Abstract
Soluble CD4 (sCD4), anti-CD4 antibody, and anti-gp120 antibody have long been regarded as entry inhibitors in human immunodeficiency virus (HIV) therapy. However, the interactions between these HIV entry inhibitors and corresponding target molecules are still poorly understood. In this study, atomic force microscopy (AFM) was utilized to investigate the interaction forces among them. We found that the unbinding forces of sCD4-gp120 interaction, CD4 antigen-antibody interaction, and gp120 antigen-antibody interaction were 25.45 ± 20.46 pN, 51.22 ± 34.64 pN, and 89.87 ± 44.63 pN, respectively, which may provide important mechanical information for understanding the effects of viral entry inhibitors on HIV infection. Moreover, we found that the functionalization of an interaction pair on AFM tip or substrate significantly influenced the results, implying that we must perform AFM force measurement and analyze the data with more caution.
Keywords: atomic force microscopy (AFM), human immunodeficiency virus (HIV), entry inhibitor, soluble CD4 (sCD4), gp120, neutralizing antibody
Introduction
HIV infection of target cells is a multi-stage process involving the entry, replication, and budding of virus. Presently, many strategies have been developed for HIV therapy depending on distinct stages of the process, among which blocking HIV entry is a well-known, important one. Many entry inhibitors or drugs have been developed in the treatment of HIV infection by blocking the interactions of HIV envelop glyprotein gp120 with cell-surface CD4 or coreceptors, or HIV envelop protein gp41-mediated membrane fusion [1–4]. However, the interactions of the inhibitors or drugs with CD4 or gp120 or gp41 remain poorly understood.
Soluble CD4 (sCD4) and CD4-mimetic compounds are well known to inhibit HIV entry in vitro or in vivo [5–8]. It has been reported that sCD4 selectively inhibited HIV replication and syncytium formation [9] or inactivated HIV by inducing the release of gp120 [10, 11]. Recently, sCD4 and CD4 mimics were found to inhibit HIV infection by inducing a short-lived activated state of gp120 and spontaneously and irreversibly transforming gp120 into a non-functional conformation from the relatively long-lived activated intermediate induced by cell-associated CD4 [12]. Neutralizing antibodies against gp120 or cell-associated CD4 are also well-known HIV entry inhibitors and antibody-based vaccines [13–15]. These antibodies inactivate or neutralize or block the invading HIV virus by interacting with gp120 on viral surface or cell-surface CD4 on CD4+ lymphocytes.
Until now, unfortunately, no safe, effective vaccine against HIV-1/AIDS has been found [16, 17]. Therefore, the development of safe, effective vaccines is a top priority in HIV/AIDS research field. Accordingly, to investigate the interactions between HIV vaccines or inhibitors and their corresponding target molecules is very important for understanding the antiviral mechanisms of vaccines or inhibitors.
Recently, atomic force microscopy (AFM) has been widely applied in biological and viral studies [18–20]. AFM also has been used to image HIV viral particles and HIV-infected lymphocytes [21, 22]. Chang et al. investigated the HIV-1 gp120-receptor interactions in living cells [23]. More recently, the kinetics of gp41 (HIV fusion protein) interaction with lipid membranes was detected by AFM [24]. To date, however, there are no reported AFM studies on interaction forces between HIV inhibitors (e.g. sCD4, anti-CD4 or anti-gp120 antibody, etc.) and their target molecules. In this study, we recruited AFM force measurement to detect the sCD4-gp120 interaction and gp120 or CD4 antigen-antibody interaction.
Materials and methods
Reagents
Human soluble CD4 (Affinity BioReagents, Golden, CO), mouse monoclonal IgG1 against CD4 (Ab-2, clone 1F6; NeoMarkers, Inc, Fremont, CA), recombinant HIV-1MN envelope glycoprotein gp120 (Advanced Biotechnologies Inc., Columbia, MD), and mouse monoclonal IgG1 against HIV-1 gp120 (Clone ED8.D4; Abcam, Cambridge, MA) were purchased from different companies. 3-Aminopropyltriethoxysilane (APTES), Bovine Serum Albumin (BSA), glutaraldehyde, and others were from Sigma.
Functionalization of AFM tips and substrates
The method for tip and sample functionalization was modified from previous studies [25–27]. Briefly, all Silicon Nitride tips and freshly cleaved micas were incubated in 1% (v/v) 3-Aminopropyltriethoxysilane (APTES; Sigma) in toluene for 2 h, and rinsed in toluene for 5 min. Subsequently, they were incubated with 0.2% (v/v) glutaraldehyde in distilled water for 30 min, and then rinsed with distilled water for 5 min. In above steps, all tips and micas were always functionalized simultaneously in the same solutions. When functionalized with different proteins, these tips and micas were modified separately in 50 ul and 10 ul protein solution for 1 h, respectively. 1mg/ml sCD4, 1 mg/ml or 0.01mg/ml HIV-1 gp120, 1mg/ml anti-CD4 or anti-gp120 antibody, 1mg/ml monkey serum or 1 mg/ml BSA, or various mixtures of these proteins in distilled water were used for modification. After protein modification, all tips or micas were treated with glycine to block free aldehyde groups. All of them were rinsed with distilled water and then incubated in pH7.4 PBS buffer for use (generally within 12 h).
Force measurements by AFM
AFM data were collected using an Explorer AFM (Veeco, Santa Barbara, CA). The spring constants of the Si3N4 cantilevers were 0.01–0.03 N/m. All force measurements of antigen-antibody or ligand-receptor unbinding interaction were performed in 100 μl PBS buffer (pH 7.4) at room temperature.
During AFM measurements, we found that the experimental results may change dramatically with alternations of tips, substrates, sites, points, and even different times of measurement on the same point. To make the results more objective and accurate, we established the following guide lines in our AFM experiments: a) only one force-distance cycle was done at each point; b) around 20 force measurements were performed randomly in each 5 μm × 5 μm site; c) more than 10 sites at intervals of < 100 μm were measured following an detecting path (For each independent experiment, more than 10 sites were observed on a protein-immobilized substrate in a from-center-to-edge-and-then-back-to-center manner.); d) more than 2 functionalized AFM tips were used to detect proteins on a same substrate, and more than 2 substrates were measured for each pair of force interaction; e) data shown in each histogram were obtained using more than 3 different AFM probes and independent samples.
Data processing and statistics
The data obtained for the unbinding force was used to generate a frequency distribution. This was then analyzed by fitting multi-Gaussian distributions. The validity of distinct unbinding distributions was tested and confirmed using one-way ANOVA analysis of statistical variance, and by Student t-test.
Results
AFM force measurements detected the specific interactions between sCD4 and gp120
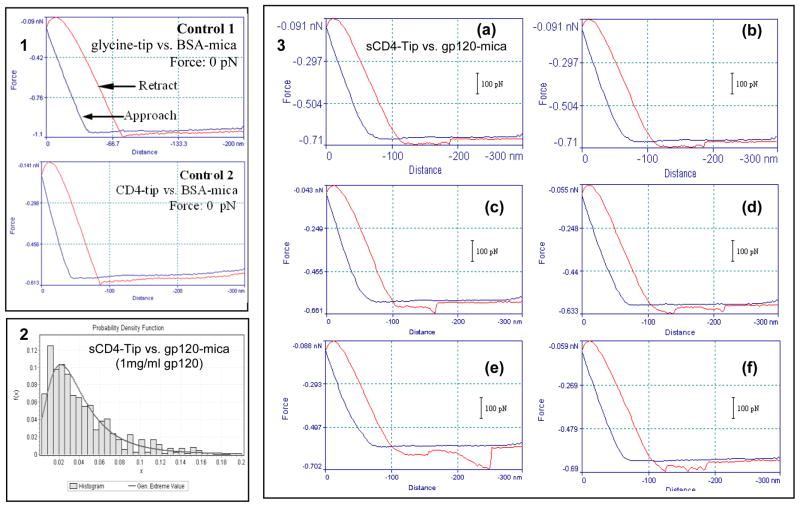
Prior to the force measurements of ligand-receptor and antigen-antibody specific interactions, we detected the non-specific interactions between glutaraldehyde- or glycine- or sCD4-functionalized AFM tips and glutaraldehyde- or glycine- or BSA (Bovine Serum Albumin)-modified micas. We found that most of the control non-specific interactions have a force of 0 pN (Panel 1 of Fig. 1 and data not shown) except for the non-specific interactions of glutaraldehyde-functionalized tip or mica with other molecules. For instance, the mean value of the non-specific interaction between the glutaraldehyde-functionalized tip and the glutaraldehyde-modified mica was 432.54 ± 233.98 pN (n=735).
Fig. 1. AFM force measurements of the sCD4-gp120 interaction.
Panel 1: representatives of force-distance curves for control no specific interactions. Most glycine-BSA interactions (upper) and sCD4-BSA interactions (bottom) have a force of 0 pN as shown here; Panel 2: force histogram for interaction between sCD4 and gp120 (1 mg/ml). Generalized Extreme Value: 25.45 ± 20.46 pN; Mean value: 42.25 ± 34.14 pN (n=705). Panel 3: representatives of force-distance curves for sCD4-gp120 interaction. (a and b) the interaction force is 26 pN; (c) the interaction force is 55 pN which indicates the simultaneous unbinding of two sCD4-gp120 specific interactions; (d) there are two peaks (the force is 24 pN for the first peak and 28 pN for the other peak) displaying double interaction points which indicates the sequent unbinding of two sCD4-gp120 specific interactions; (e) the interaction force is 105 pN which indicates the simultaneous unbinding of 3–4 sCD4-gp120 specific interactions; (f) there are three peaks (the forces are 31, 21, and 35 pN, respectively) displaying three interaction points which indicate the sequent unbinding of three sCD4-gp120 specific interactions. All these force-distance curves are representative examples from a total of thousands of curves.
Then, the sCD4-functionalized AFM tips were utilized to detect gp120 molecules (1mg/ml in PBS) on micas. The histogram in panel 2 of Fig. 1 showed a narrow distribution of sCD4-gp120 interaction forces with a generalized extreme value at 25.45 ± 20.46 pN and with a mean value at 42.25 ± 34.14 pN (n=705). We found that most force-distance curves for sCD4-gp120 interaction have only one peak of ~30 pN which represent the unbinding force between sCD4 and gp120 (Panel 3a,3b of Fig. 1). Occasionally, simultaneous unbinding of two or more sCD4-gp120 specific interactions (Panel 3c and 3e of Fig. 1, respectively) and sequent unbinding of two or more sCD4-gp120 specific interactions (Panel 3d and 3f of Fig. 1, respectively) were evident in an individual force-distance curves.
We also used the gp120-functionalized AFM tips to detect sCD4 molecules on micas. The data showed that the generalized extreme value and the mean value of sCD4-gp120 interaction force were 49.13 ± 32.41 pN and 69.23 ± 66.17 pN (n=331), respectively, which were larger than those obtained by sCD4-functionalized tip and gp120-modified mica.
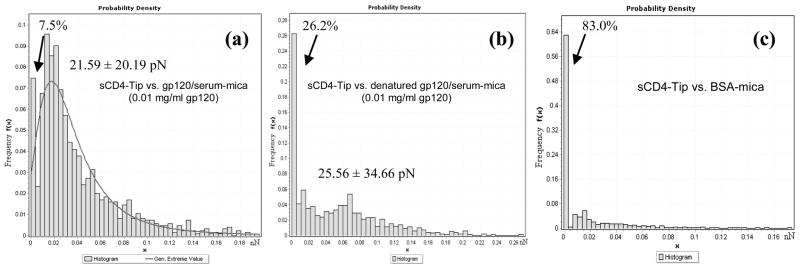
To further investigate the specificity of sCD4-gp120 interactions and the sensitivity of AFM force measurements, low-abundant (0.01mg/ml) gp120 molecules mixed with 1mg/ml monkey serum were measured by sCD4-functionalized tips (Fig. 2a). The data indicated that only 7.5% measurements displayed no interactions and that the generalized extreme and mean values of specific sCD4-gp120 interaction were 21.59 ± 20.19 pN and 39.11± 35.78 pN (n=1243), respectively. 0.01mg/ml denatured gp120 (heated at 100°C for 3 min) mixed with 1mg/ml monkey serum were also measured by sCD4-functionalized tips (Fig. 2b). As expected, the force measurements without specific interactions increased to 26.2%. However, the generalized extreme and mean values (25.56 ± 34.66 pN and 49.88 ± 49.78 pN, respectively; n=1000) of detectable sCD4-gp120 interaction forces were similar to those in Fig. 2a and panel 2 of Fig. 1. Additionally, ~83.0% measurements displayed no interactions for gp120-free BSA samples detected by sCD4-functionalized tips (Fig. 2c).
Fig. 2. Soluble CD4-modified AFM tips detected low-abundant gp120 proteins mixed with highly concentrated monkey serum proteins.
(a) Interaction force histogram obtained in PBS between sCD4 tips and the mixture of 0.01 mg/ml (low-abundant) gp120 and 1 mg/ml (high-abundant) serum. The histogram obtained from 1243 force-distance curves (n=1243) reveals a distribution of interaction forces with a generalized extreme value at 21.59 ± 20.19 pN (mean value: 39.11± 35.78 pN). The arrow indicates the curves without interaction force (~7.5%). (b) Interaction force histogram (n=1000) obtained in PBS using the same molecule pairs under the same condition as Fig. 2a, except that the gp120 proteins were denatured by heat at 100°C for 3 min prior to the mixing of gp120 with serum. Generalized extreme value: 25.56 ± 34.66 pN; mean value: 49.88 ± 49.78 pN. The arrow indicates the curves without interaction force (~26.2%). (c) Interaction force histogram (n=1000) between sCD4 tips and BSA (1 mg/ml). The arrow indicates the curves without interaction force (~83.0%).
AFM force measurements detected the specific interactions between CD4 or gp120 and anti-CD4 or anti-gp120 antibody
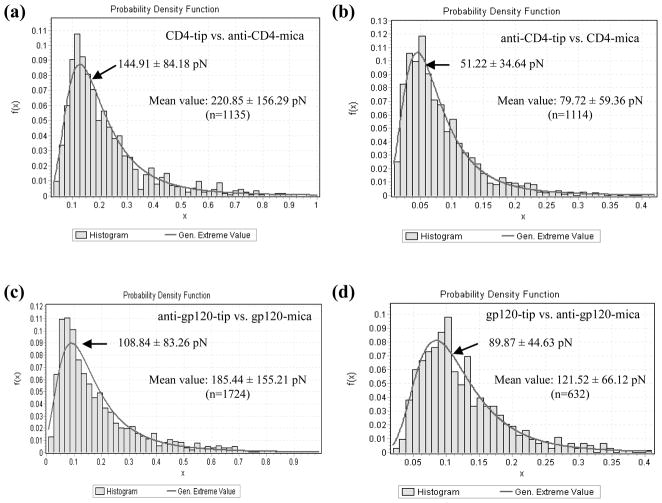
Subsequently, we functionalized the AFM tips with CD4 molecules and the micas with anti-CD4 antibodies, and then performed AFM force measurements in liquid. The data showed that the generalized extreme and mean values of specific interaction between CD4-tip and anti-CD4 antibody-mica were 144.91 ± 84.18 pN and 220.85 ± 156.29 pN (n=1135), respectively (Fig. 3a). At the same time, anti-CD4 antibody-modified tips were used to obtain the interaction forces again by detecting CD4 molecules modified on micas. The statistical analysis indicated that the generalized extreme and mean values of specific interaction between anti-CD4 antibody-tip and CD4-mica were 51.22 ± 34.64 pN and 79.72 ± 59.36 pN (n=1114), respectively (Fig. 3b).
Fig. 3. Interaction force histograms obtained in PBS between CD4 and anti-CD4 antibody (a, b) or between gp120 and anti-gp120 antibody (c, d).
Various functionalization strategies were used: (a) CD4-tip vs. anti-CD4-mica (generalized extreme value: 144.91 ± 84.18 pN; mean value: 220.85 ± 156.29 pN; n=1135); (b) anti-CD4-tip vs. CD4-mica (generalized extreme value: 51.22 ± 34.64 pN; mean value:79.72 ± 59.36 pN; n=1114); (c) anti-gp120-tip vs. gp120-mica (generalized extreme value: 108.84 ± 83.26 pN; mean value: 185.44 ± 155.21 pN; n=1724); (d) gp120-tip vs. anti-gp120-mica (generalized extreme value: 89.87 ± 44.63 pN; mean value: 121.52 ± 66.12 pN; n=632).
Similarly, we found that the generalized extreme and mean values of specific interaction between anti-gp120 antibody-tip and gp120-mica were 108.84 ± 83.26 pN and 185.44 ± 155.21 pN (n=1724), respectively (Fig. 3c), whereas the generalized extreme and mean values of specific interaction between gp120-tip and anti-gp120 antibody-mica were 89.87 ± 44.63 pN and 121.52 ± 66.12 pN (n=632), respectively (Fig. 3d). The representatives of force-distance curves for the antigen-antibody interactions are not shown here.
AFM force measurements detected the interactions between CD4 and the mixture of gp120 and anti-CD4 antibody
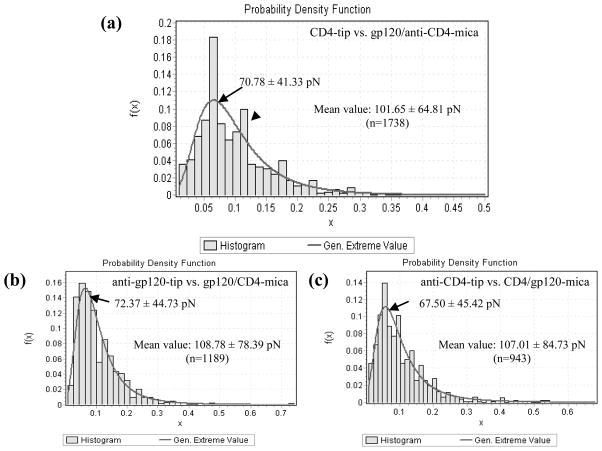
In order to study whether a functionalized AFM tip can be used to distinguish various molecule types on a substrate, we mixed gp120 and anti-CD4 antibody solutions together and then recruited CD4-functionalized AFM tips to detect the mixture. The interaction force histogram (Fig. 4a) showed two peaks at ~70 pN (arrow) and ~120 pN (arrow head), respectively, which potentially represented the CD4-gp120 interaction and the interaction between CD4 and anti-CD4 antibody. The value (70.78 ± 41.33 pN) of the first peak is much larger than the mean value (42.25 ± 34.14 pN) of CD4-gp120 interaction forces measured above (Panel 2 of Fig. 1), which was perhaps attributed to the fraction of relatively weak interaction forces between CD4-tip and anti-CD4 antibody-mica. The mean value of the force histogram was 101.65 ± 64.81 pN (n=1738).
Fig. 4. Interaction force histograms obtained in PBS between CD4 tips and the mixtures of gp120 and anti-CD4 antibody (a) or between anti-gp120-(b) or anti-CD4-(c) functionalized tips and the mixtures of gp120 and CD4.
In the histogram of Fig. 4a, the arrow and arrowhead indicate two peaks at ~70 pN (arrow) and ~120 pN (arrow head), respectively.
AFM force measurements detected the interactions between anti-gp120 or anti-CD4 antibody and gp120-CD4 complex
To detect the interactions of anti-gp120 or anti-CD4 antibody with gp120-CD4 complex, gp120 and CD4 molecules were mixed to form gp120-CD4 complexes prior to modification on micas. Then, AFM force measurements were performed to acquire the interaction forces using AFM tips functionalized with anti-gp120 or anti-CD4 antibody. As shown in Fig. 4b, the generalized extreme and mean values of specific interaction between anti-gp120 antibody and gp120-CD4 complex were 72.37 ± 44.73 pN and 108.78 ± 78.39 pN (n=1189), respectively. The generalized extreme and mean values of specific interaction between anti-CD4 antibody and gp120-CD4 complex were a little smaller, which were 67.50 ± 45.42 pN and 107.01 ± 84.73 pN (n=943), respectively (Fig. 4c).
Discussion
Functionalizing on AFM tip or substrate may cause different results
While performing AFM force measurements, researchers generally paid major attention to using a best tip/substrate-functionalizing method to acquire high-fidelity force information of an interaction pair (e.g. legend vs. receptor, antigen vs. antibody, etc.). Surprisingly, we found that the result measured by functionalizing one party of an interaction pair on AFM tip and its counterpart on substrate may significantly differ from that measured by functionalizing it on substrate and its counterpart on AFM tip. In our study, the mean value (42.25 ± 34.14 pN) of the interaction force between sCD4-tip and gp120-mica was less than that (69.23 ± 66.17 pN) between gp120-tip and sCD4-mica; the mean value (220.85 ± 156.29 pN) of the interaction force between CD4-tip and anti-CD4 antibody-mica was much larger than that (79.72 ± 59.36 pN) between anti-CD4 antibody-tip and CD4-mica; whereas, the mean value (121.52 ± 66.12 pN) of the interaction force between gp120-tip and anti-gp120 antibody-mica was a little smaller than that (185.44 ± 155.21 pN) between anti-gp120 antibody-tip and gp120-mica.
The amount of molecules functionalized on the top of an AFM tip potentially contributed to the significant differences in force measurements, since two or more molecules on the top of an AFM tip could interact simultaneously with multiple molecules on the substrate resulting in the enlargement of AFM-measured interaction forces as shown in panel 3 of Fig. 1. The molecule size is a major factor influencing the amount of the molecules functionalized on the top of an AFM tip. In general, the curvature radius of the top of a commercial AFM tip is 10 nm. It is possible that the amount of small molecules attached on the top of an AFM tip is more than that of larger molecules. The two antigen-recognizing arms of antibody might also contribute to the enlargement of AFM-measured interaction forces, as well as the aggregation of functionalized molecules, the method for tip or substrate modification, and others.
The data imply that we must carefully consider this situation while performing AFM force measurement and analyzing AFM force data.
The interaction forces between various HIV inhibitors and their target molecules are significantly different
Therefore, there were two data sets of interaction force for an interaction pair, one of which was measured by functionalizing one party of an interaction pair on AFM tip and its counterpart on substrate, and the other one of which was measured by functionalizing it on substrate and its counterpart on AFM tip. We think that the data set with lower value might reflect the truth since the reasons mentioned above (the amount of molecule, two arms of antibody, among others) would cause over-measurement of the interaction force in the data set with larger value although presently we have no evidence. On the other hand, the generalized extreme value might reflect the real interaction force between an interaction pair better than the mean value since the mean value consisted of a relatively broad distribution of interaction forces ranging up to 500 pN or 1 nN which must be from simultaneous unbinding of many interaction pairs in an individual AFM force measurement event.
Accordingly, the unbinding forces of sCD4-gp120 interaction, CD4 antigen-antibody interaction, and gp120 antigen-antibody interaction were determined in our study as 25.45 ± 20.46 pN, 51.22 ± 34.64 pN, and 89.87 ± 44.63 pN, respectively, although the values might be still a little overestimated. Obviously, the interaction forces between various HIV inhibitors and their target molecules were significantly different. We also determined the unbinding forces between anti-CD4 or anti-gp120 antibody and gp120-CD4 complex as 67.50 ± 45.42 pN and 72.37 ± 44.73 pN, respectively. It was possible that both the sCD4-gp120 interaction and the CD4 or gp120 antigen-antibody interaction contributed to the larger unbinding forces of anti-CD4 or anti-gp120 antibody versus gp120-CD4 complex than that versus gp120 or CD4 alone.
Hu et al. detected the alternation in specific binding force of CD4 antigen-antibody interaction on resting and PHA-activated CD4+ T cells, however, no exact values for the interaction force was reported in their study [28]. Chang et al. detected the interaction of gp120 with CD4 or CD4/CCR5 on living cells. They found that the gp120-CD4 and gp120-CD4/CCR5 complex on living cells could withstand mean forces of up to 26 pN and 34 pN before rupture, respectively [23]. Our gp120-sCD4 interaction force data (25.45 ± 20.46 pN) was similar to, and the CD4 or gp120 antigen-antibody interaction force data (51.22 ± 34.64 pN and 89.87 ± 44.63 pN, respectively) were much larger than, the reported interaction force between gp120 and cell-surface CD4 (~26 pN) [23]. It partially elucidates the mechanical reason why HIV entry inhibitors (such as sCD4, anti-CD4 antibody, anti-gp120 antibody, and others) can inhibit HIV infection.
Acknowledgments
This work was supported by National Institutes of Health R01 grants HL64560 (Z.W.C.) and RR13601 (Z.W.C.) and National Natural Science Foundation of China 30900340 (Y.C.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lin P, Kadow J, Alexander L. Inhibitors that target gp120-CD4 interactions. In: Parnham MJ, Bruinvels J, editors. Entry inhibitors in HIV therapy. 2007. pp. 49–62. [Google Scholar]
- 2.Strizki JM, Mosier DE. Inhibitors that target gp120 interactions with coreceptor. In: Parnham MJ, Bruinvels J, editors. Entry inhibitors in HIV therapy. 2007. pp. 63–78. [Google Scholar]
- 3.Wang W, Weiss CD. Inhibitors that target fusion. In: Parnham MJ, Bruinvels J, editors. Entry inhibitors in HIV therapy. 2007. pp. 79–98. [Google Scholar]
- 4.Kuritzkes DR. HIV-1 entry inhibitors: an overview. Curr Opin HIV AIDS. 2009;4:82–87. doi: 10.1097/COH.0b013e328322402e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fisher RA, Bertonis JM, Meier W, Johnson VA, Costopoulos DS, Liu T, Tizard R, Walker BD, Hirsch MS, Schooley RT, Flavell RA. HIV infection is blocked in vitro by recombinant soluble CD4. Nature. 1988;331:76–78. doi: 10.1038/331076a0. [DOI] [PubMed] [Google Scholar]
- 6.Traunecker A, Luke W, Karjalainen K. Soluble CD4 molecules neutralize human immunodeficiency virus type 1. Nature. 1988;331 :84–86. doi: 10.1038/331084a0. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe M, Reimann KA, Delong PA, Liu T, Fisher RA, Letvin NL. Effect of recombinant soluble CD4 in rhesus monkeys infected with simian immunodeficiency virus of macaques. Nature. 1989;337:267–270. doi: 10.1038/337267a0. [DOI] [PubMed] [Google Scholar]
- 8.Tilton JC, Doms RW. Entry inhibitors in the treatment of HIV-1 infection. Antivir Res. 2010;85:91–100. doi: 10.1016/j.antiviral.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 9.Hussey RE, Richardson NE, Kowalski M, Brown NR, Chang H, Siliciano RF, Dorfman T, Walker B, Sodroski J, Reinherz EL. A soluble CD4 protein selectively inhibits HIV replication and syncytium formation. Nature. 1988;331:78–81. doi: 10.1038/331078a0. [DOI] [PubMed] [Google Scholar]
- 10.Moore JP, McKeating JA, Weiss RA, Sattentau QJ. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science. 1990;250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 11.Hart TK, Kirsh R, Ellens H, Sweet RW, Lambert DM, Petteway SR, Leary J, Jr, Bugelski PJ. Binding of soluble CD4 proteins to human immunodeficiency virus type 1 and infected cells induces release of envelop glycoprotein gp120. PNAS. 1991;88:2189–2193. doi: 10.1073/pnas.88.6.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haim H, Si Z, Madani N, Wang L, Courter JR, Princiotto A, Kassa A, DeGrace M, McGee-Estrada K, Mefford M, Gabuzda D, Smith AB, III, Sodroski J. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog. 2009;5:e1000360. doi: 10.1371/journal.ppat.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montefiori D, Sattentau Q, Flores J, Esparza J, Mascola J. Antibody-based HIV-1 vaccines: recent developments and future directions. PLoS Med. 2007;4:e348. doi: 10.1371/journal.pmed.0040348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopalco L, Magnani Z, Confetti C, Brianza M, Saracco A, Ferraris G, Lillo F, Vegni C, Lazzarin A, Siccardi AG, Burastero SE. Anti-CD4 antibodies in exposed seronegative adults and in newborns of HIV type 1-seropositive mothers: a follow-up study. AIDS Res Human Retroviruses. 1999;15:1079–1085. doi: 10.1089/088922299310377. [DOI] [PubMed] [Google Scholar]
- 15.Song R, Franco D, Kao CY, Yu F, Huang Y, Ho DD. Epitope mapping of Ibalizumab, a humanized anti-CD4 monoclonal antibody with anti-HIV-1 activity in infected patients. J Virol. 2010;84:6935–6942. doi: 10.1128/JVI.00453-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rowland-Jones S. A winding road towards and HIV vaccine. Eur J Immunol. 2008;38:13–14. doi: 10.1002/eji.200790061. [DOI] [PubMed] [Google Scholar]
- 17.Walker BD, Burton DR. Toward an AIDS vaccine. Science. 2008;320:760–764. doi: 10.1126/science.1152622. [DOI] [PubMed] [Google Scholar]
- 18.Gaczynska M, Osmulski PA. AFM of biological complexes: What can we learn? Curr Opin Coll Interface Sci. 2008;13:351–367. doi: 10.1016/j.cocis.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caygill RL, Blair GE, Millner PA. A review on viral biosensors to detect human pathogens. Analytica Chimica Acta. 2010;681:8–15. doi: 10.1016/j.aca.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 20.Baclayon M, Wuite GJL, Roos WH. Imaging and manipulation of single viruses by atomic force microscopy. 2010;6:5273–5285. [Google Scholar]
- 21.Kuznetsov YG, Victoria JG, Robinson WE, McPherson A. Atomic force microscopy investigation of human immunodeficiency virus (HIV) and HIV-infected lymphocytes. J Virol. 2003;77:11896–11909. doi: 10.1128/JVI.77.22.11896-11909.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuznetsov YG, Victoria JG, Low A, Robinson WE, Fan H, McPherson A. Atomic force microscopy imaging of retroviruses: Human immunodeficiency virus and murine leukemia. Scanning. 2004;26:209–216. doi: 10.1002/sca.4950260409. [DOI] [PubMed] [Google Scholar]
- 23.Chang MI, Panorchan P, Dobrowsky TM, Tseng Y, Wirtz D. Single-molecule analysis of human immunodeficiency virus type 1 gp120-receptor interactions in living cells. J Virol. 2005;79:14748–14755. doi: 10.1128/JVI.79.23.14748-14755.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bitler A, Lev N, Fridmann-Sirkis Y, Blank L, Cohen SR, Shai Y. Kinetics of interaction of HIV fusion protein (gp41) with lipid membranes studied by real-time AFM imaging. Ultramicroscopy. 2010;110:694–700. doi: 10.1016/j.ultramic.2010.02.034. [DOI] [PubMed] [Google Scholar]
- 25.Allen S, Chen XY, Davies J, Davies MC, Dawkes AC, Edwards JC, Roberts CJ, Sefton J, Tendler SJB, Williams PM. Detection of antigen-antibody binding events with the atomic force microscope. Biochem. 1997;36:7457–7463. doi: 10.1021/bi962531z. [DOI] [PubMed] [Google Scholar]
- 26.Chtcheglova LA, Shubeita GT, Sekatskii SK, Dietler G. Force spectroscopy with a small dithering of AFM tip: a method of direct and continuous measurement of the spring constant of single molecules and molecular complexes. Biophys J. 2004;86:1177–1184. doi: 10.1016/S0006-3495(04)74192-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McAllister C, Karymov MA, Kawano Y, Lushnikov AY, Mikheikin A, Uversky VN, Lyubchenko YL. Protein interactions and misfolding analyzed by AFM force spectroscopy. J Mol Biol. 2005;354:1028–1042. doi: 10.1016/j.jmb.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 28.Hu M, Wang J, Cai J, Wu Y, Wang X. Nanostructure and force spectroscopy analysis of human peripheral blood CD4+ T cells using atomic force microscopy. Biochem Biophys Res Co. 2008;374:90–94. doi: 10.1016/j.bbrc.2008.06.107. [DOI] [PubMed] [Google Scholar]