAM homologue was the second most highly expressed gene in the testes of S. mansoni (Table 4, Table S5). Its expressed product plays a role in fertility by helping sperm-egg adhesion. Other than fertilisation, ADAMs are also involved in neurogenesis, myogenesis and inflammatory processes [36], [40]. Among the 2171 genes enriched 2-fold or greater in the testes were molecules specifically related to testicular function, including the testis development protein nyd-sp29, testis specific protein, sperm-associated antigen 6, spermatogenesis-associated protein, and nuclear autoantigenic sperm protein (nasp) (Table 4 and Table S5).
Schistosomes are known to use well-defined signalling pathways for development, and for between-gender and host-parasite cross-talk [41], [42]. Females need the presence of mature male worms for their development, and oogenesis and vitellogenesis [41], [42]. Our data revealed enrichment of transmembrane receptors involved in protein phosphorylation, major components of signaling pathways, including serine threonine kinase and tyrosine kinase, in gonadal tissues of males and females (Table S3, Table S5, Table S6). Furthermore, all key molecules of the TGFβ-related pathway, a pathway important for female reproductive activity [42], [43], were enriched in the ovary, including transforming growth factor-beta receptors (TGFβR) and putative Smad proteins including Smad4 (Smp_033950) and Smad 1, 5, and 8 (Smp_160650.2) (Table 2). Smads were also enriched in vitellaria, whereas TGFβR1 was up-regulated in the testes.
We performed relatively crude dissections to assess gene expression in different regions of adult S. mansoni. The anatomy of schistosomes is highly ordered with specific tissues located in different regions of the parasites (Table S1). We reasoned that this crude dataset could serve as a means for further fine-tuning sites of expression of genes that are expressed in tissues difficult to isolate with current technologies. These data have been presented here with the caveat that localisation will need to be further confirmed with other localisation methods including in situ hybridization and further LMM.
Of particular interest was the expression profile of the female head region. This region contains the oesophageal gland, the tissue likely to express enzymes initiating haemoglobinolysis and red cell lysis. Many proteases that were not common to the gastrodermal tissue were up-regulated in the head region. Genes encoding proteins associated with neuronal tissue were also up-regulated in this region (Table S7), reflecting the location of the circum-oesophageal ganglia. Abundant molecules with neuronal function in the head region included neurotransmitter gated ion channel proteins (Smp_096480 Smp_176310 Smp_037960) and a vesicular acetylcholine transporter (Smp_046120). Cerebellin (Smp_105050), a neuropeptide known to be found mainly in central nervous system related to neuromodulatory function [44], and stomatin (Smp_162440), involved in signal transduction from mechanoreceptors [45], were among some of the other enriched genes. Kuzbanian (Smp_160620.1), a molecule related to neural development in mammals [1], was up-regulated in the head region of adult females.
Chalmers and colleagues [46] have described 28 S. mansoni venom allergen-like proteins (SmVALs) which contain a sperm coating protein domain. No specific function has been ascribed to these proteins, but they may have diverse biological roles including counteracting the host immune response, host invasion and host parasite interactions. We found SmVAL- 6 (Smp_124050.1) to be 31-fold enriched in the female head region compared with the female control, although this molecule was previously reported to be predominately associated with adult males [46]. SmVAL6 was recently identified as a microexon gene [1], [47] and is one of at least 45 S. mansoni genes thought to undergo alternative splicing, resulting possibly in the generation of multiple gene products [47]. Another distinct VAL, SmVAL-13 (Smp_124060) was enriched 14-fold head region of males (Table S7 and Table S10).
In summary, this is the second study of tissue specific transcriptomics of the digestive and reproductive systems in schistosomes and the first such study of S. mansoni which, additionally, includes an expanded tissue analysis of the male reproductive tissues. In addition to established genes associated with digestive function, we have provided localisation data for new putative transporters, receptors, cytoskeletal elements and motor proteins. Furthermore, many other genes without any formal annotation were prominent transcripts in the gastrodermis. Similarly, many known genes involved in DNA fidelity, oocyte development and egg shell formation were identified in the reproductive tissues along with numerous unidentified molecules. Some of these as yet unknown genes may be involved in transport processes, signal transduction and receptor function, and include cytoskeletal elements and motor proteins. The regulation of the cell cycle and gene expression is another important function that must be maintained in the reproductive tissues of schistosomes. The findings of this study provide a valuable resource for the parasitology community and add a new dimension to the already available stage-specific transcriptomic data for S. mansoni. The presented data represent a resource that could be used effectively in reverse vaccinology allowing the targeting of hidden antigens in the gastrodermis that are exposed to the blood meal, a strategy used effectively for other blood feeding parasites [48], [49], [50], [51], and in the search for new anti-schistosome drug target candidates targeting the control of fertility and fecundity.
Supporting Information
Venn diagram showing genes up-regulated in microdissected male and female reproductive tissues of S. mansoni.
(1.01 MB TIF)
Gene Ontology distribution for male and female microdissected tissues. GO distribution for Biological Process level 2 (left) or Molecular Function level 2 (right) for S. mansoni male and female microdissected tissues. The number of genes in each category is in brackets.
(2.16 MB TIF)
Top 10 Gene Ontology annotations by gene number for Biological Process and Molecular function in S. mansoni male and female microdissected tissues. The categories are listed, with the number of sequences in each category.
(2.69 MB TIF)
Real-time PCR validation of microarray. Validation is shown using a subset of differentially expressed genes in S. mansoni female gastrodermis, ovary, vitelline tissues compared to the female control and testes compared to the male control tissue. The real-time PCR data, expressed as fold changes (normalised to control tissues, either male or female all tissues), are presented as bar graphs, while the corresponding microarray data (fold changes) are shown below in numbers.
(0.41 MB TIF)
Major tissue components of female macro-dissected regions.
(0.03 MB DOC)
Comparison of the microarray used in the S. japonicum transcriptomic study [11] and the microarray used in the current study using BioEdit BLASTn. Column A: Name of EST from either S. japonicum (ContigXXX) or S. mansoni (TCXXX) used from the study of Gobert et al (2009) [11]. Column B: Length of the EST from Column A. Column C: % of the length from EST of Column A that was homologous to the BLASTn from ESTs on the microarray from the current study. Column D: EST result from BLASTn results % nucleotide identity to the current study's microarray platform. Column E: % nucleotide identity, the extent to which two sequences were invariant. Column F: Nucleotide alignment length. Column G: The number of nucleotides that did not match. Column H: A space introduced into an alignment to compensate for insertions and deletions in one sequence relative to another. Columns I–J: The input sequence with which all of the entries in a database were to be compared. Column I: Coordinate of the query start. Column J: Coordinate of the query end. Columns K–L: The database which the input sequence was compared with. Column K: Coordinate of the subject start. Column L: Coordinate of the subject end. Column M: Expectation value. The number of different alignments with scores equivalent to or better than S that are expected to occur in a database search by chance. Column N: Bit Score, The value S′ is derived from the raw alignment score S in which the statistical properties of the scoring system used have been taken into account.
(3.41 MB XLS)
BlastX results of the complete list of differentially upregulated genes in S. mansoni female and male microdissected tissues (2 fold or higher than the microdissected whole female control for ovary, vitelline and gastrodermis and 2 fold or higher than the microdissected male control for testes). Column A: EST name from current study microarray platform [14]. Column B: BLASTx description. Column C: Length of the EST. Column D: Number of homologous matchs from BLASTn. Column E: eValue- Expectation value. The number of different alignments with scores equivalent to or better than S that are expected to occur in a database search by chance. Column F: mean % sequence identity, the extent to which two sequences are invariant. Columns G–M: Top homologous hit details. Column G: Accession numbers for the homologous protein description. Column H: Homologous hit accession number. Column I: eValue- Expectation value. Column J: % sequence identity, the extent to which two sequences are invariant. Column K: Bit Score, The value S′ is derived from the raw alignment score S in which the statistical properties of the scoring system used have been taken into account. Column L: Alignment length between the query and the top homologous hit. Column M: Number of positive sequences. Column N: Number of Gene Ontology categories associated with the top homologous hit. Column O: Associated gene ontology codes. Column P: Associated gene ontology names. Column Q: Enzyme codes. Column R: Enzyme pathways. Column S: InterProScan codes. Column T: Additional InterProScan output information.
(5.71 MB XLSX)
List of oligonucleotides used in real time PCR to validate the microarray data.
(0.03 MB DOC)
Complete list of differentially upregulated genes in S. mansoni female and male microdissected tissues. Worksheet 1–3: Up-regulated genes in the female parasite sorted for each tissue type based on descending signal intensity, Gastrodermis, Ovary and Vitelline tissues, relative to the entire LMM-obtained control female tissues. Worksheet 4: Up-regulated genes in the male parasite Testes tissues, relative to the entire LMM male tissues. Column A: Systemic Name, unique identifier. Column B: Normalised signal intensity relative to control (all tissues) tissue. Column C: Normalised signal intensity of gastrodermis tissue. (Testes in worksheet 4). Column D: Normalised signal intensity of ovary tissue. (Blank in worksheet 4). Column E: Normalised signal intensity of vitelline tissue. (Blank in worksheet 4). Column F: Synonyms for unique identifier. Column G: Contig name. Column H: Homologue in Gobert et al, 2009 [11] study (genes found upregulated in same tissue between both [current] studies are highlight in blue). Columns I–M: Class, Annotation from Blast against GenBank on Jan 2008, Probe Sequence, SOURCE_ID and Description; all from original publication [14]. Columns N–AF: New Descriptions from BlastX performed on 20/10/2010. Column N: BLASTx description. Column O: Length of the EST. Column P: Number of homologous matchs from BLASTn. Column Q: eValue- Expectation value. The number of different alignments with scores equivalent to or better than S that are expected to occur in a database search by chance. Column R: mean % sequence identity, the extent to which two sequences are invariant. Columns S–Y: Top homologous hit details. Column S: Accession numbers for the homologous protein description. Column T: Homologous hit accession number. Column U: eValue- Expectation value. Column V: % sequence identity, the extent to which two sequences are invariant. Column W: Bit Score, The value S′ is derived from the raw alignment score S in which the statistical properties of the scoring system used have been taken into account. Column X: Alignment length between the query and the top homologous hit. Column Y: Number of positive sequences. Column Z: Number of Gene Ontology categories associated with the top homologous hit. Column AA: Associated gene ontology codes. Column AB: Associated gene ontology names. Column AC: Enzyme codes. Column AD: Enzyme pathways. Column AE: InterProScan codes. Column AF: Additional InterProScan output information.
(6.83 MB XLS)
List of differentially upregulated genes common to S. mansoni male and female reproductive tissues (ovary, vitelline tissue and testes).
(0.11 MB XLS)
Complete list of 1402 differentially upregulated genes (2 fold or higher than the whole female control) in the S. mansoni female head region.
(2.50 MB XLS)
Complete list of 870 differentially upregulated genes (2 fold or higher than the whole female control) in the S. mansoni female middle region.
(1.75 MB XLS)
Complete list of 152 differentially upregulated genes (2 fold or higher than the whole female control) in the S. mansoni female hind region.
(0.31 MB XLS)
Complete list of 1120 differentially upregulated genes (2 fold or higher than the whole male control) in the S. mansoni male head region.
(2.66 MB XLS)
Complete list of 91 differentially upregulated genes (2 fold or higher than the male control) in the S. mansoni male hind region.
(0.19 MB XLS)
Acknowledgments
The authors would like to thank Clay Winterford and the staff of the Histotechnology Group of the Queensland Institute of Medical Research, for assisting with cryosectioning and Ms Mary Duke for the maintenance of the parasite lifecycle. We thank Dr D.L.O. Dissabandara for assistance with Figure 3.
Footnotes
The authors have declared that no competing interests exist.
The authors gratefully acknowledge funding from the Australian Research Council (DP1093471, awarded to MKJ and GNG), National Health and Medical Research Council of Australia (MKJ and DPM), the Sandler Centre for Basic Research into Parasitic Disease (MKJ), and the Tropical Diseases Research Programme of the World Health Organization (MKJ and GNG). SSKN is supported by a scholarship from the Fellowship Fund Branch of Australian Federation of University Women (Queensland). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Berriman M, Haas BJ, LoVerde PT, Wilson RA, Dillon GP, et al. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460:352–358. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schistosoma japonicum Genome Sequencing and Functional Analysis Consortium. The Schistosoma japonicum genome reveals features of host-parasite interplay. Nature. 2009;460:345–351. doi: 10.1038/nature08140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han ZG, Brindley PJ, Wang SY, Chen Z. Schistosoma genomics: new perspectives on schistosome biology and host-parasite interaction. Annu Rev Genomics Hum Genet. 2009;10:211–240. doi: 10.1146/annurev-genom-082908-150036. [DOI] [PubMed] [Google Scholar]
- 4.Wilson RA, Ashton PD, Braschi S, Dillon GP, Berriman M, et al. ‘Oming in on schistosomes: prospects and limitations for post-genomics. Trends Parasitol. 2007;23:14–20. doi: 10.1016/j.pt.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Hu W, Yan Q, Shen DK, Liu F, Zhu ZD, et al. Evolutionary and biomedical implications of a Schistosoma japonicum complementary DNA resource. Nat Genet. 2003;35:139–147. doi: 10.1038/ng1236. [DOI] [PubMed] [Google Scholar]
- 6.Verjovski-Almeida S, DeMarco R, Martins EA, Guimaraes PE, Ojopi EP, et al. Transcriptome analysis of the acoelomate human parasite Schistosoma mansoni. Nat Genet. 2003;35:148–157. doi: 10.1038/ng1237. [DOI] [PubMed] [Google Scholar]
- 7.Gobert GN, Moertel L, Brindley PJ, McManus DP. Developmental gene expression profiles of the human pathogen Schistosoma japonicum. BMC Genomics. 2009;10:128. doi: 10.1186/1471-2164-10-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mulvenna J, Moertel L, Jones MK, Nawaratna S, Lovas EM, et al. Exposed proteins of the Schistosoma japonicum tegument. Int J Parasitol. 2009;40:543–554. doi: 10.1016/j.ijpara.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Gobert GN. Applications for profiling the schistosome transcriptome. Trends Parasitol. 2010;26:434–439. doi: 10.1016/j.pt.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Jones MK, Higgins T, Stenzel DJ, Gobert GN. Towards tissue specific transcriptomics and expression pattern analysis in schistosomes using laser microdissection microscopy. Exp Parasitol. 2007;117:259–266. doi: 10.1016/j.exppara.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 11.Gobert GN, McManus DP, Nawaratna S, Moertel L, Mulvenna J, et al. Tissue specific profiling of females of Schistosoma japonicum by integrated laser microdissection microscopy and microarray analysis. PLoS Negl Trop Dis. 2009;3:e469. doi: 10.1371/journal.pntd.0000469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dillon GP, Illes JC, Isaacs HV, Wilson RA. Patterns of gene expression in schistosomes: localization by whole mount in situ hybridization. Parasitology. 2007;134:1589–1597. doi: 10.1017/S0031182007002995. [DOI] [PubMed] [Google Scholar]
- 13.Jones MK, Randall LM, McManus DP, Engwerda CR. Laser microdissection microscopy in parasitology: microscopes meet thermocyclers. Trends Parasitol. 2004;20:502–506. doi: 10.1016/j.pt.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 14.Verjovski-Almeida S, Venancio TM, Oliveira KC, Almeida GT, DeMarco R. Use of a 44k oligoarray to explore the transcriptome of Schistosoma mansoni adult worms. Exp Parasitol. 2007;117:236–245. doi: 10.1016/j.exppara.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 16.Gobert GN, Tran MH, Moertel L, Mulvenna J, Jones MK, et al. Transcriptional Changes in Schistosoma mansoni during early schistosomula development and in the presence of erythrocytes. PLoS Negl Trop Dis. 2010;4:e600. doi: 10.1371/journal.pntd.0000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patterson TA, Lobenhofer EK, Fulmer-Smentek SB, Collins PJ, Chu TM, et al. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat Biotechnol. 2006;24:1140–1150. doi: 10.1038/nbt1242. [DOI] [PubMed] [Google Scholar]
- 18.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 19.Moertel L, Gobert GN, McManus DP. Comparative real-time PCR and enzyme analysis of selected gender-associated molecules in Schistosoma japonicum. Parasitology. 2008;135:575–583. doi: 10.1017/S0031182008004174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morey JS, Ryan JC, Van Dolah FM. Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol Proced Online. 2006;8:175–193. doi: 10.1251/bpo126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bos DH, Mayfield C, Minchella DJ. Analysis of regulatory protease sequences identified through bioinformatic data mining of the Schistosoma mansoni genome. BMC Genomics. 2009;10:488. doi: 10.1186/1471-2164-10-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delcroix M, Sajid M, Caffrey CR, Lim KC, Dvorak J, et al. A multienzyme network functions in intestinal protein digestion by a platyhelminth parasite. J Biol Chem. 2006;281:39316–39329. doi: 10.1074/jbc.M607128200. [DOI] [PubMed] [Google Scholar]
- 23.Brindley PJ, Kalinna BH, Wong JY, Bogitsh BJ, King LT, et al. Proteolysis of human hemoglobin by schistosome cathepsin D. Mol Biochem Parasitol. 2001;112:103–112. doi: 10.1016/s0166-6851(00)00351-0. [DOI] [PubMed] [Google Scholar]
- 24.Caffrey CR, McKerrow JH, Salter JP, Sajid M. Blood ‘n’ guts: an update on schistosome digestive peptidases. Trends Parasitol. 2004;20:241–248. doi: 10.1016/j.pt.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Halton DW. Nutritional adaptations to parasitism within the platyhelminthes. Int J Parasitol. 1997;27:693–704. doi: 10.1016/s0020-7519(97)00011-8. [DOI] [PubMed] [Google Scholar]
- 26.Cass CL, Johnson JR, Califf LL, Xu T, Hernandez HJ, et al. Proteomic analysis of Schistosoma mansoni egg secretions. Mol Biochem Parasitol. 2007;155:84–93. doi: 10.1016/j.molbiopara.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Infante RE, Wang ML, Radhakrishnan A, Kwon HJ, Brown MS, et al. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc Natl Acad Sci U S A. 2008;105:15287–15292. doi: 10.1073/pnas.0807328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Storch J, Xu Z. Niemann-Pick C2 (NPC2) and intracellular cholesterol trafficking. Biochim Biophys Acta. 2009;1791:671–678. doi: 10.1016/j.bbalip.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu LL, Zhou XF. Huntingtin associated protein 1 and its functions. Cell Adh Migr. 2009;3:71–76. doi: 10.4161/cam.3.1.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albertti LA, Macedo AM, Chiari E, Andrews NW, Andrade LO. Role of host lysosomal associated membrane protein (LAMP) in Trypanosoma cruzi invasion and intracellular development. Microbes Infect. 2010;12:784–789. doi: 10.1016/j.micinf.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howe CL, Granger BL, Hull M, Green SA, Gabel CA, et al. Derived protein sequence, oligosaccharides, and membrane insertion of the 120-kDa lysosomal membrane glycoprotein (lgp120): identification of a highly conserved family of lysosomal membrane glycoproteins. Proc Natl Acad Sci U S A. 1988;85:7577–7581. doi: 10.1073/pnas.85.20.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z, Chang H, Li Y, Zhang T, Zou J, et al. MicroRNAs: potential regulators involved in human anencephaly. Int J Biochem Cell Biol. 2010;42:367–374. doi: 10.1016/j.biocel.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Tompkins K, Garrigues J, Snead ML, Gibson CW, et al. Full length amelogenin binds to cell surface LAMP-1 on tooth root/periodontium associated cells. Arch Oral Biol. 2010;55:417–425. doi: 10.1016/j.archoralbio.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomson SA, Baldwin WS, Wang YH, Kwon G, Leblanc GA. Annotation, phylogenetics, and expression of the nuclear receptors in Daphnia pulex. BMC Genomics. 2009;10:500. doi: 10.1186/1471-2164-10-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.King-Jones K, Charles JP, Lam G, Thummel CS. The ecdysone-induced DHR4 orphan nuclear receptor coordinates growth and maturation in Drosophila. Cell. 2005;121:773–784. doi: 10.1016/j.cell.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 36.Primakoff P, Myles DG. The ADAM gene family: surface proteins with adhesion and protease activity. Trends Genet. 2000;16:83–87. doi: 10.1016/s0168-9525(99)01926-5. [DOI] [PubMed] [Google Scholar]
- 37.Almeida EA, Huovila AP, Sutherland AE, Stephens LE, Calarco PG, et al. Mouse egg integrin alpha 6 beta 1 functions as a sperm receptor. Cell. 1995;81:1095–1104. doi: 10.1016/s0092-8674(05)80014-5. [DOI] [PubMed] [Google Scholar]
- 38.Krueger DA, Warner EA, Dowd DR. Involvement of thyrotroph embryonic factor in calcium-mediated regulation of gene expression. J Biol Chem. 2000;275:14524–14531. doi: 10.1074/jbc.275.19.14524. [DOI] [PubMed] [Google Scholar]
- 39.Vermeulen K, Van Bockstaele DR, Berneman ZN. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003;36:131–149. doi: 10.1046/j.1365-2184.2003.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med. 2008;29:258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loverde PT, Osman A, Hinck A. Schistosoma mansoni: TGF-beta signaling pathways. Exp Parasitol. 2007;117:304–317. doi: 10.1016/j.exppara.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.LoVerde PT, Andrade LF, Oliveira G. Signal transduction regulates schistosome reproductive biology. Curr Opin Microbiol. 2009;12:422–428. doi: 10.1016/j.mib.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beckmann S, Quack T, Burmeister C, Buro C, Long T, et al. Schistosoma mansoni: signal transduction processes during the development of the reproductive organs. Parasitology. 2010;137:497–520. doi: 10.1017/S0031182010000053. [DOI] [PubMed] [Google Scholar]
- 44.Albertin G, Malendowicz LK, Macchi C, Markowska A, Nussdorfer GG. Cerebellin stimulates the secretory activity of the rat adrenal gland: in vitro and in vivo studies. Neuropeptides. 2000;34:7–11. doi: 10.1054/npep.1999.0779. [DOI] [PubMed] [Google Scholar]
- 45.Martinez-Salgado C, Benckendorff AG, Chiang LY, Wang R, Milenkovic N, et al. Stomatin and sensory neuron mechanotransduction. J Neurophysiol. 2007;98:3802–3808. doi: 10.1152/jn.00860.2007. [DOI] [PubMed] [Google Scholar]
- 46.Chalmers IW, McArdle AJ, Coulson RM, Wagner MA, Schmid R, et al. Developmentally regulated expression, alternative splicing and distinct sub-groupings in members of the Schistosoma mansoni venom allergen-like (SmVAL) gene family. BMC Genomics. 2008;9:89. doi: 10.1186/1471-2164-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeMarco R, Mathieson W, Manuel SJ, Dillon GP, Curwen RS, et al. Protein variation in blood-dwelling schistosome worms generated by differential splicing of micro-exon gene transcripts. Genome Res. 2010;20:1112–1121. doi: 10.1101/gr.100099.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Munn EA, Smith TS, Smith H, James FM, Smith FC, et al. Vaccination against Haemonchus contortus with denatured forms of the protective antigen H11. Parasite Immunol. 1997;19:243–248. doi: 10.1046/j.1365-3024.1997.d01-205.x. [DOI] [PubMed] [Google Scholar]
- 49.Knox DP, Redmond DL, Newlands GF, Skuce PJ, Pettit D, et al. The nature and prospects for gut membrane proteins as vaccine candidates for Haemonchus contortus and other ruminant trichostrongyloids. Int J Parasitol. 2003;33:1129–1137. doi: 10.1016/s0020-7519(03)00167-x. [DOI] [PubMed] [Google Scholar]
- 50.Newton SE, Munn EA. The development of vaccines against gastrointestinal nematode parasites, particularly Haemonchus contortus. Parasitol Today. 1999;15:116–122. doi: 10.1016/s0169-4758(99)01399-x. [DOI] [PubMed] [Google Scholar]
- 51.Willadsen P, Bird P, Cobon GS, Hungerford J. Commercialisation of a recombinant vaccine against Boophilus microplus. Parasitology. 1995;110(Suppl):S43–50. doi: 10.1017/s0031182000001487. [DOI] [PubMed] [Google Scholar]
- 52.Morris GP. Fine structure of the gut epithelium of Schistosoma mansoni. Experientia. 1968;24:480–482. doi: 10.1007/BF02144405. [DOI] [PubMed] [Google Scholar]
- 53.Toh SQ, Glanfield A, Gobert GN, Jones MK. Heme and blood-feeding parasites: friends or foes? Parasit Vectors. 2010;3:108. doi: 10.1186/1756-3305-3-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Venn diagram showing genes up-regulated in microdissected male and female reproductive tissues of S. mansoni.
(1.01 MB TIF)
Gene Ontology distribution for male and female microdissected tissues. GO distribution for Biological Process level 2 (left) or Molecular Function level 2 (right) for S. mansoni male and female microdissected tissues. The number of genes in each category is in brackets.
(2.16 MB TIF)
Top 10 Gene Ontology annotations by gene number for Biological Process and Molecular function in S. mansoni male and female microdissected tissues. The categories are listed, with the number of sequences in each category.
(2.69 MB TIF)
Real-time PCR validation of microarray. Validation is shown using a subset of differentially expressed genes in S. mansoni female gastrodermis, ovary, vitelline tissues compared to the female control and testes compared to the male control tissue. The real-time PCR data, expressed as fold changes (normalised to control tissues, either male or female all tissues), are presented as bar graphs, while the corresponding microarray data (fold changes) are shown below in numbers.
(0.41 MB TIF)
Major tissue components of female macro-dissected regions.
(0.03 MB DOC)
Comparison of the microarray used in the S. japonicum transcriptomic study [11] and the microarray used in the current study using BioEdit BLASTn. Column A: Name of EST from either S. japonicum (ContigXXX) or S. mansoni (TCXXX) used from the study of Gobert et al (2009) [11]. Column B: Length of the EST from Column A. Column C: % of the length from EST of Column A that was homologous to the BLASTn from ESTs on the microarray from the current study. Column D: EST result from BLASTn results % nucleotide identity to the current study's microarray platform. Column E: % nucleotide identity, the extent to which two sequences were invariant. Column F: Nucleotide alignment length. Column G: The number of nucleotides that did not match. Column H: A space introduced into an alignment to compensate for insertions and deletions in one sequence relative to another. Columns I–J: The input sequence with which all of the entries in a database were to be compared. Column I: Coordinate of the query start. Column J: Coordinate of the query end. Columns K–L: The database which the input sequence was compared with. Column K: Coordinate of the subject start. Column L: Coordinate of the subject end. Column M: Expectation value. The number of different alignments with scores equivalent to or better than S that are expected to occur in a database search by chance. Column N: Bit Score, The value S′ is derived from the raw alignment score S in which the statistical properties of the scoring system used have been taken into account.
(3.41 MB XLS)
BlastX results of the complete list of differentially upregulated genes in S. mansoni female and male microdissected tissues (2 fold or higher than the microdissected whole female control for ovary, vitelline and gastrodermis and 2 fold or higher than the microdissected male control for testes). Column A: EST name from current study microarray platform [14]. Column B: BLASTx description. Column C: Length of the EST. Column D: Number of homologous matchs from BLASTn. Column E: eValue- Expectation value. The number of different alignments with scores equivalent to or better than S that are expected to occur in a database search by chance. Column F: mean % sequence identity, the extent to which two sequences are invariant. Columns G–M: Top homologous hit details. Column G: Accession numbers for the homologous protein description. Column H: Homologous hit accession number. Column I: eValue- Expectation value. Column J: % sequence identity, the extent to which two sequences are invariant. Column K: Bit Score, The value S′ is derived from the raw alignment score S in which the statistical properties of the scoring system used have been taken into account. Column L: Alignment length between the query and the top homologous hit. Column M: Number of positive sequences. Column N: Number of Gene Ontology categories associated with the top homologous hit. Column O: Associated gene ontology codes. Column P: Associated gene ontology names. Column Q: Enzyme codes. Column R: Enzyme pathways. Column S: InterProScan codes. Column T: Additional InterProScan output information.
(5.71 MB XLSX)
List of oligonucleotides used in real time PCR to validate the microarray data.
(0.03 MB DOC)
Complete list of differentially upregulated genes in S. mansoni female and male microdissected tissues. Worksheet 1–3: Up-regulated genes in the female parasite sorted for each tissue type based on descending signal intensity, Gastrodermis, Ovary and Vitelline tissues, relative to the entire LMM-obtained control female tissues. Worksheet 4: Up-regulated genes in the male parasite Testes tissues, relative to the entire LMM male tissues. Column A: Systemic Name, unique identifier. Column B: Normalised signal intensity relative to control (all tissues) tissue. Column C: Normalised signal intensity of gastrodermis tissue. (Testes in worksheet 4). Column D: Normalised signal intensity of ovary tissue. (Blank in worksheet 4). Column E: Normalised signal intensity of vitelline tissue. (Blank in worksheet 4). Column F: Synonyms for unique identifier. Column G: Contig name. Column H: Homologue in Gobert et al, 2009 [11] study (genes found upregulated in same tissue between both [current] studies are highlight in blue). Columns I–M: Class, Annotation from Blast against GenBank on Jan 2008, Probe Sequence, SOURCE_ID and Description; all from original publication [14]. Columns N–AF: New Descriptions from BlastX performed on 20/10/2010. Column N: BLASTx description. Column O: Length of the EST. Column P: Number of homologous matchs from BLASTn. Column Q: eValue- Expectation value. The number of different alignments with scores equivalent to or better than S that are expected to occur in a database search by chance. Column R: mean % sequence identity, the extent to which two sequences are invariant. Columns S–Y: Top homologous hit details. Column S: Accession numbers for the homologous protein description. Column T: Homologous hit accession number. Column U: eValue- Expectation value. Column V: % sequence identity, the extent to which two sequences are invariant. Column W: Bit Score, The value S′ is derived from the raw alignment score S in which the statistical properties of the scoring system used have been taken into account. Column X: Alignment length between the query and the top homologous hit. Column Y: Number of positive sequences. Column Z: Number of Gene Ontology categories associated with the top homologous hit. Column AA: Associated gene ontology codes. Column AB: Associated gene ontology names. Column AC: Enzyme codes. Column AD: Enzyme pathways. Column AE: InterProScan codes. Column AF: Additional InterProScan output information.
(6.83 MB XLS)
List of differentially upregulated genes common to S. mansoni male and female reproductive tissues (ovary, vitelline tissue and testes).
(0.11 MB XLS)
Complete list of 1402 differentially upregulated genes (2 fold or higher than the whole female control) in the S. mansoni female head region.
(2.50 MB XLS)
Complete list of 870 differentially upregulated genes (2 fold or higher than the whole female control) in the S. mansoni female middle region.
(1.75 MB XLS)
Complete list of 152 differentially upregulated genes (2 fold or higher than the whole female control) in the S. mansoni female hind region.
(0.31 MB XLS)
Complete list of 1120 differentially upregulated genes (2 fold or higher than the whole male control) in the S. mansoni male head region.
(2.66 MB XLS)
Complete list of 91 differentially upregulated genes (2 fold or higher than the male control) in the S. mansoni male hind region.
(0.19 MB XLS)