Abstract
Quinine (QN) continues to be an important treatment option for severe malaria, however resistance to this drug has emerged in field isolates of the etiologic agent Plasmodium falciparum. Quantitative trait loci investigations of QN resistance have mapped three loci of this complex trait. Two coincide with pfcrt and pfmdr1, involved in resistance to chloroquine (CQ) and other quinoline-based antimalarials. A third locus on chromosome 13 contains the sodium-proton exchanger (pfnhe) gene. Previous studies have associated pfnhe polymorphisms with reduced QN sensitivity in culture-adapted field isolates. Here, we provide direct evidence supporting the hypothesis that pfnhe contributes to QN resistance. Using allelic exchange, we reduced pfnhe expression by introducing a truncated 3’ untranslated region (UTR) from pfcrt into the endogenous pfnhe 3’UTR. Transfections were performed with 1BB5 and 3BA6 (both CQ- and QN-resistant) as well as GC03 (CQ- and QN-sensitive), all progeny of the HB3×Dd2 genetic cross. RNA and protein analyses of the ensuing recombinant clones demonstrated a ~50% decrease in pfnhe expression levels. A statistically significant 30% decrease in QN IC50 values was associated with these decreased expression levels in 1BB5 and 3BA6 but not in GC03. CQ, mefloquine and lumefantrine IC50 values were unaltered. Cytosolic pH values were similar in all parental lines and recombinant clones. Our observations support a role for pfnhe in QN resistance in a strain-dependent manner, which might be contingent on pre-existing resistance to CQ and/or QN. These data bolster observations that QN resistance is a complex trait requiring the contribution of multiple transporter proteins.
Keywords: Malaria, quinine, multidrug resistance, transfection, knockdown
1. Introduction
For centuries quinine (QN) has been a critical component of the antimalarial pharmacopoeia. In many malaria-endemic regions it remains a principal drug of choice for the treatment of severe malaria [1]. Naturally present in the bark of cinchona trees, QN was first introduced into Europe from South America in the 17th century for the treatment of malaria-like fevers (“ague”) [2]. By 1820, QN had been isolated, leading to its standardized use for the treatment of severe and/or CQ-resistant malaria [3]. Like the 4-aminoquinoline drugs such as chloroquine (CQ), this quinoline derivative is thought to act on Plasmodium falciparum asexual blood stages by interfering with their ability to detoxify heme via its biomineralization into inert hemozoin [4].
P. falciparum, the most virulent of the plasmodial species, is increasingly difficult to treat because of the spread of parasite strains resistant to the former first line antimalarials, CQ and sulfadoxine-pyrimethamine. Reduced susceptibility to QN has also been observed, notably in Southeast Asia and South America as well as the Pacific region and sub-Saharan Africa. In areas where reduced susceptibility occurs, standard treatment regimens may no longer be adequate, and high concentrations of QN, often combined with tetracycline or doxycycline, are implemented to achieve clinical cure. QN resistance has been slow to evolve and disseminate in P. falciparum [5]. This slow rate suggests a multifactorial basis of resistance. Interestingly, culture-adapted parasite lines harboring QN resistance phenotypes can display complex patterns of cross-resistance with CQ and/or mefloquine (MFQ), indicating some overlap in mechanisms of resistance to these distinct drugs [6]. Recent studies of the genetic basis of QN resistance have identified contributions from the P. falciparum genes pfcrt and pfmdr1 that encode the transporters PfCRT and PfMDR-1, respectively. These transporters reside on the membrane of the parasite digestive vacuole, inside which heme is detoxified. In the case of the primary CQ resistance determinant pfcrt, introduction of mutant alleles conferring CQ resistance into a CQ-sensitive line was found to increase QN sensitivity, whereas introduction of the wild type K76 residue in the place of the PfCRT K76T mutation in CQ-resistant lines led to increased susceptibility to both CQ and QN [7; 8]. These seemingly paradoxical results suggest that the effect of PfCRT mutations on QN resistance depends on the particular mutant allele as well as the genetic background of the host strain. An association between elevated QN resistance and point mutations in PfMDR1 has also been reported, notably N1042D and D1246Y [8; 9]. However, none of the aforementioned studies account for high-level QN resistance (with in vitro IC50 values typically defined as 450 nM or higher [9]).
Investigations into the inheritance of QN resistance in the progeny of a P. falciparum genetic cross [10] (between the CQ-resistant and low-level QN-resistant Dd2 clone from Indochina and the drug-sensitive HB3 clone from Honduras) used quantitative trait loci analysis to dissect contributions of individual loci [11]. These studies associated low-level QN resistance with mutations in pfcrt and pfmdr1 (present on chromosomes 7 and 5, respectively) and an additional locus that mapped to a 380 kb segment of chromosome 13. This segment includes the P. falciparum sodium-proton exchanger gene, pfnhe, which in Dd2 harbors point mutations and a DNNND repeat unit polymorphism (termed ms4760-1) that associate with elevated QN IC50 values in the progeny receiving this allele [11]. pfnhe sequence analysis of a set of 71 culture-adapted isolates from around the globe provided additional support for an association between Dd2-type PfNHE DNNND microsatellite repeat lengths (present in the C-terminal predicted cytoplasmic domain) and increased QN IC50 values. This was corroborated by a recent analysis of parasites collected from 5 regions in India that found an increased number of the previously identified DNNND repeats in parasites from multi-drug resistant regions [12].
Previous studies have observed a change in antimalarial susceptibility upon alteration of the expression levels of either pfcrt or pfmdr1 [13; 14]. Based upon those findings, we hypothesized that if pfnhe is a component of QN resistance, then alteration of its expression levels might affect parasite susceptibility to this drug. We tested this hypothesis by generating “knockdown” parasites with ~50% reduced pfnhe expression, using an experimental approach similar to that of Waller et al. [13]. Here we report the findings from this experiment.
2. Materials and methods
2.1. DNA constructs
The pfnhe allelic exchange fragment (corresponding to the 3’ 1.15 kb end of the full-length 5.76 kb coding sequence) was PCR amplified from 3D7 genomic DNA using primers p1 and p2 (Table 1; Fig. 1A). The 148 bp pfcrt 3’ untranslated region (UTR) was PCR amplified from the same genomic DNA using primers p3 and p4. Primers p2 and p3 shared overlapping sequence and the two products were fused together by the technique of splicing by overlap extension by PCR [15], with amplification of the complete 1.3 kb pfhne-pfcrtΔ3’UTR fusion using primers p1 and p4. The resulting product was subcloned into the pCam_bsd plasmid, which uses the blasticidin S deaminase gene (bsd) as a selectable marker flanked by the 5’UTR of calmodulin and the 3’UTR of hrp2 [16]. The resulting 5.8 kb transfection plasmid, denoted pCam_bsd/pfnheΔ3’UTR, was electroporated into ring stage parasites of the three cloned HB3×Dd2 progeny 1BB5, 3BA6 and GCO3.
Table 1.
Oligonucleotide primer sequences
| Oligomers | Sequence | Origin | Restriction site |
|---|---|---|---|
| p1 | 5’-CAAggatccCACAGCAGATTTTACACCACAGAGA | pfnhe gene | Bam HI site in lower case |
| p2 | 5’-ATAAAAAAATAAATGATATATTTTCATTTTATAAACCTTTTTAG | pfnhe gene | pfcrt-specific 3’UTR overhang underlined |
| p3 | 5’-CTAAAAAGGTTTATAAAATGAAAATATATCATTTATTTTTTTATATT | pfcrt 3’UTR | pfcrt-specific 3’UTR overhang underlined |
| p4 | 5’-CAAgcggccgcATTCCTTATAAAGTGTAATGCGATAG | pfcrt 3’UTR | NotI site in lower case |
| p5 | 5’-GAAATTAACCCTCACTAAAG | plasmid-specific | None |
| p6 | 5’-CAAggatccGAAGGATATGATGGTAACTCATG | pfnhe gene | Bam HI site in lower case |
| p7 | 5’-AATGTCGGAATGTGGTATATGC | pfnhe 3’UTR | None |
| p8 | 5’-TGTTCGGAAATATAAAGACGTGG | pfnhe gene | None |
| p9 | 5’-TTATAAAGTGTAATGCGATAGC | pfcrt 3’UTR | None |
Fig. 1.
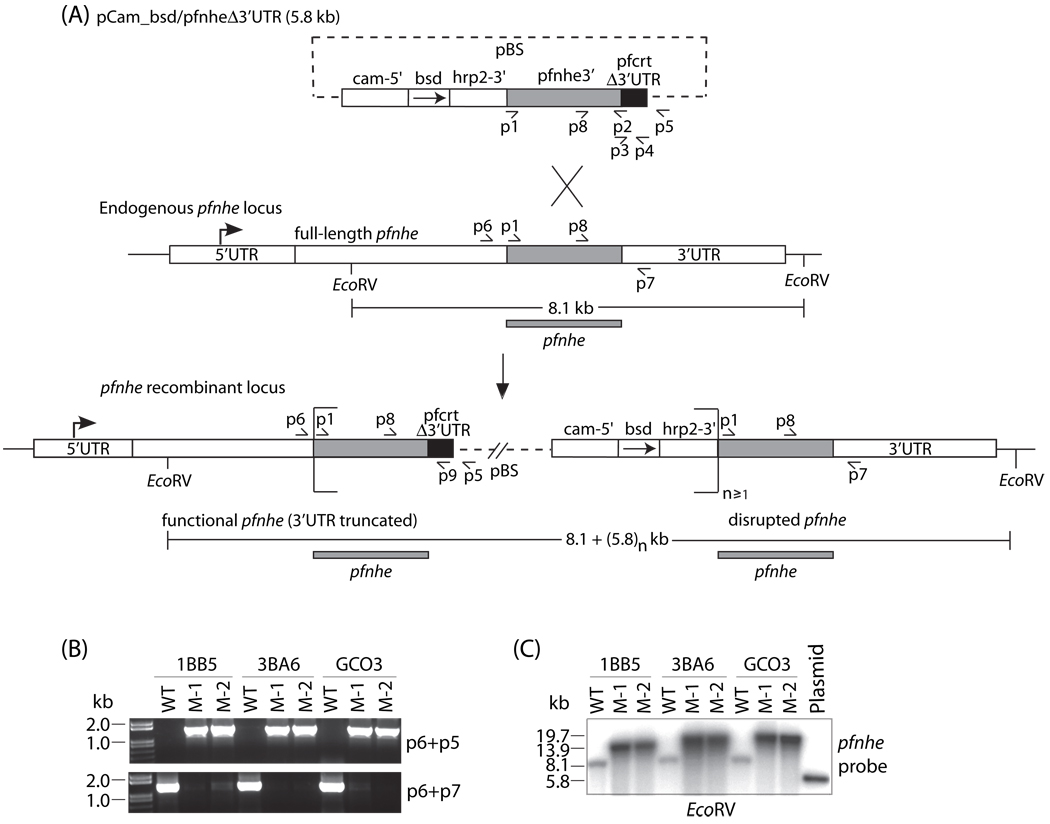
Generation of pfnhe recombinant knockdown clones. (A) Schematic representation of pCam_bsd/pfnheΔ3’UTR plasmid integration into the pfnhe locus via homologous recombination and single-crossover to replace the pfnhe endogenous 3’UTR with the truncated pfcrt 3’UTR. This resulted in the generation of an upstream functional recombinant pfnhe gene possessing the endogenous pfnhe promoter, the full-length coding sequence and the 150 bp truncated 3’UTR of pfcrt. The downstream integrated blasticidin S deaminase selection marker (bsd), driven by the calmodulin (cam) 5’UTR and the hrp2 3’UTR, is followed immediately by a nonfunctional pfnhe remnant. This downstream, disrupted locus lacks 4.6 kb of 5’ pfnhe coding sequence and its promoter and retains the endogenous 3’UTR. (B) PCR detection of the recombinant functional pfnhe locus with primers p6+p5 (top panel), or the endogenous undisrupted pfnhe with primers p6+p7 (bottom panel), for the pfnhe parental lines (WT) and their respective knockdown clones (listed as M-1 and M-2 for each genetic background). (C) Southern hybridization of EcoRV-digested genomic DNA from recombinant and parental lines probed with a pfnhe 3’ coding fragment.
2.2. Parasite transfections and cloning
P. falciparum parasites were cultured in vitro in freshly-drawn human red blood cells (RBC), transfected as described previously [17], and selected by adding 2.5 µg/ml blasticidin hydrochloride to the culture medium. Plasmid rescue confirmed that the parasites were transfected with the appropriate plasmid. This was performed by electroporating plasmid-containing parasite genomic DNA into Escherichia coli, selecting with 50 µg/ml ampicillin, and confirming plasmid identity by restriction mapping. Recombinant parasites were cloned by limiting dilution and identified using the Malstat assay that detects expression of P. falciparum lactate dehydrogenase [18].
2.3. Nucleic acid and protein analyses
Parasites that had undergone plasmid integration into the pfnhe locus (PlasmoDB identifier: PF13_0019) via homologous recombination and single-site crossover were identified by PCR using the primer pair p6+p5 that was specific for upstream integration events (Fig. 1A). Southern blot analysis was used to confirm replacement of the endogenous pfnhe locus by a locus harboring the truncated pfcrt 3’UTR, and employed a pfnhe 3’ coding sequence probe amplified using the primers p1 and p2. Northern, reverse-transcriptase PCR and Western blot analyses were performed on saponin-lysed parasite preparations from sorbitol-synchronized cultures, as described [7; 19]. For Western blot analysis, a multiple antigen peptide (MAP4) directed against PfNHE amino acids 1127–1142 (LIEHATHLPKTLSDNL) was synthesized by Bio-Synthesis Inc., Lewisville, TX. Anti-peptide sera were produced (by Spring Valley Laboratories, Inc.) by immunizing a rabbit with 250 µg peptide in complete Freund’s adjuvant on day 0 followed by boosts in incomplete Freund’s adjuvant on days 21, 42 and 63. Rabbit antiserum to PfERD2 was obtained through the MR4 (MRA-1, deposited by Dr. John Adams).
2.4. Drug susceptibility assays and statistical analyses
P. falciparum lines were typed for their antimalarial susceptibility profiles using [3H]-hypoxanthine incorporation assays, as described [17]. Assays were performed using a range of 2-fold dilutions, with each dilution tested in duplicate. The final data set consisted of results from assays performed on 5 to 9 separate occasions for each line with each drug. Unpaired two tailed t tests were used to screen for statistically significant differences between recombinant mutant and control lines.
2.5. Parasite preparations for microscopy
All cytosolic pH measurements were performed on sorbitol-synchronized trophozoite stage parasites. A strong magnet (MidiMACS, Miltenyi Biotec, Auburn, CA) [20] was used to enrich parasite cultures to greater than 80% infected RBCs (iRBCs). MACS LD columns placed within the magnet were washed twice with RPMI 1640 (Invitrogen, Carlsbad, CA) lacking the pH indicator phenol red and containing 2% BSA (RPMI+BSA). Three to five ml of parasite culture was applied to the column, followed by four 1 ml washes with RPMI+BSA. The MACS column was removed from the magnet and iRBCs were eluted with 1 ml of RPMI+BSA. The entire magnet purification procedure was performed in a 37°C incubator. The magnet-enriched iRBC eluate was centrifuged at 200×g for 4 minutes, and the pellet was resuspended in 1 ml Na+ E1 buffer (140 mM NaCl, 2.8 mM KCl, 2 mM MgCl2, 1 mM CaCl2, 10 mM dextrose, 10 mM HEPES, pH 7.4).
2.6. Cytosolic pH measurements
The steady state cytosolic pH of intra-erythrocytic parasites was measured using the ratiometric pH sensitive dye BCPCF-AM (Invitrogen) [21]. For this, iRBCs were incubated for 10 minutes at 37°C with Na+ E1 buffer containing 10 µM BCPCF-acetoxymethyl ester, in an environment of 5% O2/5% CO2/90% N2. iRBCs were then washed 3 times with Na+ E1 buffer. BCPCF loaded iRBCs were attached to poly-D-lysine coated coverslips and perfused on the microscope stage with Na+ E1 buffer at 37°C. After a 5 minute equilibration, the RBC membrane was selectively permeabilized by adding 0.01% saponin to the perfusion medium. RBC membrane permeabilization occurred within approximately 30 seconds of introducing saponin, as evidenced by extracellular leakage of hemoglobin. The ratiometric BCPCF response was measured for many individual parasites, and the BCPCF probe was calibrated in each individual parasite by perfusion with high K+ E1 buffer (2.8 mM NaCl, 140 mM KCl, 2 mM MgCl2, 1 mM CaCl2, 10 mM dextrose, 10 mM HEPES, pH 7.4) containing 1 µM nigericin at various pH values (6.7, 7.0, 7.3, 7.6). All measurements were carried out within 5 minutes of saponin treatment.
2.7. Nigericin/BSA pH clamp assays
In an attempt to examine parasite NHE activity, the nigericin/BSA pH clamp method was used [22; 23]. Following BCPCF loading and saponin permeabilization (by perfusion on the microscope stage as described above), the parasites were perfused with pH 7.5 high K+ E1 buffer. After 2–3 minutes, 1 µM nigericin was added to the perfusate and the cytosolic BCPCF probe signal was allowed to equilibrate. The perfusion pH was then shifted to pH 6.0 and the cytoplasmic BCPCF signal was again allowed to equilibrate. Nigericin was then removed from the perfusate and 5 mg/ml BSA was added to adsorb the remaining nigericin, clamping the cytoplasmic pH at 6.0. The perfusion pH was then returned to 7.4, either in Na+ E1 buffer or in a Na+ free NMDG+ buffer (140 mM N-Methyl-D-glucamine (NMDG), 2.8 mM KCl, 2 mM MgCl2, 1 mM CaCl2, 10 mM dextrose, 10 mM HEPES, pH 7.4).
2.8. Microscopy and image collections
Microscopy was performed using a Nikon Eclipse TE2000-U inverted microscope with a 100× S-Fluor 1.3 NA lens. Excitation light of 440 and 495 nm was produced by a PTI DeltaRAM V monochromator (Photon Technology International, Birmingham, NJ), with a slit width of 2 nm. Emission was collected through a 505 nm long pass dichroic mirror (Chroma Technology, Rockingham, VT) with a CoolSnap Fx CCD camera (Photometrics, Tucson, AZ). Exposure time was 0.5 seconds at both 440 and 495 nm wavelengths. Image analysis was performed using Imaging Workbench 5.2 Software (Indec Biosystems, Santa Clara, CA) as described previously [24]. Regions of interest were defined around individual parasites within a single field of view. All ratio values were derived from background-subtracted images, with background defined as the average intensity in a region of interest containing no RBCs. The threshold for ratio images was set at 50 counts per pixel for both 440 and 495 nm excitation, thereby eliminating noise from low intensity fluorescence in border regions. Ratio values were determined for each individual parasite and converted to a pH value based on the calibration ratios for that parasite. Each field of view contained multiple parasites. For each parasite line, pH was measured in at least three separate batches of parasites on separate days.
3. Results
3.1. Generation of recombinant pfnhe-modified lines
To generate P. falciparum lines that under-express pfnhe, an allelic exchange strategy was designed to replace the endogenous full-length 2.0 kb 3’UTR of pfnhe with a truncated 0.15 kb region from the pfcrt 3’UTR. This strategy was used in an earlier study in which the full length (1.9 kb) pfcrt 3’UTR had been replaced with this same truncated 0.15 kb version, causing a 52–55% reduction in pfcrt steady state mRNA levels and a 30–40% reduction in protein expression levels [13]. The decrease in pfcrt expression levels was attributed to reduced mRNA stability resulting from the shorter 3’UTR. Recombinant pfcrt knockdown lines, produced in the CQ-resistant 7G8 line, had demonstrated a 40% reduction in CQ mean IC50 levels. To achieve a similar transcriptional knockdown of pfnhe, the pCam_bsd/pfnheΔ3’UTR construct was electroporated into the lines 1BB5, 3BA6 and GC03, all cloned progeny of the HB3×Dd2 genetic cross [25]. 1BB5 and 3BA6 were chosen because they are both QN- and CQ-resistant and carry the Dd2 mutant pfcrt allele that is the primary determinant of CQ resistance. By contrast, GC03 is sensitive to QN and CQ and carries the CQ-sensitive HB3 wild-type pfcrt allele (Table 2). All three lines carry the HB3-type pfmdr1 allele, which encodes the N1042D mutation that was recently found (in 1BB5 and 3BA6 parasites) to augment QN IC50 values, without significantly altering levels of CQ susceptibility [8]. These three lines also carry the Dd2-type pfnhe allele harboring the ms4760-1 copy number variant of the DNNND repeat that previously had been indirectly associated with resistance to QN [11].
Table 2.
Genetic and drug susceptibility parameters of lines chosen for pfnhe modification
| Allelic type | Susceptibility status | ||||
|---|---|---|---|---|---|
| Linea | pfcrtb | pfmdr1c | pfnhe | CQ | QN |
| IBB5 | Dd2 | HB3 | Dd2 | resistant | resistant(low) |
| 3BA6 | Dd2 | HB3 | Dd2 | resistant | resistant(low) |
| GC03 | HB3 | HB3 | Dd2 | senstitive | senstitive |
All three clones are progeny of the HB3×Dd2 genetic cross.
The Dd2-type pfcrt allele carries the following mutations associated with CQR: M741/N75E/K76T/A220S/Q271E/N326S/I356T/R371I.
The HB3-type pfmdr1 allele carries the N1042D mutation associated with increased resistance to QN and increased susceptibility to MFQ.
Transfection of the three lines (1BB5, 3BA6 and GC03) with the pfnhe knockdown construct produced episomally transformed lines that were maintained by selection with blasticidin hydrochloride. After 12 weeks, these lines were found by PCR to have undergone integration into the pfnhe locus and were subsequently cloned by limiting dilution. Two independent pfnhe-modified recombinant clones from each parental genetic background were randomly chosen for molecular and phenotypic analyses. These clones were designated M-11BB5, M-21BB5, M-13BA6, M-23BA6, M-1GC03 and M-2GC03. PCR results were consistent with plasmid integration via homologous recombination and single crossover into the 3’ end of the pfnhe coding sequence, resulting in a functional recombinant allele regulated by its endogenous promoter and the pfcrt truncated 3’UTR knockdown terminator (Fig. 1A). This locus was followed by the plasmid backbone (pBluescript) and a downstream, disrupted pfnhe locus that lacked the bulk of the coding sequence and a promoter. PCR assays with the primer pair p6+p5, specific for the upstream junction of integration, yielded a 1.5 kb band from all recombinant clones, which was absent in the parental (nontransfected) lines (Fig. 1B). Similarly, PCR assays with primers p6+p7, specific for the undisrupted endogenous pfnhe locus, produced a 1.4 kb band from the parental lines but not from the cloned recombinant parasites. Southern blot hybridization with a pfnhe probe confirmed complete allelic replacement of the endogenous pfnhe 3’UTR with the pfcrt truncated 3’UTR in all recombinant clones (Fig. 1C). EcoRV digestion, which cleaves sites flanking the pfnhe region of homologous recombination, revealed replacement of the 8.1 kb wild type band with a 13.9 kb band in the 1BB5 knockdown clones and a 19.7 kb band in 3BA6 and GCO3 knockdown clones. This suggested integration of a single plasmid in the 1BB5 knockdown clones, as opposed to tandem integration of a concatemer of 2 plasmid copies in the 3BA6 and GC03 clones.
3.2. Analysis of pfnhe expression levels in knockdown recombinant lines
Reverse transcriptase-PCR with primers p8+p9 (see Fig. 1A), which flank a 0.15 kb intron present in the 3’ end of the pfnhe coding region, generated the expected 0.4 kb cDNA product in all pfnhe-modified clones, but not in the parental lines (Fig. 2A). This confirmed that the recombinant pfnhe locus was functional in the knockdown clones, producing a pfnhe transcript with the truncated pfcrt 3’UTR. No product was generated from control reactions lacking reverse transcriptase, nor was any 0.6 kb product observed that would have derived from genomic DNA, indicating lack of DNA contamination of the RNA preparations (data not shown).
Fig. 2.
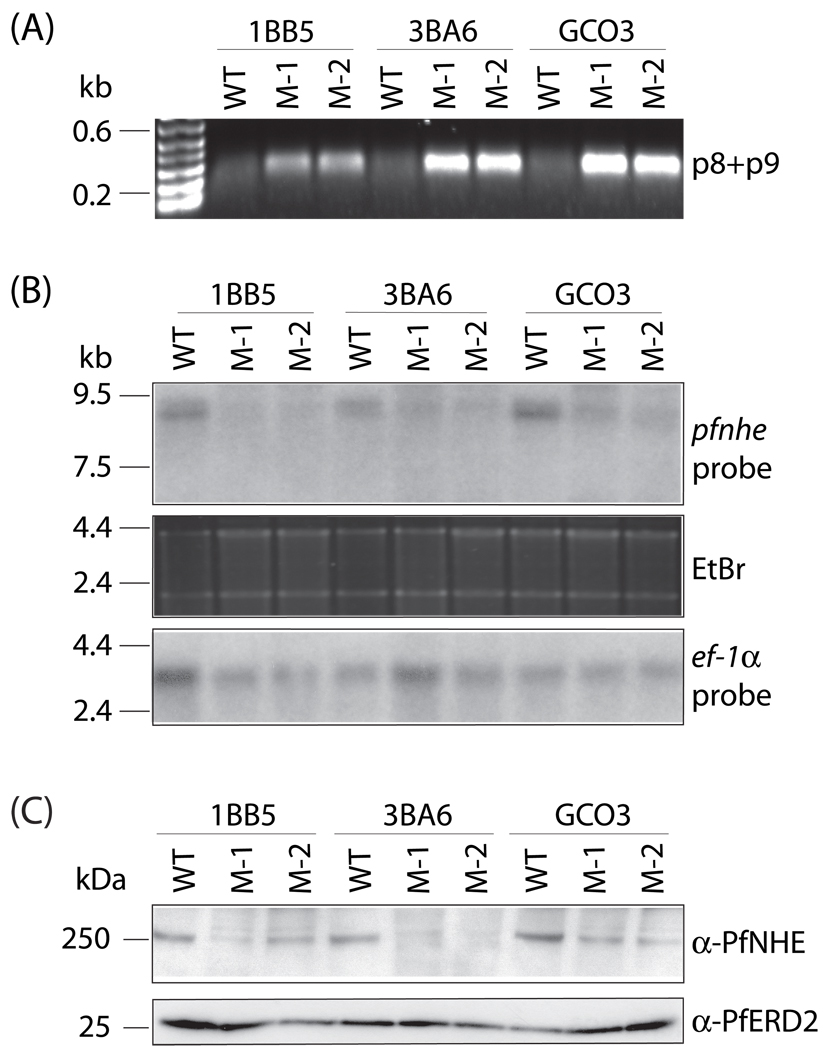
Analysis of pfnhe expression levels in recombinant clones. (A) RT-PCR analysis with primers p8+p9, specific for the functional recombinant pfnhe allele, reveals a 0.4 kb transcript from the knockdown clones. (B) Northern hybridization of total RNA from synchronized schizont stage parasites probed with a pfnhe fragment (top panel) reveals substantially reduced steady-state pfnhe transcription in the recombinant clones compared to their parental lines. Loading controls are shown in the middle panel (ethidium bromide (EtBr) stained gel), and the bottom panel (same blot rehybridized with an ef-1α probe). (C) Western blot of total protein samples from recombinant and parental lines probed with antibodies to PfNHE or the P. falciparum Golgi marker PfERD2.
To assess the impact of the truncated pfcrt 3’UTR on pfnhe expression, we performed Northern blots on RNA samples collected from synchronized cultures of schizonts, the stage in the intraerythrocytic developmental cycle when pfnhe transcription is maximal (as determined by the DeRisi and Winzeler microarray datasets available on [www.plasmodb.org]). This showed a significant reduction in the levels of pfnhe transcript in the recombinant clones relative to the parental lines, in all three parasite genetic backgrounds (Fig. 2B). P. falciparum rRNAs and ef-1α mRNAs were quantified and used to normalize sample loading for each lane. Densitometric analysis, comparing ratios of detected pfnhe to ef-1α mRNAs for each line, revealed a 40–70% reduction in steady-state pfnhe transcript levels in the knockdown clones compared to the parental lines.
To compare PfNHE protein expression levels between individual lines, immunoblots were performed with protein extracts of synchronized merozoite and early ring-stage parasites (which gave the highest levels of detection). These were probed with anti-PfNHE IgG antibodies, which were purified from the antiserum of a rabbit immunized against a 16 amino acid PfNHE multiple antigen peptide (representing amino acids 1127 – 1142 of the 1920 amino acid full-length protein). These anti-PfNHE antibodies recognized a protein of approximately 250 kDa in all lines (Fig. 2C). This is close to the 226 kDa molecular weight predicted from the pfnhe nucleotide sequence. Identical immunoblots were probed with rabbit antiserum specific to the ER-Golgi marker PfERD2 [26]. This antiserum detected a band of expected size (26 kDa) and permitted normalization of protein loadings between samples (Fig. 2C). Densitometric analysis of the PfNHE and PfERD2 signals predicted a 50–80% decrease in PfNHE protein expression levels in the recombinant clones compared to their parental lines (Fig. 2C). Analysis of several Western blots prepared on different occasions and probed with these two antisera revealed equivalent levels of reduced expression across all three genetic backgrounds (data not shown). This result was in accordance with the estimated 40–70% decrease in pfnhe transcript levels in the recombinant clones (Fig. 2B). This demonstrated that the decrease in mRNA levels resulted in decreased PfNHE protein expression levels in the recombinant clones as compared to the parental lines.
3.3. Evidence of reduced QN resistance in PfNHE-under-expressing clones
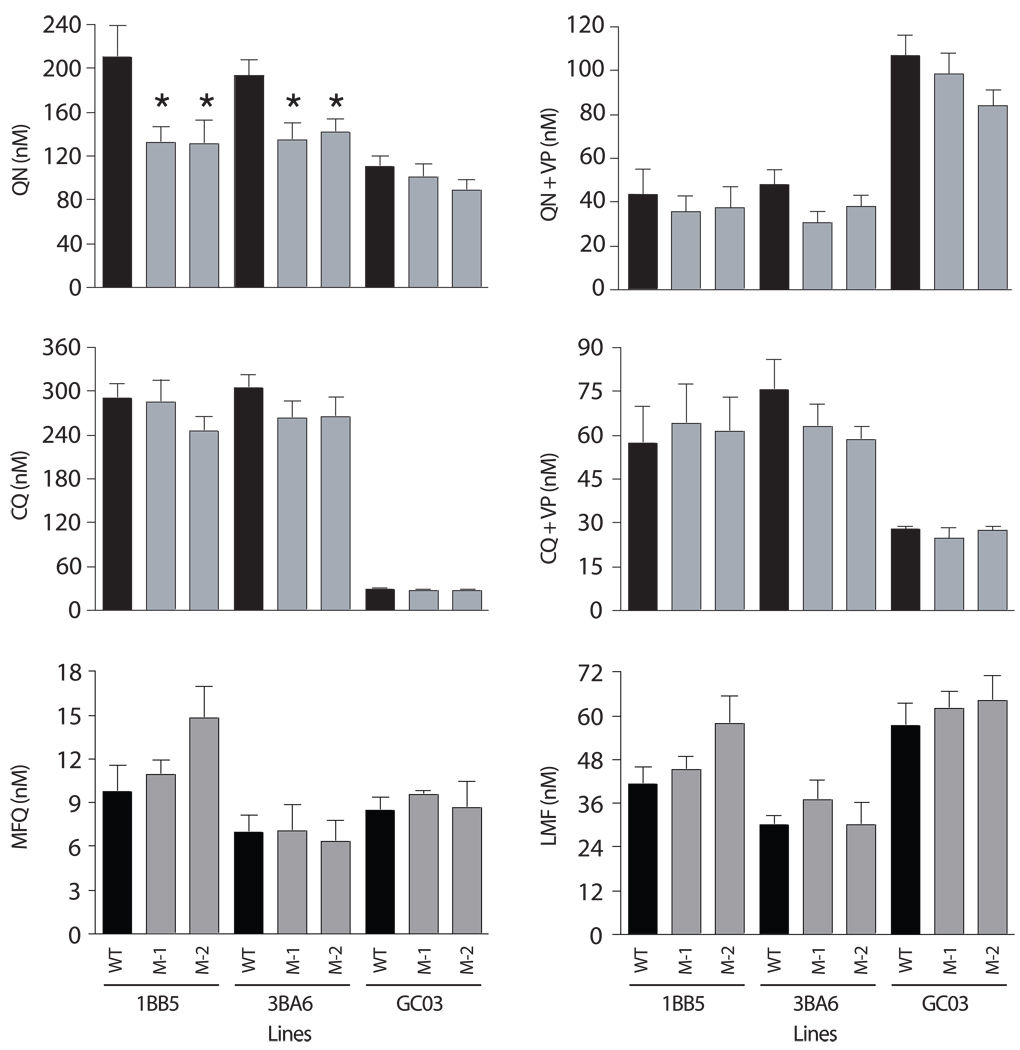
The drug susceptibility profiles of parental and pfnhe knockdown clones were determined by [3H]-hypoxanthine uptake assays with cultured iRBCs. Results showed QN mean IC50 values in the range of 131 to 142 nM (± SEM of 12 to 21 nM) for the 1BB5 and 3BA6 knockdown transformants, versus 210±29 and 194±14 nM for the untransformed parental lines (Table 3; Fig. 3). These values reflected 27% to 37% decreases in the QN mean IC50 values of the 1BB5 and 3BA6 pfnhe knockdown clones, which in all cases attained statistical significance (P< 0.05). With the GC03 knockdown transformants, the QN mean±SEM IC50 values were 101±13 and 89±9 nM for M-1GC03 and M-2GC03 respectively, which was not significantly different from the mean±SEM IC50 of 111±9 nM observed for the untransformed GC03 line. We note that in our study, GC03 is defined as QN sensitive, whereas 1BB5 and 3BA6 are defined as having low-level resistance to QN.
Table 3.
Summary of QN and CQ IC50 values of pfnhe-modified and parental clones
| 1BB5 | M-11BB5 | M-21BB5 | 3BA6 | M-13BA6 | M-23BA6 | GC03 | M-1GC03 | M-2GC03 | |
|---|---|---|---|---|---|---|---|---|---|
| QN mean±SEM | 210.2±29.2 | 132.5±14.2 | 131.4±20.8 | 193.5±14.0 | 134.2±16.2 | 142.1±11.8 | 110.6±9.3 | 101.1±12.5 | 89.1±9.4 |
| # assays | 9 | 9 | 9 | 9 | 9 | 9 | 7 | 7 | 7 |
| t test vs. parent: P value | 0.034* | 0.048* | 0.014* | 0.013* | 0.553 | 0.130 | |||
| QN+VP mean±SEM | 43.1±11.4 | 35.2±7.5 | 37.5±9.4 | 47.9±7.4 | 30.7±4.7 | 38.1±4.8 | 106.9±9.5 | 98.4±9.9 | 84.2±7.4 |
| # assays | 6 | 6 | 6 | 6 | 6 | 6 | 7 | 7 | 7 |
| t test vs. parent: P value | 0.577 | 0.716 | 0.077 | 0.290 | 0.547 | 0.083 | |||
| CQ mean±SEM | 290.8±19.4 | 285.0±29.5 | 245.1±19.9 | 304.0±17.8 | 262.5±23.5 | 264.7±26.4 | 29.3±0.9 | 27.6±1.5 | 27.1±2.1 |
| # assays | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| t test vs. parent: P value | 0.873 | 0.138 | 0.198 | 0.252 | 0.383 | 0.379 | |||
| CQ+VP mean±SEM | 57.0±12.8 | 63.9±13.5 | 61.1±11.9 | 75.5±10.3 | 63.1±7.6 | 58.6±4.3 | 27.8±1.2 | 25.0±3.2 | 27.4±1.4 |
| # assays | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| t test vs. parent: P value | 0.721 | 0.823 | 0.365 | 0.171 | 0.512 | 0.921 | |||
CQ, chloroquine; QN, quinine; VP, verapamil (used at 0.8 µM).
All IC50 values are listed in nM. Those that differ significantly between mutant and parental lines are indicated with an asterisk (P<0.05).
P values were calculated from two-tailed t tests.
Fig. 3.
Antimalarial susceptibility profiles of pfnhe-modified clones and their parental lines. This is shown as a graphical representation of IC50 mean±SEM values for Quinine (QN)±Verapamil (VP), Chloroquine (CQ)±VP, Mefloquine (MFQ), and Lumefantrine (LMF) for the various lines. Values (listed in Table 3) were calculated from 5–9 separate assays performed in duplicate (*, P<0.05).
Verapamil (VP) chemosensitizes the CQ as well as the QN response of CQ-resistant but not CQ-sensitive P. falciparum strains [7; 27]. In accordance with these observations, 0.8 µM VP reduced the QN IC50 values of the CQ-resistant 1BB5 and 3BA6 lines and their respective knockdown lines by 71–80% (Table 3; Fig. 3). In contrast, VP produced no detectable effect on the QN IC50 of either the CQ-sensitive GC03 line or its knockdown transformants (Table 3). We note that VP reversibility has been attributed to CQ-resistant alleles of pfcrt, including the Dd2 allele present in 1BB5 and 3BA6 but not in GC03 [8]. This is consistent with our finding here that in all our 1BB5 and 3BA6 lines, VP reduced the QN mean IC50 values to levels below the QN mean IC50 values of the GC03 parental and knockdown lines (Table 3; Fig. 3). The data indicate that PfNHE expression levels do not affect VP reversibility of the QN response.
In contrast to the results with QN, we found no significant differences between the knockdown and parental lines in the CQ mean IC50 values for all three strains investigated in this study. This was consistent both in the absence or presence of VP (Fig. 3, Table 3) and provided evidence against a role for PfNHE expression levels in influencing the degree of CQ susceptibility.
Parasite resistance to the quinoline-methanol mefloquine (MFQ) has been strongly associated with mutations or copy number variations of pfmdr1 [28; 29]. The parental lines used in this study were all sensitive to MFQ and the pfnhe knockdown mutants showed no significant change in MFQ mean IC50 values compared to the parental lines (Fig. 3). Similarly, the pfnhe knockdown mutants and parental lines, irrespective of their genetic background, showed equivalent mean IC50 values for the aryl amino alcohol lumefantrine, which can share patterns of cross-resistance with MFQ (Fig. 3; [14]). Thus, our data suggest that the ability of PfNHE expression levels to influence QN susceptibility does not extend to other antimalarials whose mode of action is also believed to involve inhibition of heme detoxification.
3.4. Examination of parasite cytosolic pH in parental and PfNHE knockdown parasites
In light of a recent report associating PfNHE variants with alterations in cytosolic pH [23], we sought to determine this cellular parameter in our parental and PfNHE knockdown lines. This was investigated using live cell ratiometric imaging of trophozoite stage parasites with the pH dependent fluorescent probe BCPCF-AM. Incubation of parasites in medium containing BCPCF-AM resulted in accumulation of the probe within the parasite cytoplasm, while only small amounts of BCPCF accumulated in the enveloping erythrocyte (Fig. 4A–C). To ensure that the measured pH values were derived strictly from the parasite, all extra-parasitic BCPCF was released through a brief application of 0.01% saponin, which selectively permeabilizes the RBC and parasitophorous vacuole membranes but not the parasite plasma membrane.
Fig. 4.
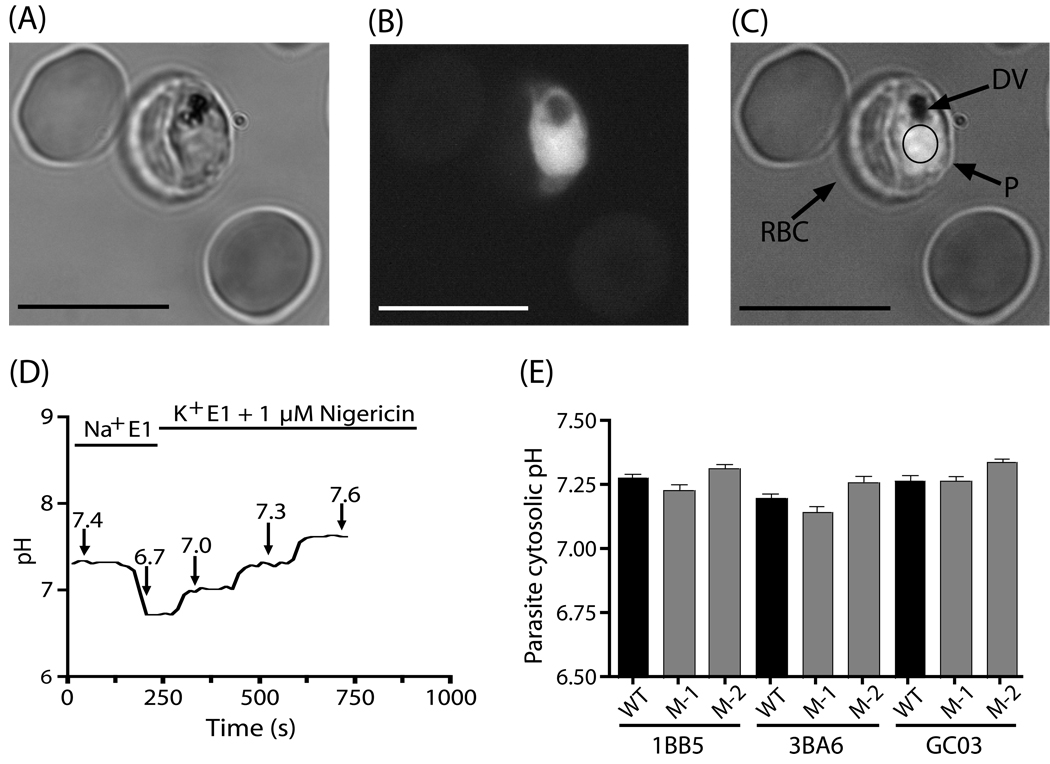
Fluorimetric pH measurement of the P. falciparum cytosol. Bright field image (A) of infected and uninfected RBCs loaded with the pH sensitive dye BCPCF-AM. A fluorescence image obtained following excitation at 495 nm is shown in (B), and panels A and B are merged in (C). The membranes of the RBC, parasite (P), and digestive vacuole (DV) are indicated. All pH measurements were made in individual parasites by examining a region of interest within the parasite cytoplasm (illustrated here as a circle). The scale bar is 10 µm. (D) BCPCF loaded parasitized erythrocytes were treated with 0.01% saponin to permeabilize the RBC membrane, and then exposed to rapidly alternating 495 nm and 440 nm excitation light. The pH within the parasite cytoplasm was derived from the ratio of the fluorescent emission derived from 495/440 nm excitation. The BCPCF probe was calibrated within each individual parasite by exchanging the perfusate with an iso-osmotic high K+ solution in the presence of 1 µM nigericin, over a range of pre-measured pH values (6.7, 7.0, 7.3, 7.6). (E) The average cytosolic pH values of the strains examined in this study are shown. For each of the parental strains (GC03, 3BA6, 1BB5), two pfnhe underexpressing strains were examined. The cytosolic pH value of each line (shown as the mean±SEM) represents data from 39–72 individual parasites, acquired on 3 separate days.
Following saponin treatment, the measured parasite pH was stable for minutes and was distinct from the pH of the extracellular buffer solution (Fig. 4D). This suggests that saponin treatment did not disrupt the parasite’s plasma membrane or its physiologic pH maintenance mechanisms. In addition, the cytosolic pH of parasites was found to be the same in iRBCs whether or not they were treated with 0.01% saponin (data not shown). Our studies revealed similar cytosolic pH values between all three unmodified parental strains. Mean±SEM pH values were 7.28 ± 0.01, 7.20 ± 0.01 and 7.27 ± 0.02 and for 1BB5, 3BA6 and GC03 respectively. In all three genetic backgrounds, we also observed no evident change in cytosolic pH in the PfNHE under expressing lines compared to parental controls (Fig. 4E; Table 4).
Table 4.
Summary of cytosolic pH data
| 1BB5 | 3BA6 | GC03 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| pH values | WT | M-1 | M-2 | WT | M-1 | M-2 | WT | M-1 | M-2 |
| Mean | 7.28 | 7.23 | 7.32 | 7.20 | 7.15 | 7.27 | 7.27 | 7.27 | 7.34 |
| SEM | 0.01 | 0.02 | 0.01 | 0.01 | 0.02 | 0.02 | 0.02 | 0.02 | 0.01 |
| N | 51 | 72 | 55 | 64 | 63 | 55 | 51 | 39 | 51 |
3.5. Attempted measurements of parasite NHE activity
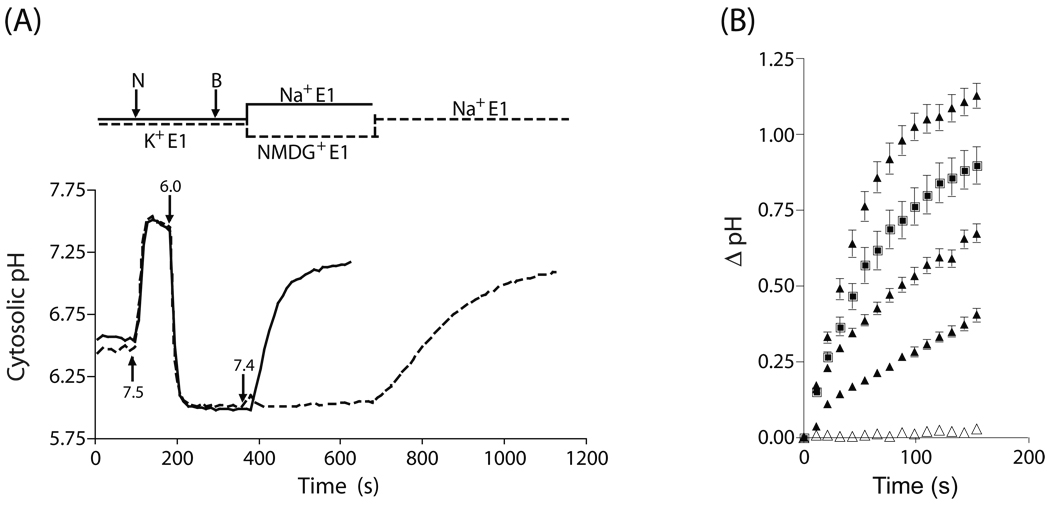
In further experiments, we explored whether PfNHE activity might be decreased in the pfnhe knockdown clones. To measure this activity, we employed the nigericin/BSA “pH clamp” technique, initially reported by Ginsburg and colleagues to be an effective measure of PfNHE-dependent cytosolic alkalinization in parasites [22]. For these experiments, saponin-lysed trophozoite stage parasites were first perfused with Na+ E1 buffer at pH 7.4, as described above for cytosolic pH measurements. Nigericin is a K+-H+ exchanger but it can also exchange other small monovalent cations [30]. To clamp the cytosolic pH of the parasites at an acidic pH, the perfusate was switched to an equimolar high K+ E1 buffer containing 1 µM nigericin, first at pH 7.5, and then at pH 6.0. In the presence of the pH 6.0 buffer, nigericin was removed from the perfusate and 5 mg/ml BSA was added to adsorb any remaining nigericin, thus clamping the cytosolic pH at 6.0. The parasites were returned to Na+ E1 buffer at pH 7.4 and rapid cytosolic alkalinization was observed (Fig. 5A, solid line). In contrast, return of parasites to (Na+ free) pH 7.4 NMDG+ buffer did not show similar alkalinization (Fig. 5A, dashed line). Subsequent perfusion with Na+ E1 buffer at pH 7.4 resulted in cytosolic alkalinization. This demonstrated that in this assay the cytosolic alkalinization required the presence of both extracellular Na+ and pH 7.4.
Fig. 5.
Nigericin/BSA pH clamp method applied to PfNHE recombinant clones and their parental lines. (A) Data were obtained using the nigericin/BSA pH clamp method that has been reported to measure NHE activity. Saponin-permeabilized iRBCs were perfused with high K+ containing E1 (K+ E1) buffer at pH 7.5. 1 µM nigericin was added to the perfusate at the downward arrow marked (N), first at pH 7.5 and then at pH 6.0. After equilibration of the cytosolic BCPCF probe signal at pH 6.0, nigericin was removed and 5 mg/ml BSA was added to the perfusate at the downward arrow marked (B) to adsorb the remaining nigericin, clamping the cytoplasmic pH at 6.0. The perfusion pH was then returned to 7.4, either in the presence of Na+ (solid trace) or in a Na+ free NMDG+ buffer (dashed trace). Each trace represents the average of over 10 individually analyzed parasites within one microscopic field. (B) Prior to the time zero in these traces the full pH clamp experiment was performed as in panel (A) up to 400 seconds, where the buffer pH was returned to 7.4. Data from three separate experiments with 3BA6 parasites (closed triangles) in which the return to pH 7.4 Na+ E1 buffer is shown on an expanded time scale (i.e., from ~400 seconds to end in panel A). Parental 3BA6 parasites exhibit significant variability in their realkalinization rate upon return to Na+ E1 (closed triangles), but show very little alkalinization upon return to NMDG+ E1 (open triangles). A 3BA6 NHE under expressing strain (closed squares) exhibits a rate of alkalinization within the range of the parental strain responses. Traces are representative of many experiments, conducted on multiple days. Each set of points represents the average and SEM of at least 10 parasites in one microscopic field of view. As noted in the text, in a given experiment all parasites in a microscopic field of view alkalinized with a similar rate but the rates varied significantly from trial to trial.
While Na+ dependent cytosolic alkalinization was observed for both parental and pfnhe knockdown strains, our quantitative measurements of the rate of alkalinization proved to be highly variable between independent assays. In a given experiment, all parasites within the field of view alkalinized at a similar rate. However, there was significant variation in the alkalinization rate following reintroduction of Na+ between experimental trials, even with the same batch of parasites used in consecutive experiments (Fig. 5B, filled triangles). Despite extensive, rigorous efforts to control multiple parameters including the temperature, rate of perfusion, and the duration of reagent applications, the rate of alkalinization after return to high Na+ buffer at pH 7.4 exhibited a significant degree of variability from experiment to experiment (Fig. 5B). The variability observed was present both in parental and pfnhe knockdown lines. Consequently, we were unable to test for any decrease in NHE activity commensurate with the decreased levels of protein.
A recent study suggested that Na+ dependent alkalinization observed in this assay is due to the presence of residual amounts of nigericin in the cell membrane [30]. This occurs because the BSA wash fails to completely remove the nigericin, which then exchanges Na+ and H+, albeit with lower efficiency than K+ and H+. The variability that we observed between experimental trials could be due to differing residual amounts of nigericin in the parasite plasma membrane. This would explain why all parasites in a given experiment and microscopic field of view alkalinized with a similar rate, but the rate varied from trial to trial (Fig. 5B).
4. Discussion
Earlier studies into the genetic basis of QN resistance point to this being governed by a complex multigenic trait, which includes contributions from pfcrt, pfmdr1 and a third locus residing on a 380 kb segment of P. falciparum chromosome 13 [11]. From the ~100 predicted genes within this chromosomal segment, pfnhe was identified as a possible mediator of QN resistance because of the association between QN resistance and inheritance of the Dd2 pfnhe allele (harboring microsatellite repeat variants and point mutations that contrasted with the HB3 allele) in the progeny of the HB3×Dd2 cross, and also because the Dd2 type microsatellite variant of the DNND repeats (known as ms4760-1; [11]) was associated with decreased QN susceptibility in field isolates from geographically disparate regions [11]. More recently, an independent investigation into PfNHE in the HB3×Dd2 progeny reported activity measurements that supported a role for this sodium-proton exchanger in QN resistance [23]. The experimental basis for this conclusion however has recently been questioned in a separate report [30]. In the present work, we employed an allelic exchange strategy to reduce the level of expression of pfnhe in an isogenic background and directly assess its effect on the parasite response to QN.
Under-expression of pfnhe in the CQ-resistant backgrounds of the 1BB5 and 3BA6 parasites caused a statistically significant ~30% reduction in QN IC50 values. This effect was not observed in the CQ-sensitive background of the GC03 parasite. The absence of an effect on QN IC50 in recombinant GC03 clones suggests that the extent to which pfnhe can affect QN susceptibility is strain-specific and dependent on its interplay with additional genetic factors. One plausible candidate is pfcrt (see below), which was present in its wild type HB3 form in GC03 and in its Dd2 allelic form in 1BB5 and 3BA6. The other known determinant, pfmdr1, would not appear to contribute to this different effect of reducing pfnhe expression in these distinct progeny as the HB3 type allele was present in all three progeny (Table 1).
QN resistance has been found to display a complex relationship with CQ, and as stated earlier the contribution of the primary CQ resistance determinant PfCRT to QN response appears to depend on its haplotype and the parasite genetic background. For example, in vitro CQ pressure on the CQ-sensitive 106/1 parasite line selected a PfCRT K76I mutation, which decreased susceptibility to CQ by 12-fold but also increased susceptibility to QN by 18-fold. In contrast, a K76N PfCRT mutant line selected in parallel displayed increased resistance to both CQ and QN [31]. In a subsequent study, separate mutations in PfCRT were identified in parasites that had been selected for QN resistance and that were observed to have lost their resistance to CQ [32]. Another study employing an allelic exchange strategy (similar to the approach used in the current work) in the CQ-resistant 7G8 strain showed that reduced PfCRT expression levels were associated with an increase in CQ sensitivity but had no effect on QN or mefloquine sensitivity [13]. We also note that introduction of the Dd2 pfcrt allele into recombinant GC03 parasites rendered them more susceptible to QN [7]. Yet paradoxically the Dd2 pfcrt allele is present in 1BB5 and 3BA6 parasites that are less susceptible than GC03 to QN. All three progeny share the same pfnhe and pfmdr1 haplotypes (Table 1). These data argue that parasite proteins in addition to PfCRT, PfMDR1 and PfNHE are an important component of the QN response. One additional candidate comes from pair-wise scans of quantitative trait loci association, which recently identified a potential interaction between the chromosome 13 segment harboring pfnhe and a separate locus on chromosome 9 [11]. This interaction was also reportedly linked with alterations in cytosolic pH, or the pH gradient across the digestive vacuole membrane, that were proposed to associate with QN resistance in some HB3 ×Dd2 progeny [23]. Detailed biochemical and genetic studies will be required in an array of genetically distinct backgrounds to elucidate how these proteins might interact to regulate QN access to its intracellular targets and modulate its potency.
While our data corroborate a link between PfNHE and QN resistance, the subcellular localization of this transporter, harboring 12 predicted transmembrane domains and a putative signal peptide, and its function in P. falciparum physiology are not established. In most eukaryotic cells, NHE activity has an important role in proton export from the cytoplasm, however most mammalian NHEs are minimally active at normal cytosolic pH [33]. An early report proposed that a P. falciparum NHE might be active at resting cytosolic pH because amiloride and EIPA induced cytoplasmic acidification, albeit at higher concentrations than needed to inhibit most mammalian NHEs [22]. Subsequent experiments showed that the cytoplasmic acidification following the application of amiloride and EIPA also occurred in Na+-free media, indicating that the effects were not mediated by NHE inhibition [34]. The recent study of PfNHE in 13 HB3×Dd2 progeny reported higher NHE activity in the progeny with reduced sensitivity to QN, regardless of their CQ status [23]. It should be noted, however, that a recent report by Kirk and coworkers raised questions regarding the validity of the nigericin/BSA pH clamp assay used to measure NHE activity due to technical difficulties in completely removing nigericin from the cell preparations during these assays [30]. From their data, it appears that the observed alkalinization resulted from the residual nigericin rather than from NHE activity [30]. The difficulty in completely removing nigericin provides a likely explanation for our inability to obtain reproducible rates of alkalinization in the present work using the nigericin/BSA pH clamp assay (Fig. 5B). Thus, we were unable to confirm earlier reports of PfNHE activity in the P. falciparum plasma membrane [22; 23].
The study of the 13 HB3×Dd2 progeny also reported a linear relationship between cytosolic pH and QN resistance that coincided with allelic combinations of pfnhe and the chromosome 9 partner [23]. In contrast, in the current work we found no significant differences in cytoplasmic pH amongst the three parental parasite lines or in any of the PfNHE knockdown lines. Our interpretation of the present results leads us to suggest that variations in PfNHE expression levels do not significantly affect cytosolic pH, nor does cytosolic pH correlate with QN resistance. We think that it is unlikely that parasites might have a significant excess NHE capacity for proton export such that a lower rate of sodium-H+ exchange resulting from reduced expression of PfNHE does not measurably alter cytosolic pH in the knockdown lines. This is consistent with the view PfNHE does not play a significant role in H+ extrusion and regulation of cytoplasmic pH [30]. Proton transport is multi-faceted and its control is likely to be determined by partner pathways that regulate pH balance in the cell. For example, a study of P. falciparum pH regulation found that the parasite plasma membrane V-type H+ ATPase is a major determinant of cytosolic pH maintenance [34]. In another example, much of the proton load produced by glycolysis can be removed from the parasite cytoplasm during lactate export via the P. falciparum H+/monocarboxylate symporter [35].
Historically, reliable studies of cytosolic pH in P. falciparum have proven difficult in part because of the photodynamic damage to the digestive vacuole produced by high excitation light intensity [36], the compartmentalization of reporter dye within extra-cytosolic locations, and dye overloading. These factors can combine to clamp the pH to the pKa of the reporter dye [37; 38]. Such problems have led different groups to report significantly different results using related but not identical methodologies. These confounding factors led us to strive for an experimental method that minimizes their impact. The method we report herein allowed us to measure cytosolic pH values that differed from the extracellular pH and from the pKa of the BCPCF fluorescent dye (pKa = 7.0) [21]. Furthermore, our cytosolic pH measurements were stable for over 10 minutes under the illumination intensity conditions used, indicating that photodynamic damage was not a significant factor. In contrast to Bennett et al. [23], we found no difference between CQ-sensitive and -resistant lines in terms of their cytosolic pH. Our data agree with a recent study that employed the pH-sensitive green fluorescent protein pHluorin and observed no difference in cytosolic pH between the CQ-sensitive HB3 and CQ-resistant Dd2 lines [38].
QN is a monoprotic weak base that accumulates within the low pH environment of the parasite digestive vacuole. A principle mode of its action is thought to be interference with the detoxification of heme produced during hemoglobin degradation, with resultant buildup of toxic degradation byproducts [39]. While this initial framework for understanding the toxicity of the quinoline antimalarial drugs for parasites is widely accepted, it has become clear that the mechanisms of resistance to QN and related quinolines can differ in important ways. Our study provides evidence that PfNHE expression levels can influence QN sensitivity, although this appears to be strain dependent. Furthermore, our data suggest that this effect might depend on the CQ resistance status of the host parasite strain, even though reduced pfnhe expression levels did not alter the CQ response of the strains investigated herein. Additional transfection studies are now required to define the contribution of known PfNHE polymorphisms to QN resistance in culture-adapted laboratory lines and to test their association with QN treatment outcomes. The successful use of QN for centuries illustrates the difficulties that the Plasmodium parasite faces in establishing resistance and disseminating this phenotype across malarial regions. Delineating the contribution of PfNHE and other parasite genes to QN resistance will be increasingly important as this drug continues to be relied upon for the treatment of severe malarial infections that are resistant to other antimalarial agents.
Acknowledgements
We thank Dr. Marcus Lee and Celeste D. Li for their kind help with the preparation of this manuscript. Funding for this work was provided in part by an Investigator in Pathogenesis Award in Infectious Diseases from the Burroughs Wellcome Fund (to D.A.F.) and from the NIH (to M.H.A.). T.E.W. is supported by the Intramural Research Program of the NIH, NIAID.
References
- 1.Winstanley P. Modern chemotherapeutic options for malaria. Lancet Infect Dis. 2001;1:242–250. doi: 10.1016/S1473-3099(01)00119-0. [DOI] [PubMed] [Google Scholar]
- 2.Meshnick SR. From quinine to qinghaosu: historical perspectives. In: Sherman I, editor. Malaria: parasite biology, pathogenesis and protection. Washington, DC: ASM Press; 1998. pp. 341–354. [Google Scholar]
- 3.Smith DC. Quinine and fever: The development of the effective dosage. J Hist Med Allied Sci. 1976;31:343–367. doi: 10.1093/jhmas/xxxi.3.343. [DOI] [PubMed] [Google Scholar]
- 4.Foley M, Tilley L. Quinoline antimalarials: mechanisms of action and resistance and prospects for new agents. Pharmacol Ther. 1998;79:55–87. doi: 10.1016/s0163-7258(98)00012-6. [DOI] [PubMed] [Google Scholar]
- 5.White NJ. Antimalarial drug resistance: the pace quickens. J Antimicrob Chemother. 1992;30:571–585. doi: 10.1093/jac/30.5.571. [DOI] [PubMed] [Google Scholar]
- 6.Uhlemann AC, Krishna S. Antimalarial multi-drug resistance in Asia: mechanisms and assessment. Curr Top Microbiol Immuno. 2005;295:39–53. doi: 10.1007/3-540-29088-5_2. [DOI] [PubMed] [Google Scholar]
- 7.Sidhu ABS, Verdier-Pinard D, Fidock DA. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science. 2002;298:210–213. doi: 10.1126/science.1074045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lakshmanan V, Bray PG, Verdier-Pinard D, et al. A critical role for PfCRT K76T in Plasmodium falciparum verapamil-reversible chloroquine resistance. EMBO J. 2005;24:2294–2305. doi: 10.1038/sj.emboj.7600681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reed MB, Saliba KJ, Caruana SR, et al. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- 10.Wellems TE, Panton LJ, Gluzman IY, et al. Chloroquine resistance not linked to mdr-like genes in a Plasmodium falciparum cross. Nature. 1990;345:253–255. doi: 10.1038/345253a0. [DOI] [PubMed] [Google Scholar]
- 11.Ferdig MT, Cooper RA, Mu J, et al. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol Microbiol. 2004;52:985–997. doi: 10.1111/j.1365-2958.2004.04035.x. [DOI] [PubMed] [Google Scholar]
- 12.Vinayak S, Alam MT, Upadhyay M, et al. Extensive genetic diversity in the Plasmodium falciparum Na+/H+ exchanger 1 transporter protein implicated in quinine resistance. Antimicrob Agents Chemother. 2007;51:4508–4511. doi: 10.1128/AAC.00317-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waller KL, Muhle RA, Ursos LM, et al. Chloroquine resistance modulated in vitro by expression levels of the Plasmodium falciparum chloroquine resistance transporter. J Biol Chem. 2003;278:33593–33601. doi: 10.1074/jbc.M302215200. [DOI] [PubMed] [Google Scholar]
- 14.Sidhu AB, Uhlemann AC, Valderramos SG, et al. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J Infect Dis. 2006;194:528–535. doi: 10.1086/507115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horton RM, Hunt HD, Ho SN, et al. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 16.Sidhu AB, Valderramos SG, Fidock DA. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol Microbiol. 2005;57:913–926. doi: 10.1111/j.1365-2958.2005.04729.x. [DOI] [PubMed] [Google Scholar]
- 17.Fidock DA, Nomura T, Wellems TE. Cycloguanil and its parent compound proguanil demonstrate distinct activities against Plasmodium falciparum malaria parasites transformed with human dihydrofolate reductase. Mol Pharmacol. 1998;54:1140–1147. doi: 10.1124/mol.54.6.1140. [DOI] [PubMed] [Google Scholar]
- 18.Goodyer ID, Taraschi TF. Plasmodium falciparum: a simple, rapid method for detecting parasite clones in microtiter plates. Exp Parasitol. 1997;86:158–160. doi: 10.1006/expr.1997.4156. [DOI] [PubMed] [Google Scholar]
- 19.Kyes S, Pinches R, Newbold C. A simple RNA analysis method shows var and rif multigene family expression patterns in Plasmodium falciparum. Mol Biochem Parasitol. 2000;105:311–315. doi: 10.1016/s0166-6851(99)00193-0. [DOI] [PubMed] [Google Scholar]
- 20.Staalsoe T, Giha HA, Dodoo D, et al. Detection of antibodies to variant antigens on Plasmodium falciparum-infected erythrocytes by flow cytometry. Cytometry. 1999;35:329–336. doi: 10.1002/(sici)1097-0320(19990401)35:4<329::aid-cyto5>3.3.co;2-p. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Diwu Z, Klaubert DH. Fluorescent molecular probes III. 2',7'-bis-(3-carboxypropyl)-5-(and-6)-carboxyfluorescein (BCPCF): A new polar dual-excitation and dual-emission pH indicator with a pKa of 7.0. Bioorg & Med Chem Lett. 1997;7:3069–3072. [Google Scholar]
- 22.Bosia A, Ghigo D, Turrini F, et al. Kinetic characterization of Na+/H+ antiport of Plasmodium falciparum membrane. J Cell Physiol. 1993;154:527–534. doi: 10.1002/jcp.1041540311. [DOI] [PubMed] [Google Scholar]
- 23.Bennett TN, Patel J, Ferdig MT, et al. Plasmodium falciparum Na+/H+ exchanger activity and quinine resistance. Mol Biochem Parasitol. 2007;153:48–58. doi: 10.1016/j.molbiopara.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reeves DC, Liebelt DA, Lakshmanan V, et al. Chloroquine-resistant isoforms of the Plasmodium falciparum chloroquine resistance transporter acidify lysosomal pH in HEK293 cells more than chloroquine-sensitive isoforms. Mol Biochem Parasitol. 2006;150:288–299. doi: 10.1016/j.molbiopara.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su X, Ferdig MT, Huang Y, et al. A genetic map and recombination parameters of the human malaria parasite Plasmodium falciparum. Science. 1999;286:1351–1353. doi: 10.1126/science.286.5443.1351. [DOI] [PubMed] [Google Scholar]
- 26.Elmendorf HG, Haldar K. Identification and localization of ERD2 in the malaria parasite Plasmodium falciparum: separation from sites of sphingomyelin synthesis and implications for organization of the Golgi. Embo J. 1993;12:4763–4773. doi: 10.1002/j.1460-2075.1993.tb06165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin SK, Oduola AM, Milhous WK. Reversal of chloroquine resistance in Plasmodium falciparum by verapamil. Science. 1987;235:899–901. doi: 10.1126/science.3544220. [DOI] [PubMed] [Google Scholar]
- 28.Price RN, Uhlemann AC, Brockman A, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valderramos SG, Fidock DA. Transporters involved in resistance to antimalarial drugs. Trends Pharmacol Sci. 2006;27:594–601. doi: 10.1016/j.tips.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spillman NJ, Allen RJ, Kirk K. Acid extrusion from the intraerythrocytic malaria parasite is not via a Na(+)/H(+) exchanger. Mol Biochem Parasitol. 2008;162:96–99. doi: 10.1016/j.molbiopara.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 31.Cooper RA, Ferdig MT, Su X-Z, et al. Alternative mutations at position 76 of the vacuolar transmembrane protein PfCRT are associated with chloroquine resistance and unique stereospecific quinine and quinidine responses in Plasmodium falciparum. Mol Pharmacol. 2002;61:35–42. doi: 10.1124/mol.61.1.35. [DOI] [PubMed] [Google Scholar]
- 32.Cooper RA, Lane KD, Deng B, et al. Mutations in transmembrane domains 1, 4 and 9 of the Plasmodium falciparum chloroquine resistance transporter alter susceptibility to chloroquine, quinine and quinidine. Mol Microbiol. 2007;63:270–282. doi: 10.1111/j.1365-2958.2006.05511.x. [DOI] [PubMed] [Google Scholar]
- 33.Orlowski J, Grinstein S. Na+/H+ exchangers of mammalian cells. J Biol Chem. 1997;272:22373–22376. doi: 10.1074/jbc.272.36.22373. [DOI] [PubMed] [Google Scholar]
- 34.Saliba KJ, Kirk K. pH regulation in the intracellular malaria parasite, Plasmodium falciparum. H(+) extrusion via a v-type h(+)-atpase. J Biol Chem. 1999;274:33213–33219. doi: 10.1074/jbc.274.47.33213. [DOI] [PubMed] [Google Scholar]
- 35.Elliott JL, Saliba KJ, Kirk K. Transport of lactate and pyruvate in the intraerythrocytic malaria parasite, Plasmodium falciparum. Biochem J. 2001;355:733–739. doi: 10.1042/bj3550733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wissing F, Sanchez CP, Rohrbach P, et al. Illumination of the malaria parasite Plasmodium falciparum alters intracellular pH. Implications for live cell imaging. J Biol Chem. 2002;277:37747–37755. doi: 10.1074/jbc.M204845200. [DOI] [PubMed] [Google Scholar]
- 37.Hayward R, Saliba KJ, Kirk K. The pH of the digestive vacuole of Plasmodium falciparum is not associated with chloroquine resistance. J Cell Sci. 2006;119:1016–1025. doi: 10.1242/jcs.02795. [DOI] [PubMed] [Google Scholar]
- 38.Kuhn Y, Rohrbach P, Lanzer M. Quantitative pH measurements in Plasmodium falciparum-infected erythrocytes using pHluorin. Cell Microbiol. 2007;9:1004–1013. doi: 10.1111/j.1462-5822.2006.00847.x. [DOI] [PubMed] [Google Scholar]
- 39.Hawley SR, Bray PG, Mungthin M, et al. Relationship between antimalarial drug activity, accumulation, and inhibition of heme polymerization in Plasmodium falciparum in vitro. Antimicrob Agents Chemother. 1998;42:682–686. doi: 10.1128/aac.42.3.682. [DOI] [PMC free article] [PubMed] [Google Scholar]