Abstract
The future of personalized medicine will hinge on effective management of patient genetic profiles. Molecular diagnostic testing laboratories need to track knowledge surrounding an increasingly large number of genetic variants, incorporate this knowledge into interpretative reports and keep ordering clinicians up to date as this knowledge evolves. Treating clinicians need to track which variants have been identified in each of their patients along with the significance of these variants. The GeneInsightSM Suite assists in these areas. The suite also provides a basis for interconnecting laboratories and clinicians in a manner that increases the scalability of personalized medicine processes.
Keywords: Variation, Database, Patient Genetic Profile, Laboratory Software, Knowledgebase
Introduction
Clinical laboratories that offer sequencing-based tests face unique and rapidly-growing challenges. These laboratories must not only identify variants present in the regions of patient DNA targeted by their tests but must also write reports explaining what is known about the clinical significance of any variants that are found. The quality of these reports is often a key determinant of the effectiveness of genetic medicine. High-quality reports concisely explain the state of knowledge surrounding the variants identified in a patient and their clinical relevance, so clinicians can include this information in patient care decisions. Because these reports must be specific to the variants found and the patient’s indication for testing in order to be useful, producing them is often labor-intensive and therefore expensive. In addition to the cost issues, there are a limited number of laboratory professionals trained in genetics who can sign these reports. Ultimately, the lack of trained geneticists could impair the fundamental scalability of personalized medicine. Furthermore, the interpretative process should not end when the report is sent. More information often emerges about variants over time. There is a need for laboratories to keep treating clinicians updated as new, clinically significant information is learned about variants previously identified in their patients. However, most laboratories lack the capability to effectively update reports, thereby making it very difficult for clinicians to ensure that they are making decisions based on the most recent information about each patient’s genetic profile. These issues are increasing in severity as exponentially decreasing sequencing costs enable much larger regions of patient DNA to be clinically assayed in an ever-growing number of patients.
We have developed a series of software applications, collectively called the GeneInsight (GI) Suite (Table 1), that focus on addressing these challenges. The GI Suite consists of a laboratory/knowledge management application (GI Lab), a provider application (GI Clinic) and networking infrastructure that enables organizations using GI Lab to communicate with organizations using GI Clinic. GI can be interfaced to both laboratory information management systems and electronic health records. GI Lab and GI Clinic have already been implemented at several sites.
Table 1.
GeneInsight Suite Components
| Component | Target Organizations | Functions |
|---|---|---|
| GI Lab - Knowledgebase | Laboratories and other curators | Record genetic knowledge relevant to genetic/genomic test reporting |
| GI Lab - Reporting Tool | Laboratories | Leverage knowledgebase to draft informative reports on variants identified in individual patients |
| GI Clinic | Treating clinicians | Enable clinicians to access their patients’ genetic reports and receive updates on the variants detected in their patients |
| Network Interconnects | All | Facilitate inter-organizational communication of identified variants, knowledge surrounding these variants and clinical annotations related to specific cases |
GI Lab
The combination of a genetic knowledgebase and a highly flexible report-generation tool can streamline both report generation and patient-specific knowledge distribution. The knowledgebase can centralize the knowledge maintenance process so that a single action by a geneticist both updates future reports and generates alerts for clinicians treating patients previously known to harbor a variant. Automating report drafting itself can save considerable time for geneticists and genetic counselors and increase quality and reporting consistency across geneticists as a laboratory grows.
The GI Lab application is the product of more than seven years of iterative development focused on meeting these needs. The system has supported operations of the Partners Center for Personalized Genetic Medicine’s Laboratory for Molecular Medicine for approximately six years, and in 2010 began supporting both the Genetic Sequencing Laboratory Section at ARUP Laboratories and the Massachusetts General Hospital (MGH) Molecular Pathology Laboratory.
The genetic knowledgebase built into GI Lab is the core of the entire GI Suite. We have found the knowledgebase must store information on the tests that a laboratory offers, the genes and reference sequences covered by those tests, the variants known to exist in those genes and the state of knowledge linking these variants to diseases, drug efficacies and drug responses. These represent high-level information categories, each requiring its own fields and interrelationships to the other categories. The associations between variants and diseases and/or pharmacogenomic effects are particularly important for enabling report drafting and knowledge updates to clinicians. The system allows geneticists to associate genes with one or more diseases and/or pharmacogenomic properties. Variants within genes can then be classified to specify the likelihood that the variant will cause the effect. For example, variants in inherited diseases can be classified as benign, likely benign, unknown significance, likely pathogenic or pathogenic. Clinical alerts are driven off of changes to these associations and the evidence behind each change can be recorded and distributed. Permissions can be granted to those individuals who can validate mutation classification, and for each change the system maintains a record of the time, operator, and notes/comments about the new classification.
Laboratories can specify alternate variant classification terminology if they choose. While we see the benefits that would arise from restricting laboratories to a single standardized terminology, in our experience, the field is not ready to accept this yet. By enabling customization we increase adoption. At the same time, we have found that the system can highlight terminology differences between laboratories. Delineating these differences has already led to discussions that have incrementally moved the participants towards standardized vocabularies.
The system currently supports reporting on a wide variety of disease areas including tests for cardiomyopathy, cancer, hearing loss, Medium Chain Acyl-CoA Dehydrogenase, hereditary hemorrhagic telangiectasia and numerous genetic syndromes
GI Lab also tracks the laboratory’s case history and uses a family number to link together families that have been tested by the laboratory. The goal of the knowledgebase is to track as much information as possible that could be useful in drafting a report or providing information about a variant to a clinician. Building this knowledgebase is a task that will never be truly completed; it will always evolve.
Literature references can be imported directly from PubMed and associated with disease/pharmacogenomic effects, tests, genes or variants. Each reference is imported and annotated by the end users of the system and can be marked as to whether it should be included on genetic reports. In addition to tracking literature references, the system also tracks the reference sequences comprising each test. This assists with variant validation and also specifies which regions of patient DNA were assayed in each test. The accuracy and consistency of reference sequences is ensured through automated input using downloads from NCBI Entrez Nucleotide database, though modification is allowed if the users identify errors in publically available reference sequences.
The system drafts reports based on order information and the variants, or lack of variants, identified in a particular patient. When GI Lab is interfaced with a laboratory information-management system, it can receive this information automatically. When it is not interfaced, the information can be entered manually through the application’s user interface. Currently the system has supported tests that include up to several hundred variants per patient. However, it has the capacity to accept much more data per patient. We believe the system’s underlying data structures will, with minor modifications, scale to support whole exomes. We will be testing this capability later this year. Immediately after validating the ability to handle whole exomes, we plan to begin extending the system to support whole genomes. We are currently exploring enhancements to enable capture of not only the variants that are identified in a patient, but also test quality information including confidence/quality scores for the variants identified and coverage information to define areas of a large test (gene panel, exome, genome) that may or may not have been covered fully.
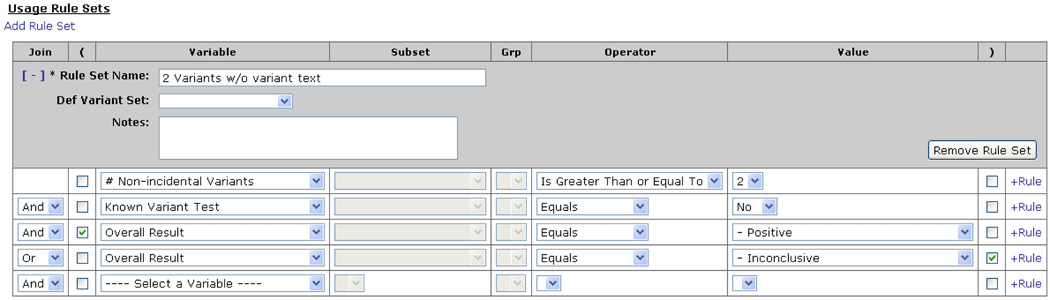
Once GI Lab has the patient and variant information, it proposes an overall test result and drafts a report based on rules and templates maintained by the geneticist and genetic counselor users of the system. The body of each report is composed of five sections: non-incidental (pathogenic, likely pathogenic, or unknown) DNA variants, interpretation, recommendation, comments, and incidental variants (benign or likely benign). Each text section is driven by templates that are maintained by end-users at the disease/pharmacogenomic effect level. Users, typically geneticists or genetic counselors, can create an unlimited number of templates. We have found that it is important for end-users to be able to create and maintain these templates without the involvement of IT personnel. GI provides screens that enable laboratory personnel to create and maintain rules and templates. Rules are attached to each template created. (Figure 1) These rules enable the system to determine which template should be applied to each case. The rules can be driven off order information, test information, or variant information. As of this writing, 38 variables are available for use in template selection rules. Complex rules can be constructed by using logical And/Or operators to concatenate multiple conditions. For example, a rule might specify that a template should only be used for a positive known familial variant test, or another may select a template based on the number of non-incidental variants identified, a positive overall result, and a certain ethnic background. The system also holds fields for defining patients' symptoms that range from structured fields to enter general categories (affected or unaffected) or specific disease diagnoses as well as a free text field to enter the patient specific clinical features.
Figure 1.
Specifying Selection Rules Associated with a Template. This functionality enables users to specify conditions that must be met for a template to be included in a report.
End-users also maintain the content of each template. (Figure 2) This content consists of text that may include dynamic variables that are replaced with a specific variant, test, or patient information associated with each case. For example, when setting up a template, the end-user might insert a dynamic variable that is replaced with the patient’s race when a report is drafted. Another dynamic variable may be replaced with the amino acid change of the first non-incidental variant. There is also a dynamic variable that enables users to embed other templates with a template, which simplifies the template maintenance process.
Figure 2.
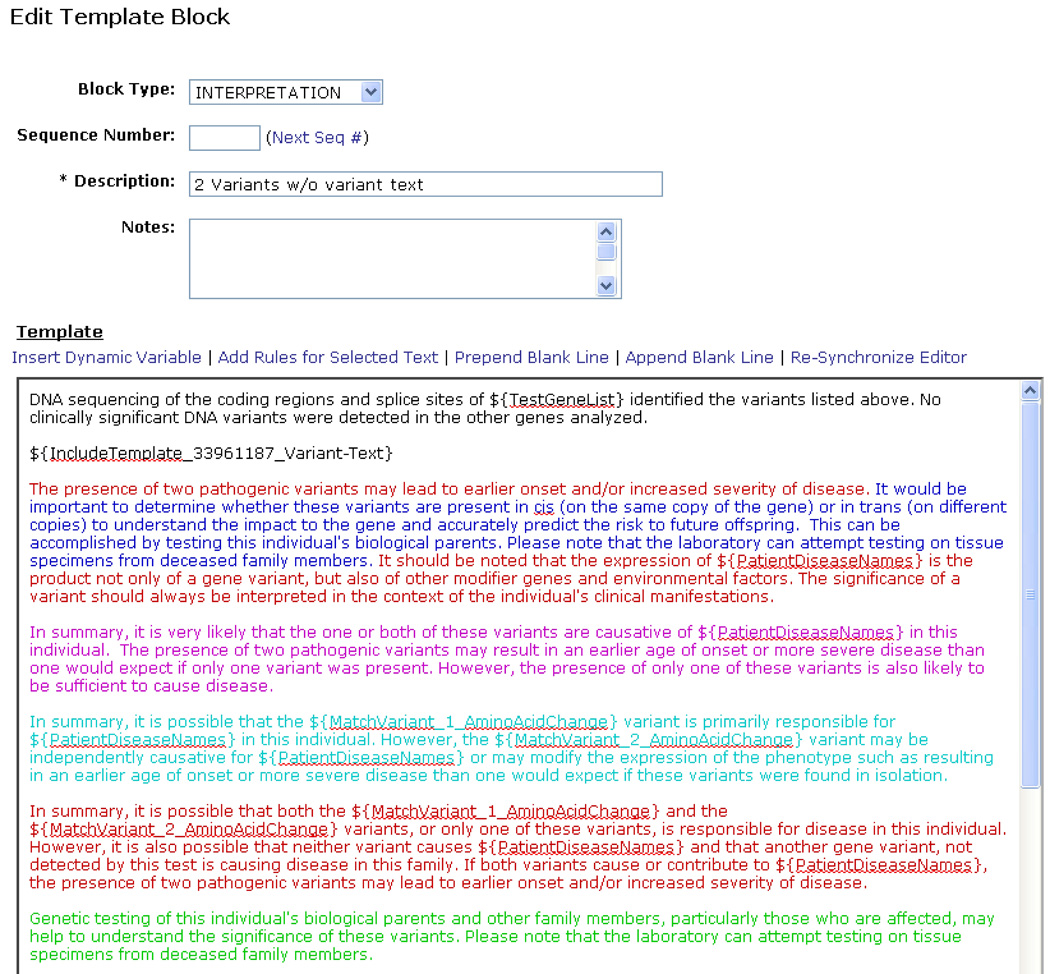
An example template. Dynamic variables within the text block appear as ${VariableName}. These are replaced with case specific information when a report is drafted. Colored text is only included if an associated rule is met. A means of underlining text associated with a rule is provided to enable color blind individuals to operate the system. ${IncludeTemplate…} is an example of a link to another template which generates text to support the variant interpretation.
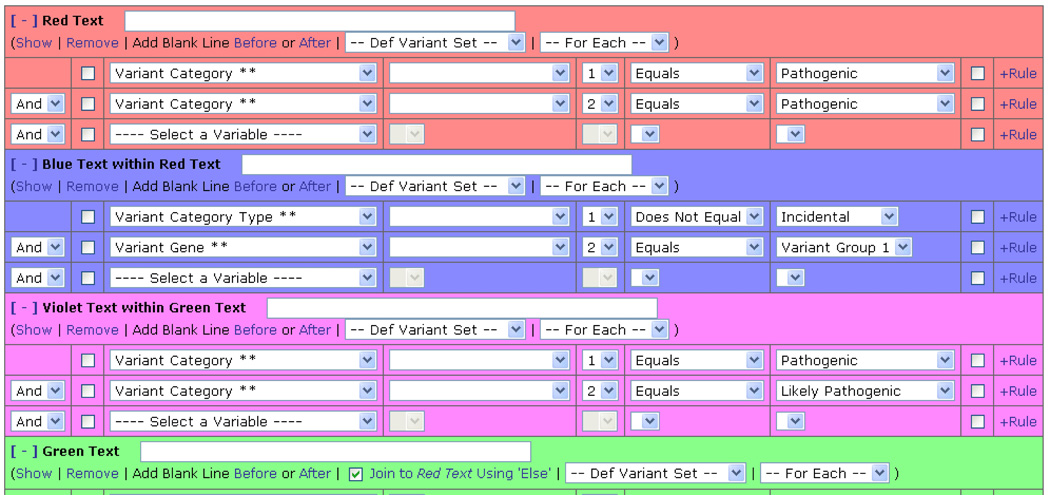
Users can also make text within a template conditional. (Figure 3) This is done by highlighting text within a template and then associating a rule with that text. We have found that users will typically apply these rules to whole paragraphs within templates. But at times they also restrict a rule to an individual sentence or even an individual word.
Figure 3.
Examples of conditional rules associated with template text. These rules are applied to specific text within a template. The text in the template is color coded to match the rule. The “Show” link can also be used to underline the associated text for color blind individuals. These rules are evaluated for each case and the associated text is only included in the report if the conditions are met.
Reports automatically drafted through this process are reviewed by a geneticist prior to signout if the system is supporting sequencing tests or complex genotyping tests. This provides an opportunity to review the output of the templates and make modifications as required. Once a report is signed out, it is sent to connected GI Clinics as appropriate. An example of a test report is shown in Supp. Figure S1. Changes to variant interpretations can also be forwarded at future time-points.
While GI Lab was originally designed for, and is currently only used by, reporting laboratories, the system could also support curators of genetic knowledge who do not run laboratory tests. There are many situations where this may be helpful, including efforts under consideration to create centrally-managed, clinically-validated public knowledgebases, commercial enterprises interested in offering curated knowledge on a subscription basis, and treating clinicians willing to distribute their knowledge about best practices for treating patients with specific variants. Enhancements would be required to optimize the system to support these usages, but the core required functionality is present.
GI Clinic
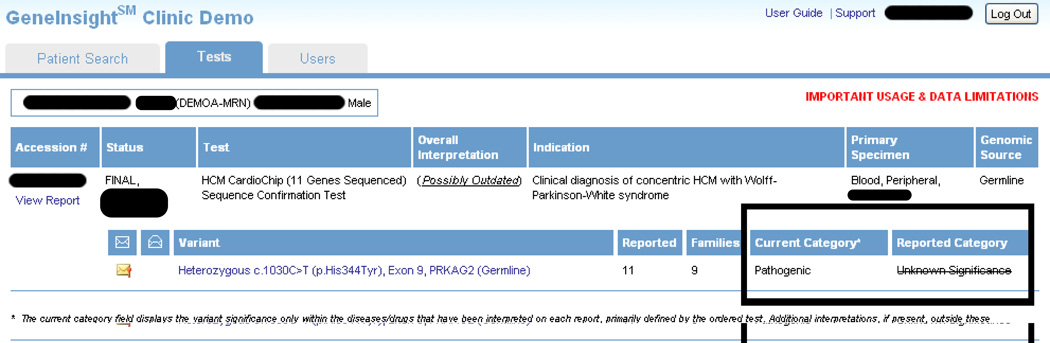
GI Clinic has functionality designed to help treating clinicians manage their patient genetic profiles and stay up-to-date through proactive alerts when new information emerges on a variant previously detected in one or more of their patients. The system enables clinicians to search for patients using a variety of different filters. Once a patient is identified, they can view genetic tests that have been received for that patient. (Figure 4) When a category is updated in a connected GI Lab, this information is used to generate both email and screen updates for all cases where the laboratory identified the variant. As of this writing, GI Clinics are in use at Brigham and Women’s Hospital, Massachusetts General Hospital, and the University of Michigan.
Figure 4.
The GeneInsight Clinic Patient Screen. Treating providers can use this screen to review genetic reports associated with a patient. The box highlights a knowledge change.
Each GI Clinic instance only displays results for tests ordered by that institution. Currently, each authorized user has access to all results housed within their institution’s GI Clinic. However, in our next release, currently scheduled for April 2011, we will include functionality that by default restricts a clinician’s access to reports where they are specifically referenced as an associated provider. Clinicians will have to provide a reason for searching for the reports where they are not listed.
We hope to add functionality to a future release of GI Clinic that will enable users to enter clinical annotations, including clinical symptoms, on patients. Our intention is to bundle this functionality with order entry support. Our goal will be to combine these two functions in a way that will streamline treating clinician workflow while enabling the efficient capture of this additional information.
Building a Network
At present, providers who do significant amounts of genetic testing are likely to order tests from multiple laboratories. In order for these providers to maintain complete patient genetic profiles, they either need to establish connections to all of these laboratories or manually re-enter results from laboratories to which they are not connected. Manual re-entry is suboptimal because it is labor-intensive and can increase the risk of error relative to electronic interfaces. In addition, as the scope of DNA assayed in genetic tests continues to grow, it will become more difficult for each individual laboratory to fully interpret all the variants their tests might identify. Ultimately, it is unlikely that any institution will be able to fully interpret whole exome results, much less whole genome results.
We are collaborating closely with the Partners HealthCare Longitudinal Medical Record team, ARUP, and Intermountain HealthCare to build the interfaces required to enable high-throughput delivery of genetic results into the electronic health record (EHR) environment through GI Clinic.
Our goal in creating GI is to establish the networking components needed to cost-effectively and securely link together providers, laboratories, and other sources of curated genetic knowledge. Our hope is to create a federated network that will enable full clinical curation of the genome by linking a critical mass of institutions capable of annotating different diseases and genomic regions. Currently, GI Clinics can be established for each GI Lab customer. More work is needed, and actively underway, to deepen this capability to fully support the federated network vision.
We currently deploy GI Lab and Clinic instances through a hosted model. Each GI Lab and GI Clinic instance is fully separated from the others. As a result, users can only access patient data associated with their own laboratory or clinic. The physical servers that support this model are housed in a Partners HealthCare data center. We have offered the option of installing GI instances at end user sites but thus far economic and support considerations have always favored the hosted model.
Thus far we have been able to setup and maintain the basic GI Clinic service without charging a fee to end users. We intend to maintain this policy as long as possible but do not know whether we will be able to do so indefinitely. We will need to charge for extended services based on these GI Clinic instances such as interface projects, manual entry of reports from unconnected laboratories and expanded clinical decision support functionality. We charge a hosting fee for setting up and maintaining instances of GI Lab.
Regulatory Status
We have registered GI with the FDA as a class I exempt medical device. This means it is subject to inspection and must comply with quality regulations but does not involve premarket approval.
Supplementary Material
Acknowledgments
The funding for development of the GeneInsight Suite was provided by Partners HealthCare. The rollout of GeneInsight Clinic modules was supported, in part, by grant number RC1LM010526 from the National Library of Medicine and the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Library of Medicine or the National Institutes of Health. License fees for GeneInsight Laboratory also supported this effort.
Footnotes
Supporting Information for this preprint is available from the Human Mutation editorial office upon request (humu@wiley.com)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.