Abstract
Molecular chaperone proteins play a pivotal role in the protozoan parasite Leishmania donovani, controlling cell fate and ensuring intracellular survival. In higher eukaryotes, the so-called co-chaperone proteins are required for client protein recognition and proper function of chaperones, among them the small glutamine-rich tetratricopeptide repeat proteins (SGT) which interact with both HSP70 and HSP90 chaperones. An atypical SGT homolog is found in the L. donovani genome, encoding a protein lacking the C-terminal glutamine-rich region, normally typical for SGT family members. The gene is expressed constitutively during the life cycle and is essential for survival and/or growth of the parasites. LdSGT forms large, stable complexes that also include another putative co-chaperone, HSC70 interacting protein (HIP). The gene product forms cytoplasmic clusters, matching the subcellular distribution of HIP and partly that of the major cytoplasmic chaperones, HSP70 and HSP90, reflecting a direct molecular interaction with both chaperones.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-009-0160-7) contains supplementary material, which is available to authorized users.
Keywords: Leishmania, Tetratricopeptide repeat, SGT, Foldosome complex, Co-chaperone
Introduction
Parasites of the genus Leishmania cause a variety of human disease syndromes, ranging from self-healing skin lesions to lethal, generalized infections that are difficult to control. The parasites are found on four continents, with 12 million actively infected humans and another 350 million more at risk. Transmitted by sandflies as flagellated promastigotes, the parasites differentiate into nonmotile amastigotes inside mammalian macrophages. The concomitant destruction of macrophages and the spread within the reticuloendothelial system are responsible for the pathology of Leishmania infections. The conversion from the promastigote to the amastigote stage is pivotal for parasite survival and linked to the parasite's heat shock response (Clos and Krobitsch 1999; Clos 2007).
Among the leishmaniae, Leishmania donovani stands out for its ability to undergo life cycle stage conversion in vitro, induced by changes in culture conditions. Simplified, an increase of cultivation temperature and a subsequent acidification suffice to trigger promastigote-to-amastigote conversion under axenic culture conditions. This allows the study of stage conversion and of the so-called axenic amastigotes in a biochemically well-defined context (Zilberstein and Shapira 1994; Barak et al. 2005; Clos 2007).
The chaperone HSP90 (HSP83) was found to play a crucial role in the stage conversion. Pharmacological inactivation of HSP90 by a variety of inhibitors such as geldanamycin, radicicol, and also taxol triggers the increased expression of heat shock proteins, a reduced growth, and the formation of amastigote-like culture forms, similar to those induced by heat shock and acidic shift (Wiesgigl and Clos 2001). This result indicates a pivotal role for HSP90 homeostasis in the regulation of cell fate and stage-specific gene regulation.
HSP90 is an essential protein in all eukaryotes where it is found in functionally distinct forms. HSP90 homodimers may serve as basic chaperones, binding to non-native proteins in cells exposed to unfavorable growth conditions. Higher-order complexes of HSP90 consist of the HSP90 dimer, HSP70, HSP40, and an assortment of so-called co-chaperones. Co-chaperones mediate the presentation of client proteins, regulate the ATPase activity of chaperones, change the fate of clients, and induce the formation of large chaperone complexes (Pratt and Toft 2003; Tobaben et al. 2003; Wandinger et al. 2008). These large complexes are also known as foldosomes and are crucial for the proper folding of post-translationally regulated proteins, such as transcription factors, cyclin-dependent kinases, MAP kinases, and steroid hormone receptors (Scheibel and Buchner 1998; Buchner 1999; Jackson et al. 2004).
Only limited information exists with regard to possible foldosome complexes in the leishmaniae (Webb et al. 1997). To further our knowledge of the HSP90 complexes, we screened the L. infantum Genome Project database for co-chaperone candidate genes. With a few notable exceptions, putative homologs to several co-chaperone families could be identified. Most of the identified co-chaperones belong to the tetratricopeptide repeat (TPR) protein family.
Among the identified genes, we focused on homologs of established foldosome components. A Sti-1/HOP homolog which interacts with HSP90 was previously described in Leishmania major (Webb et al. 1997). The gene product comprises three TPR domains, containing nine TPR motifs. Another gene related to Sti-1/HOP, dubbed HOP-2, and a gene encoding a protein similar to HSC70 interacting protein (HIP) were recently shown to be dispensable for growth of L. donovani promastigotes, for axenic amastigote differentiation, and for infectiousness towards cultured macrophage-like cells (Ommen et al. 2009). Small glutamine-rich TPR proteins (SGT) were first described as co-chaperones of the HSC70 chaperones and as possible negative regulators of the protein refolding activity of the HSP70/HSP40 complex (Liu et al. 1999; Angeletti et al. 2002). They are now known to be involved in a variety of cellular functions, including, but not restricted to, participation in endocytosis, interaction with beta-amyloid proteins and growth hormone receptors, and control of mitosis and apoptosis (Fonte et al. 2002; Schantl et al. 2003; Wang et al. 2005; Winnefeld et al. 2006; Yin et al. 2006; Buchanan et al. 2007).
As implied by their name, the SGT protein family is characterized by three characteristic sequence motifs: (1) a TPR domain, an all-helical structural motif affording protein–protein interactions, (2) an N′-terminal coiled-coil motif required for dimerization, and (3) a glutamine-rich region at the C terminus. The latter region shows a clustering of glutamine, asparagine, and proline residues and are thought to interact with short peptide segments composed of consecutive nonpolar amino acids, while the N′-terminal coiled-coil motif is thought to support the formation of SGT homodimers (Tobaben et al. 2003; Liou and Wang 2005; Worrall et al. 2008).
In L. donovani, as in other leishmaniae, only an atypical SGT family member (LinJ30_V3.2740) could be identified, lacking the characteristic glutamine-rich region. Given our interest in the co-chaperones, we characterized this SGT homolog and the encoded protein by reverse genetics, expression analyses, subcellular localization, and coprecipitation studies. We found the gene to be essential for promastigote growth and expressed in both life cycle stages. The protein localizes to clusters within the cytoplasm, strongly resembling the localization of another putative co-chaperone, HIP. Moreover, a molecular interaction with cytoplasmic chaperones is observed, further supporting a role of LdSGT in the formation of leishmanial foldosome equivalents.
Materials and methods
Sequence analysis
The genome databases of two Leishmania species (http://www.genedb.org) were used to screen for sequence homologs of known HSP90 co-chaperone proteins. The protein sequence of human co-chaperones was used in a direct BLAST search against the Leishmania genome databases (http://www.genedb.org/genedb/leish/blast.jsp) to select candidates with the best match.
The sequence of the L. infantum SGT ortholog (LinJ30_V3.2740) was entered into a BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to identify the closest matches from various phylogenetic entities. These amino acid sequences were then subjected to a ClustalW analysis using human HIP, another TPR protein, as root. Default settings of the MacVector 10.6 analysis suite were applied.
We searched for possible interaction partners for LdSGT using the Simple Modular Architecture Research Tool (SMART) algorithm made available at the EMBL website (http://smart.embl-heidelberg.de).
Leishmania donovani strain and cultivation
Promastigote stages of L. donovani 1SR (Rosenzweig et al. 2007) were grown at 25°C in supplemented M199 medium (Hubel et al. 1997). After electroporation, recombinant parasites were cultivated in supplemented M199 medium with the appropriate selection antibiotics. For routine cultivation, cells were grown to late log phase in order to maintain exponential growth. Cell density was determined using a Schaerfe System CASY Cell Counter.
In vitro stage differentiation
Axenic amastigotes of L. donovani 1SR were differentiated as previously described (Saar et al. 1998). Briefly, differentiation of promastigote towards axenic amastigote was induced by a combination of 37°C cultivation and a time-coordinated acidification of the culture medium to pH 5.5 (Doyle et al. 1991; Castilla et al. 1995; Gupta et al. 1996; Saar et al. 1998; Barak et al. 2005). Axenic amastigotes were cultured at 37°C with 5% CO2. For amastigote quantification, the CASY Cell Counter was run in cumulative cell volume mode to include cell clusters. Equal cell volume aliquots were applied to sodium dodecyl sulfate (SDS)-mediated lysis and reduction with dithiothreitol (DTT) and were then subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot.
Western blot analysis
For immunization, chaperone and co-chaperone genes were amplified and cloned into the expression vector pJC45 (Schlueter et al. 2000). The proteins were then expressed in Escherichia coli BL21(DE3) [pAPlacIQ] cells and purified using a Novagen His•Bind Resin as described (Clos and Brandau 1994). The purified proteins were then used to immunize NMRI mice. After boostering, sera were collected and tested in Western blots against the recombinantly expressed proteins and against L. donovani whole-cell lysate. Production of SDS cell lysates, discontinuous SDS-PAGE, and Western blot were preformed according to standard protocols. Briefly, membranes were treated with blocking solution (5% milk powder and 0.1% Tween 20 in Tris-buffered saline) before they were probed using the polyclonal antiserum (1:1,000 in blocking solution), followed by incubation with an antimouse IgG-alkaline phosphatase conjugate (1:5,000) as secondary antibody. Blots were developed using nitro blue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl phosphate.
Animal care and experimentation were performed in accordance with the German Federal Animal Protection Laws, in particular subsections 4, 7, and 10a, in the Animal Facility of the Bernhard Nocht Institute. Animals were euthanized in CO2 prior to final bleeds.
Construction and preparation of recombinant DNA
Approximately 1,000 bp of 5′ untranslated region (UTR) and 3′ UTR of the LdSGT gene were amplified enzymatically from genomic L. donovani DNA with primers that added EcoRI and Acc65I sites (5′ UTR) or BamHI and HindIII sites (3′ UTR) to upstream and downstream ends. SwaI sites were introduced to flank the constructs. The 5′ UTR amplificates were digested with EcoRI and Acc65I and ligated into pUC19 plasmid, digested with the same enzymes. The resulting plasmids were digested with BamHI and HindIII, and the 3′ UTR amplificates, cut with the same enzymes, were ligated between those sites. Afterwards, plasmid pUC19-SGT-5′-3′ UTR was opened with Acc65I and BamHI. Bleomycin resistance and puromycin resistance genes were amplified with primers that added Acc65I and BamHI sites to their 5′ and 3′ ends. Cut with those enzymes, the resistance markers were ligated into pUC19-SGT-5′-3′ UTR to yield pSGT5′bleo3′ and pSGT5′puro3′. After construction, the plasmids were amplified in E. coli, purified by cesium chloride density gradient ultracentrifugation (Sambrook et al. 1989), and linearized with SwaI to yield the constructs depicted in Fig. 3a. The fragments containing the recombination cassette were separated by agarose gel electrophoresis and purified using the Machery&Nagel NucleoSpin Extract II Kit.
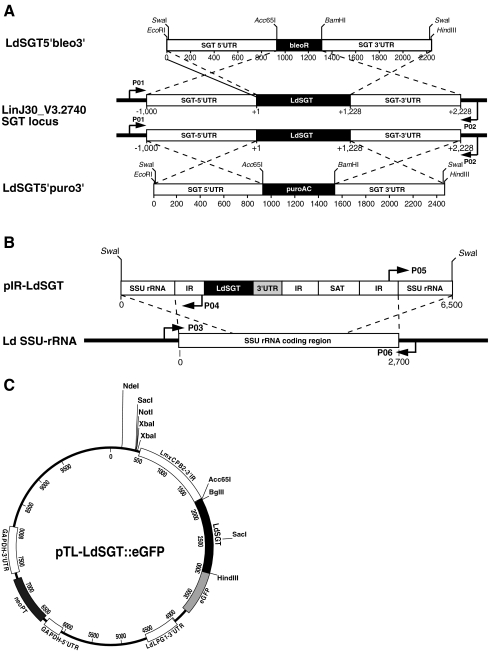
Fig. 3.
Schematic representation of the DNA constructs and PCR primers used in this study. a The drawing shows the L. infantum SGT gene locus with 5′ and 3′ UTR (open boxes) and ORF (closed box). The arrows symbolize the annealing regions for PCR primers used in Fig. 4a. The rulers show the length of the DNA in base pairs with each tick representing 200 bp. The top and bottom drawings show the two gene-replacement constructs. Cut sites for Acc65I, BamHI, EcoRI, HindIII, and SwaI are marked. b The drawing shows the add back construct, pIR-LdSGT, and an SSU rRNA-coding region. Arrows symbolize primer annealing regions, as used in Fig. 4b. SAT streptomycin acetyl transferase, 3′ UTR LdSGT 3′ untranslated region. c The map shows the pTL-LdSGT::eGFP expression vector. The construction is described in detail in Supplementary Fig. 8. LdSGT is fused to eGFP and flanked by the L. mexicana cysteine peptidase 2 intergenic region (IR) and the L. donovani LPG1 IR. The neomycin phosphotransferase selection marker gene is controlled by the flanking sequences of the Trypanosoma cruzi glyceraldehyde 3′-phosphate dehydrogenase (GAPDH) gene. Relevant restriction sites are marked
Construction and preparation of overexpression systems
A construct for the integration of an exogenous SGT gene copy into the 18S rRNA gene was generated by amplification of the open reading frame (ORF) of LdSGT, together with 1,000 bp of its natural 3′ UTR, using specific primers that added XbaI and BamHI sites to the 5′ and 3′ ends, respectively. The polymerase chain reaction (PCR) product was then ligated between the XbaI and BglII sites of the integration vector pIRmcs3+ (Hoyer et al. 2004). After construction, the plasmids were amplified in E. coli, purified by cesium chloride density gradient ultracentrifugation, and linearized with SwaI (Fig. 3b). The fragments containing the exogenous gene copy were separated by agarose gel electrophoresis and purified prior to electrotransfection.
For localization studies, an LdSGT::eGFP expression vector was constructed (Supplementary Fig. 8). The eGFP coding sequence was amplified with Acc65I and BamHI sites at the 5′ and 3′ ends and fused between the Acc65I and BglII sites of pIRmcs3+ (Hoyer et al. 2004) to yield pIReGFP. A 3,506-bp SacI–SalI fragment from pcosTL (Kelly et al. 1994) was cloned between the SacI and SalI sites of pUC19 (Yanisch-Perron et al. 1985). The product, pTL.v2, lacks the cos sequences of pcosTL and the single-strand origin of replication. The single HindIII site had to be deleted by HindIII digest, Klenow enzyme fill-in, and religation, to yield pTL.v3, to allow the use of HindIII at the gene insertion site. The 3,154-bp BamHI fragment from pIReGFP, containing the entire GFP expression cassette, was then ligated into the BamHI site of pTL.v3 to yield the vector pTL.v3-eGFP which allows the expression of eGFP and 3′-terminal eGFP fusion proteins from an episome. The coding sequence of LdSGT minus the stop codon was then amplified with Acc65I and HindIII sites at its 5′ and 3′ ends, digested with these enzymes, and fused in-frame between the Acc65I and HindIII sites of pTL.v3-eGFP to yield pTL-LdSGT::eGFP (Fig. 3c).
Electrotransfection and selection
Electrotransfection of Leishmania promastigotes was carried out essentially as described (Krobitsch et al. 1998). Leishmania parasites were taken from late log phase cultures (1–2 × 107 ml−1), sedimented (720 × g, 8 min, 4°C), and washed once with ice-cold phosphate-buffered saline (PBS) and once in prechilled electroporation buffer (Laban and Wirth 1989). Cells were resuspended in electroporation buffer at a density of 1 × 108 ml−1. Aliquots of 0.4 ml were mixed either with 2 µg of linearized DNA for homologous recombination or with 50 µg of circular DNA for stable episomal transfection. Electroporation was carried out in prechilled 4-mm electroporation cuvettes in a Bio-Rad Gene Pulser with three pulses at 1.5 kV (3,750 V/cm), 25 µF, and 200 Ω. A mock transfection was performed in identical fashion, but without DNA, to provide negative control strains for the antibiotic selection.
Following electroporation, cells were kept on ice for 10 min before they were transferred to 10 ml drug-free medium. Bleomycin (5 µg ml−1), puromycin (25 µg ml−1), or nourseothricin (150 µg ml−1) was added after 24 h and cultivation was continued until the mock transfection population had succumbed to the antibiotic pressure. For cloning, promastigotes were seeded in supplemented M199 medium at 0.5 cells per well in microtiter plates. After 10–14 days, wells positive for promastigote growth were identified and their content transferred to culture flasks.
Preparation of genomic DNA and PCR
Mutant genotypes were verified by PCR analysis of genomic DNA. For the preparation of genomic DNA, the Gentra Systems Puregene Tissue Core Kit A (Qiagen) was used, following the manufacturer's instructions.
Correct integration of gene-replacement constructs was verified by PCR using the primers listed below. Their relative positions are shown schematically in Fig. 3a, b. Products were separated by agarose gel electrophoresis and ethidium bromide staining. Primers used are as follows: P01 (GGCCAAGCGAGTCATGGAG), P02 (CATACATGTGTACGCATGC), P03 (ATTACATCAGACGTAATCTG), P04 (TCGGGAAGCCTTGAAAGTGAG), P05 (GTACGCCTTCCTTGACTTGC), and P06 (GAAAATGATCCAGCTGCAGG).
Expression profiling
Semiquantitative real-time reverse transcription polymerase chain reaction (RT-PCR) was performed essentially as described (Choudhury et al. 2008). SGT-specific primers were SGT-fwd (TGGGAACGGCTCTCTTCTACCAGG) and SGT-rev (TTGTGGGTGGCATTGTCGGG). SGT mRNA abundance was calculated relative to the actin signal.
Immunofluorescence and confocal microscopy
Log phase promastigotes (1 × 107 cells) were sedimented by gentle centrifugation (750 × g), washed twice with PBS, and suspended in 1 ml PBS. Aliquots (2 × 105 cells) of the suspension were applied on poly-l-lysine-coated microscopic slides. After fixing the cells for 10 min in ice-cold acetone, the slides were air-dried for 20 min. Nonadherent cells were removed by a gentle wash (0.1% Triton-X 100 in PBS) followed by incubation in blocking solution (2% bovine serum albumin, 0.1% Triton-X 100 in PBS). Slides were then incubated for 1 h with mouse antisera diluted 1:500 to 1:1,000 in blocking solution. Cells were washed thrice and then incubated for 1 h with antimouse (goat) secondary antibody coupled to Alexa 594 (Invitrogen #A11005; 1:2,500) and, simultaneously, with 4′,6-diamidino-2-phenylindole (DAPI; 1:50). After washing the slides thrice, Mowiol and cover slips were applied and the slides were left to harden for 24 h at 4°C.
Fluorescence microscopy was carried out on an Olympus FluoView1000 confocal microscope (SIM scanner and spectral detection).
Native electrophoresis
Extraction of nondenatured Leishmania proteins and native gradient gel electrophoresis was performed largely according to Westwood et al. (1991). Promastigotes (1 × 107) were harvested by centrifugation, washed twice with PBS, and resuspended in 40 µl extraction buffer (15% glycerol, 0.5 mM 1,10-phenanthroline, 10 mM Tris–HCl pH 8,0, 70 mM KCl). After four freeze and thaw cycles, cell lysates were cleared by centrifugation. The supernatant, containing the soluble protein fraction, was mixed (5:1, v/v) with loading buffer (50% glycerol, 0.1% bromophenol blue).
The samples were run alongside a high-molecular-weight protein marker for native gels (Amersham) on a 4–18% polyacrylamide (2.5–6% glycerol) gradient gel in 0.5× TBE buffer. Electrophoresis was allowed to proceed for 24 h at 4°C, at 20 V/cm. This long duration of electrophoresis was necessary for all proteins to migrate to their exclusion limit regardless of their net charge (Andersson et al. 1972; Clos et al. 1990). After that, the gel was incubated at 60°C in transfer buffer (48 mM Tris, 39 mM glycine, 0.5% SDS, 10 mM DTT) for 30 min, followed by Western blot analysis. A control gel was stained with Coomassie Blue R-250 to verify successful separation (data not shown).
Immune precipitation
Promastigote cells (1 × 108) of L. donovani [pTL-LdSGT::eGFP] or of wild-type L. donovani were washed twice in prechilled PBS and homogenized in 200 µl of cell lysis buffer (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.5 mM ethylenediaminetetraacetic acid [EDTA], 0.5% [v/v] NP-40, 1 mM phenylmethylsulphonyl fluoride [PMSF], 0.5 mM 1,10-phenanthrolin). After incubation on ice for 30 min, lysates were centrifuged (20,000 × g, 4°C, 10 min), and the supernatant was adjusted to a volume of 1 ml with dilution buffer (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA,1 mM PMSF, 0.5 mM 1,10-phenanthrolin). Aliquots of this fraction (50 µl) were mixed at 1:1 ratio (v/v) with 2× SDS sample buffer (“input” fraction). For pull down of immune complexes, 25 µl of GFP-Trap® Beads (Chromotek, Germany) were washed twice with ice-cold dilution buffer, mixed with cell lysate, and incubated for 2 h at 4°C under rotation. After a centrifugation step (2,000 × g, 4°C, 2 min), 50 µl of supernatant was mixed with 2× SDS sample buffer (“flow-through” fraction). The GFP-Trap beads were washed twice in dilution buffer, suspended in 1× SDS sample buffer and heated to 95°C for 10 min. Beads were sedimented by centrifugation (2,700 × g, 4°C, 2 min), and SDS-PAGE was performed with the supernatant (“bound” fraction) along with the “input” and “flow-through” fractions. Separated proteins were then stained either by Coomassie Brilliant Blue (loading control) or by immune blot.
Results
The Leishmania SGT is atypical
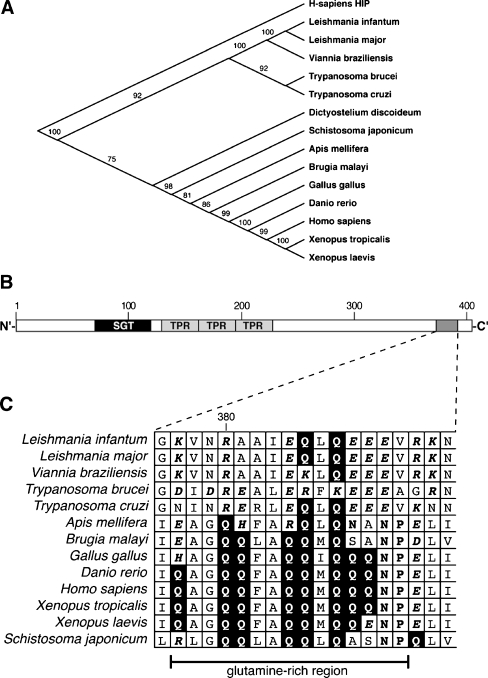
A sequence comparison of SGT family members from vertebrates, insects, nematodes, platyhelminths, and various protozoa shows the Kinetoplastida orthologs as separate branch (Fig. 1a), reflecting the phylogenetic distance of this order from the crown group Eukaryota. Normally, SGT is organized into three structural units with distinct functions: an N-terminal region for dimerization (SGT), a conserved TPR domain, and a C-terminal glutamine-rich region (Fig. 1b). In particular, we find that the C-terminal glutamine-rich sequence typical for SGT proteins of Metazoa is notably absent in the Kinetoplastida orthologs and replaced with a region rich in charged residues, in particular glutamate residues (Fig. 1c). The SGT homologs of the Kinetoplastida are, therefore, atypical. The replacement of a noncharged, hydrophilic domain with a highly charged domain may hint at a diverged function.
Fig. 1.
Phylogenetic analysis. a Using ClustalW, SGT amino acid sequences from the species indicated were aligned and a phylogenetic tree was assembled using the human HIP sequence as outgroup. Bootstrap values are indicated. b Schematic representation of the LdSGT primary structure. SGT the N′-terminal coiled-coil structure, TPR tetratricopeptide repeat. The shaded box near the C terminus represents a region rich in charged residues which is enlarged in c. c Alignment of the SGT sequences from a, showing the region equivalent to the glutamine-rich region of mammalian SGTs. Glutamine residues are shown in inverse print, asparagine and proline amino acids are in bold print and shaded, charged amino acids are in bold italic
Of note is that BLAST searches of the genomes of Apicomplexa, including Plasmodium falciparum and Toxoplasma gondii, did not reveal any putative SGT family members in those important parasites. Neither does Entamoeba histolytica harbor an SGT homolog.
Expression of LdSGT
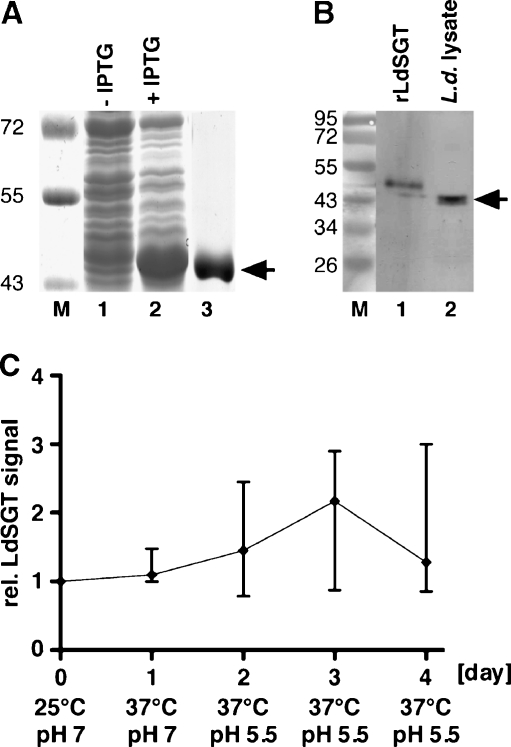
The L. donovani SGT gene was expressed with an N-terminal (His)10 tag in E. coli and purified by metal chelate chromatography (Fig. 2a). NMRI mice were immunized with the recombinantly expressed protein (rLdSGT) and polyclonal sera were obtained. The mouse antisera recognize the rLdSGT faithfully (Fig. 2b, lane 1). Moreover, the antisera also recognize a protein band of ~45 kD in a lysate of L. donovani promastigotes (Fig. 2b, lane 2), in keeping with the theoretical molecular mass of 45,800. The rLdSGT has an apparent molecular mass 2 kD larger than the natural protein due to a 20-amino-acid N-terminal His-Tag sequence (Clos and Brandau 1994).
Fig. 2.
Expression of LdSGT. a Bacterial expression and purification of LdSGT. E. coli BL21 (DE3)[pAPlacIQ] transformed with pJC45-LdSGT before (lane 1) and after (lane 2) induction with IPTG. The rLdSGT was purified using His•Bind resin (lane 3). The arrow points at the ~48 kD band representing the rLdSGT. The M.M. of protein marker bands (×1,000) are given to the left. b Immunoblot using anti-LdSGT mouse serum. Lane 1 rLdSGT, lane 2 L. donovani promastigote (5 × 106 cells) lysate. The arrow points at the ~46 kD band representing LdSGT. The M.M. of protein marker bands (×1,000) are given to the left. c Expression kinetic of LdSGT during promastigote-to-amastigote conversion. L. donovani promastigotes grown at 25°C and pH 7.0 were subjected to elevated temperature (37°C) and a subsequent acidification (pH 5.5). Cell volume equivalents equaling 5 × 106 cells were collected daily, lysed in SDS sample buffer, subjected to SDS-PAGE and Western blotting, and probed with anti-LdSGT serum. Identical control gels were stained with Coomassie Brilliant Blue. Western blots and Coomassie Blue-stained gels were digitalized. The anti-LdSGT bands were quantified using the ImageJ software and normalized against general Coomassie Blue staining on the lanes. The graph shows the statistical means of the relative LdSGT signal intensities (with SD) from four repeat experiments
Lysates from a promastigote-to-amastigote differentiation time course (Fig. 2c) show that expression of LdSGT is only marginally induced during amastigote stage conversion. The approximately twofold transient increase observed at day 3 is not statistically significant.
The anti-LdSGT antisera are, therefore, specific for the protein and reveal a constitutive expression of the protein in both in vitro life cycle stages.
Replacement of SGT in L. donovani
To unravel the function and role of LdSGT, we undertook to replace both alleles of the gene with selection marker genes. Constructs were generated in which the bleomycin resistance gene and the puromycin acetylase gene, respectively, are flanked by 1,000 bp each of the 5′ and 3′ UTR from the LdSGT gene (Fig. 3a). Also, a LdSGT add back construct was generated, with the LdSGT ORF and 1,000 bp of the LdSGT 3′ UTR fused into the pIRmcs3+ vector (Fig. 3b). The lack of the LdSGT 5′ UTR precludes the integration of the add back construct into the LdSGT gene locus.
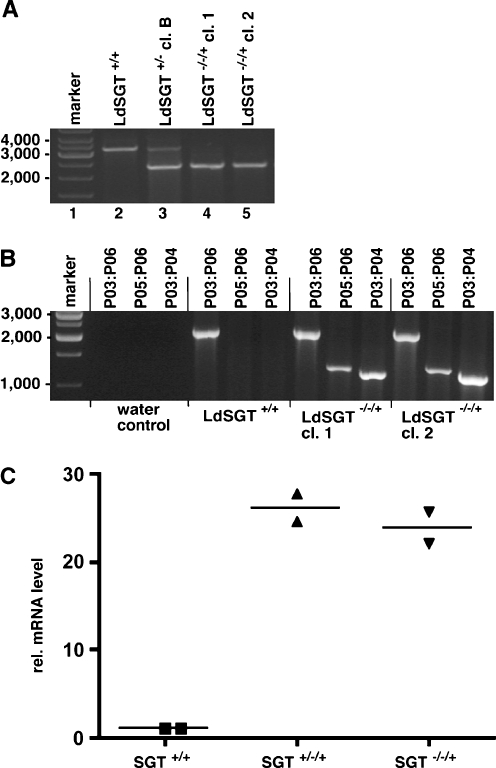
By transfecting wild-type L. donovani 1SR strain with the LdSGT5′bleo3′ construct, we were able to achieve single-allele gene replacement to yield a LdSGT+/− mutant (data not shown). Single-allele gene replacement was verified by amplification of the gene locus using a pair of primers that anneal on either side of the 5′ and 3′ UTRs (Fig. 3a). We observe a shortening of approximately one half of the PCR products, indicating a single-allele gene-replacement event.
We repeatedly attempted to replace the second SGT allele by transfection of the LdSGT+/− clones with the LdSGT5′puro3′ construct and a subsequent double selection under bleomycin and puromycin. Although we obtained several double-resistant clones, none of them were functional null mutants. This is a problem frequently encountered when essential genes or genes affecting promastigote growth are targeted. We, therefore, suspected LdSGT to be important for promastigote growth in culture.
To exclude the possibility that our gene-replacement constructs were faulty, we used the pIR-SGT add back construct (Fig. 3b) to introduce the LdSGT ORF and 3′ UTR into an 18S rRNA-coding region. The resulting LdSGT+/−/+ strain was tested for LdSGT overexpression by semiquantitative real-time RT-PCR (Fig. 4c). The add back gene in the 18S rRNA locus shows a 25-fold higher LdSGT RNA level compared with LdSGT+/+. This is in keeping with previous results with the pIRmcs3+ integration vector (Hoyer et al. 2004).
Fig. 4.
PCR analysis. a Verification of LdSGT gene replacement. Genomic DNA derived from wild-type L. donovani (LdSGT+/+), a single-allele replacement clone (LdSGT+/− clone B) and two putative LdSGT−/−/+ clones was used as template for PCR. Amplification of the SGT locus using primers P01 and P02 (Fig. 3). Primers anneal to sequences outside the region homologous to the gene-replacement constructs, yielding a predicted PCR product of 3,219 bp. Replacement of the SGT ORF with BleoR and PuroAC, respectively, results in PCR products of 2,368 and 2,591 bp. b Test for correct integration of the add back gene. Genomic DNA from LdSGT+/+ and two LdSGT−/−/+ clones was used as template for PCR. The primer pair P03:P04 or P05:P06 (Fig. 3), as indicated above the lanes, only allow the amplification of 1,156 and 1,247 bp products if the LdSGT add back construct are correctly integrated into an 18S rRNA gene unit. The combination of primer P03 and P06 was used as positive control. c Semiquantitative real-time RT-PCR analysis of LdSGT mRNA. RNA isolated from wild-type L. donovani LdSGT+/+, from a SGT single-allele mutant with add back LdSGT+/−/+, and from a LdSGT−/−/+ clone was reversely transcribed. The resulting cDNAs were used for semiquantitative real-time PCR using specific primers for LdSGT. For normalization of the results, actin mRNA was amplified, too. The analysis was performed in duplicate from each sample; the horizontal bars mark the mean of two results. The Y-axis shows the relative mRNA levels calculated relative to the actin signal
The add back strain LdSGT+/−/+ was then subjected to second-allele gene replacement using the LdSGT5′puro3′ construct. Triple selection under bleomycin, nourseothricin, and puromycin was applied. At the third attempt, the triple selection yielded proliferating promastigotes.
Amplification of the LdSGT gene locus yielded no natural length products, indicating that both natural alleles were replaced by selection marker genes (Fig. 4a). Moreover, the correct integration of the add back gene (Fig. 4b) and the integration of the selection marker genes into the LdSGT locus (data not shown) were verified by PCR analysis. This is evidence that the gene-replacement constructs can readily recombine with the target gene.
Neither the monoallelic gene-replacement mutant nor the double-allele gene-replacement mutant with add back show overt phenotypes in vitro under standard culture conditions.
Our results strongly suggest that replacement of both LdSGT alleles is not compatible with in vitro promastigote growth. LdSGT is probably an essential gene of L. donovani, at least during the promastigote stage.
Subcellular localization studies
For indirect immune fluorescence, we first ascertained the specificity of the antisera raised against chaperones and co-chaperones. L. donovani lysates were separated by SDS-PAGE, subjected to Western blot, and probed with the antisera. All antisera faithfully recognized protein bands at the expected position, and no cross-specificity to other co-chaperones or chaperones could be detected, except for the anti-HOP-2 antiserum which showed a weak reactivity with L. donovani Sti-1/HOP (data not shown).
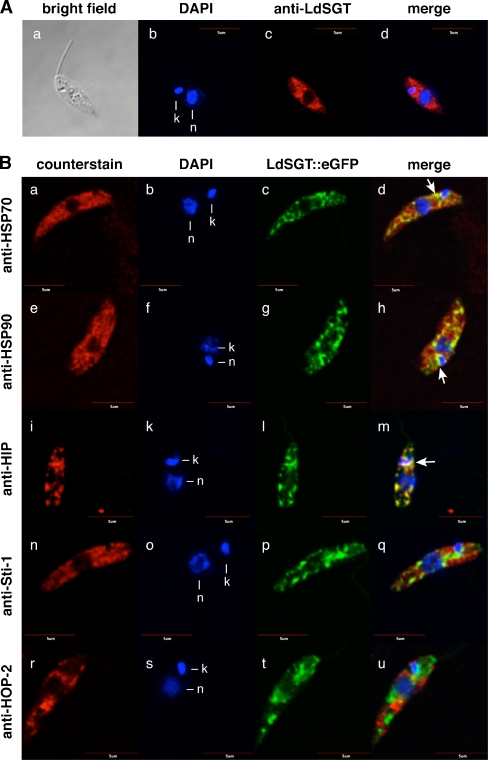
Indirect immune fluorescence using the anti-LdSGT antisera stains promastigotes in a nonhomogenous, cytoplasmic pattern (Fig. 5a a–d). Neither the nucleus nor the DNA-containing kinetoplast shows anti-LdSGT staining. The antiserum appears to react preferentially with the anterior part of the parasite.
Fig. 5.
Colocalization studies by confocal microscopy. a Subcellular LdSGT localization in promastigotes by indirect fluorescence microscopy. Wild-type promastigotes were fixed and stained with DNA dye DAPI (b) and with anti-LdSGT (c) before visualizing by indirect immunofluorescence. A bright field image of the cell was obtained (a), the merged image (d) exclude the bright field figure. Bar = 5 µm. b Colocalization studies of LdSGT::eGFP with several chaperones and putative co-chaperones in fixed promastigotes. Wild-type promastigotes transfected with pTL-LdSGT::eGFP were fixed and stained with DAPI (b, f, k, o, s) and either anti-HSP70 (a), anti-HSP90 (e), anti-HIP (i), anti-Sti-1 (n), or anti-HOP-2 (r) and visualized by indirect immunofluorescence. C-terminal GFP-tagged LdSGT fusion protein was revealed by direct fluorescence (c, g, l, p, t). Overlays of direct GFP fluorescence, DAPI, and indirect immunofluorescence images are shown in bd, h, m, q, and u. The figure shows representative results of several independent experiments. Yellow color is indicative of colocalization. Arrows indicate clusters where the fluorescent patterns colocalize. Bar = 5 µm
We also introduced an episomal LdSGT::eGFP chimeric gene in L. donovani. Green fluorescence is again observed in nonhomogenous pattern in the anterior part of the cytoplasm, mostly close to the nucleus (Fig. 5b c, g, l, p, t).
Making use of the LdSGT::eGFP chimera, we next attempted to compare the subcellular localization of LdSGT to that of a number of chaperones and putative co-chaperones. Using mouse sera against HSP70 (Fig. 5b a–d), HSP90 (Fig. 5b e–h), HIP (Fig. 5b i–m), Sti-1/HOP (Fig. 5b n–q), and HOP-2 (Fig. 5b r–u), we compared the fluorescence patterns. No significant overlap was found for Sti-1/HOP or HOP-2 co-chaperones. However, we found a partial overlap for the fluorescence patterns of LdSGT::eGFP and the major chaperones, HSP70 and HSP90, indicating that LdSGT may colocalize partly with these heat shock proteins.
LdSGT shows a highly congruent fluorescence pattern to another TPR domain protein, the putative HIP homolog. This protein is encoded by a nonessential gene that may be replaced readily (Ommen et al. 2009). The LdSGT::eGFP chimera and the immunofluorescent stain with anti-HIP sera highlight identical subcellular patterns in the cytoplasm (Fig. 5b i–m), strongly indicating colocalization.
Native gradient gel electrophoresis
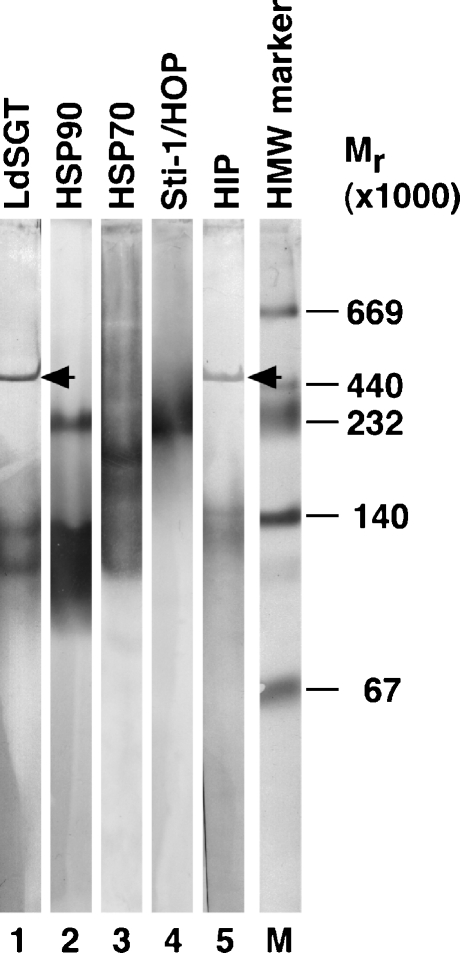
Coimmune precipitation experiments yielded ambiguous results only (data not shown) and did not allow us to draw conclusions as to direct interactions between LdSGT and other co-chaperones or chaperones. Therefore, we resorted to an analysis of the native pore exclusion sizes of chaperone and co-chaperone complexes in a native gradient gel electrophoresis. We produced lysates from L. donovani promastigotes and separated the native, soluble proteins in a nondenaturing polyacrylamide gradient gel. After equilibrium electrophoresis, protein complexes were denatured, reduced, and blotted onto a polyvinylidene fluoride (PVDF) membrane for detection with antisera. Antisera against LdSGT recognize defined protein bands, representing complexes of 470, 130, and 110 kD (Fig. 6, lane 1). The LdSGT complexes do not comigrate with the native forms of either HSP90 (230 and 90–140 kD, lane 2) or HSP70 (200 kD, lane 3). The Sti-1/HOP complex comigrates with HSP90 (230 kD, lane 4), but not with LdSGT. By contrast, the anti-HIP serum recognizes bands at identical position to those bound by anti-LdSGT (460, 140, and 130 kD, lane 5), suggesting that both proteins are part of the same 470-kD multiprotein complex. Thus, the two putative co-chaperones, LdSGT and HIP, remain associated during 24 h of electrophoresis, indicating a strong, stable interaction.
Fig. 6.
Nondenatured size of protein complexes formed by LdSGT and several other putative chaperones and co-chaperones by native gradient gel electrophoresis. Nondenatured proteins from 1 × 107L. donovani wild-type promastigotes were separated in a 0.5× TBE-buffered polyacrylamide gradient gel. The gel was run for 24 h for the complexes to reach their size exclusion limit. The native protein complexes were then denatured, reduced, and blotted onto a PVDF membrane for immunoblot analysis. The blot was probed with antisera against LdSGT (lane 1), HSP90 (lane 2), HSP70 (lane 3), Sti-1 (lane 4), and HIP (lane 5). Protein bands were visualized by alkaline phosphatase reaction. The sizes of marker proteins are indicated to the right of the respective panel (lane M). The arrowheads point at bands at 470 kD. The lanes are derived from one representative immunoblot out of four independent experiments
GFP-Trap coprecipitation experiments
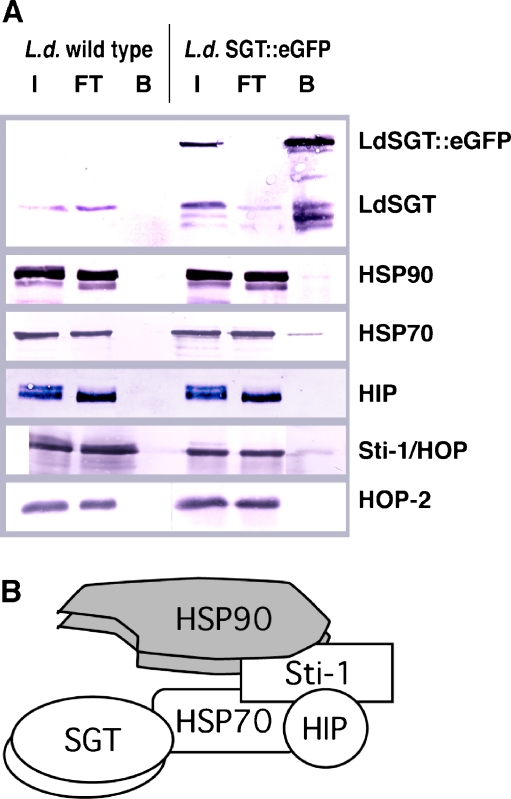
In order to identify direct molecular interactions of LdSGT with heat shock proteins and co-chaperones, we performed coprecipitation experiments using the GFP-Trap technology and LdSGT::eGFP chimera. Probing of the coprecipitated fractions indicate multiple interactions of LdSGT::eGFP (Fig. 7a). Firstly, the product of the endogenous LdSGT alleles is coprecipitated. This indicates that LdSGT is present in duplicate in native protein complexes. Secondly, we also detect HSP90 and HSP70 in the coprecipitates with LdSGT::eGFP. This finding establishes LdSGT as a bona fide co-chaperone of HSP70 and HSP90. Moreover, we find a signal for Sti-1/HOP in the coprecipitates. HOP-2 was not found in the “bound” fraction, as was expected from the previous results. By contrast, the absence of HIP from the coprecipitate was unexpected and will be discussed below. No heat shock proteins or co-chaperones could be precipitated from lysates of wild-type L. donovani, confirming the specificity of the GFP-Trap resin.
Fig. 7.
a GFP-Trap experiments. Lysates from L. donovani wild-type or L. donovani [pTL-LdSGT::eGFP] cells were bound to GFP-Trap-A resin and eluted. Fractions (I input, FT flow-through, B bound) were subjected to SDS-PAGE and Western blotting with mouse antisera against selected chaperones and co-chaperones as indicated to the right. Note that staining intensity does not necessarily reflect quantities due to different development parameters. b Schematic drawing of SGT-containing, mammalian foldosome complex redrawn from Buchanan et al. (2007)
Use of prediction algorithms
To test whether in silico prediction algorithms may support our experimental findings, we used the SMART algorithm (http://smart.embl-heidelberg.de) to predict possible interaction partners for LdSGT. The SMART algorithm at EMBL, Heidelberg, predicted an interaction of LdSGT with L. infantum HOP-2 with a score of 0.8. This is not confirmed by our experimental data, at least during the promastigote stage. Our colocalization and coprecipitation studies contradict such an interaction, as both proteins localize to different areas in the cytosol (Fig. 5b r–u).
Discussion
We have analyzed a probable L. donovani ortholog of the human SGT protein (LdSGT). The Leishmania protein is atypical in its structure, harboring a highly charged amino acid sequence in place of the glutamine-rich region found in metazoan SGTs. The charged sequence appears to be special to the Kinetoplastida, hinting at an evolved function specific to that order. Interestingly, other lineages of parasitic protozoa seem to lack SGT homologs, among them important genera such as Plasmodium spp., causative agents of human and mammalian malaria, or Entamoeba spp.
Is the Leishmania “SGT” a true homolog of the mammalian SGTs? Judging by its primary structure, LdSGT is indeed the only candidate found in the Leishmania Genome Project databases. Running the LdSGT sequence against the Human Genome database also identifies human SGT as its closest structural match. LdSGT lacks the name-giving glutamine-rich sequence, but possesses both a TPR domain and the N′-terminal coiled-coil motif typical for the SGT family. Its interaction with HSP70 also argues in favor of LdSGT being a member of this family. It should be mentioned that the “HSP70” of the leishmaniae shows highest sequence similarity to mammalian HSC70 proteins and not to the heat-inducible HSP70 family members.
Gene-replacement analysis reveals that the LdSGT is an essential gene for proliferation and/or survival of the L. donovani promastigote. In spite of the successful integration of two marker genes, the resulting viable parasites had retained functional copies of LdSGT. Such expansion of gene copy number is a rare event and likely to occur only under strong survival pressure. Therefore, this is considered a criterion for the essentiality of a gene (Beverley 2003). Furthermore, the successful replacement of both wild-type alleles in the presence of an add back construct, but not in its absence, is also strong proof of a gene's importance for growth and/or viability. In the absence of RNA interference strategies for Old World leishmaniae (Robinson and Beverley 2003), the LdSGT gene is not accessible to further genetic characterization.
This finding is in agreement with earlier reports on the essentiality of the human SGT protein for cell proliferation (Winnefeld et al. 2006). RNA interference-mediated knockdown of SGT in HeLa cells caused a mitotic arrest in the metaphase. Currently, we have no knowledge whether the gene deletion in L. donovani causes similar defects as in human cells due to the lack of an RNAi system in Old World Leishmania spp. The constitutive expression in both morphological forms, promastigote and amastigote, would suggest that LdSGT is essential for the amastigote, too. Thus, LdSGT fulfills the criteria for a potential therapeutic target (Barrett et al. 1999), in particular due to its structural differences to mammalian SGTs.
In Mammalia, SGT forms a dimer bound to a foldosome complex (Fig. 7b) comprising two subunits of HSP90 and one each of HSP70, Sti-1/HOP, and HIP (Liou and Wang 2005; Buchanan et al. 2007). For Leishmania, our coprecipitation studies confirm interaction with HSP70, HSP90, and Sti-1/HOP, but not with HIP. However, two lines of evidence point at a probable interaction between LdSGT and the leishmanial HIP ortholog: (1) both proteins are found in stable complexes of ~470 kD, a size range that is not recognized by any of the other antichaperone sera we used and (2) the fluorescence of LdSGT::eGFP and the indirect immune fluorescence by anti-HIP in L. donovani promastigotes show virtually identical patterns, proving at least colocalization. Moreover, the native size of LdSGT-containing complexes of 470 kD is consistent with a hypothetical foldosome complex of LdSGT(2×), HSP90(2×), HSP70(1×), Sti-1/HOP(1×), and HIP(1×). The combined molecular mass of such a complex, using the hypothetical polypeptide masses, is 424,000, compared with the experimentally obtained 470,000. The fact that HSP90, HSP70, and Sti-1/HOP are heavily phosphorylated in L. donovani (Morales et al. 2008) may explain this difference.
It is peculiar that the ~470-kD complexes are not recognized by the HSP70 and HSP90 antisera. An explanation for this may be that these complexes constitute only a minor fraction of HSP70- and HSP90-containing complexes and are thus underrepresented in the native gel electrophoresis. By contrast, the coprecipitation enriches these complexes and may thus lead to detectable signals.
By contrast, LdSGT does not, in all likelihood, interact stably with the HOP-2 gene product. Antibodies against this putative co-chaperone indicate clearly different subcellular localization compared with the LdSGT::eGFP. This is further supported by the coprecipitation studies and contradicts the in silico search results for interaction partners.
Chaperone proteins in L. donovani may serve different functions than in eukaryotic model organisms. For instance, the HSP100 (ClpB) does not mediate temperature tolerance, nor does it form homohexameric complexes (Krobitsch et al. 1998; Krobitsch and Clos 1999) as does its counterpart in yeast (Sanchez and Lindquist 1990; Sanchez et al. 1992; Parsell and Lindquist 1994). We are only beginning to understand the functional variability of the chaperones and co-chaperones between different phyla and how this affects the operative range of client proteins that are activated by molecular chaperones (Johnson and Brown 2009). Therefore, caution should be exercised in the functional interpretation of structural similarities of genes and proteins from kinetoplastid protozoa to those of the other crown group Eukaryota.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Cloning strategy of pTL-LdSGT::eGFP, an expression vector for stable expression of LdSGT::eGFP fusion proteins in Leishmania. The figure illustrates the materials and methods, construction, and preparation of overexpression systems (PDF 91 kb)
Acknowledgements
The authors acknowledge the technical assistance of Dorothea Zander, Manfred Krömer (died 2009), and Stefanie Pflichtbeil. Doreen Gutzke created the pTL.v3-eGFP plasmid. M.C. was a fellow of the Vereinigung der Freunde des Tropeninstituts e.V., Hamburg. We also thank Wai-Lok Yau for the helpful suggestions.
Footnotes
Gabi Ommen and Mareike Chrobak contributed equally to this study.
References
- Andersson LO, Borg H, Mikaelson M. Molecular weight estimations of proteins by electrophoresis in polyacrylamide gels of graded porosity. FEBS Lett. 1972;20:199–201. doi: 10.1016/0014-5793(72)80793-2. [DOI] [PubMed] [Google Scholar]
- Angeletti PC, Walker D, Panganiban AT. Small glutamine-rich protein/viral protein U-binding protein is a novel cochaperone that affects heat shock protein 70 activity. Cell Stress Chaperones. 2002;7:258–268. doi: 10.1379/1466-1268(2002)007<0258:SGRPVP>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak E, Amin-Spector S, Gerliak E, Goyard S, Holland N, Zilberstein D. Differentiation of Leishmania donovani in host-free system: analysis of signal perception and response. Mol Biochem Parasitol. 2005;141:99–108. doi: 10.1016/j.molbiopara.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Barrett MP, Mottram JC, Coombs GH. Recent advances in identifying and validating drug targets in trypanosomes and leishmanias. Trends Microbiol. 1999;7:82–88. doi: 10.1016/S0966-842X(98)01433-4. [DOI] [PubMed] [Google Scholar]
- Beverley S. Genetic and genomic approaches to the analysis of Leishmania virulence. In: Marr JJ, Nilsen TW, Komuniecki R, editors. Molecular medical parasitology. New York: Academic; 2003. pp. 111–122. [Google Scholar]
- Buchanan G, Ricciardelli C, Harris JM, Prescott J, Yu ZC, Jia L, Butler LM, Marshall VR, Scher HI, Gerald WL, Coetzee GA, Tilley WD. Control of androgen receptor signaling in prostate cancer by the cochaperone small glutamine rich tetratricopeptide repeat containing protein alpha. Cancer Res. 2007;67:10087–10096. doi: 10.1158/0008-5472.CAN-07-1646. [DOI] [PubMed] [Google Scholar]
- Buchner J. Hsp90 & Co.—a holding for folding. Trends Biochem Sci. 1999;24:136–141. doi: 10.1016/S0968-0004(99)01373-0. [DOI] [PubMed] [Google Scholar]
- Castilla JJ, Sanchez-Moreno M, Mesa C, Osuna A. Leishmania donovani: in vitro culture and [1H] NMR characterization of amastigote-like forms. Mol Cell Biochem. 1995;142:89–97. doi: 10.1007/BF00928929. [DOI] [PubMed] [Google Scholar]
- Choudhury K, Zander D, Kube M, Reinhardt R, Clos J. Identification of a Leishmania infantum gene mediating resistance to miltefosine and SbIII. Int J Parasitol. 2008;38:1411–1423. doi: 10.1016/j.ijpara.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Clos J. The heat shock response in Leishmania spp. In: Radons J, Multhoff G, editors. Heat shock proteins in biology and disease. Kerala: Research Signpost; 2007. pp. 421–448. [Google Scholar]
- Clos J, Brandau S. pJC20 and pJC40—two high-copy-number vectors for T7 RNA polymerase-dependent expression of recombinant genes in Escherichia coli. Protein Expr Purif. 1994;5:133–137. doi: 10.1006/prep.1994.1020. [DOI] [PubMed] [Google Scholar]
- Clos J, Krobitsch S. Heat shock as a regular feature of the life cycle of Leishmania parasites. Am Zool. 1999;39:848–856. [Google Scholar]
- Clos J, Westwood JT, Becker PB, Wilson S, Lambert K, Wu C. Molecular cloning and expression of a hexameric Drosophila heat shock factor subject to negative regulation. Cell. 1990;63(5):1085–1097. doi: 10.1016/0092-8674(90)90511-C. [DOI] [PubMed] [Google Scholar]
- Doyle PS, Engel JC, Pimenta PF, Silva PP, Dwyer DM. Leishmania donovani: long-term culture of axenic amastigotes at 37 degrees C. Exp Parasitol. 1991;73:326–334. doi: 10.1016/0014-4894(91)90104-5. [DOI] [PubMed] [Google Scholar]
- Fonte V, Kapulkin V, Taft A, Fluet A, Friedman D, Link CD. Interaction of intracellular beta amyloid peptide with chaperone proteins. Proc Natl Acad Sci USA. 2002;99:9439–9444. doi: 10.1073/pnas.152313999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N, Goyal N, Kumar R, Agrawal AK, Seth PK, Rastogi AK. Membrane characterization of amastigote-like forms of Leishmania donovani. Trop Med Int Health. 1996;1:495–502. doi: 10.1046/j.1365-3156.1996.d01-90.x. [DOI] [PubMed] [Google Scholar]
- Hoyer C, Zander D, Fleischer S, Schilhabel M, Kroener M, Platzer M, Clos J. A Leishmania donovani gene that confers accelerated recovery from stationary phase growth arrest. Int J Parasitol. 2004;34:803–811. doi: 10.1016/j.ijpara.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Hubel A, Krobitsch S, Horauf A, Clos J. Leishmania major Hsp100 is required chiefly in the mammalian stage of the parasite. Mol Cell Biol. 1997;17:5987–5995. doi: 10.1128/mcb.17.10.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SE, Queitsch C, Toft D. Hsp90: from structure to phenotype. Nat Struct Mol Biol. 2004;11:1152–1155. doi: 10.1038/nsmb1204-1152. [DOI] [PubMed] [Google Scholar]
- Johnson JL, Brown C. Plasticity of the Hsp90 chaperone machine in divergent eukaryotic organisms. Cell Stress Chaperones. 2009;14:83–94. doi: 10.1007/s12192-008-0058-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JM, Das P, Tomás AM. An approach to functional complementation by introduction of large DNA fragments into Trypanosoma cruzi and Leishmania donovani using a cosmid shuttle vector. Mol Biochem Parasitol. 1994;65:51–62. doi: 10.1016/0166-6851(94)90114-7. [DOI] [PubMed] [Google Scholar]
- Krobitsch S, Clos J. A novel role for 100 kD heat shock proteins in the parasite Leishmania donovani. Cell Stress Chaperones. 1999;4:191–198. doi: 10.1379/1466-1268(1999)004<0191:ANRFKH>2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krobitsch S, Brandau S, Hoyer C, Schmetz C, Hübel A, Clos J. Leishmania donovani heat shock protein 100: characterization and function in amastigote stage differentiation. J Biol Chem. 1998;273:6488–6494. doi: 10.1074/jbc.273.11.6488. [DOI] [PubMed] [Google Scholar]
- Laban A, Wirth DF. Transfection of Leishmania enriettii and expression of chloramphenicol acetyltransferase gene. Proc Natl Acad Sci USA. 1989;86:9119–9123. doi: 10.1073/pnas.86.23.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou ST, Wang C. Small glutamine-rich tetratricopeptide repeat-containing protein is composed of three structural units with distinct functions. Arch Biochem Biophys. 2005;435:253–263. doi: 10.1016/j.abb.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Liu FH, Wu SJ, Hu SM, Hsiao CD, Wang C. Specific interaction of the 70-kDa heat shock cognate protein with the tetratricopeptide repeats. J Biol Chem. 1999;274:34425–34432. doi: 10.1074/jbc.274.48.34425. [DOI] [PubMed] [Google Scholar]
- Morales MA, Watanabe R, Laurent C, Lenormand P, Rousselle JC, Namane A, Spath GF. Phosphoproteomic analysis of Leishmania donovani pro- and amastigote stages. Proteomics. 2008;8:350–363. doi: 10.1002/pmic.200700697. [DOI] [PubMed] [Google Scholar]
- Ommen G, Lorenz S, Clos J. One-step generation of double-allele gene replacement mutants in Leishmania donovani. Int J Parasitol. 2009;39:541–546. doi: 10.1016/j.ijpara.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Lindquist S. Heat shock proteins and stress tolerance. In: Morimoto RI, Tissières A, Georgopoulos C, editors. The biology of heat shock proteins and molecular chaperones. Plainview: Cold Spring Harbor Laboratory Press; 1994. pp. 457–494. [Google Scholar]
- Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Medicine (Maywood, NJ) 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- Robinson KA, Beverley SM. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol Biochem Parasitol. 2003;128:217–228. doi: 10.1016/S0166-6851(03)00079-3. [DOI] [PubMed] [Google Scholar]
- Rosenzweig D, Smith D, Opperdoes F, Stern S, Olafson RW, Zilberstein D. Retooling Leishmania metabolism: from sand fly gut to human macrophage. FASEB J. 2007;22(2):590–602. doi: 10.1096/fj.07-9254com. [DOI] [PubMed] [Google Scholar]
- Saar Y, Ransford A, Waldman E, Mazareb S, Amin-Spector S, Plumblee J, Turco SJ, Zilberstein D. Characterization of developmentally-regulated activities in axenic amastigotes of Leishmania donovani. Mol Biochem Parasitol. 1998;95:9–20. doi: 10.1016/S0166-6851(98)00062-0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. Plainview: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanchez Y, Lindquist SL. Hsp104 required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Taulien J, Borkovich KA, Lindquist S. Hsp104 is required for tolerance to many forms of stress. EMBO J. 1992;11:2357–2364. doi: 10.1002/j.1460-2075.1992.tb05295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schantl JA, Roza M, Jong AP, Strous GJ. Small glutamine-rich tetratricopeptide repeat-containing protein (SGT) interacts with the ubiquitin-dependent endocytosis (UbE) motif of the growth hormone receptor. Biochem J. 2003;373:855–863. doi: 10.1042/BJ20021591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibel T, Buchner J. The Hsp90 complex—a super-chaperone machine as a novel drug target. Biochem Pharmacol. 1998;56:675–682. doi: 10.1016/S0006-2952(98)00120-8. [DOI] [PubMed] [Google Scholar]
- Schlueter A, Wiesgigl M, Hoyer C, Fleischer S, Klaholz L, Schmetz C, Clos J. Expression and subcellular localization of cpn60 protein family members in Leishmania donovani. Biochim Biophys Acta. 2000;1491:65–74. doi: 10.1016/s0167-4781(00)00028-2. [DOI] [PubMed] [Google Scholar]
- Tobaben S, Varoqueaux F, Brose N, Stahl B, Meyer G. A brain-specific isoform of small glutamine-rich tetratricopeptide repeat-containing protein binds to Hsc70 and the cysteine string protein. J Biol Chem. 2003;278:38376–38383. doi: 10.1074/jbc.M301558200. [DOI] [PubMed] [Google Scholar]
- Wandinger SK, Richter K, Buchner J. The Hsp90 chaperone machinery. J Biol Chem. 2008;283:18473–18477. doi: 10.1074/jbc.R800007200. [DOI] [PubMed] [Google Scholar]
- Wang H, Shen H, Wang Y, Li Z, Yin H, Zong H, Jiang J, Gu J. Overexpression of small glutamine-rich TPR-containing protein promotes apoptosis in 7721 cells. FEBS Lett. 2005;579:1279–1284. doi: 10.1016/j.febslet.2004.12.092. [DOI] [PubMed] [Google Scholar]
- Webb JR, Campos-Neto A, Skeiky YA, Reed SG. Molecular characterization of the heat-inducible LmSTI1 protein of Leishmania major. Mol Biochem Parasitol. 1997;89:179–193. doi: 10.1016/S0166-6851(97)00115-1. [DOI] [PubMed] [Google Scholar]
- Westwood JT, Clos J, Wu C. Stress-induced oligomerization and chromosomal relocalization of heat-shock factor. Nature. 1991;353:822–827. doi: 10.1038/353822a0. [DOI] [PubMed] [Google Scholar]
- Wiesgigl M, Clos J. Heat shock protein 90 homeostasis controls stage differentiation in Leishmania donovani. Mol Biol Cell. 2001;12:3307–3316. doi: 10.1091/mbc.12.11.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnefeld M, Grewenig A, Schnolzer M, Spring H, Knoch TA, Gan EC, Rommelaere J, Cziepluch C. Human SGT interacts with Bag-6/Bat-3/Scythe and cells with reduced levels of either protein display persistence of few misaligned chromosomes and mitotic arrest. Exp Cell Res. 2006;312:2500–2514. doi: 10.1016/j.yexcr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Worrall LJ, Wear MA, Page AP, Walkinshaw MD. Cloning, purification and characterization of the Caenorhabditis elegans small glutamine-rich tetratricopeptide repeat-containing protein. Biochim Biophys Acta. 2008;1784:496–503. doi: 10.1016/j.bbapap.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yin H, Wang H, Zong H, Chen X, Wang Y, Yun X, Wu Y, Wang J, Gu J. SGT, a Hsp90beta binding partner, is accumulated in the nucleus during cell apoptosis. Biochem Biophys Res Commun. 2006;343:1153–1158. doi: 10.1016/j.bbrc.2006.03.090. [DOI] [PubMed] [Google Scholar]
- Zilberstein D, Shapira M. The role of pH and temperature in the development of Leishmania parasites. Annu Rev Microbiol. 1994;48:449–470. doi: 10.1146/annurev.mi.48.100194.002313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
Cloning strategy of pTL-LdSGT::eGFP, an expression vector for stable expression of LdSGT::eGFP fusion proteins in Leishmania. The figure illustrates the materials and methods, construction, and preparation of overexpression systems (PDF 91 kb)