Abstract
Background
The TolC outer membrane channel is a key component of several multidrug resistance (MDR) efflux pumps driven by H+ transport in Escherichia coli. While tolC expression is under the regulation of the EvgA-Gad acid resistance regulon, the role of TolC in growth at low pH and extreme-acid survival is unknown.
Methods and Principal Findings
TolC was required for extreme-acid survival (pH 2) of strain W3110 grown aerobically to stationary phase. A tolC deletion decreased extreme-acid survival (acid resistance) of aerated pH 7.0-grown cells by 105-fold and of pH 5.5-grown cells by 10-fold. The requirement was specific for acid resistance since a tolC defect had no effect on aerobic survival in extreme base (pH 10). TolC was required for expression of glutamate decarboxylase (GadA, GadB), a key component of glutamate-dependent acid resistance (Gad). TolC was also required for maximal exponential growth of E. coli K-12 W3110, in LBK medium buffered at pH 4.5–6.0, but not at pH 6.5–8.5. The TolC growth requirement in moderate acid was independent of Gad. TolC-associated pump components EmrB and MdtB contributed to survival in extreme acid (pH 2), but were not required for growth at pH 5. A mutant lacking the known TolC-associated efflux pumps (acrB, acrD, emrB, emrY, macB, mdtC, mdtF, acrEF) showed no growth defect at acidic pH and a relatively small decrease in extreme-acid survival when pre-grown at pH 5.5.
Conclusions
TolC and proton-driven MDR efflux pump components EmrB and MdtB contribute to E. coli survival in extreme acid and TolC is required for maximal growth rates below pH 6.5. The TolC enhancement of extreme-acid survival includes Gad induction, but TolC-dependent growth rates below pH 6.5 do not involve Gad. That MDR resistance can enhance growth and survival in acid is an important consideration for enteric organisms passing through the acidic stomach.
Introduction
Escherichia coli expresses a large number of multi-drug resistance (MDR) efflux pumps for the expulsion of antibiotics and metabolic wastes. An important group of inner membrane efflux pumps interacts with the outer membrane channel TolC proteins to form complexes that traverse the inner membrane, periplasm, and outer membrane. These complexes efficiently pump the materials outside of the cell [1]–[5]. The other components of these TolC-dependent tripartite efflux systems consist of an inner membrane bound transporter such as the “resistance nodulation division” (RND) family transporter AcrB or the major facilitator superfamily (MFS) transporter EmrB, both driven by H+ influx, or the ABC-superfamily transporter MacB driven by ATP hydrolysis [6]. Stabilizing the transporter-channel interaction is a cognate periplasmic membrane fusion protein (MFP) such as AcrA, EmrA and MacA. Homologs of the E. coli tolC are important in virulence for pathogens such as Salmonella typhimurium [7], Legionella pneumophila [8], Francisella tularensis [9], and Xylella fastidiosa [10]. The TolC-dependent efflux system is responsible not only for expulsion of toxic compounds but also for export of intracellular metabolites, such as enterobactin, porphyrin, and excess cysteine [4], [11], [12].
Several pieces of evidence link tolC expression to acid pH resistance. TolC shows acid-enhanced expression in the E. coli proteome [13]. In E. coli, tolC is a member of the EvgA acid resistance regulon [14], [15] and, in F. tularensis, the tolC homolog is expressed in the same operon with gad (glutamate decarboxylase) [9], an important acid resistance factor (reviewed by [16], [17]). The Gad acid resistance system (AR2) is active in stationary-phase cells grown at pH 7 or pH 5.5, in contrast to the glucose-repressed CRP system (AR1) which requires induction in acid, pH 5.5 [16]. Furthermore, assembly of TolC into efflux complexes requires low pH [18]. The acid-dependent expression and MDR assembly have been suggested to explain the increased sensitivity of bacteria to many antibiotics above pH 7 [18].
Nevertheless, the role of MDR pumps in E. coli acid growth and survival has not been tested. For comparison, at high pH, overexpression of the drug resistance pump MdfA has been shown to increase survival, and actually extends the E. coli growth range to pH 10 [19]. Since enteric pathogens must pass through the stomach, it is important to know whether MDR pumps have a role in growth or survival in acid. Here we report the contributions of tolC, emrB, and mdtB to extreme-acid survival (viability of cells following exposure to pH 2), the requirement of TolC for normal exponential growth at moderately low external pH (pH 4.5–6.0), and the requirement of TolC for Gad expression and induction at low pH.
Results
Extreme-acid survival of tolC, emrB, and mdtB
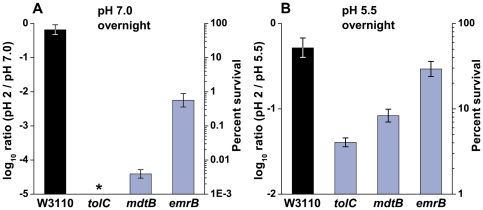
TolC associates with at least nine different inner-membrane protein complexes (such as EmrAB or MdtABC) to form a connected efflux pump system [6]; several of these in the RND and MFS families, are driven by H+ influx. The growth and survival phenotypes of tolC defect strains may result directly from the absence of TolC or from the combined inactivation of several inner-membrane efflux pumps. Therefore, we investigated whether these RND and MFS transporter pump components played a role in extreme acid survival. Of the strains tested, only tolC, emrB, and mdtB deletions showed a significant effect on extreme-acid survival of aerobic cultures (Fig. 1). MDR deletion strains acrB, emrY, and mdtF showed survival levels comparable to the wild-type (data not shown). Survival was tested first for overnight cultures grown at external pH 7, where the Gad system is available but not the acid-inducible CRP system [16]. Extreme-acid survival (exposure at pH 2 for 2 hrs) was over 105-fold lower for tolC, 104-fold lower for mdtB, and 100-fold lower for emrB compared to wild-type strain W3110 (Fig. 1A). There was no increase or decrease in survival for a marR defective strain in which TolC expression is upregulated (data not shown) [20].
Figure 1. TolC, EmrB, and MdtB are required for extreme-acid survival.
Strains W3110 (K-12 parent strain), JLS1015 (W3110 tolC::kan), JLS1027 (W3110 emrB::kan), and JLS1024 (mdtB::kan) were diluted into LBK pH 2 and exposed with rotation for 2 hours at 37°C after overnight growth to stationary phase in buffered LBK at A) pH 7.0 for non-acid-adapted cells and B) pH 5.5 for acid-adapted cells. Error bars = SEM, n = 6. *Below the level of detection.
Acid survival was also tested for bacteria cultured overnight at pH 5.5, where RpoS- and CRP-dependent acid resistance systems are expressed [16]. Cultures grown at external pH 5.5 showed a 13-fold decrease in survival of tolC compared to W3110 (Fig. 1B). Thus, the TolC requirement was much greater for cells grown at pH 7 than for cells grown at pH 5.5. Complementation of tolC with plasmid pMX, which produces a functional TolC, grown at pH 5.5 and challenged at pH 2 restored the strain's acid survival comparable to that of the wild-type (data not shown). Strains defective for mdtB and emrB showed only a 6-fold and 2-fold decrease in survival under these conditions, respectively.
In extreme base (pH 10), the tolC strain (cultured aerobically to stationary phase at pH 8) showed comparable survival to the wild-type (data not shown). Thus, the pH sensitivity of tolC mutants was limited to acidic pH.
TolC is required for expression of the glutamate-dependent acid resistance system
A major contribution to acid resistance can result from the glutamate decarboxylase (Gad) system encoded by gadA and gadBC. The gadA and gadB genes encode isoforms of glutamate decarboxylase and gadC encodes the glutamate/γ-aminobutyric acid antiporter [16]. Survival of strains MG1655 (wild-type) and MG1655T (tolC::Tn10) grown in LBK at pH 5.5 (100 mM MES) overnight and exposed to low external pH (pH 2.5) was tested in M9-glucose medium supplemented with 1.5 mM L-glutamic acid. After 30 min, survival of the tolC strain was decreased 10-fold relative to the wild type; and after 60 min, survival of the tolC strain dropped to nearly 20-fold below the wild-type (data not shown). This is comparable to the acid-survival seen in complex LB medium (Fig. 1B) and suggests that the tolC strain is unable to utilize the glutamate-dependent acid resistance system, which is expressed during the overnight growth before exposure to pH 2.
Cultures of MG1655 and MG1655T (tolC::Tn10) were also assayed for activity and expression of the Gad system. Glutamate decarboxylase activity was assessed using the pH indicator dye bromocresol green; the dye changes from yellow to blue upon pH increase in the reaction mixture, following decarboxylation of L-glutamate (Fig. 2A). The wild-type strain behaved as expected with no decarboxylation at pH 7.5 and very clear evidence of glutamate decarboxylation at pH 5.5. The tolC strain, however, showed almost no Gad activity at pH 5.5. The gadA mRNA transcription was observed in wild-type but not in tolC cultures at pH 5.5, whereas the mRNA transcript of the lysine-dependent acid resistance system (cadA) was present in both wild-type and tolC strains (Fig. 2B). In the tolC strain at pH 5.5, the cadA mRNA transcript showed decreased expression compared to the wild-type, which may result from decreased regulation by GadE [21]. Both GadA and GadB proteins were absent in the tolC strain at pH 5.5 (Fig. 2C). Without TolC, no gadA or gadB expression could be detected.
Figure 2. TolC is required for expression of the glutamate-dependent acid resistance system.
E. coli wild-type MG1655 and its TolC-deficient derivative MG1655T (tolC::Tn10) were assayed for glutamate decarboxylase activity, gadA mRNA expression, and GadA/B expression. A) Both strains were tested for glutamate decarboxylase activity at pH 7.5 and pH 5.5 using the GAD reagent as described in Materials and Methods. The pH indicator dye bromocresol green changes from yellow to blue when L-glutamic acid is decarboxylated. There was almost no change in color with the tolC::Tn10 strain at pH 5.5. B) Northern analysis of gadA and cadA mRNA at pH 7.5 and pH 5.5 with the wild-type and tolC::Tn10 strains; 16S rRNA bands are provided as a control. C) A 53 kDa protein band (indicated by an arrow) was observed by SDS-PAGE only in wild-type cultures at pH 5.5; this band was identified as a mixture of GadA and GadB proteins by mass spectrometry analysis (MALDI-TOF).
Gad expression was restored by the pMX plasmid carrying the wild-type tolC gene (data not shown). Thus, TolC is required for expression of gadA mRNA and GadA and GadB proteins, as well as for activity of glutamate decarboxylases. Furthermore, in a tolC defective strain, plasmids expressing either GadB-C (pMF565) or the positive regulator GadE (pQEgadE) each restored extreme-acid survival at pH 2. This complementation confirms the role for Gad in the TolC requirement for extreme-acid survival.
Extreme-acid survival in a multiple-MDR efflux pump mutant
TolC acts as the outer-membrane conduit for export by several inner-membrane efflux pump complexes [5]. We investigated whether loss of several TolC-dependent pump complexes would affect acid resistance in a manner comparable to loss of TolC. Extreme-acid survival was tested for strain M6293, which is defective for the inner-membrane pumps of eight known TolC-dependent MDR efflux complexes (AcrAB, AcrAD, AcrEF, EmrAB, EmrKY, MacAB, MdtABC, MdtEF). Acid survival of the multi-pump defective strain was compared to its parent strain N7829, and to strains deleted for tolC grown to stationary phase at pH 5.5 (Fig. 3). The parent strain N7829 survived the acid challenge as well as other wild-type E. coli K-12 strains. M6293, the strain lacking the TolC-associated efflux pumps, including EmrB and MdtC, showed a 6-fold decrease in survival versus the parent strain N7829. This result is comparable to the survival percentages seen with strains lacking MdtB (6-fold) or EmrB (2-fold) in Fig. 1B. When tolC was also disrupted in these two strains, survival was decreased to below 1%. A strain with defects in both the EmrAB and MdtABC complexes (W3110 emrB::frt mdtB::kan) showed 4- to 10-fold decrease in survival (data not shown).
Figure 3. TolC is required for extreme-acid survival in a strain defective for eight MDR efflux inner membrane pumps.
Strains N7829 (K-12 derivative), M5567 (N7829 tolC::kan), M6293 (N7829 acrB::frt acrD::frt emrB::frt emrY::frt macB::frt mdtC::frt mdtF::frt acrEF::spc), and JLS1042 (M6293 tolC::kan) were grown overnight to stationary phase in LBK buffered at pH 5.5 (100 mM MES), diluted into LBK pH 2, and exposed for 2 hours at 37°C. Grey bars represent the parent strain listed with the additional kanamycin resistance insertion in tolC. Error bars = SEM, n = 6.
pH-dependent growth of tolC defective strains
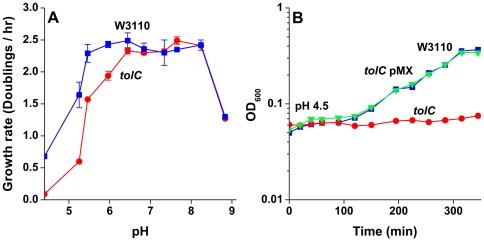
While numerous genes are known to affect survival at pH 2, relatively few affect exponential growth at moderately low pH. The best studied case is the triple potassium transport deletion which results in K+-dependent growth at low pH [22]. To determine the role of TolC in acidic growth, we assessed the ability of a strain defective for tolC to grow in LBK with an external pH range of 4.5–9.0 (Fig. 4A). At pH 4.5, the growth rate of tolC was near zero. Over the range of pH 4.5–6.0, the tolC strain grew at a slower rate than the parent. Over the range of external pH 6.5–9.0, there was no significant difference in growth; thus, the effect of the deletion of tolC on growth rate was limited to the range of acid stress. Wild-type growth at pH 4.5 was restored by complementation of the tolC strain with the tolC-carrying plasmid pMX (Fig. 4B). The tolC defect had no effect on cytoplasmic pH when cultures were suspended at pH 4.5 to pH 6.0 (data not shown), using GFPmut3b fluorimetry as described previously [22], [23].
Figure 4. TolC is required for acid growth.
A) Strains were grown from pH 4.5–pH 9.0 using the appropriate buffer in half unit increments at 37°C. W3110 (blue) and tolC::kan (red) growth rates calculated in early log-phase (OD600 0.1 to 0.3) are depicted as a function of pH. B) Successful complementation of the tolC strain with pMX, a low-copy plasmid that carries the functional tolC gene (green), in LBK at pH 4.5 (100 mM HOMOPIPES) restored the acid growth capability to that of the wild type W3110 (blue). Cultures were maintained as described in the Materials and Methods. Error bars = SEM (n = 3) and absent when smaller than the symbol.
The pH-dependent growth of the tolC strain could be caused by a defect in the channel connection to one or more of its associated inner-membrane efflux complexes [24]. Growth at pH 5 was tested for a mutant deleted for the known TolC-dependent efflux porters (N7829 acrB::frt acrD::frt emrB::frt emrY::frt macB::frt mdtC::frt mdtF::frt acrEF::spc). No difference was seen between the growth of mutant and parent (data not shown).
Given that a tolC strain does not appear to express the glutamate-dependent acid resistance system, we assessed the contribution of the Gad system to the pH-sensitive growth of a tolC mutant. The antiporter GadC imports glutamate in exchange for the decarboxylation product [16]. When a gadC tolC double mutant (JLS1048) was grown in early log-phase at pH 5, growth rates similar to the tolC strain were observed, while the wild-type W3110 and a strain lacking gadC had comparable growth rates that were higher than the tolC strains (data not shown). No growth defect was seen in a gadC strain at pH 5.0; this finding is consistent with previous studies showing that the Gad regulon is needed only for extreme acid survival, not for growth in moderate acid [16]. Furthermore, growth rates in tolC cultures grown at pH 5 were unaffected by expression of GadE or GadB-C produced by plasmids pQEgadE and pMF565, respectively (data not shown), indicating that even over-expression of Gad system components on a low-copy plasmid could not restore wild-type growth rates. Thus, the poor growth at moderately low pH in tolC strains must involve a mechanism other than TolC-mediated induction of the glutamate-dependent acid resistance system.
Discussion
TolC is a component of several MDR efflux complexes that enable E. coli to expel both toxins and metabolic wastes across the periplasm and outer membrane, driven by H+ antiport [5]. Expression of TolC is regulated by MarA, SoxS and Rob [20] and by the EvgA acid resistance regulon [14], which suggests that TolC may function in acid adaptation. Consistent with acid adaptation, TolC-associated drug efflux of toxins is more active at low pH [25]. Our finding that TolC contributes to acid resistance is the first report of an MDR pump component that enhances acid adaptation. For comparison, at high pH over-expression of MDR pump MdfA confers alkali-tolerance and extends the upper pH range for growth [19]. Our results suggest a general possibility that when antibiotics select for gain of MDR pumps by enteric bacteria, the bacteria may show increased resistance to stomach acid.
Additionally, two TolC-associated inner-membrane MDR efflux pump components, EmrB and MdtB, contributed to extreme-acid survival. MdtABC comprises RND-family efflux pumps and a membrane fusion protein which, along with tolC, is under the regulation of the BaeR regulon [26]–[28]. The BaeSR two-component regulatory system is an envelope stress signaling pathway that responds to extracytoplasmic stress, which may include acidic pH [27], [29]. EmrB is a major facilitator superfamily MDR efflux pump that is induced by permeant weak acids, such as salicylate [30]. Other TolC-associated inner membrane pumps that were tested for extreme-acid survival, such as AcrB, EmrY, and MdtF, showed comparable survival to that of the wild-type.
Of all MDR efflux pump components tested, TolC contributes the most to extreme-acid survival at pH 2, even in a background strain that lacks eight TolC-dependent inner membrane pumps (Fig. 3). While the multiple MDR efflux pump mutant pre-grown at pH 5.5 showed 6-fold decreased survival at pH 2 compared to its wild-type, a tolC deletion in either the parent strain or the MDR mutant showed a much larger decrease (45- and 12-fold, respectively; Fig. 3). Thus, either TolC itself plays a major role in extreme-acid survival independent of its associated inner-membrane pumps, or else an unidentified TolC-dependent pump is involved.
The mechanism of the TolC effect in acid survival was shown to include regulation of the Gad system. The decarboxylation of glutamate by GadA and GadB is one of the main pH homeostasis mechanisms active at low external pH and in stationary phase; cells lacking this system are unable to maintain cytoplasmic pH and perform poorly when challenged in acidic media [16]. In a strain lacking TolC grown at pH 5.5, we identified almost no glutamate decarboxylase activity (Fig. 2A), observed no gadA mRNA among total RNA isolated (Fig. 2B), and detected no GadA and GadB protein expression (Fig. 2C). Thus, TolC is required for induction of the glutamate-dependent acid-resistance system. The restoration of pH 2 survival by GadB-C or by GadE provided on a plasmid confirms that the TolC requirement involved Gad regulation. This may explain why the TolC requirement for acid survival was greatest for cells grown at pH 7 (Fig. 1A) where the acid-induced AR1 system thatinvolves CRP is unavailable, and thus Gad offers the main mechanism of acid resistance [16].
On the other hand, the requirement for TolC for exponential growth in moderate acid (pH 4.5–6.0) was shown to be independent of Gad. Deletion of gadC significantly reduces extreme-acid survival (pH 2.5) [31], [32], verifying that a gadC deletion inactivates Gad activity. Nevertheless, a gadC deletion strain did not exhibit a growth defect during exponential growth in moderate acid and expression of the positive regulator GadE or GadB-C from plasmids did not restore wild-type growth rates in a tolC mutant strain (LBK, pH 5.0; data not shown). Thus, while the role of TolC in Gad regulation may be the reason TolC is required for extreme-acid survival, the decreased growth rates of the tolC strain only in acidic conditions are not the result of a lack of Gad activity. TolC was needed for maximal growth below pH 6.5, where cytoplasmic pH is less than optimal (pH 7.4–7.8) (Fig. 4).
The growth defect and the acid resistance defect of the tolC strain were both complemented with a complete tolC gene on a low copy plasmid. The complementing plasmid pMX does not carry the ygiABC genes downstream from tolC that may be in the tolC operon [33]. Thus, the low pH growth effect is not due to YgiABC activity. Complementation confirms that tolC, and not adjacent genes, contributes to acid resistance.
Our findings suggest a novel physiological role for TolC in pH homeostasis in acidic conditions. Previous reports demonstrate no growth defects in LB medium, but find impaired cell division and growth in minimal glucose medium [33]. The requirement of TolC for growth at low pH is surprising because TolC resides in the outer membrane, mediating exchange of the external medium with the periplasm; and the periplasmic pH generally equals the external pH [23]. Thus it is hard to see why cytoplasmic pH homeostasis would require an outer-membrane channel. A possibility is that products excreted during metabolism at low external pH accumulate in the periplasm, if they cannot be removed without the TolC channel.
The mechanism of TolC may or may not involve its interactions with the inner membrane efflux pumps [24]. The fact that deletion of eight major inner-membrane efflux pumps has no effect on growth and a relatively modest effect on extreme-acid survival of pH 5.5-grown cultures (Fig. 3) suggests that the significant reduction of extreme-acid survival in tolC deletion strains is independent of the channel's association with these pumps. The proton motive force from the periplasm to the cytoplasm drives the functioning of many multidrug efflux transporters [34]. In addition to functioning as an outer membrane pore for many MDR pumps, TolC may also play a physiological role in pH homeostasis through an interaction with the proton motive force that drives efflux. The original function of TolC may have been to provide the cell with a pH homeostasis mechanism in acidic conditions that later was co-opted to function as a common outer membrane porin in multidrug resistance.
As we completed our manuscript, we became aware of an unpublished plate screen showing that E. coli colony growth at low pH requires several envelope and inner membrane components besides TolC, such as TolB and TolR; the report has since been published [35]. We have since confirmed with quantitative growth curves and survival assays the low-pH specific growth requirements for TolB and TolR (G. Garduque and J. Slonczewski, unpublished). It will be of interest to determine how all these envelope components relate to pH homeostasis.
Materials and Methods
Bacterial strains, media, and growth conditions
The E. coli K-12 strains used here are described in Table 1. W3110 [36] was used as the wild-type strain unless indicated otherwise. Deletion strain M6293 (N7829 acrB::frt acrD::frt emrB::frt emrY::frt macB::frt mdtC::frt mdtF::frt acrEF::spc) was compared to parental strain N7829 (GC4468). Deletion alleles containing a kanamycin resistance insertion (KmR) were transduced from the Keio collection [37], obtained from the Coli Genetic Stock Center (Yale University), into the wild-type strain by P1 phage transduction. “frt” is the designation for the “scar” sequence remaining at the site of the cured Keio kan insertion. Deletion strains were maintained with 50 µg/ml kanamycin. Plasmid pMX carrying the wild-type tolC gene on a low-copy-number vector pMW119 (derived from pSC101) was transformed into JLS1015 (W3110 tolC::kan) for complementation experiments [4]. Strains containing plasmid pMX were maintained with 50 µg/ml ampicillin in overnight cultures and 20 µg/ml ampicillin in growth cultures.
Table 1. E. coli K-12 strains and plasmids used in this study.
| Strain or plasmid | Genotype | Source |
| W3110 | K-12 (F− λ−) | [36] |
| JLS9318 | MC4100 gadC::Tn10 | [31] |
| JLS1015 | W3110 tolC732::kan (Keio JW5503) | This work |
| JLS1023 | W3110 acrB747::kan (Keio JW0451) | This work |
| JLS1024 | W3110 mdtB774::kan (Keio JW2060) | This work |
| JLS1025 | W3110 mdtF769::kan (Keio JW3482) | This work |
| JLS1026 | W3110 emrY776::kan (Keio JW2364) | This work |
| JLS1027 | W3110 emrB767::kan (Keio JW2661) | This work |
| JLS1036 | W3110 tolC732::kan pMX | This work |
| JLS1039 | W3110 marR751::kan (Keio JW5248) | This work |
| JLS1048 | W3110 tolC732::kan gadC::Tn10 | This work |
| JLS1050 | W3110 emrB767::frt mdtB774::kan | This work |
| MG1655 | K-12 | M. Wachi |
| MG1655T | MG1655 tolC::Tn10 | This work |
| N7829 | K-12; GC4458 | J.L. Rosner |
| M5567 | N7829 tolC732::kan | J.L. Rosner |
| M6293 | N7829 acrB747::frt acrD790::frt emrB767::frt emrY776::frt macB780::frt mdtC775::frt mdtF769::frt acrEF::spc | J.L. Rosner |
| JLS1042 | M6293 tolC732::kan | This work |
| Plasmids | ||
| pMX | pMW119 carrying the tolC gene | [41] |
| pQEgadE | pQE80L carrying the gadE gene | [44] |
| pMF565 | pQE80L [gadBC]his-gadB ApR | J.W. Foster |
Bacteria were cultured in LBK medium (10 g/l tryptone, 5 g/l yeast extract, and 7.45 g/l KCl) supplemented with pH buffers as needed [38]. Overnight cultures of deletion strains were maintained with kanamycin (50 µg/ml). Media were buffered with 100 mM Homopiperazine-N, N′-bis-2-(ethanesulfonic acid) (HOMOPIPES; pKa = 4.55), 2-(N-morpholino) ethanesulfonic acid (MES; pKa = 5.96), 1,4-Piperazinebis(ethanesulfonic acid) (PIPES; pKa = 6.66), 3-(N-morpholino)propanesulfonic acid (MOPS; pKa = 7.01), N-Tris(hydroxymethyl)methyl-3-aminopropanesulfonic acid (TAPS; pKa = 8.11), 3-[(1,1-Dimethyl-2-hydroxyethyl)amino]-2-hydroxypropanesulfonic acid (AMPSO; pKa = 9.10), or 3-(Cyclohexylamino)-1-propanesulfonic acid (CAPS; pKa = 10.08). At the end of the experiments, the pH of the cultures was checked to ensure that it was within 0.2 pH units of the original uninoculated medium.
Acid and base resistance assays
The conditions for testing acid resistance (survival in extreme acid) of aerated cultures were based on those previously described, with modifications [39], [40]. Cells were cultured with rotary aeration overnight (16–18 hr at 37°C) to stationary phase in LBK pH 5.5 (100 mM MES) or LBK pH 7 (100 mM MOPS). Overnight cultures were diluted 200-fold into LBK pH 2 and incubated with rotation at 37°C. Following a 2 hr exposure, cultures were serially diluted and plated on LBK-agar. Overnight cultures were also diluted 200-fold into LBK 100 mM MOPS, pH 7 and immediately serially diluted and plated onto LBK-agar. Plates were incubated overnight at 30°C.
Percent survival was calculated as follows: since acid survival represents an exponential death curve, colony counts of surviving cells and control plates were log10-transformed to provide a normal distribution of the data. The mean of the unexposed controls was then subtracted from the mean of exposed pH 2 colony counts, resulting in a log10 ratio that correlates to percent survival. All errors stated are the standard error of the mean (SEM). Each experimental condition consisted of six biological replicates from the same overnight culture. Each entire experiment was conducted at least twice.
For base resistance (survival in extreme base), bacteria were cultured with aeration in LBK pH 8.0 (100 mM TAPS) and diluted into LBK pH 10 (100 mM CAPS). Survival was measured and calculated as for acid resistance.
Glutamic acid decarboxylase assays
E. coli K-12 derivative strains MG1655 and MG1655T (tolC::Tn10), transduced by P1-phage from JA300T [41], were used in the assessment of glutamic acid decarboxylase activity. Glutamic acid decarboxylase activity was assessed using the GAD reagent (1 g/l L-glutamic acid, 0.05 g/l bromocresol green, 90 g/l NaCl, 3 ml/l Triton X-100) with minor modifications [42]. MG1655 and MG1655T cultures were grown for 1 hr in LB (pH 7.5±0.2) or LB buffered with 100 mM MES (pH 5.5). Cells were harvested, washed with saline (0.85% NaCl), and suspended in the same solution. An aliquot of cell suspension (9×108 cells) was transferred to a new tube, and 1 ml of the GAD reagent was added. The reaction mixtures were incubated for 1 hr at 35°C and then evaluated for decarboxylase activity by a color change from yellow to blue.
The presence of gadA and cadA mRNA in both MG1655 and MG1655T at pH 7.5 and pH 5.5 was assessed using Northern analysis. Total cellular RNA was isolated using the RNeasy kit (Qiagen) and separated by formaldehyde-agarose gel electrophoresis. Hybridization was done with the DIG luminescent detection kit (Roche Dignostics).
To assess the presence of GadA and GadB proteins, cultures of MG1655 and MG1655T were grown in LB medium at pH 5.5 (100 mM MES) and pH 7.5. Cells were harvested, suspended in a 50 mM sodium phosphate buffer (pH 7.0), and disrupted by sonication. After unbroken cells were removed, lysate proteins were separated by SDS-polyacrylamide gel electrophoresis (10% acrylamide) and stained with Coomassie Brilliant Blue. The 53-kDa protein band was cut out and analyzed by mass spectrometry (MALDI-TOF/TOF ultrafleXtreme, Bruker Daltonics).
Glutamate-dependent extreme-acid resistance was tested with overnight cultures grown in LB buffered with 100 mM MES pH 5.5, then diluted into warmed M9 medium (6.8 g/l Na2HPO4, 3.0 g/l KH2PO4, 0.5 g/l NaCl, 1.0 g/l NH4Cl, 2 mM MgSO4, 0.1 mM CaCl2, and 0.4% glucose) supplemented with 1.5 mM L-glutamic acid and adjusted to pH 2.5. Surviving cells were counted after 30 and 60 min of acid challenge as previously described [43].
Acid growth assays
To test acid growth, cells were cultured with aeration to stationary phase (16–18 hr, 37°C) in unbuffered LBK. Overnight cultures were diluted 100-fold into LBK pH 4.5–9.0 (in half unit increments) including 100 mM of the pH-appropriate buffer, and rotated at 37°C until cultures reached stationary phase. OD600 was measured at regular intervals after the initial dilution. Growth rates were calculated as doublings per hour over the region of exponential growth (approximately OD600 = 0.1 to 0.3). The wild-type strain W3110 and its tolC derivative (JLS1015) were also tested for loss of cytoplasmic pH homeostasis at low pH as described previously [22], [23].
Acknowledgments
We thank J. W. Foster for generously providing plasmids pMF565 and pQEgadE.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grant MCB-1050080 to JLS from the National Science Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Piddock LJV. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev. 2006;19:382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Piddock LJV, Garvey MI, Rahman MM, Gibbons S. Natural and synthetic compounds such as trimethoprim behave as inhibitors of efflux in Gram-negative bacteria. J Antimicrob Chemother. 2010;65:1215–1223. doi: 10.1093/jac/dkq079. [DOI] [PubMed] [Google Scholar]
- 3.Rosner JL, Martin RG. An excretory function for the Escherichia coli outer membrane pore TolC: upregulation of marA and soxS transcription and Rob activity due to metabolites accumulated in tolC mutants. J Bacteriol. 2009;191:5283–5292. doi: 10.1128/JB.00507-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tatsumi R, Wachi M. TolC-dependent exclusion of porphyrins in Escherichia coli. J Bacteriol. 2008;190:6228–6233. doi: 10.1128/JB.00595-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koronakis V, Eswaran J, Hughes C. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu Rev Biochem. 2004;73:467–489. doi: 10.1146/annurev.biochem.73.011303.074104. [DOI] [PubMed] [Google Scholar]
- 6.Piddock LJV. Multidrug-resistance efflux pumps - not just for resistance. Nat Rev Microbiol. 2006;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 7.Buckley AM, Webber MA, Cooles S, Randall LP, La Ragione RM, et al. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell Microbiol. 2006;8:847–856. doi: 10.1111/j.1462-5822.2005.00671.x. [DOI] [PubMed] [Google Scholar]
- 8.Ferhat M, Atlan D, Vianney A, Lazzaroni J-C, Doublet P, et al. The TolC protein of Legionella pneumophila plays a major role in multi-drug resistance and the early steps of host invasion. PLoS ONE. 2009;4:e7732. doi: 10.1371/journal.pone.0007732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gil H, Platz GJ, Forestal CA, Monfett M, Bakshi CS, et al. Deletion of TolC orthologs in Francisella tularensis identifies roles in multidrug resistance and virulence. Proc Natl Acad Sci USA. 2006;103:12897–12902. doi: 10.1073/pnas.0602582103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reddy JD, Reddy SL, Hopkins DL, Gabriel DW. TolC is required for pathogenicity of Xylella fastidiosa in Vitis vinifera grapevines. Mol Plant Microbe Interact. 2007;20:403–410. doi: 10.1094/MPMI-20-4-0403. [DOI] [PubMed] [Google Scholar]
- 11.Bleuel C, Große C, Taudte N, Scherer J, Wesenberg D, et al. TolC is involved in enterobactin efflux across the outer membrane of Escherichia coli. J Bacteriol. 2005;187:6701–6707. doi: 10.1128/JB.187.19.6701-6707.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiriyathanawudhiwong N, Ohtsu I, Li Z-D, Mori H, Takagi H. The outer membrane TolC is involved in cysteine tolerance and overproduction in Escherichia coli. Appl Microbiol Biotechnol. 2009;81:903–913. doi: 10.1007/s00253-008-1686-9. [DOI] [PubMed] [Google Scholar]
- 13.Yohannes E, Barnhart M, Slonczewski JL. pH-dependent catabolic protein expression during anaerobic growth of Escherichia coli K-12. J Bacteriol. 2004;186:192–199. doi: 10.1128/JB.186.1.192-199.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masuda N, Church GM. Escherichia coli gene expression responsive to levels of the response regulator EvgA. J Bacteriol. 2002;184:6225–6234. doi: 10.1128/JB.184.22.6225-6234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eguchi Y, Oshima T, Mori H, Aono R, Yamamoto K, et al. Transcriptional regulation of drug efflux genes by EvgAS, a two-component system in Escherichia coli. Microbiology. 2003;149:2819–2828. doi: 10.1099/mic.0.26460-0. [DOI] [PubMed] [Google Scholar]
- 16.Foster JW. Escherichia coli acid resistance: tales of an amateur acidophile. Nat Rev Microbiol. 2004;2:898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- 17.Slonczewski JL, Fujisawa M, Dopson M, Krulwich TA. Cytoplasmic pH measurement and homeostasis in bacteria and archaea. Adv Microb Physiol. 2009;55:1–79. doi: 10.1016/S0065-2911(09)05501-5. [DOI] [PubMed] [Google Scholar]
- 18.Tikhonova EB, Dastidar V, Rybenkov VV, Zgurskaya HI. Kinetic control of TolC recruitment by multidrug efflux complexes. Proc Natl Acad Sci USA. 2009;106:16416–16421. doi: 10.1073/pnas.0906601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewinson O, Padan E, Bibi E. Alkalitolerance: a biological function for a multidrug transporter in pH homeostasis. Proc Natl Acad Sci USA. 2004;101:14073–14078. doi: 10.1073/pnas.0405375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang A, Rosner JL, Martin RG. Transcriptional activation by MarA, SoxS and Rob of two tolC promoters using one binding site: a complex promoter configuration for tolC in Escherichia coli. Mol Microbiol. 2008;69:1450–1455. doi: 10.1111/j.1365-2958.2008.06371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krin E, Danchin A, Soutourina O. RcsB plays a central role in H-NS-dependent regulation of motility and acid stress resistance in Escherichia coli. Res Microbiol. 2010;161:363–371. doi: 10.1016/j.resmic.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Kitko RD, Wilks JC, Garduque GM, Slonczewski JL. Osmolytes contribute to pH homeostasis of Escherichia coli. PLoS ONE. 2010;5:e10078. doi: 10.1371/journal.pone.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilks JC, Slonczewski JL. pH of the cytoplasm and periplasm of Escherichia coli: rapid measurement by green fluorescent protein fluorimetry. J Bacteriol. 2007;189:5601–5607. doi: 10.1128/JB.00615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tal N, Schuldiner S. A coordinated network of transporters with overlapping specificities provides a robust survival strategy. Proc Natl Acad Sci USA. 2009;106:9051–9056. doi: 10.1073/pnas.0902400106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martins A, Spengler G, Rodrigues L, Viveiros M, Ramos J, et al. pH modulation of efflux pump activity of multi-drug resistant Escherichia coli: protection during its passage and eventual colonization of the colon. PLoS ONE. 2009;4:e6656. doi: 10.1371/journal.pone.0006656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagakubo S, Nishino K, Hirata T, Yamaguchi A. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J Bacteriol. 2002;184:4161–4167. doi: 10.1128/JB.184.15.4161-4167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bury-Moné S, Nomane Y, Reymond N, Barbet R, Jacquet E, et al. Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet. 2009;5:e1000651. doi: 10.1371/journal.pgen.1000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishino K, Honda T, Yamaguchi A. Genome-wide analyses of Escherichia coli gene expression responsive to the BaeSR two-component regulatory system. J Bacteriol. 2005;187:1763–1772. doi: 10.1128/JB.187.5.1763-1772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raffa RG, Raivio TL. A third envelope stress signal transduction pathway in Escherichia coli. Mol Microbiol. 2002;45:1599–1611. doi: 10.1046/j.1365-2958.2002.03112.x. [DOI] [PubMed] [Google Scholar]
- 30.Lomovskaya O, Lewis K, Matin A. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J Bacteriol. 1995;177:2328–2334. doi: 10.1128/jb.177.9.2328-2334.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hersh BM, Farooq FT, Barstad DN, Blankenhorn D, Slonczewski JL. A glutamate-dependent acid resistance gene in Escherichia coli. J Bacteriol. 1996;178:3978–3981. doi: 10.1128/jb.178.13.3978-3981.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castanié-Cornet M-P, Penfound TA, Smith D, Elliott JF, Foster JW. Control of acid resistance in Escherichia coli. J Bacteriol. 1999;181:3525–3535. doi: 10.1128/jb.181.11.3525-3535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dhamdhere G, Zgurskaya HI. Metabolic shutdown in Escherichia coli cells lacking the outer membrane channel TolC. Mol Microbiol. 2010;77:743–754. doi: 10.1111/j.1365-2958.2010.07245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharff A, Fanutti C, Shi J, Calladine C, Luisi B. The role of the TolC family in protein transport and multidrug efflux. Eur J Biochem. 2001;268:5011–5026. doi: 10.1046/j.0014-2956.2001.02442.x. [DOI] [PubMed] [Google Scholar]
- 35.Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, et al. Phenotypic landscape of a bacterial cell. Cell. 2011;144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith MW, Neidhardt FC. Proteins induced by anaerobiosis in Escherichia coli. J Bacteriol. 1983;154:336–343. doi: 10.1128/jb.154.1.336-343.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maurer LM, Yohannes E, BonDurant SS, Radmacher M, Slonczewski JL. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J Bacteriol. 2005;187:304–319. doi: 10.1128/JB.187.1.304-319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorden J, Small PLC. Acid resistance in enteric bacteria. Infect Immun. 1993;61:364–367. doi: 10.1128/iai.61.1.364-367.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noguchi K, Riggins DP, Eldahan KC, Kitko RD, Slonczewski JL. Hydrogenase-3 contributes to anaerobic acid resistance of Escherichia coli. PLoS ONE. 2010;5:e10132. doi: 10.1371/journal.pone.0010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aono R, Tsukagoshi N, Yamamoto M. Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance of Escherichia coli K-12. J Bacteriol. 1998;180:938–944. doi: 10.1128/jb.180.4.938-944.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rice EW, Johnson CH, Dunnigan ME, Reasoner DJ. Rapid glutamate decarboxylase assay for detection of Escherichia coli. Appl Environ Microbiol. 1993;59 doi: 10.1128/aem.59.12.4347-4349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin J, Lee IS, Frey J, Slonczewski JL, Foster JW. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J Bacteriol. 1995;177:4097–4104. doi: 10.1128/jb.177.14.4097-4104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masuda N, Church GM. Regulatory network of acid resistance genes in Escherichia coli. Mol Microbiol. 2003;48:699–712. doi: 10.1046/j.1365-2958.2003.03477.x. [DOI] [PubMed] [Google Scholar]