Abstract
The Glutaminase Interacting Protein (GIP) is composed of a single PDZ domain that interacts with a growing list of partner proteins, including Glutaminase L, that are involved in a number of cell signaling and cancer pathways. Therefore, GIP makes a good target for structure-based drug design. Here we report the solution structures of both free GIP and GIP bound to the C-terminal peptide analog of Glutaminase L. This is the first reported NMR structure of GIP in a complex with one of its binding partners. Our analysis of both free GIP and GIP complexed with the Glutaminase L peptide provides important insights into how a promiscuous binding domain can have affinity for multiple binding partners. Through a detailed chemical shift perturbation analysis and backbone dynamics studies, we demonstrate here that the binding of the Glutaminase L peptide to GIP is an allosteric event. Taken together, the insights reported here lay the groundwork for the future development of a specific inhibitor for GIP.
Protein interaction networks are essential to maintain order and direct the flow of traffic within cells. These networks are mediated by proteins composed of one or more protein-protein interaction domains such as SH2 (1), SH3 (2), PH (3), PDZ (4) and others (5). PDZ domains are named for the three founding members: Post Synaptic Density 95 (PSD-95), Discs Large (Dlg) and Zonula Occludentes (ZO-1) (6). PDZ domains are small protein-protein interaction motifs that contain 80-100 amino acid residues. They form a compact domain primarily composed of 1-2 α-helices and 5-6 β-strands, and can be found in animal species, with homologous domains also observed in yeast and plants (7, 8). Proteins containing PDZ domains are often involved in signaling pathways and act as scaffolds in the organization of multimeric complexes, frequently in tandem with other protein-protein interaction modules (5). As specialized protein interaction motifs, PDZ domains are best known for binding the unstructured C-terminal tails of their binding partners. However, rarely PDZ domains can bind to internal motifs that structurally mimic a C-terminus (9, 10). Based on the sequence specificity of the interacting proteins, PDZ domains are broken down into a number of different classes: class I (X-S/T-X-Φ-COOH), class II (X-Φ-X-Φ-COOH) (5), class III (X-E/D-X-Φ-COOH) (11) and various other minor classes (12) where Φ is any hydrophobic residue and X is any residue.
One such PDZ domain containing protein that intersects a number of important biological pathways is known as either Glutaminase Interacting Protein GIP (13) or Tax Interacting Protein TIP-1 (14) based on the bait proteins used to find interacting partners. Human GIP is a 124-residue protein that is highly unusual among PDZ domain containing proteins in that it is comprised almost exclusively of a single PDZ domain rather than one of many domains as part of a larger protein (5). To date GIP has been shown to interact with a number of different proteins including: Glutaminase L (13), β-Catenin (15, 16), FAS (17, 18), HTLV Tax (14), HPV E6 (19), Rhotekin (20) and Kir 2.3 (21, 22). β-Catenin and Rhotekin are important in the Wnt and Rho signaling pathways, respectively. FAS is a member of the Tumor Necrosis Factor (TNF) family of receptors, while HTLV Tax and HPV E6 are both viral proteins from oncogenic viruses. Finally, the inward rectified potassium channel Kir 2.3 is known to be regulated by GIP in renal epithelial cells. These proteins all contain the PDZ class I (X-S/T-X-I/L/V-COOH) binding motif. Thus, in addition to its regulation of Glutaminase L, which has been shown to be up-regulated in various cancers (23-25), GIP has been shown to be involved in a variety of different cancer and cell signaling pathways with its numerous binding-partner proteins. Sequence alignment of all of these currently known binding-partners reveals the optimal consensus sequence for GIP binding to be E-S-X-V-COOH (Table 1).
Table 1.
Sequential Alignment of C-terminal Binding Partners of GIP
| Binding Partner | Position | |||||||
|---|---|---|---|---|---|---|---|---|
| −7 | −6 | −5 | −4 | −3 | −2 | −1 | 0 | |
| Glutaminase L | K | E | N | L | E | S | M | V |
| Kir 2.3 | S | Y | R | R | E | S | A | I |
| HTLV Tax | K | H | F | R | E | T | E | V |
| HPV E6 | R | Q | A | T | E | S | T | V |
| Rhotekin | R | T | W | L | Q | S | P | V |
| β-Catenin | L | A | W | F | D | T | D | L |
| FAS | R | N | E | I | Q | S | L | V |
| Consensus | X | X | X | X | E | S | X | V |
The importance of GIP as a scaffolding protein in the mammalian brain has been implicated by the demonstration of the presence of both GIP and Glutaminase L in astrocytes and neurons (26). Glutaminase, which is activated by inorganic phosphate, catalyzes an important energy generation reaction in mammalian tissues using glutamine as a substrate to produce glutamate and ammonia (27). Glutaminase is also involved in synaptic transmission, hepatic ureagenesis, renal ammoniagenesis and regulation of cerebral concentrations of glutamine and glutamate (13, 28). There are two isoforms of the Glutaminase enzyme; one is the kidney-type (K) isozyme, which is encoded by a gene located in chromosome 2 and the other is the liver-type (L) isozyme, whose coding gene locus is in chromosome 12 (29). Immunostaining demonstrated that Glutaminase L localizes to neuronal nuclei and Glutaminase K to mitochondria, suggesting a role of GIP in determining the subcellular distribution of Glutaminase L as well as potential interactions with other nuclear proteins (30). In both tumor cells and normally dividing cells, glutamine catabolism has been shown to be a key pathway in the process of bioenergetics (31-33). The C-terminus of Glutaminase L has been reported to interact with several PDZ domain-containing proteins such as alpha-1-syntrophin (SNT) and GIP. The C-terminal end of Glutaminase L contains a class I binding motif (ESMV-COOH) whereas its K-counterpart lacks this motif, allowing the two isoforms to be differentially regulated and spatially localized, even if they are present in the same tissue (29).
Here we report the first NMR solution structure of a complex formed between GIP and the C-terminal peptide analog of Glutaminase L that has the sequence KENLESMV hereinafter referred to as the Glutaminase L peptide. We also demonstrate that ligand binding perturbs both NMR chemical shifts and backbone dynamics within GIP providing important insights into the binding mechanism. The comparative structural analysis of the free and bound states of GIP provides specific knowledge about how this protein interacts with the Glutaminase L peptide. Furthermore, our analysis also gives insight into the way GIP can interact with multiple different binding partners. Finally, it sets the groundwork for the design of a small molecule inhibitor for GIP that would have specificity to a number of class I PDZ domains due to their promiscuity for many binding partners.
Materials and Methods
Cloning, over-expression and purification of 15N, 13C-labeled GIP
Following the procedure as described previously (17), the recombinant pET-3c/GIP plasmid was transformed into Escherichia coli BL21DE3pLysS cells, and expression was performed in M9 minimal media containing 15N-labeled ammonium chloride and 13C-labeled glucose. An overnight culture was diluted 1:25, (v/v) in minimal media and grown at 37 °C to an OD600 of 0.4-0.5. Expression was then induced with 1 mM IPTG at 30° C, and after 15 h of incubation, the cells were harvested by centrifugation. The harvested cells were lysed by sonication using lysis buffer, which contains 50 mM phosphate buffer at pH 8 as well as 200 mM NaCl, 4 mM EDTA, 4% glycerol, and 1 mM PMSF. After centrifugation of the lysed cells, the supernatant was retained for further purification. 15N, 13C-labeled GIP was purified and the NMR sample was prepared following the protocol as described previously (17).
NMR Data collection
All NMR data were collected on a Bruker Avance 600 MHz spectrometer with a triple resonance 1H/13C/15N TCI cryoprobe equipped with z-axis pulsed field gradients at either the Department of Chemistry and Biochemistry, Auburn University, Auburn, AL, Bruker BioSpin Corporation, Billerica, MA, or the New York Structural Biology Center, New York, NY. The data were processed using NMRPipe (34) and analyzed using NMRView (35) or Sparky (36). For structure determination, samples between 500 μM and 1 mM of uniformly 15N/13C-labeled GIP in 50 mM phosphate buffer containing 5% D2O pH 6.5, 1 mM EDTA and 0.01% (w/v) NaN3 were prepared either with or without addition of the Glutaminase L peptide (Chi Scientific, Maynard, MA, USA) at a 1:3 protein to peptide ratio. All NMR experiments were performed at 298 K. To determine the 15N T1 values, NMR spectra were recorded with relaxation delays of 10, 600, 50, 500, 100, 400, 200, 300 and 10 ms. To determine 15N T2 values, NMR spectra were recorded with delays of 17, 153, 34, 17, 136, 51, 119, 68, 102, 85 and 34 ms. The relaxation times were randomized and some points repeated in order to avoid any systematic errors that may arise when the data are collected sequentially. The relaxation rates were calculated by least squares fitting of peak heights versus relaxation delay to a single exponential decay. Steady state 1H-15N NOE values were calculated from the ratio of peak heights in a pair of NMR spectra acquired with and without proton saturation. For backbone and side-chain assignments of both free GIP and the GIP-Glutaminase L peptide complex the following spectra were recorded at 298 K: 2D 1H,15N-HSQC (37), 3D HNCACB (38), 3D CC(CO)NH (39), 3D CBCA(CO)NH (38), 3D 15N-edited HSQC-TOCSY (40, 41) with an 80 ms mixing time, 3D HC(CO)NH (39), 3D HNHA (42), 3D HNCO (39) and 3D HN(CA)CO (43). NOE distance restraints were collected from 3D 15N-edited HSQC-NOESY (40, 41, 44) and 3D 13C-edited HSQC-NOESY (40, 41, 44) with the 13C carrier frequency in the aliphatic (44 ppm) and aromatic (125 ppm) regions and mixing times of 140 for 15N and 110 ms for 13C, respectively. For complex structure determination of GIP with the Glutaminase L peptide, selectively filtered 2D NOESY(45) with a mixing time of 100 ms, 3D 15N-filtered and 3D 13C-filtered NOESY experiments, each with mixing times of 120 ms, were performed (46). The backbone and side-chain assignments of the Glutaminase L peptide were obtained with an unlabeled peptide sample (~4mM) from the following spectra: 2D 1H,15N-HSQC, 2D 1H-13C-HMQC, homonuclear 2D TOCSY (47) and ROESY (48) each with a mixing time of 60 ms.
Chemical shift perturbation analysis
The combined chemical shift perturbation (ΔHN) is given by the equation ΔHN={(Hf−Hb)2+((Nf−Nb)/10)2}½. A scaling factor of 10 was used to normalize the differences in the 1H and 15N spectral widths. Hf, Hb, Nf and Nb are the chemical shifts of each residue's amide 1H and 15N in the free (GIP alone) and bound (GIP-Glutaminase L peptide complex) states, respectively. The combined chemical shift perturbation (ΔHC) is given by the equation ΔHC={(Hf−Hb)2+((Cf2−Cb/4)2}½. A scaling factor of 4 was used to normalize the differences in the 1H and 13C spectral widths. Hf, Hb, Cf and Cb are the chemical shifts of each residue's alpha 1H and 13C in the free (GIP alone) and bound (GIP-Glutaminase L peptide complex) states, respectively.
Analysis of dynamics data
Measured relaxation parameters R1, R2 and the steady-state 1H-15N NOE for each residue were used as inputs in the Modelfree 4.15 program developed by Palmer et al (49, 50) to analyze 15N-backbone dynamics. The τc values for both free GIP and the GIP-Glutaminase L peptide complex were calculated using the program Tensor2 for the core region A11-Q112 (51, 52). Of five different models, the best one was chosen according to the selection criteria (49) to get the order parameter (S2) that represents the degree of spatial restriction within the 1H-15N bond vector. These values range from zero for completely isotropic internal motions to unity for totally restricted motion and represent dynamics on the picosecond to nanosecond time scale.
Structure calculation and refinement
A total of 4303 and 2866 NOE cross peaks were assigned manually using Sparky (36) for free GIP and the GIP-Glutaminase L peptide complex, respectively. The assignments were corrected or confirmed with the NOEASSIGN module of CYANA 2.1 (53), using the standard protocol of eight iterative cycles of NOE assignment and structure calculation. Alternately, the CANDID module of CYANA 1.0.6 was used on the complex to initially fit the Glutaminase L peptide into the binding pocket of GIP because it allowed the intermolecular assignments to be fixed separately from the intramolecular assignments. To calculate the complex structure, 36 glycine residues were added as a flexible linker between the protein and the peptide. A total of 118 dihedral angles restrains were derived from the TALOS (54) program based on the chemical shift index (CSI) and primary sequence of GIP for both free protein and protein-peptide complex calculations. Additionally, a total of 64 and 66 hydrogen bond distance restraints (two restraints per bond) for the free protein and the protein-peptide complex, respectively, were derived from the CSI by TALOS. During the iterative NOE assignments, a total of 1134 and 490 assignments for free GIP and the GIP-Glutaminase L peptide complex were removed due to overlap, redundancy, or unresolved ambiguity that resulted from low stringency in the initial peak picking phase and high stringency in the final assignments. The final assignments averaged over 25, 18 and 12 NOEs per residue for free protein, protein in the complex, and for the peptide in the complex, respectively. Final refinement of the 100 lowest energy structures of the 200 total calculated structures was performed with the water refinement protocol implemented in ARIA (55). The 20 structures with the lowest potential energy and best Ramachandran statistics as assessed by PROCHECK (56) were selected for analysis. The structures were visualized with VMD and figures were created using Pymol (57, 58). Table 2 shows the complete structural statistics for both structures.
Table 2.
NMR Structural Statistics for the 20 Selected Lowest Energy Structures of Free GIP and the GIP-Glutaminase L Peptide Complex
| Assignments | Free GIP | GIP-Glutaminase L |
|---|---|---|
| Sequential |i−j|=1 | 871 | 718 |
| Medium 2≤|i−j|≤4 | 331 | 241 |
| Long |i−j|>4 | 622 | 360 |
| Intermolecular | 0 | 37 |
| Hydrogen Bonds a | 64 | 66 |
| Dihedral Constraints b | 118 | 118 |
| Ensemble Average c | ||
| Total energy | −3625 ± 125 | −4816 ± 175 |
| NOE energy | 1131 ± 189 | 1586 ± 302 |
| VDW energy | −937 ± 75 | −1096 ± 67 |
| Bonds energy | 85 ± 5 | 170 ± 8 |
| Dihedral energy | 657 ± 10 | 749 ± 13 |
| Angle energy | 318 ± 22 | 434 ± 26 |
| Improper energy | 963 ± 78 | 1009 ± 89 |
| Electrostatic energy | −4712 ± 67 | −6082 ± 123 |
| Ramachandran Plot d | ||
| Favorable | 68.6 | 71.2 |
| Additionally Allowed | 26.6 | 24.3 |
| Generously Allowed | 3.4 | 2.7 |
| Disallowed | 1.5 | 1.8 |
| RMSD (Å) e | ||
| Well-ordered Backbone | 0.45 | 0.67 |
| Well-ordered Sidechain | 0.92 | 1.28 |
Hydrogen bonds were defined by a set of two distance restraints per bond for residues of predicted secondary structure based on TALOS (54) predictions from CSI.
Dihedral constraints were derived from TALOS (54) predictions from CSI.
Energy terms were calculated by the water refinement module of ARIA 1.2 (55).
Ramachandran plot statistics were calculated by PROCHECK (56).
Well ordered regions included residues 11-19, 29-36 and 54-112.
Results
NMR Structure determination of free GIP and the GIP-Glutaminase L peptide complex
We have previously reported the backbone assignments of free GIP (17), which is a crucial initial rate-limiting step prior to full structural determination. Subsequently, the NMR structure of free GIP was reported by Durney et al (59). We find good agreement between both structures of free GIP. We had previously examined the binding of several peptides representing the C-termini of various proteins, including β-catenin, FAS, and Glutaminase L (17). To understand the molecular mechanism of ligand binding to GIP, we have initiated a detailed structural characterization of not only the free protein, but also GIP in complex with its various binding partners, starting with the Glutaminase L peptide. While titrating the Glutaminase L peptide to GIP, it was possible to track the movements of individual peaks within the (1H, 15N)-HSQC spectra, since most of the protein resonances are in the fast-exchange time scale. However, the peak intensities of the amino acid residues I18, L21, I28-G35, Q39, D40, Q43, N44, E48, I55, E62, A66, E67, A69 and R96 are either greatly diminished or completely disappear below the noise threshold, presumably due to intermediate to slow exchange. Once GIP approaches saturation and the bound state predominates, these resonances that had disappeared earlier reappear, often in remote regions of the HSQC spectrum relative to their original positions. This engendered considerable uncertainty as to the assignments of many of the residues predicted to be critical to complex formation, such as I28-E48 and R96, which are located within the β2 strand, the β2-β3 loop and the α2 helix. This evidence points to the interaction between the protein and peptide to primarily proceed through the β-strand addition mechanism (7) rather than a direct interaction with the α2 helix. The observation of intermediate to slow exchange also suggests that there are some long-range allosteric interactions within the protein due to ligand binding, as this phenomenon was also seen for a number of residues that were not directly a part of the predicted binding region, such as I18, I55, and E62-A69, which belong to the β1 and β3 strands and the α1 helix, respectively.
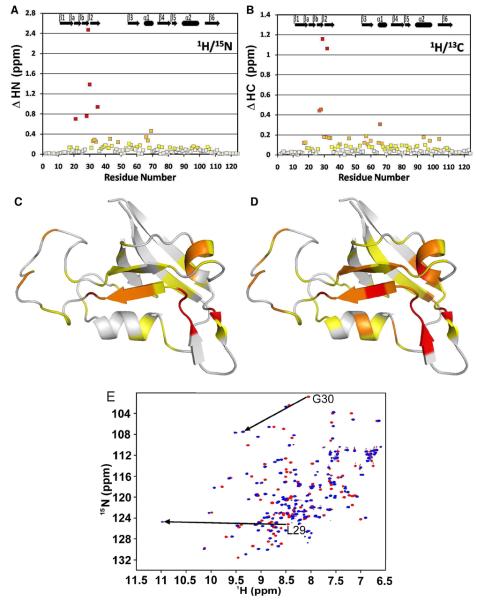
Likewise, the (1H, 13C)-HSQC spectra were remarkably different between free GIP and the GIP-Glutaminase L peptide complex. Due to the severe overlap of carbon and proton chemical shifts and the large chemical shift perturbations within GIP assignments, there was considerable uncertainty about the identity of a number of key protein side-chain assignments based solely on the previously assigned free GIP. Therefore, to determine the structure of the GIP-Glutaminase L peptide complex, we assigned all 15N, 13C and 1H resonances from GIP both in the free and complexed states using the following 3D experiments: HNCACB, CBCA(CO)NH, HCC(CO)NH, CC(CO)NH, HSQC-TOCSY and HCCH-TOCSY. These experiments were used to fully re-assign the protein in the complexed state including residues that had disappeared and re-appeared in remote locations during the course of the titration of GIP with the Glutaminase L peptide, such as L27 – G35. The re-assignment of the protein in the complex proved essential as nearly all of the resonances, both backbone and side-chain, shifted either slightly or dramatically (Figure 1 A & B). Generally speaking, the amide region is the most sensitive to chemical shift perturbation, but with so many overlapping 1H and 13C resonances, each assignment must be individually sorted out before structure calculation could proceed.
Figure 1.
(A) Combined 1H and 15N backbone amide chemical shift perturbations (ΔHN) are plotted as a function of residue number for GIP. (B) Combined HA and CA backbone chemical shift perturbations (ΔHC) are plotted as a function of residue number for GIP. (C) The magnitudes of HN presented in (A) are represented in different colors on a ribbon diagram of free GIP. White is < 0.1 ppm, yellow is < 0.2 ppm, orange is < 0.5 ppm and red is > 0.5 ppm. (D) The magnitudes of HC presented in (B) are represented as different colors on a ribbon diagram of free GIP. White is < 0.05 ppm, yellow is < 0.1 ppm, orange is < 0.2 ppm, red-orange is < 0.5 ppm and red is > 0.5 ppm. For (C) and (D) residues A11-Q112 are shown. Residues M1-T10 and A113-S124 are not shown since they are highly disordered and have chemical shifts perturbations of < 0.05 ppm. (E) An overlay of the HSQC spectra for free GIP (red) and the GIP-Glutaminase L peptide complex (blue). Arrows indicate the dramatic chemical shift perturbations of L29 and G30.
NMR Structure determination of the GIP-Glutaminase L peptide complex
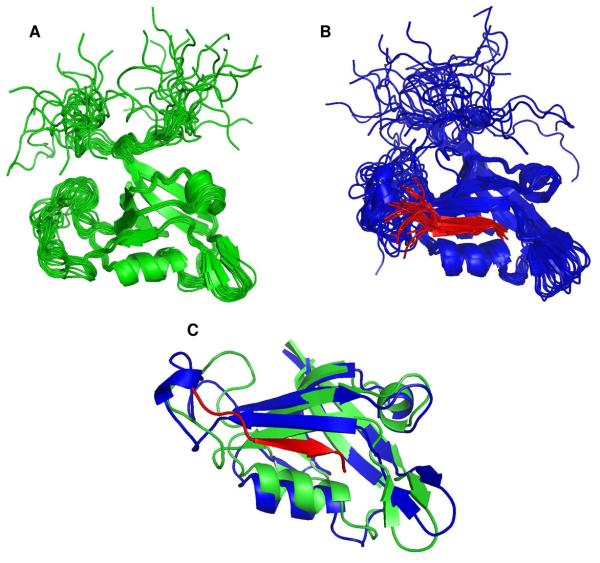
A number of key experiments were recorded with the goal of calculating the structure of the GIP-Glutaminase L peptide complex. First, we used a 2D selectively filtered NOESY that can be separated into four 2D NOESY spectra (45). This experiment selectively filters NOEs that originate from protons attached to either 12C/14N (peptide) or 13C/15N (protein). One of the four filtered experiments, which shows NOEs from only protons attached to 12C or 14N, allowed us to re-assign the resonances of the Glutaminase L peptide in the bound conformation by comparing them to the resonances of the unbound form. It also helped us to determine the structure of the Glutaminase L peptide in the bound state. Second, F1-filtered/F3-selected NOESY experiments with both 15N/14N and 13C/12C filtering methods were used to identify intermolecular NOEs between the unlabeled peptide and the 13C, 15N-labeled protein in the complex. In addition, we identified intermolecular NOEs from the traditional 3D 15N- and 13C-edited NOESY experiments. Third, the selective formation of specific hydrogen bonds between the negatively charged C-terminal Val carboxyl oxygens from the Glutaminase L peptide to the amide protons of L29 and G30 from GIP could be directly identified from their very large induced chemical shift perturbations (60). These hydrogen bonds greatly enhanced the iterative assignment process in fitting the Glutaminase L peptide into the structure of GIP. The GIP- Glutaminase L peptide complex differs from most other complexes of PDZ domains because far fewer NOEs were observed using only the standard 13C-filtered NOESY that is most frequently used to determine the structure of a complex. We believe that the lack of observable NOEs is due to line broadening resulting from intermediate to slow exchange of residues in the entire β2 strand. Thus only the strongest NOEs were observable as a result of the reduced intensity of residues critical to the binding interaction. Initial assignments of intermolecular NOEs arising from the traditional 3D (unfiltered) NOESY were ambiguous. However, once the peptide's relative position in the binding site was established, many of the initial ambiguities could be sorted out, which helped to add to the total number of intermolecular NOEs used for the final structure calculation. The ensemble of structures of both free GIP (Figure 2A) and the GIP-Glutaminase L peptide complex (Figure 2B) were each calculated independently using completely different NOE data sets.
Figure 2.
Ribbon diagrams of the ensemble of the 20 superimposed lowest energy structures. (A) Free GIP is shown in green. (B) Complexed GIP is in blue and the Glutaminase L peptide is in red. (C) Overlays of free GIP and complexed GIP are shown in green and blue, respectively, with the Glutaminase L peptide in red.
Comparison of free GIP with the GIP-Glutaminase L peptide complex
Generally, the architecture of a PDZ domain is characterized by a six-stranded β-roll (β1- β6) and two α-helices (α1 - α2) (60). Although there is general agreement between the structures of free GIP and the GIP-Glutaminase L peptide complex; the protein does adjust to accommodate the additional β-strand in an allosteric manner. Both contain the same overall fold and architecture with the peptide binding via α-strand addition (7). Upon binding, the α2 helix moves away from β2 by 0.95 Å to accommodate the additional β-strand (Figure 2C). The β2-β3 loop is largely unstructured in both free GIP and the complex. However, in the complex we observe a few NOEs between this loop and the Glutaminase L peptide. It has been previously reported that GIP interacts with the C-terminal β-catenin peptide through its PFS loop (residues 45-47) (16). The above observations clearly demonstrate that GIP interacts with different binding partners with specificity. We observe substantial chemical shift perturbations in the α1 helix due to its relative proximity to the binding site (Figure 1). However, these chemical shift changes do not translate into the 3-dimensional structure in the complex (Figure 2). The significant chemical shift perturbations in residues not lining the binding pocket illustrate that without complete structure determination, NMR titration data can potentially be misinterpreted as indication of direct protein-ligand interactions. It is also difficult to determine through a side-by-side structural comparison which specific interactions led to the relatively large changes in chemical shifts that we observed in regions of the protein located away from the binding-site.
Glutaminase L peptide binding and site specificity
The binding pocket of GIP is formed from a groove located between the α2 helix and β2 strand. The C-terminus of the interacting protein binds to this groove as an additional antiparallel β-sheet to β2 through β-strand addition (7). The sequence specificity of PDZ domains for the interacting partner is broken down into several classes usually of 4 residues numbered as positions −3, −2, −1 and 0 starting with the C-terminal residue as P0. In particular, the consensus GLGF loop located at the beginning of the β2 strand forms a series of hydrogen bonds between the backbone amides of the protein and the COO- of the C-terminal peptide. Furthermore, this loop provides a hydrophobic pocket that helps with the sequence selectivity for the C-terminal residue of the substrate peptide.
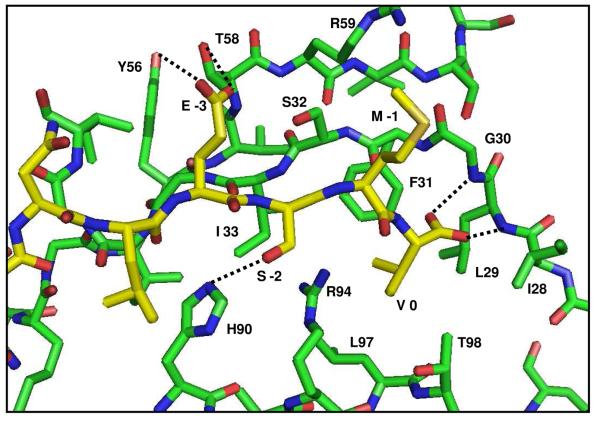
Although both free and the Glutaminase L bound complex of GIP share the same general fold typical of PDZ domains, there are several notable exceptions. Between the canonical β1 and β2 strands, GIP contains an additional short β-hairpin composed of strands βa and βb (16, 59, 61). Importantly, the final residue in βb is I28. The residue I28 forms the ILGF motif for GIP, which deviates from the canonical GLGF motif of PDZ domains. This suggests that while G28 is the consensus amino acid for the binding motif for PDZ domains, the mutation to Ile is tolerated perhaps due to the structural role it plays in forming the βa-βb hairpin. In contrast, G30 is the only amino acid that can accommodate the geometry needed for the formation of hydrogen bonds from L29 and G30 of GIP to the COO- from the C-terminal peptide. We believe that G30 is absolutely required, as it provides a structural framework for the C-terminal specificity. In the Glutaminase L peptide, the charged carboxyl group from the C-terminal Val (P0) forms two hydrogen bonds to the backbone amide protons of L29 and G30 from GIP. Meanwhile, the hydrophobic side-chain of Val (P0) buries itself into the hydrophobic pocket created by L29, F31, L97 and I33 as well as T98 at the periphery (Figure 3). The above two hydrogen bonds cause unusually large chemical shift changes of up to 2.5 ppm for the amides of L29 and G30 in the 1H, 15N-HSQC spectra upon binding to the peptide (Figure 1E). The chemical shift perturbations of both HN/N and HA/CA pairs show the effect of the Glutaminase L peptide binding to GIP. When these chemical shift changes are mapped onto the structure of GIP (Figure 1), one can clearly see that while regions near the binding sight including β2, α2 and the β2-β3 loop are generally the most perturbed, α1, which does not appear to be directly involved in the binding is also significantly affected. This clearly demonstrates that the mechanism of peptide binding to GIP is allosteric. The residue H90 at the beginning of α2 (α2:1 in PDZ nomenclature) is oriented into the binding pocket and makes a specific hydrogen bond with the Ser at P−2 of the peptide (Figure 3). This is a general feature of class I PDZ domains as the residue at position α2:1 provides the sequence selectivity that distinguishes between different classes (12). Generally, there is no specificity at P−1 (Table 1). The Glutaminase L peptide has Met at P−1, which is oriented away from the binding pocket toward the solvent. Some class I PDZ domains have specificity towards E/D or a small amino acid at P−3 (12). This interaction comes from hydrogen bonds between E at P−3 from the Glutaminase L peptide with Y56 and T58 of GIP. Alternately, a transient salt-bridge could potentially exist, but does not appear to be formed with R59 (Figure 3) of GIP. This particular salt-bridge has been observed in the crystal structures of GIP with β-catenin (16) and Kir 2.3 (61). However, we did not find any observable NOEs to support the formation of a salt bridge between E at P−3 of the Glutaminase L peptide with R59 of GIP. This is due to the dynamic flexibility of the protein side chains in solution, in contrast to the static nature of a crystal environment. It could be possible that the flexibility of these side chains would allow them to come close enough to form a transient salt-bridge. However, our results demonstrate that both E at P−3 and R59 are solvent-exposed, thus decreasing the strength of such an interaction in solution. This demonstrates that the salt-bridges observed in the two crystal structures are likely due to packing artifacts of crystallization while the true nature of the salt-bridge in solution is more dynamic.
Figure 3.
A close-up view of the binding site of GIP (green) with the Glutaminase L peptide (yellow). Specified individual residues are labeled and hydrogen bonds are shown as dashed lines.
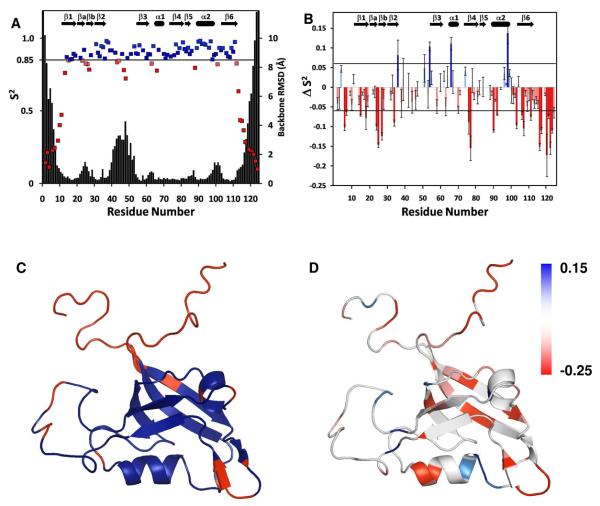
Dynamics of free GIP and the GIP-Glutaminase L peptide complex from 15N relaxation measurements
To further elucidate the binding mechanism of the Glutaminase L peptide to GIP, we carried out backbone dynamics studies. The order parameters (S2) from both free and complexed forms of GIP were calculated using steady-state 1H-15N NOE intensities, R1 and R2 relaxation rates using the model-free analysis based on the Lipari-Szabo formalism (62). Overlapped peaks and residues that could not be characterized due to low intensity or absence in the HSQC spectra were excluded from our data analysis. Residues M1, P5, P8, V12, L21, P41, P45, K50, D52, P65, D75 and V80 were excluded from both free and bound states. Residues K20, L29, G30, V57, R59, I68, A69, I73, M87, K95, V105 and V118 were excluded only in the free state. Residues V13, N26, F31, G35, I37, D40, Q43, E48, D49, Y56, S61, Q72, W83, M85, T86 and A93 were excluded only in the bound state. Of these, L29, G30, F31, G35, D40, Q43, E48, D49 are from residues that form part of the binding pocket including the ILGF motif (canonical GLGF) and the β2-β3 loop, could not be measured as a result of being too close to the intermediate exchange regime to provide sufficient intensity required for observation in the NMR dynamics data. While the S2 values could not be determined for every residue in both the free and complexed states due to either spectral overlap or line broadening, we could determine S2 for 100 and 96 of 118 residues (excluding the N-terminus and 5 prolines) for the free and bound forms of GIP, respectively. Additionally, ΔS2 values between bound and free states were determined for 84 residues. The generalized order parameters, S2, are broadly similar for both the free and complexed states, but exhibit certain differences as explained below. The residues located in well-defined secondary structures that show relatively restricted mobility of 0.85 or above are highlighted in blue (Figure 4A & C). The residues at both termini of the protein and various loops including the βa-βb hairpin, the β2-β3 loop and a few other short loops between secondary structural elements exhibit greater flexibility as shown in red (Figure 4A & C). We observe a correlation between our order parameters and the overall RMSD. The modelfree analysis of free GIP yielded generally high values of S2 with an average value of 0.89 for the core region (residues A11-Q112), indicative of the restricted backbone mobility of a well folded protein on the sub-nanosecond timescale which drops off precipitously for residues at both unstructured termini (Figure 4A). Likewise, for the GIP-Glutaminase L peptide complex, the average S2 value is 0.87 for the core region. This demonstrates that in general the core of the protein maintains its structure and flexibility upon binding to the Glutaminase L peptide. However, a closer examination of the changes in S2 reveals that there are specific residues that exhibit either an increase or decrease in flexibility. For residues where we could calculate ΔS2, the following showed a substantial (ΔS2 > 0.06) decrease in flexibility: G36, G54, A66 and T98. Furthermore, residues I4, T51, G74, R96 and K99 showed smaller but still significant increases in S2 (0.03 < ΔS2 < 0.06) where the average variance in S2 was ±0.015 for all measured residues. Twelve other residues showed positive, but statistically insignificant increases in S2. Likewise twenty-four residues showed statistically insignificant decreases in S2 upon binding. However, residues Q14, H19, I28, D38, N44, F46, T58, G63, G70, D91 and V109 showed a small but statistically significant (−0.03 > S2 > −0.06) increase in flexibility. Additionally, residues R15, I18, G24, E25, L27, G34, K76, I77, H90, Q92, E103, R106, L107, R111 and many of the measured residues in the unstructured termini (M1-T10, S113-S124) showed a substantial increase in flexibility (ΔS2 < −0.06) as shown in Figure 4B. When these residues are mapped onto the structure of free GIP (Figure 4D) they demonstrate that the biggest decreases in flexibility are in residues at the C-terminal end of the α2 helix near the binding site and at the hinge points of the β2-β3 loop. However, the biggest increases in backbone flexibility occur for residues located either on the β4 and β6 strands that are distal to the binding site or in the flexible loops such as the βa-βb hairpin and the β2-β3 loop.
Figure 4.
(A) The S2 values derived using the modelfree analysis from the steady state 1H-15N NOE, R1 and R2 relaxation times of free GIP for each non-overlapping well defined residue. Residues with order parameters above the threshold 0.85 are colored in blue while those below are colored in red. The backbone RMSD of free GIP for each residue is overlaid on this plot in black. (B) A plot of ΔS2 as a function of residue number where ΔS2 refers to S2 of the GIP-Glutaminase L peptide complex minus that of free GIP. Positive values are indicated with increasing blue intensity while negative values are indicated with increasing red intensity. (C) Residues with S2 values below the threshold of 0.85 are mapped in red onto the structure of free GIP colored blue. (D) The magnitude of ΔS2 upon binding to the Glutaminase L peptide is mapped onto the structure of free GIP and is indicated by darker intensity for red (increased flexibility) or blue (decreased flexibility). Residues were colored white for one of the following reasons: they could not be measured in both structures due to overlap, they had ΔS2 values between the threshold values 0.06 and −0.06, or the residue was a proline.
Intermediate exchange within GIP caused by the binding of the Glutaminase L peptide
The chemical shift perturbations for most of the residues of GIP are in the fast exchange regime as demonstrated by the titration experiments with the peptide. However, the residues lining the binding pocket appear to be in intermediate exchange. As GIP approaches saturation, residues L27, I28, L29, G30, F31, S32, I33, G34 and G35, which had disappeared or greatly diminished due to intermediate to slow exchange at a low protein to peptide ratio, reappeared in new locations in the 1H, 15N-HSQC spectra. Additionally, both in the free and complexed state of GIP, residues L29 and G30 have lower intensity in the 2D 1H, 15N-HSQC spectra compared to all other residues of the protein due to line broadening caused by intermediate exchange. It is noteworthy that L27-G35 are the residues that comprise the ILGF loop and most of the β2 strand that line the binding pocket of GIP. Along with significant chemical shift perturbations, there are substantial changes in the measurable order parameters for different regions within the binding pocket. It appears that both ends of the binding pocket experience opposite effects in S2 values. One end of the binding pocket that is near the C-terminus of the peptide is composed of the IGLF loop and the C-terminal half of the α2 helix (K95-R100). The residues L29, G30 and F31 of the IGLF loop are in intermediate exchange, which precludes the measurement of ΔS2. The residues R96, T98 and K99 from the α2 helix) experience a decreased flexibility upon binding the C-terminal end of the peptide. However, at the opposite end of the binding pocket, residues from both β2 and α2 (G34, H90 and Q92) experience an increase in flexibility. This is reflected in the relatively high RMSD for the N-terminal end of the Glutaminase L peptide. The above observation of increased or decreased flexibility in the binding pocket of the protein and that of the peptide suggests that the substrate specificity is limited to the C-terminal four residues of Glutaminase L.
Discussion
Binding specificity of the Glutaminase L peptide with GIP
The specificity of GIP toward the target protein C-terminal sequence i.e. E-S/T-X-I/L/V-COOH [Table-1] of Glutaminase L peptide comes from various interactions. The amide protons of residues L29 and G30 in the ILGF loop are uniquely positioned in such a way that allows them to form a pair of hydrogen bonds to both carboxyl oxygen atoms of V at P0 from the Glutaminase L peptide (Figure 3). These interactions are characterized by the very large chemical shift perturbations observed for the L29 and G30 amide groups (Figure 1A & E). Although the structure is not significantly affected, the chemical environment is dramatically different due to the proximity to the negatively charged carboxyl oxygens from the C-terminus of the peptide. The peptide binding also dramatically affects the dynamics of GIP, so much so that residues L27-G35 disappear during the course of the titration into the intermediate to slow exchange regime and do not reappear until after the binding-site becomes saturated.
The specificity for a hydrophobic residue at P0 comes from the hydrophobic pocket created by L29, F31, I33 and L97. Val seems to be preferred at P0 more than Leu or Ile. The steric nature of these hydrophobic interactions can be investigated through point mutation of one or more of the following residues in the binding pocket of GIP: L29V, L97V or T98A. Residue L97, located at position α2:8, is highly conserved across class I PDZ domains and is known to confer specificity at P0 (12). The side-chains of L29 and L97 interact to form the majority of the surface area of this hydrophobic pocket. These mutations would likely change the selectivity at P0 from Val to Ile, Leu or potentially a larger hydrophobic amino acid currently not allowed such as Phe or Trp.
Specificity for S/T at P−2 is due to H90 at position α2:1 of GIP. In contrast, the lack of specificity at P−1 appears to be two-fold. The residue G30 is highly conserved across PDZ domains. The geometry of G30 is a steric requirement for the binding to the C-terminus of the target protein. The absence of a side chain at this position most likely serves as an evolutionary trade-off between specificity for the C-terminus and sequence specificity at P−1. Second, because the binding occurs as β-strand addition, alternating amino acids are oriented away from the binding site.
By comparing the first NMR structure of the GIP -Glutaminase L peptide complex to the crystal structures of GIP bound to other target proteins, we can identify and distinguish between common and unique features of binding for each ligand. We have shown here that the mode of binding between GIP and each of its ligands is unique and specific. We show that only a few interactions occur between the β2-β3 loop of GIP and the Glutaminase L peptide, unlike the specific interactions seen between the PFS loop of GIP with β-catenin (16). Additionally, the E at P−3 of the Glutaminase L peptide makes specific hydrogen bonds to Y56 and T58 of GIP rather than the salt-bridge observed between the D or E at P−3 of β-catenin or Kir 2.3 respectively with R59 of GIP that we believe to be crystallization artifacts (16, 61). In comparison to the observed changes in chemical shifts for other ligands of GIP such as β-catenin and FAS that we have previously reported (17), the unique mode of binding and chemical shift perturbation patterns observed for GIP with the Glutaminase L peptide means that it is necessary to experimentally determine the structure of GIP in complex with each of its known ligands.
By maximizing the common features and taking advantage of the unique features of ligand binding, we should be able to efficiently design a competitive inhibitor with higher affinity than any of the natural ligands. When designing a target site interacting partner a good choice at P−1 would likely be hydrophilic and specifically negatively charged such as D or E to potentially form a salt-bridge with R59 (16) rather than hydrophobic since it is solvent exposed. Specificity for E at P−3 of the peptide is due to the formation of a hydrogen bond with Y56 and/or T58 of GIP. Since Y56 and T58 can each act both as hydrogen bond donors or acceptors, this explains why P−3 can also accommodate multiple sidechains. Furthermore, GIP has three glycines in a row: G34, G35 and G36. The lack of side-chains for these residues can explain the binding of GIP to various binding partners. Finally, GIP has specificity to β-catenin that is not present for the Glutaminase L peptide, at positions beyond P−3. An aromatic residue at P−5 or P−6 (Table 1) could provide additional specificity to GIP for any future drug design effort.
The effects of the Glutaminase L peptide binding on the dynamics of GIP
Where S2 could be measured in both free GIP and the GIP-Glutaminase L complex, some residues show either substantial increases or decreases in backbone flexibility. Generally speaking, residues at the binding site tend to become more ordered, while residues peripheral to the binding site in GIP become more disordered, with a few exceptions. G34 is part of the β2 strand that forms an antiparallel β-sheet with the Glutaminase L peptide, and thus should be more stabilized, yet it actually becomes more disordered. While it is part of the binding site, it is located on the opposite end of the β2-strand from the ILGF binding loop and is near the hinge-point between the β2 strand and the β2-β3 loop (residues G36-G54). Additionally, H90, D91 and Q92 show increased flexibility. While H90 makes a direct H-bond to the S at P−2, (Figure 3) the specificity of the Glutaminase L peptide is limited to the four C-terminal residues, while the N-terminal four residues are disordered with higher RMSD values. However, the general trend is that in the regions of GIP where the peptide directly interacts, the structure becomes more rigid, which is offset by an increase in flexibility that is distributed throughout the rest of the protein. The decrease in the flexibility of the binding pocket is offset by an increase in flexibility distal to the binding site in the core of the protein including the β1, β4 and β6 strands. Furthermore, the increase in flexibility throughout GIP also includes the flexible regions of the protein such as the ba-bb hairpin and β2-β3 loops as well as both termini.
Comparison to other GIP-peptide complex structures
It is worth noting that both the N-terminal (M1-T10) and C-terminal (S113-S124) regions of GIP are completely unstructured both in the free-state and in the complex with very few observed NOEs and correspondingly high RMSDs in our structural ensembles (Figure 2A & B). This is further supported by our dynamics data, which also show these regions to be completely unstructured (Figure 4A). It has been previously reported that C-terminal truncation of GIP leads to a decreased affinity for full length β-catenin in vivo (15). We observed general similarity in the binding modes of the β-catenin and Glutaminase L peptides to GIP (17). Therefore, it appears unlikely that the reported decrease in full length β-catenin affinity to a C-terminally truncated GIP is due to an interaction between the canonical C-terminal binding motif of β-catenin and the C-terminus (113-124) of GIP. We previously observed very little change in the chemical shifts of the C-terminal region of GIP upon ligand binding, regardless of whether the ligand is the Glutaminase L, β-catenin or FAS peptide (17). Therefore, we propose a possible explanation for the observed decrease in the affinity for the full length β-catenin upon C-terminal truncation of GIP. The C-terminus of GIP most likely interacts with either a different region of full length β-catenin or another interacting partner protein in vivo. This hypothesis is supported by the in vivo 2-hybrid interaction studies between various deletion mutants for both GIP and β-catenin (15). These studies showed a central core region of β-catenin (173-483) lacking the class I C-terminus still maintained some affinity for GIP (15). However, we believe that the best explanation of the above results, in light of our structural and dynamics characterization, is that it is the central core region of β-catenin that interacts directly and specifically with the C-terminus of GIP. Taken together, it appears that β-catenin and GIP each bind to the other protein's C-terminus.
Comparison of NMR and crystal structures of free GIP
A solution NMR structure provides insight into the dynamic nature of a protein. By comparing the solution structure of free GIP with the previously solved crystal structures, we show that while there is good agreement between NMR and crystallographic methods, there are a few key differences. First, in each of the NMR structures, both the N- and C-termini (regions 1-10 and 113-124) are highly dynamic and unstructured. This is in contrast to the crystal structure of free GIP where the C-terminus forms a helix, which is most likely an artifact of crystallization. Second, the β2-β3 loop from G36-G54 has a defined structure in the crystal structures (16, 61), but it is considerably more flexible in our NMR structure. Our dynamics data also indicates that this region had significantly lower order parameters compared to the rest of the central core region. Additionally, relatively few NOEs were observed compared to other regions of the protein. All of the observed NOEs were medium range (|i−j|<5) or shorter, but there were no unambiguously defined long-range NOEs (|i−j|>5). We found this to be the case for both free GIP as well as the GIP-Glutaminase L peptide complex. However, there were some intermolecular NOEs between the loop and the peptide, which indicates that this flexible loop may undergo a conformational change upon binding. This conformational change is observed from the decrease in flexibility of G36 and G54 near the hinge-point of the β2-β3 loop while flexibility increases on either side of the hinge point. We do observe a distinct conformational change (Figure 2C), but the loop remains relatively unstructured compared to the rest of the core protein in both free and bound states. Third, the non-canonical β-hairpin formed by residues L21-I28 has a higher relative backbone RMSD of around 0.85 Å in the free form of GIP compared to the rest of the core structured portion of the protein at 0.45 Å. In the GIP-Glutaminase L complex the corresponding RMSD values are 2.73 Å and 0.67 Å. As was the case for the β2-β3 loop, there are NOEs that define the β-hairpin. However, since it is exposed to the solvent, it does not make as many contacts with the rest of the protein and is therefore relatively unconstrained during the structural calculation. The increase in RMSD for this hairpin loop within the complex is due to relatively more long-range NOEs observed for free GIP than for the GIP-Glutaminase L complex. It is further supported by the increases in flexibility for the complex seen for residues G24, E25, L27 and I28. This hairpin loop may play a role in how GIP might discriminate between canonical class I C-terminal sequences and C-terminal structural mimics presented by internal motifs.
Potential for targeted drug design
Taken together, the elements of specificity within GIP for certain types of molecular interactions do make it a tempting target for drug design. Because cells contain literally hundreds of PDZ domains, a potential drug would either have to be designed to interact very specifically with only the PDZ domain within GIP or to broadly target and disrupt the function of other PDZ domains that may share the same specificity as GIP. Based on the structural insights shown here, targeting GIP could lead to promising anticancer therapeutics.
Acknowledgements
We thank Dr. Donna M. Baldisseri of the Bruker BioSpin Corporation for some of the NMR data collection as well as Dr. Uma Katre and Dr. Janarthanan Krishnamoorthy for help with figure preparation and Prof. Peter Livant of Department of Chemistry and Biochemistry at Auburn University for critical reading of the manuscript. Dedication: The authors dedicate this article in the loving memory of Dr. Marie W. Wooten, Dean and Professor of the College of Sciences and Mathematics (COSAM) at Auburn University, who tragically passed away on the 5th of November, 2010. Dean Wooten was our mentor, colleague, friend and role-model. She will be deeply and painfully missed, yet celebrated, honored and never forgotten.
This research was financially supported by USDA PECASE Presidential Early Career Award for Scientists and Engineers award 2003-35302-12930, NSF grant IBN-0628064, and NIH grant DK082397 to Smita Mohanty.
Abbreviations
- NMR
Nuclear Magnetic Resonance
- PDZ
Post synaptic density 95, Disc-large, Zonula occludentes-1
- GIP
Glutaminase Interacting Protein
- NOE
Nuclear Overhauser Effect
Footnotes
The chemical shifts of the resonances and the atomic coordinates for free GIP as well as the GIP - Glutaminase L peptide complex have been deposited in the BioMagnetic Resonance Bank (BMRB) and the Protein Data Bank (PDB) with the following accession numbers: 17254 and 17255 for the BMRB and 2L4S and 2L4T for the PDB, respectively.
References
- 1.Neel BG. Structure and function of SH2-domain containing tyrosine phosphatases. Semin Cell Biol. 1993;4:419–432. doi: 10.1006/scel.1993.1050. [DOI] [PubMed] [Google Scholar]
- 2.Kardinal C, Posern G, Zheng J, Knudsen BS, Moarefi I, Feller SM. Rational development of cell-penetrating high affinity SH3 domain binding peptides that selectively disrupt the signal transduction of Crk family adapters. Amgen Peptide Technology Group. Ann N Y Acad Sci. 1999;886:289–292. doi: 10.1111/j.1749-6632.1999.tb09439.x. [DOI] [PubMed] [Google Scholar]
- 3.Falke JJ. Membrane Recruitment as a Cancer Mechanism: A Case Study of Akt PH Domain. Cellscience. 2007;4:25–30. [PMC free article] [PubMed] [Google Scholar]
- 4.Fanning AS, Anderson JM. Protein-protein interactions: PDZ domain networks. Curr Biol. 1996;6:1385–1388. doi: 10.1016/s0960-9822(96)00737-3. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Mata R, Burridge K. Catching a GEF by its tail. Trends Cell Biol. 2007;17:36–43. doi: 10.1016/j.tcb.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Woods DF, Bryant PJ. ZO-1, DlgA and PSD-95/SAP90: homologous proteins in tight, septate and synaptic cell junctions. Mech Dev. 1993;44:85–89. doi: 10.1016/0925-4773(93)90059-7. [DOI] [PubMed] [Google Scholar]
- 7.Jelen F, Oleksy A, Smietana K, Otlewski J. PDZ domains - common players in the cell signaling. Acta Biochim Pol. 2003;50:985–1017. [PubMed] [Google Scholar]
- 8.Spaller MR. Act globally, think locally: systems biology addresses the PDZ domain. ACS Chem Biol. 2006;1:207–210. doi: 10.1021/cb600191y. [DOI] [PubMed] [Google Scholar]
- 9.Christopherson KS, Hillier BJ, Lim WA, Bredt DS. PSD-95 assembles a ternary complex with the N-methyl-D-aspartic acid receptor and a bivalent neuronal NO synthase PDZ domain. J Biol Chem. 1999;274:27467–27473. doi: 10.1074/jbc.274.39.27467. [DOI] [PubMed] [Google Scholar]
- 10.Lemaire JF, McPherson PS. Binding of Vac14 to neuronal nitric oxide synthase: Characterisation of a new internal PDZ-recognition motif. FEBS Lett. 2006;580:6948–6954. doi: 10.1016/j.febslet.2006.11.061. [DOI] [PubMed] [Google Scholar]
- 11.Wiedemann U, Boisguerin P, Leben R, Leitner D, Krause G, Moelling K, Volkmer-Engert R, Oschkinat H. Quantification of PDZ domain specificity, prediction of ligand affinity and rational design of super-binding peptides. J Mol Biol. 2004;343:703–718. doi: 10.1016/j.jmb.2004.08.064. [DOI] [PubMed] [Google Scholar]
- 12.Tonikian R, Zhang Y, Sazinsky SL, Currell B, Yeh JH, Reva B, Held HA, Appleton BA, Evangelista M, Wu Y, Xin X, Chan AC, Seshagiri S, Lasky LA, Sander C, Boone C, Bader GD, Sidhu SS. A specificity map for the PDZ domain family. PLoS Biol. 2008;6:e239. doi: 10.1371/journal.pbio.0060239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olalla L, Aledo JC, Bannenberg G, Marquez J. The C-terminus of human glutaminase L mediates association with PDZ domain-containing proteins. FEBS Lett. 2001;488:116–122. doi: 10.1016/s0014-5793(00)02373-5. [DOI] [PubMed] [Google Scholar]
- 14.Rousset R, Fabre S, Desbois C, Bantignies F, Jalinot P. The C-terminus of the HTLV-1 Tax oncoprotein mediates interaction with the PDZ domain of cellular proteins. Oncogene. 1998;16:643–654. doi: 10.1038/sj.onc.1201567. [DOI] [PubMed] [Google Scholar]
- 15.Kanamori M, Sandy P, Marzinotto S, Benetti R, Kai C, Hayashizaki Y, Schneider C, Suzuki H. The PDZ protein tax-interacting protein-1 inhibits beta-catenin transcriptional activity and growth of colorectal cancer cells. J Biol Chem. 2003;278:38758–38764. doi: 10.1074/jbc.M306324200. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Yan X, Shi C, Yang X, Guo Y, Tian C, Long J, Shen Y. Structural basis of beta-catenin recognition by Tax-interacting protein-1. J Mol Biol. 2008;384:255–263. doi: 10.1016/j.jmb.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 17.Banerjee M, Huang C, Marquez J, Mohanty S. Probing the structure and function of human glutaminase-interacting protein: a possible target for drug design. Biochemistry. 2008;47:9208–9219. doi: 10.1021/bi800287v. [DOI] [PubMed] [Google Scholar]
- 18.Saras J, Engstrom U, Gonez LJ, Heldin CH. Characterization of the interactions between PDZ domains of the protein-tyrosine phosphatase PTPL1 and the carboxyl-terminal tail of Fas. J Biol Chem. 1997;272:20979–20981. doi: 10.1074/jbc.272.34.20979. [DOI] [PubMed] [Google Scholar]
- 19.Hampson L, Li C, Oliver AW, Kitchener HC, Hampson IN. The PDZ protein Tip-1 is a gain of function target of the HPV16 E6 oncoprotein. Int J Oncol. 2004;25:1249–1256. [PubMed] [Google Scholar]
- 20.Reynaud C, Fabre S, Jalinot P. The PDZ protein TIP-1 interacts with the Rho effector rhotekin and is involved in Rho signaling to the serum response element. J Biol Chem. 2000;275:33962–33968. doi: 10.1074/jbc.M000465200. [DOI] [PubMed] [Google Scholar]
- 21.Le Maout S, Welling PA, Brejon M, Olsen O, Merot J. Basolateral membrane expression of a K+ channel, Kir 2.3, is directed by a cytoplasmic COOH-terminal domain. Proc Natl Acad Sci U S A. 2001;98:10475–10480. doi: 10.1073/pnas.181481098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alewine C, Olsen O, Wade JB, Welling PA. TIP-1 has PDZ scaffold antagonist activity. Mol Biol Cell. 2006;17:4200–4211. doi: 10.1091/mbc.E06-02-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez-Gomez C, Campos-Sandoval JA, Alonso FJ, Segura JA, Manzanares E, Ruiz-Sanchez P, Gonzalez ME, Marquez J, Mates JM. Co-expression of glutaminase K and L isoenzymes in human tumour cells. Biochem J. 2005;386:535–542. doi: 10.1042/BJ20040996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallagher FA, Kettunen MI, Day SE, Lerche M, Brindle KM. 13C MR spectroscopy measurements of glutaminase activity in human hepatocellular carcinoma cells using hyperpolarized 13C-labeled glutamine. Magn Reson Med. 2008;60:253–257. doi: 10.1002/mrm.21650. [DOI] [PubMed] [Google Scholar]
- 26.Olalla L, Gutierrez A, Jimenez AJ, Lopez-Tellez JF, Khan ZU, Perez J, Alonso FJ, de la Rosa V, Campos-Sandoval JA, Segura JA, Aledo JC, Marquez J. Expression of the scaffolding PDZ protein glutaminase-interacting protein in mammalian brain. J Neurosci Res. 2008;86:281–292. doi: 10.1002/jnr.21505. [DOI] [PubMed] [Google Scholar]
- 27.Krebs HA. Metabolism of amino-acids: The synthesis of glutamine from glutamic acid and ammonia, and the enzymic hydrolysis of glutamine in animal tissues. Biochem J. 1935;29:1951–1969. doi: 10.1042/bj0291951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curthoys NP, Watford M. Regulation of glutaminase activity and glutamine metabolism. Annu Rev Nutr. 1995;15:133–159. doi: 10.1146/annurev.nu.15.070195.001025. [DOI] [PubMed] [Google Scholar]
- 29.Aledo JC, Gomez-Fabre PM, Olalla L, Marquez J. Identification of two human glutaminase loci and tissue-specific expression of the two related genes. Mamm Genome. 2000;11:1107–1110. doi: 10.1007/s003350010190. [DOI] [PubMed] [Google Scholar]
- 30.Olalla L, Gutierrez A, Campos JA, Khan ZU, Alonso FJ, Segura JA, Marquez J, Aledo JC. Nuclear localization of L-type glutaminase in mammalian brain. J Biol Chem. 2002;277:38939–38944. doi: 10.1074/jbc.C200373200. [DOI] [PubMed] [Google Scholar]
- 31.Medina MA, Sanchez-Jimenez F, Marquez J, Rodriguez Quesada A, Nunez de Castro I. Relevance of glutamine metabolism to tumor cell growth. Mol Cell Biochem. 1992;113:1–15. doi: 10.1007/BF00230880. [DOI] [PubMed] [Google Scholar]
- 32.Brand K. Glutamine and glucose metabolism during thymocyte proliferation. Pathways of glutamine and glutamate metabolism. Biochem J. 1985;228:353–361. doi: 10.1042/bj2280353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovacevic Z, McGivan JD. Mitochondrial metabolism of glutamine and glutamate and its physiological significance. Physiol Rev. 1983;63:547–605. doi: 10.1152/physrev.1983.63.2.547. [DOI] [PubMed] [Google Scholar]
- 34.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 35.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 36.Goddard TD, K. DG. SPARKY 3. University of California; San Francisco: [Google Scholar]
- 37.Kay L, Keifer P, Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. Journal of the American Chemical Society. 1992;114:10663–10665. [Google Scholar]
- 38.Muhandiram DR, Kay LE. Gradient-Enhanced Triple-Resonance Three-Dimensional NMR Experiments with Improved Sensitivity. Journal of Magnetic Resonance, Series B. 1994;103:203–216. [Google Scholar]
- 39.Grzesiek S, Bax A. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. Journal of the American Chemical Society. 1992;114:6291–6293. [Google Scholar]
- 40.Norwood TJ, Boyd J, Heritage JE, Soffe N, Campbell ID. Comparison of techniques for 1H-detected heteronuclear 1H---15N Spectroscopy. Journal of Magnetic Resonance. 1990;87:488–501. 1969. [Google Scholar]
- 41.Palmer AG, Cavanagh J, Wright PE, Rance M. Sensitivity improvement in proton-detected two-dimensional heteronuclear correlation NMR spectroscopy. Journal of Magnetic Resonance. 1991;93:151–170. 1969. [Google Scholar]
- 42.Vuister GW, Bax A. Quantitative J correlation: a new approach for measuring homonuclear three-bond J(HNH.alpha.) coupling constants in 15N-enriched proteins. Journal of the American Chemical Society. 1993;115:7772–7777. [Google Scholar]
- 43.Clubb RT, Thanabal V, Wagner G. A constant-time three-dimensional triple-resonance pulse scheme to correlate intraresidue 1HN, 15N, and 13C′ chemical shifts in 15N---13C-labelled proteins. Journal of Magnetic Resonance. 1992;97:213–217. 1969. [Google Scholar]
- 44.Zhang O, Kay LE, Olivier JP, Forman-Kay JD. Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J Biomol NMR. 1994;4:845–858. doi: 10.1007/BF00398413. [DOI] [PubMed] [Google Scholar]
- 45.Otting G, Wüthrich K. Extended heteronuclear editing of 2D 1H NMR spectra of isotope-labeled proteins, using the X([omega]1, [omega]2) double half filter. Journal of Magnetic Resonance. 1989;85:586–594. 1969. [Google Scholar]
- 46.Zwahlen C, Legault P, Vincent SJF, Greenblatt J, Konrat R, Kay LE. Methods for Measurement of Intermolecular NOEs by Multinuclear NMR Spectroscopy: Application to a Bacteriophage λ N-Peptide/boxB RNA Complex. Journal of the American Chemical Society. 1997;119:6711–6721. [Google Scholar]
- 47.Bax A, Davis DG. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. Journal of Magnetic Resonance. 1985;65:355–360. 1969. [Google Scholar]
- 48.Bax A, Davis DG. Practical aspects of two-dimensional transverse NOE spectroscopy. Journal of Magnetic Resonance. 1985;63:207–213. 1969. [Google Scholar]
- 49.Mandel AM, Akke M, Palmer AG., 3rd. Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J Mol Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- 50.Palmer AG, Rance M, Wright PE. Intramolecular motions of a zinc finger DNA-binding domain from Xfin characterized by proton-detected natural abundance carbon-13 heteronuclear NMR spectroscopy. Journal of the American Chemical Society. 1991;113:4371–4380. [Google Scholar]
- 51.Dosset P, Hus J-C, Blackledge M, Marion D. Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. Journal of Biomolecular NMR. 2000;16:23–28. doi: 10.1023/a:1008305808620. [DOI] [PubMed] [Google Scholar]
- 52.Tsan P, Hus J-C, Caffrey M, Marion D, Blackledge M. Rotational Diffusion Anisotropy and Local Backbone Dynamics of Carbon Monoxide-Bound Rhodobacter capsulatus Cytochrome c′. Journal of the American Chemical Society. 2000;122:5603–5612. [Google Scholar]
- 53.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 54.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 55.Linge JP, Habeck M, Rieping W, Nilges M. ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics. 2003;19:315–316. doi: 10.1093/bioinformatics/19.2.315. [DOI] [PubMed] [Google Scholar]
- 56.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 57.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27-38. [DOI] [PubMed] [Google Scholar]
- 58.Schrodinger, LLC The PyMOL Molecular Graphics System, Version 1.3r1. 2010 [Google Scholar]
- 59.Durney MA, Birrane G, Anklin C, Soni A, Ladias JA. Solution structure of the human Tax-interacting protein-1. J Biomol NMR. 2009;45:329–334. doi: 10.1007/s10858-009-9361-8. [DOI] [PubMed] [Google Scholar]
- 60.Schultz J, Hoffmuller U, Krause G, Ashurst J, Macias MJ, Schmieder P, Schneider-Mergener J, Oschkinat H. Specific interactions between the syntrophin PDZ domain and voltage-gated sodium channels. Nat Struct Biol. 1998;5:19–24. doi: 10.1038/nsb0198-19. [DOI] [PubMed] [Google Scholar]
- 61.Yan X, Zhou H, Zhang J, Shi C, Xie X, Wu Y, Tian C, Shen Y, Long J. Molecular mechanism of inward rectifier potassium channel 2.3 regulation by tax-interacting protein-1. J Mol Biol. 2009;392:967–976. doi: 10.1016/j.jmb.2009.07.060. [DOI] [PubMed] [Google Scholar]
- 62.Lipari G, Szabo A. Effect of librational motion on fluorescence depolarization and nuclear magnetic resonance relaxation in macromolecules and membranes. Biophysical Journal. 1980;30:489–506. doi: 10.1016/S0006-3495(80)85109-5. [DOI] [PMC free article] [PubMed] [Google Scholar]