Abstract
We studied the reactivity of 35 genetically engineered Cys sulfhydryl groups at different locations in Escherichia coli FepA. Modification of surface loop residues by fluorescein maleimide (FM) was strongly temperature-dependent in vivo, whereas reactivity at other sites was much less affected. Control reactions with bovine serum albumin showed that the temperature dependence of loop residue reactivity was unusually high, indicating that conformational changes in multiple loops (L2, L3, L4, L5, L7, L8, L10) transform the receptor to a more accessible form at 37 °C. At 0 °C colicin B binding impaired or blocked labeling at 8 of 10 surface loop sites, presumably by steric hindrance. Overall, colicin B adsorption decreased the reactivity of more than half of the 35 sites, in both the N - and C- domains of FepA.
However, colicin B penetration into the cell at 37 °C did not augment the chemical modification of any residues in FepA. The FM modification patterns were similarly unaffected by the tonB locus. FepA was expressed at lower levels in a tonB host strain, but when we accounted for this decrease its FM-labeling was comparable whether TonB was present or absent.
Thus we did not detect TonB-dependent structural changes in FepA, either alone or when it interacted with colicin B at 37 °C. The only changes in chemical modification were reductions from steric hindrance when the bacteriocin bound to the receptor protein. The absence of increases in the reactivity of N-domain residues argues against the idea (Devanathan and Postle, Mol. Microbiol. 65: 441–453, 2007) that the colicin B polypeptide traverses the FepA channel.
The natural function of the Escherichia coli outer membrane (OM) protein FepA is recognition and uptake of ferric enterobactin (FeEnt) (Pugsley and Reeves, 1976; McIntosh and Earhart, 1976; Wayne et al., 1976). Bacteriocins {in the case of FepA, colicins B (colB) and D (Davies and Reeves, 1975; Cao and Klebba, 2002)} and viruses {bacteriophage H8 (Rabsch et al., 2007)} parasitize bacterial solute and nutrilite uptake pathways. Colicins initially interact with an OM receptor protein, denature and unfold on the cell surface (Benedetti et al., 1992; Zakharov et al., 2008), and then transfer a functional domain that kills the cell by mechanisms like inner membrane (IM) depolarization or degradation of nucleic acids or peptidoglycan. One model of colicin uptake through the OM involves movement of the toxin polypeptide through a transmembrane porin channel (Yamashita et al., 2008; Zakharov et al., 2008). FepA and its homologs contain a C-terminal, 22-stranded porin channel (Saier, 2000) and a globular N-terminus (N-domain) inside the pore. Without structural rearrangement, the N-domain in the channel prohibits the passage of a metal chelate or a bacteriocin. This enigma may be resolved by conformational motion that forms a transient pore or moves the N-domain out of the channel (Ma et al., 2007; Gumbart et al., 2007; Devanathan and Postle, 2007).
The energetics of colicin uptake are not definitively known, but colicin activity requires the additional cell envelope proteins TonB (Wang and Newton, 1971; Guterman and Dann, 1973; Reynolds et al., 1980; Postle, 1993) or TolA (Gerding et al., 2007; Pommier et al., 2005).
TonB-dependent metal transport, and Tol-dependent cellular systems are energy dependent (Wang and Newton, 1969; Pugsley and Reeves, 1977; Bradbeer, 1993; Goemaere et al., 2007). These requirements are not fully understood: the OM cannot sustain an ion gradient because of its open channels (Nikaido and Vaara, 1985); TonB and TolA are minor cell envelope proteins whose exact functions are unknown. The N-termini of both proteins are postulated to reside in the IM, but their C-termini interact with OM proteins (Shultis et al., 2006; Pawelek et al., 2006; Cascales and Lloubes, 2004), suggesting that they span the periplasm. Consistent with this idea, the TonB C-terminus also binds peptidoglycan {PG; (Kaserer et al., 2008)}, and the C-terminus of TolA binds the PG-associated protein Pal (Bouveret et al., 1999). The requirements for energy and TonB during FepA-mediated transport may relate to the structural rearrangement of the receptor protein’s globular N-domain, noted above.
Ligands bind to FepA in biphasic reactions (Payne et al., 1997). FeEnt first adsorbs to residues in the loop extremities (Cao et al., 2000; Annamalai et al., 2004), and loops ultimately coalesce around it (Buchanan et al., 1999; Scott et al., 2002), ensconcing the metal complex in the receptor’s vestibule at binding equilibrium. Ligand binding generally elicits structural changes in the N-domain that relocate the TonB-box away from the β-barrel wall, signaling receptor occupancy at the periplasmic side of the OM (Locher et al., 1998; Ferguson et al., 1998).
However, this phenomenon does not occur during binding of ColIa to Cir (Buchanan et al., 2007) in vitro. These initial stages of ligand uptake occur with equivalent affinity and rate in energy-sufficient or -deficient, and tonB+ or tonB cells (Newton et al., 1999, Annamalai et al., 1984). In tonB+, fepA+ cells the C-terminal domain of ColB causes cell death by forming a depolarizing channel in the IM. tonB cells survive colB, presumably because its killing domain does not penetrate their OM.
To assess models of ColB transport we studied the accessibility of genetically engineered Cys side chains in FepA to covalent modification by fluorescein maleimide (FM). The reagent strongly labeled surface loop sites. These reactions were temperature-dependent, and inhibited by ColB binding to FepA. However, we did not observe increases in the accessibility of any Cys residues in FepA during ColB killing at 37 °C. Thus, we found no evidence that the ColB polypeptide passes through the FepA channel.
RESULTS
Kinetics of ColB binding and killing
We used the membrane soluble carbocyanine dye DiOC2(3) to cytometrically measure the time required for ColB-induced depolarization of E. coli cells. DiOC2(3) associates with and accumulates in bacterial cells; changes in its emission spectrum reflect alterations in cell membrane potential (Suzuki et al., 2003; Novo et al., 1999). Upon exposure of the ColB-sensitive strain BN1071 (fepA+, tonB+) to ColB and DiOC2(3), cell-associated red fluorescence increased within 5 min and reached maximum levels by 30–40 min (Fig. S1). Conversely, red fluorescence of the ColB-resistant strain OKN13 (fepA, tonB) did not increase during incubation with ColB and DiOC2(3), whereas the control depolarizing agent CCCP increased the fluorescence of both BN1071 and OKN13. On the basis of these and other findings (Kadner and McElhaney, 1978; Benedetti et al., 1992), we exposed the bacteria to excess ColB (0.28 uM; 15-fold molar excess over [FepA]; see Experimental Procedures) for 30 min at 0 or 37 °C prior to the initiation of FM labeling, which was 15 min in duration.
Temperature dependence of modification of genetically engineered Cys residues by FM
The experiments involved 25 existing Cys substitution mutations throughout FepA (Ma et al., 2007), and 10 new Cys substitutions in the N-domain (at residues 12, 39, 42, 51, 56, 59, 63, 101, 127, 135). In OKN3 (fepA, tonB+) the native fepA+ promoter on pITS47 conferred wild type expression levels for all the mutant proteins (Fig. 2s; (Ma et al., 2007)). With the exception of W101C, the mutant proteins transported FeEnt like wild-type FepA in siderophore nutrition tests, and showed normal ColB susceptibility (data not shown). W101C sometimes formed an aberrant disulfide bond with the native Cys residues in L7 (Cys 487 and 494), which was seen in non-reducing SDS-PAGE (data not shown).
We determined the accessibility of the sulfhydryl side chains to FM labeling at 0 °C and 37 °C (Fig. 1, Fig S2). From studies of the concentration- and time-dependence of fluoresceination {(Ma et al., 2007); data not shown} we employed FM at 5 uM for 15 min in PBS, pH 6.5. This protocol primarily labeled the FepA mutant proteins, and other cellular proteins at lower levels, but not wild-type FepA {Fig. S2; (Ma et al., 2007). To standardize the analysis of FM labeling we compared the fluoresceination of FepA to that of a cellular protein {band 3 in Fig. S2 (see also band 3 in Fig. 3 of (Ma et al., 2007))} that was consistently modified throughout the experiments (mean among 20 independent samples, 7.7% ± 0.4% of total fluorescence in cell lysate proteins); we report the data as the ratio of fluorescence intensities: FepA-FL/Band 3-FL (Fig. 1).
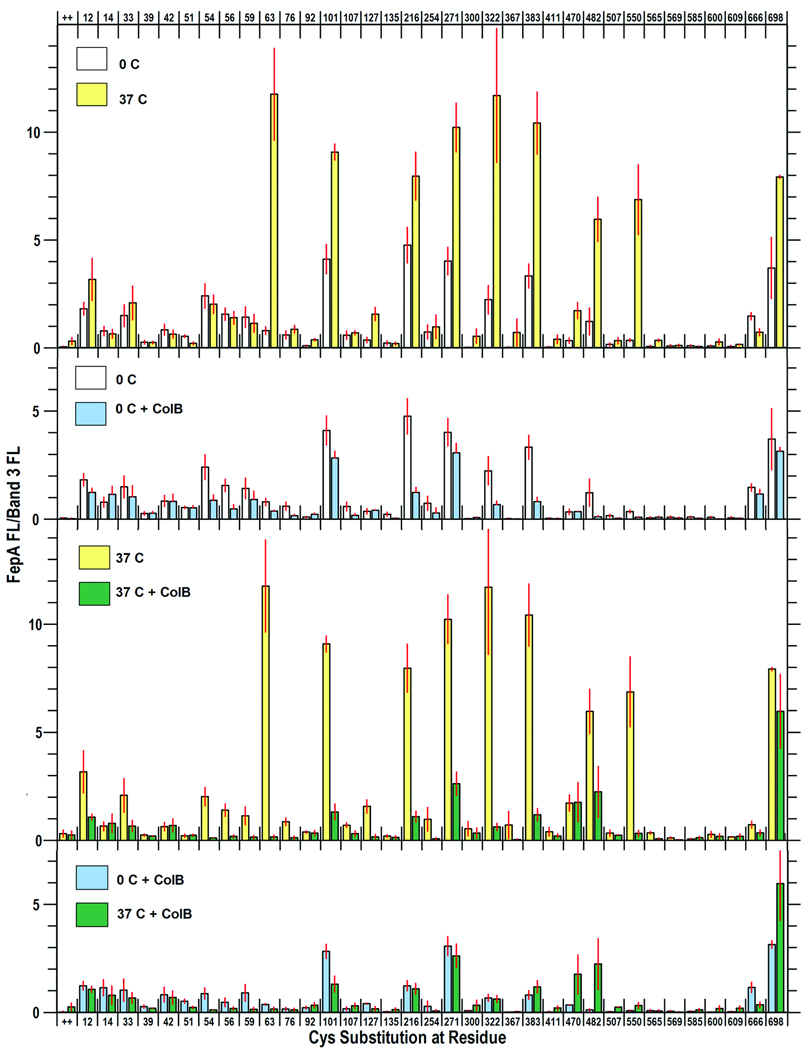
Figure 1. Accessibility of Cys residues in FepA to FM-labeling at 0 °C and 37 °C, and during ColB binding (at 0 °C) and uptake (at 37 °C).
OKN3 (fepA) harboring plasmids carrying fepA+ or its derivatives that encode Cys substitutions mutations were prepared as described in Experimental Procedures, resuspended in PBS, pH 6.5, and incubated for 30 min at 0 °C or 37 °C in the absence or presence of ColB, and then exposed to FM (5 uM) for 15 min at the same temperature. Cells lysates were resolved by SDS-PAGE and fluorescence images from the gels (Fig. S2) were analyzed by IMAGEQUANT (Molecular Dynamics). Each panel shows the mean FM-labeling of FepA (relative to band 3) from three or more independent experiments, with associated standard error. A. 0 °C vs 37 °C. White bars derive from cells labeled at 0 °C; yellow bars are from cells labeled at 37 °C. The inset shows fluoresceination of BSA at 0 °C, 25 °C and 27 °C, in the presence of 5 uM (grey bars) and 50 uM (black) FM. B. 0 °C, ± ColB. At 0 °C the cells are metabolically inactive, so ColB binds but is not transported. White bars derive from cells labeled at 0 °C in the absence of ColB; light blue bars are from cells labeled at 0 °C in the presence of ColB. C. 37 °C ± ColB. At 37 °C the cells are metabolically active, so ColB binds and kills. Yellow bars derive from cells labeled at 37 °C in the absence of ColB; green bars are from cells labeled at 37 °C in the presence of ColB. D. 0 °C vs 37 °C, + ColB. The graph compares the effects of ColB on the labeling of FepA Cys mutants at 0 °C and 37 °C. No increases in FM-labeling of N-domain residues were observed during ColB killing.
Figure 3. Evaluation of TonB-dependent conformational changes in FepA: Comparison of Cys fluoresceination in tonB+ and tonB cells.
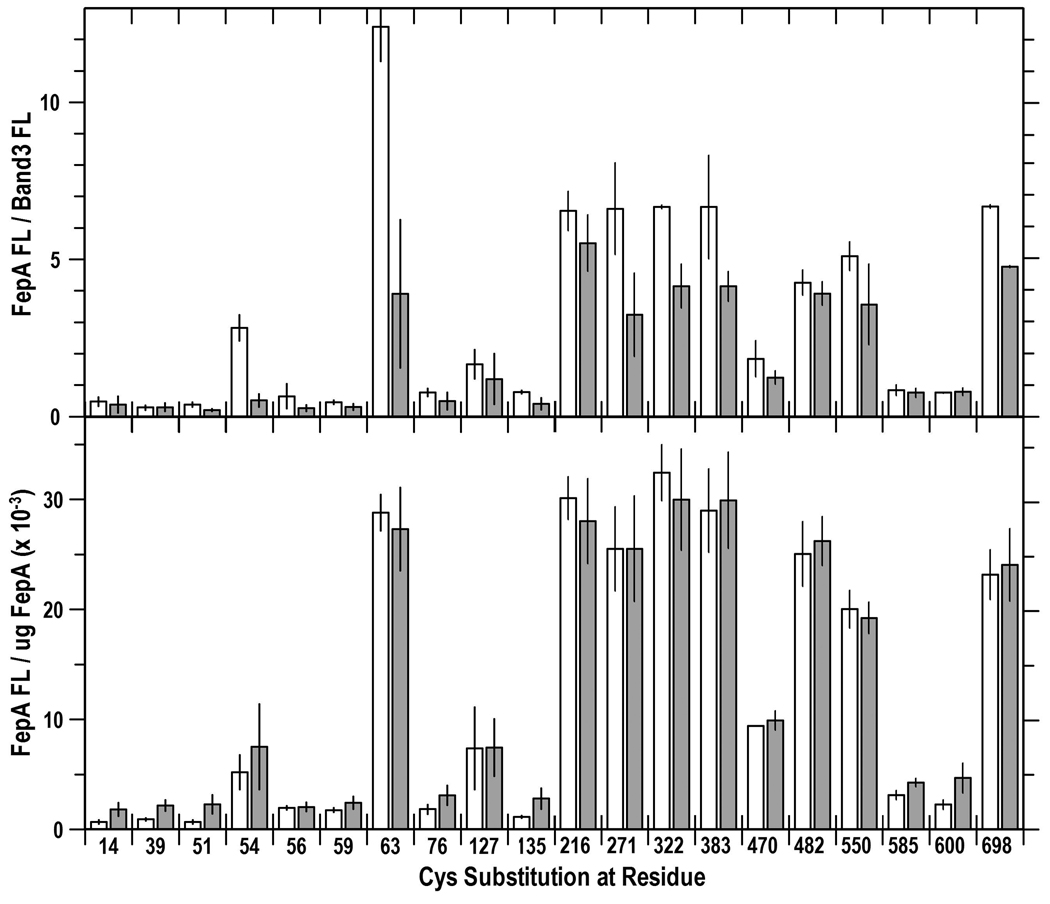
Sites in FepA that were significantly labeled by FM were re-analyzed and compared in tonB+ (white bars) and tonB (grey bars) cells. We incorporated the concentration of FepA (from anti-FepA immunoblots) into calculations to compare the relative and absolute FM-labeling levels. (Top) Relative levels of FM-labeling in tonB+ and tonB bacteria. Fluorescence images from SDS-PAGE gels of cell lysates were analyzed by IMAGEQUANT (Molecular Dynamics). Bars depict the mean FM-labeling of FepA proteins relative to band 3 in OKN3 (white) and OKN13 (grey; mean of 3 experiments, with associated standard error). FepA proteins were less fluoresceinated in the tonB strain, because they were expressed at lower levels (Fig. S4). (Bottom) Absolute levels of FM-labeling in tonB+ and tonB strains. The extent of residue labeling was corrected for the expression level of each of the mutant FepA protein, to yield the absolute labeling level (fluorescence intensity/ug FepA). The correction eliminated the differences in labeling between tonB+ and tonB cells seen in the top panel.
When exposed to FM in the absence of ligands the reactivity of many FepA Cys side chains was temperature dependent. At 0 °C FM strongly modified Cys residues in the surface loops (101, 216, 271, 322, 383, and 698), and less intensely reacted with several sulfhydryls in the N-domain (54, 56, 59, 63, 76) or at the periplasmic interface (residues 12, 14, 33, 666). Other residues in the N-domain (39, 92, 135), or on the interior (411, 565, 569, 585, 600) or exterior (367, 609) of the C-domain β-barrel were unreactive or only marginally labeled above background. Fluoresceination at many sites was temperature-independent (residues 33, 54, 59, 216, 666), but modification of cell-surface Cys residues intensified as the temperature rose from 0 °C to 37 °C: nominally 2–3 - fold (residues 101, 216, 271, 383, 698), and as much as 7–10 - fold (residues 63, 322, 470, 482, 550). These data suggested structural alterations in the loops in response to physiological temperature. Control reactions with bovine serum albumin (BSA) confirmed this inference: as temperature changed from 0 °C to 37 °C the maleimide modification of Cys in BSA only increased 26% (Fig. 1), which corresponded to a Q10 temperature coefficient of 1.09 (Table 1). Thus the 200 – 1000% increase in loop residue labeling (Q10 values of 1.15 – 2.23) did not solely accrue from diffusion-controlled enhancement of the maleimide reaction rate, but rather, from conformational changes. It was noteworthy that enhanced chemical modification occurred in 9 of the 10 surface loops that we tested (S63 in NL1; W101, NL2; T216, L2; S271, L3; A322, L4; A383, L5; S470, S482, L7; T550, L8, and A698, L11; S600 in L9 was not modified), which implied concerted conformational motion in the loops of FepA as temperature increased to physiological levels.
Table 1. Temperature-dependence of FM-modification of engineered Cys sulfhydryls in the surface loops of FepA.
| Residue | Loop | Q10 |
|---|---|---|
| BSA | 1.09 | |
| 63 | NL1 | 2.07 |
| 101 | NL2 | 1.24 |
| 216 | L2 | 1.15 |
| 271 | L3 | 1.29 |
| 322 | L4 | 1.56 |
| 383 | L5 | 1.36 |
| 470 | L7 | 1.55 |
| 482 | L7 | 1.53 |
| 550 | L8 | 2.23 |
| 698 | L11 | 1.23 |
Effect of ColB binding and uptake on the accessibility of Cys residues to FM labeling
At 0 °C ColB only binds to FepA, whereas at 37 °C it binds and kills the bacteria. We determined the susceptibility of sites within FepA to FM labeling in the presence of ColB (15-fold molar excess over [FepA]) at both temperatures. At 0 °C toxin binding reduced modification (20 – 90%) at the following residues: 54, 56, 63, 101, 216, 271, 322, 383, 482 (Fig 1, Fig S2). With the exception of G54 and N56, these sites exist within surface loops, and reduction of their modification at 0 °C presumably originated from steric hindrance when ColB adsorbed to FepA. G54 and N56 reside deeper within the N-domain, but may be labeled from the cell exterior (Ma et al., 2007), so ColB likely also obscured them by its surface binding.
When we shifted cells with bound ColB to 37 °C we observed few changes in the FM-labeling patterns (Fig 1, Fig S2). As noted above, increasing the temperature augmented FM labeling of surface loop residues, but, just as at 0 °C, incubation with ColB at 37 °C generally impaired or blocked fluoresceination of these sites. For instance, modification of S63C in NL1 and T550C in L8, which was 10-fold more intense at 37 °C, was virtually eliminated by the presence of the bacteriocin. ColB blocked all the same residues at 37 °C that it blocked at 0 °C. We looked for sites whose reactivity increased in response to incubation with ColB at 37 °C, which was previously reported to occur from passage of the toxin through the FepA channel (Devanathan and Postle, 2007) but we did not observe any increase in FM modification for the 35 sites that we surveyed, including 15 in the N-domain. We saw no changes in residues 33, 42, 51, and 92, which (Devanathan and Postle, 2007) reported to increase in accessibility as a result of ColB penetration. At 13 of the 35 positions ColB had no effect on labeling; at the remaining 22 sites it decreased reactivity with FM. These latter residues were predominantly located in surface loops, but 5 positions in the globular domain also showed decreased reactivity. None of these changes in Cys accessibility were TonB-dependent (see below), and likely resulted from steric hindrance during the interaction of the toxin with FepA.
Because we did not observe ColB-induced increases in FM labeling during 30 min incubations, as reported by (Devanathan and Postle, 2007), and because we saw membrane depolarization within 5 min of exposure to ColB, we also performed experiments in which FM labeling initiated 1 min after exposure to ColB at 37 °C, and continued for a 15 min duration. This protocol produced the same results: ColB blocked fluoresceination of the same residues, and did not increase the reactivity of any of the sites we surveyed (Fig. 2, Fig S3).
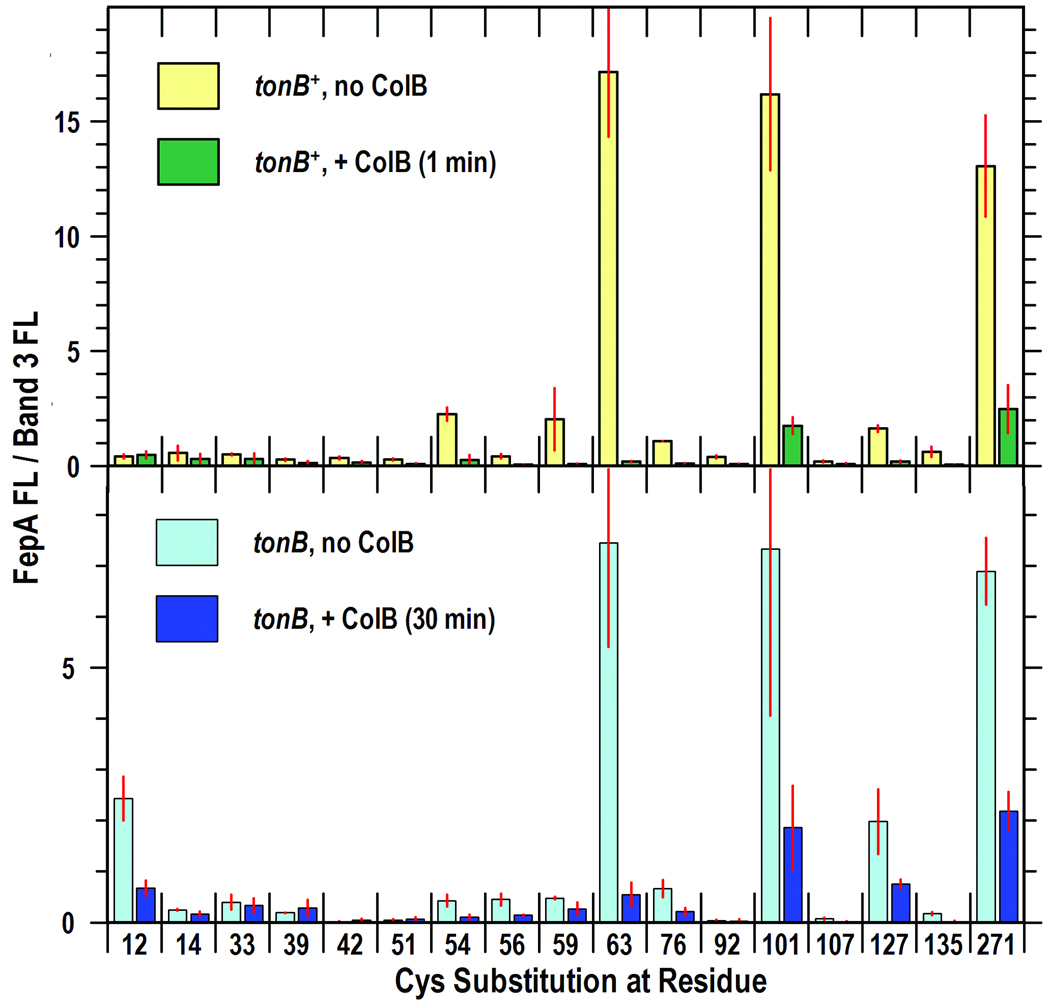
Figure 2. Effects of ColB exposure time and TonB on the fluoresceination of N-domain Cys residues in FepA.
SDS-PAGE gels of cell lysates (Fig. S3) were scanned on the fluorescence imager and analyzed by IMAGEQUANT (Molecular Dynamics). Bars show the mean FM-labeling of FepA (relative to band 3) from 2 or 3 independent experiments, with associated standard error. (Top) 1 min exposure of tonB+ cells to ColB. OKN3 harboring plasmids carrying fepA+ (lane 1) or its Cys substitution derivatives were subjected to FM-labeling as in Fig. 1, except that cells were incubated for only 1 min at 37 °C in the absence (yellow) or presence (green) of ColB, prior to labeling with FM (5 uM ; 15 min; 37 °C). (Bottom) 30 min exposure of tonB cells to ColB. OKN13 (tonB, fepA) carrying fepA+ or its Cys substitution derivatives was incubated for 30 min at 37 °C in the absence (cyan) or presence (blue) of ColB, prior to exposure to FM at 37 °C for 15 min.
Effects of TonB on FM-labeling
ColB-mediated killing of E. coli requires TonB, presumably for passage of the toxin through the OM, so we expected to see TonB-dependent structural changes associated with colicin uptake {e.g., extrusion of the N-domain from the FepA channel (Devanathan and Postle, 2007) or other mechanistic dynamics}. We measured the reactivity of Cys residues in FepA in a tonB host strain in the absence and presence of ColB, and compared them to the reactivity of the same residues in a tonB+ host strain (Fig. 1, Fig 2, Fig S2, Fig S3).
In the absence of ColB the main effect of the tonB locus was decreased fluoresceination of Cys residues in FepA (Fig. S3). However, further analysis (see below) showed that the diminution resulted from lower FepA expression in the tonB host. Interaction with ColB at 37 °C identically decreased the accessibility of Cys residues in the N-domain whether the target bacteria were killed by the toxin (tonB+) or immune to it (ΔtonB). In both strains we only saw reductions in fluoresceination associated with ColB binding, of the same magnitude and at the same sites. The absence of ColB-driven increases in the reactivities of 16 Cys substitutions in the globular domain, and the TonB-independence of the ColB-associated decreases in fluoresceination suggested that TonB-dependent structural changes do not occur during FepA-mediated ColB killing.
(James et al., 2008) reported that in the absence of ligands, binding of TonB to FhuA in vitro caused conformational changes in the surface loops of the receptor, and our fluoresceination procedures were appropriate to observe similar TonB-driven structural changes in FepA in vivo. We compared the reactivity of the panel of Cys residues in tonB+/− strains at 37 °C, but we did not find differences in their reactivity in either the N-domain or the C-domain loops of FepA (Fig. 3). Again, FepA was expressed at lower levels in the tonB host: [125I]-protein A anti-FepA immunoblots revealed a 58% reduction in mean FepA expression level in OKN13 (tonB), relative to its expression level in OKN3 (tonB+). Consequently, the engineered Cys sulfhydryls were less strongly labeled in the former strain. When we accounted for these protein concentration differences by determining the quantity of FepA (ug) in each sample (by [125I]-protein A anti-FepA immunoblots), the fluorescence intensities of the FepA bands per ug of FepA protein was the same in tonB+ or tonB host strains (Fig. 3). Hence, the relative fluorescence labeling levels of sites in both the globular domain and in the surface loops were the same whether TonB was present or absent.
DISCUSSION
The differential labeling of engineered Cys sulfhydryl groups by FM reflects their chemical accessibility. The changing chemical modification patterns document structural states in FepA: at different temperatures, during its interaction with ColB, and in the presence or absence of TonB. Enhanced fluoresceination at 37 °C reflects greater reactivity of Cys sulfhydryl groups in certain locations, and this greater chemical accessibility inferred conformational changes in surface-exposed regions of the receptor as the conditions warmed to physiological levels. Loop motion was previously seen by site-directed spectroscopy (Liu et al., 1994; Payne et al., 1997; (Jiang et al., 1997; Cao et al., 2003), by the disordered nature of L4, L5 and L7 in FepA crystal structure (Buchanan et al., 1999), and by transitions in FepA (Payne et al., 1997; Scott et al., 2002), FhuA (Locher et al., 1998; Ferguson et al., 1998) and FecA (Ferguson et al., 2002) from open forms that adsorb ligands to closed forms that subsequently transport them. Fluoresceination was highly temperature-dependent at certain sites (S63C, T550C), which had Q10 values of approximately 2 (a doubling of reaction rate for each 10 degree increment in temperature). These data imply loop conformational motion as the temperature rises (Gutfreund, 1995). Fluoresceination of BSA only slightly increased at 37 °C, and Cys sulfhydryls at other sites in FepA (residues 14, 42, 54, 56, 59, 76, 107, 254, 666) were much less, or unaffected by the change to 37 °C. Hence, the increased chemical modification of the loops at 37 °C reflects conformational dynamics that allow better permeation of the reagent. The simplest explanation is that higher temperatures open or relax the loops of FepA, increasing their permeability to small molecules. The hydrophobicity and negative charge of FM roughly mimic FeEnt: it’s an aromatic, anionic probe with slightly smaller dimensions (427 Da vs 716 Da), so ingress of the fluorophore into FepA structure may reflect the natural movement of FeEnt during transport. These experiments do not define the range of motion, because virtually any alteration of protein architecture may enhance the accessibility of a small reagent to the target sites. But, they indicate, for the first time, that as the cell warms L2, L3, L4, L5, L7, L8 and L10 undergo concerted conformational change, potentially improving receptor activity by increasing ligand interactions with relevant amino acid side chains (Cao et al., 2000; Annamalai et al., 2004). FepA has 100-fold higher affinity for FeEnt in vivo {Kd = 0.1 nM; (Newton et al., 1999; Cao et al., 2000; Annamalai et al., 2004)} than in vitro {Kd = 10 nM; (Payne et al., 1997)}, and the receptor’s conversion between open and closed forms may contribute to these differences.
The mechanism of OM penetration by colicins (assuming that they enter by a common mechanism) is unknown: do they pass the OM by transit through porin channels, or by a more obscure transport process? In their analysis of the interaction between ColB and FepA, (Devanathan and Postle, 2007) used different reagents and methodologies to conclude that the bacteriocin traverses the receptor’s transmembrane channel. Our results do not support this interpretation: we found no evidence of ColB passage through the FepA pore. Whereas (Devanathan and Postle, 2007) considered the accessibility of 12 engineered Cys residues in the N-terminal globular domain of FepA; we studied 35 sites dispersed throughout its tertiary structure: 16 in the N-domain and 19 in the C-terminal β-barrel. Four substitutions were common to both studies (at A33, A42, T51 and S92). (Devanathan and Postle, 2007) reported increased modification of the N-terminal-most 6 sites in the globular domain (T13, S29, A33 A42, S46, T51) when cells were exposed to excess ColB (4-fold) at 37 °C for 30 min (Devanathan and Postle, 2007). Conversely, we saw no increases in the modification of any FepA residues, including the 4 common sites. The discrepancies in the results may originate from the different methodologies. (Devanathan and Postle, 2007) used a catenation of reagents and procedures: after exposure to 1-biotinamido-4-[4'-(maleimidomethyl) cyclohexanecarboxamido] butane (BMCC), they lysed the cells, solubilized membrane proteins with detergent, immunoprecipitated FepA with an anti-FepA antibody, subjected the immunoprecipitates to SDS-PAGE, transferred the resolved proteins to nitrocellulose, stained them with avidin-horseradish peroxidase, and “…The extent of labeling in vivo for each FepA Cys substitution was estimated visually.” Furthermore, measurement of chemical reactivity in the periplasm depends on passage of the reagent through the OM. (Devanathan and Postle, 2007) used BMCC: its mass of 534 Da is near the exclusion limit of E. coli general porins (Nikaido and Rosenberg, 1981), and its extended molecular dimension (Stoke’s diameter of 34 Å) is 3-fold greater than the diameter of the OmpF channel (Cowan et al., 1992). These considerations make its OM transport problematical and much slower than that of FM, which is 20% smaller in mass (427 Da) and compact (Stoke’s diameter of 10 Å). FM traverses the OM via porin channels and efficiently labels sulfhydryls in the periplasm (Ma et al., 2007). On the other hand, visualization of BMCC labeling in the periplasm required high concentrations of the reagent (93 uM, about 20-fold higher than that of FM in our experiments), combined with amplified immunochemical and enzymatic procedures. We cannot fully explain the conflicting results of the two studies, but the methodological differences are likely responsible. Our experiments exposed bacteria to FM, lysed them in SDS sample buffer, subjected the lysates to SDS-PAGE, and measured fluorescence intensity on a phosphorimager. These simpler procedures and fewer manipulations created less opportunity for adventitious error, and produced better fluorescent images, whose analysis yielded statistical measures of reproducibility. We expected the methodologies to accurately detect differences in reactivity at specific sites, and the range of fluorescence labeling that we found in response to temperature and ColB binding confirmed this expectation.
Exposure to ColB at 37 °C slightly decreased the labeling of side chains in the interior or on the periplasmic surface of FepA (residues 12, 14, 54, 56, 59, 76 and 127), suggesting that during the ColB killing phase the structure of FepA remains closed and less accessible to the extrinsic fluorophore. Shorter incubation times with ColB and equivalent experiments in a tonB strain produced identical results: the same residues were labeled to the same relative extent at 37 °C in the absence and presence of ColB. Thus, our results show no indication of ColB passage through the FepA channel. We also note, however, that once excess ColB saturates FepA on the cell surface, we know neither how many colicin molecules successfully traverse the OM and depolarize the IM, nor how long thereafter ColB transport continues, before energy depletion terminates the process. The single hit-killing mechanism of bacteriocins (Luria, 1964) raises the possibility that only one or a few ColB molecules/cell breach the OM barrier, and in this case we do not anticipate gross changes in the FM-labeling pattern of FepA, which is present at a level of approximately 40,000 chromosomally encoded receptors/cell (Newton, 1999; Annamalai, 2004). Finally, we did not observe any TonB-dependent fluctuations in FM-labeling levels at the sites we analyzed in FepA. These results differ from the behavior of FepA during FeEnt transport, where TonB-dependent labeling of residue G54C from the periplasmic space implied extrusion of the N-domain during the metal transport reaction (Ma et al., 2007).
EXPERIMENTAL PROCEDURES
Bacteria and plasmids
Bacterial strains (Ma et al., 2007) OKN1 (ΔtonB), OKN3 (ΔfepA) and OKN13 (ΔtonB, ΔfepA) harbored the low copy plasmids pITS23 (Scott et al., 2002) or pITS47, which carry fepA+ or mutant fepA alleles under control of their natural Fur promoter. Existing Cys substitution mutations were carried on pITS23 (Ma et al., 2007); new constructions were on pITS47, which is identical to pITS23 except that it was generated with different restriction sites flanking the fepA+ gene. For pITS47, we amplified fepA+ and its upstream promoter region from E. coli chromosomal DNA, flanked by PstI and HindIII sites, and cloned it into PstI and HindIII -cut pHSG575 DNA (Hashimoto-Gotoh et al., 1981; Takeshita et al., 1987).
FeEnt, ColB and FepA
We formed and purified ferric enterobactin (Annamalai et al., 2004) and determined its concentration from the extinction coefficient (5.6 mM−1) at 495 nm. We purified colicin B from cultures of E. coli strain DM1187/pCLB1 (Payne et al., 1997), and determined the stoichiometry of the FepA-ColB interaction as follows: FepA is present in BN1071 at 37.3 pMol/109 cells (Kaserer et al., 2008); for labeling reactions with ColB we diluted 20 uL of purified toxin (0.8 mg/ml) into 1 mL containing 5–108 cells, resulting in final concentrations of 0.28 uM ColB and 0.019 uM FepA (i.e., 15-fold excess ColB). The Kd of the ColB-FepA binding interaction is 0.185 uM (Payne et al., 1997), so at binding equilibrium FepA was 49% saturated with ColB. Microbiological plate counts after labeling reactions in the presence of ColB showed that this protocol resulted in 99.85% mortality of the bacterial cells.
Site-directed Cys substitution mutagenesis
We used QuikChange mutagenesis (Stratagene, San Diego, Calif.) to create mutations in fepA on pITS47 that encoded Cys substitutions at A12, N39, A42, T51, N56, A59, S63, G76, W101, G127, N135. After verification by DNA sequence analysis (McLab, San Francisco, CA) we evaluated the colicin B and D sensitivity, ability to transport FeEnt, and expression levels of the mutant FepA proteins (Newton et al., 1999): except for W101C (see Results) their phenotypes were indistinguishable from wild-type FepA.
Kinetics of ColB killing
ColB kills bacterial cells by the insertion of its C-terminal pore-forming domain into the IM. We utilized the fluorescence emissions of 3,3'-diethyloxacarbocyanine {DiOC2(3); (Sims et al., 1974)}, which reflects cellular membrane potential, to observe ColB-induced depolarization. BN1071 (fepA+, tonB+) and OKN13 (ΔfepA, ΔtonB) were grown overnight in LB broth, subcultured (1%) into MOPS minimal media with glucose and nutritional supplements, and grown to late-log phase (O.D.600 nm ≈ 0.9). The bacteria were pelleted by centrifugation, resuspended in PBS plus 0.4% glucose at 5 × 108/mL, and exposed to purified ColB for varying times (5 – 30 minutes). DiOC2(3) was added to 3 uM, incubated 30 min at room temperature, and the cells were immediately analyzed on a Beckman-Coulter Epics Elite flow cytometer at 575 nm. Emission data were collected with log amplification and mean red fluorescence intensity was plotted versus time. The results were compared to those from bacteria which were not exposed to the bacteriocin, and to the same strains exposed to 5 uM carbonyl cyanide 3-chlorophenylhydrazone (CCCP).
Fluorescence labeling
We fluoresceinated Cys sulfhydryl groups in FepA in live cells (Ma et al., 2007). After growth in MOPS media to mid-log phase (4–5 × 108 cells/ml; 5–6 h), we collected the bacteria by centrifugation, and washed and resuspended them in 50 mM NaHPO4, pH 6.5 (see Results). FM was added to the cells at 5 uM for 15 min, at 0°C or 37 °C, the reactions were quenched with 100 uM cysteine, and the cells were washed with and resuspended in ice-cold TBS. After determining cell number by absorption at 600 nm, we lysed 108 bacteria by boiling in SDS-PAGE sample buffer, and subjected the lysate to SDS-PAGE.
SDS-PAGE, fluorescence imagery and immunoblots
After resolution of cell proteins by SDS-PAGE (Ames, 1974; Newton et al., 1999), we rinsed the gels with water and scanned for fluorescence on a STORMSCAN phosphorimager (Molecular Dynamics). Proteins in the gels were transferred to nitrocellulose paper, which was developed with mouse anti-FepA mAb 45 {0.5%; (Murphy et al., 1990)} and 125I-protein A (Newton et al., 1999). After exposure on an imaging screen, radioactivity was quantitated on the STORMSCAN phosphorimager.
Image analysis
Scanned images of SDS-PAGE fluorescence and [125 I] emissions were analyzed by ImageQuant 5.2 (Molecular Dynamics). For relative measurements of FepA fluorescence we determined the intensity of the FepA band in each lane of the gel, relative to the fluorescence of a cellular protein that was labeled at constant levels {Band 3; (Ma et al., 2007)}.
For absolute measurements of FepA fluorescence we ran aliquots of purified FepA in the gels, and used them as standards in the [125I]-protein A immunoblots to determine the precise quantity (ug) of FepA in each lane. From these data we calculated the fluorescence intensity of the FepA bands per ug FepA protein.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NSF grant MCB-0414694 and NIH grant GM53836 to PEK and SMCN. Thanks to Daniel Scheurch for technical assistance. This work is dedicated to the memory of J. B. Neilands, for his initial characterizations of this subject, and many helpful discussions.
REFERENCES
- Annamalai R, Jin B, Cao Z, Newton SM, Klebba PE. J Bacteriol. 2004;186:3578–3589. doi: 10.1128/JB.186.11.3578-3589.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti H, Lloubes R, Lazdunski C, Letellier L. Embo J. 1992;11:441–447. doi: 10.1002/j.1460-2075.1992.tb05073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret E, Benedetti H, Rigal A, Loret E, Lazdunski C. J Bacteriol. 1999;181:6306–6311. doi: 10.1128/jb.181.20.6306-6311.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbeer C. J Bacteriol. 1993;175:3146–3150. doi: 10.1128/jb.175.10.3146-3150.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan SK, Lukacik P, Grizot S, Ghirlando R, Ali MM, Barnard TJ, Jakes KS, Kienker PK, Esser L. Embo J. 2007;26:2594–2604. doi: 10.1038/sj.emboj.7601693. Epub 2007 Apr 2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan SK, Smith BS, Venkatramani L, Xia D, Esser L, Palnitkar M, Chakraborty R, van der Helm D, Deisenhofer J. Nat Struct Biol. 1999;6:56–63. doi: 10.1038/4931. [DOI] [PubMed] [Google Scholar]
- Cao Z, Klebba PE. Biochimie. 2002;84:399–412. doi: 10.1016/s0300-9084(02)01455-4. [DOI] [PubMed] [Google Scholar]
- Cao Z, Qi Z, Sprencel C, Newton SM, Klebba PE. Mol Microbiol. 2000;37:1306–1317. doi: 10.1046/j.1365-2958.2000.02093.x. [DOI] [PubMed] [Google Scholar]
- Cao Z, Warfel P, Newton SM, Klebba PE. J Biol Chem. 2003;278:1022–1028. doi: 10.1074/jbc.M210360200. [DOI] [PubMed] [Google Scholar]
- Cascales E, Lloubes R. Mol Microbiol. 2004;51:873–885. doi: 10.1046/j.1365-2958.2003.03881.x. [DOI] [PubMed] [Google Scholar]
- Cowan SW, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit RA, Jansonius JN, Rosenbusch JP. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- Davies JK, Reeves P. J Bacteriol. 1975;123:96–101. doi: 10.1128/jb.123.1.96-101.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devanathan S, Postle K. Mol Microbiol. 2007;65:441–453. doi: 10.1111/j.1365-2958.2007.05808.x. Epub 2007 Jun 2018. [DOI] [PubMed] [Google Scholar]
- Ferguson AD, Chakraborty R, Smith BS, Esser L, van der Helm D, Deisenhofer J. Science. 2002;295:1715–1719. doi: 10.1126/science.1067313. [DOI] [PubMed] [Google Scholar]
- Ferguson AD, Hofmann E, Coulton JW, Diederichs K, Welte W. Science. 1998;282:2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- Goemaere EL, Cascales E, Lloubes R. J Mol Biol. 2007;366:1424–1436. doi: 10.1016/j.jmb.2006.12.020. Epub 2006 Dec 1415. [DOI] [PubMed] [Google Scholar]
- Gumbart JC, Wiener MC, Tajkhorshid E. Biophys J. 2007;20:20. doi: 10.1529/biophysj.107.104158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guterman SK, Dann L. J Bacteriol. 1973;114:1225–1230. doi: 10.1128/jb.114.3.1225-1230.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutfreund H. Kinetics for the Life Sciences: Receptors, Transmitters and Catalysts. Cambridge, UK: Cambridge University Press; 1995. [Google Scholar]
- Hashimoto-Gotoh T, Franklin FC, Nordheim A, Timmis KN. Gene. 1981;16:227–235. doi: 10.1016/0378-1119(81)90079-2. [DOI] [PubMed] [Google Scholar]
- James KJ, Hancock MA, Moreau V, Molina F, Coulton JW. Protein Sci. 2008;17:1679–1688. doi: 10.1110/ps.036244.108. Epub 2008 Jul 1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Payne MA, Cao Z, Foster SB, Feix JB, Newton SM, Klebba PE. Science. 1997;276:1261–1264. doi: 10.1126/science.276.5316.1261. [DOI] [PubMed] [Google Scholar]
- Kadner RJ, McElhaney G. J Bacteriol. 1978;134:1020–1029. doi: 10.1128/jb.134.3.1020-1029.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaserer WA, Jiang X, Xiao Q, Scott DC, Bauler M, Copeland D, Newton SM, Klebba PE. J Bacteriol. 2008;190:4001–4016. doi: 10.1128/JB.00135-08. Epub 2008 Apr 4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locher KP, Rees B, Koebnik R, Mitschler A, Moulinier L, Rosenbusch JP, Moras D. Cell. 1998;95:771–778. doi: 10.1016/s0092-8674(00)81700-6. [DOI] [PubMed] [Google Scholar]
- Luria SE. Ann Inst Pasteur (Paris) 1964;107 SUPPL:67–73. [PubMed] [Google Scholar]
- Ma L, Kaserer W, Annamalai R, Scott DC, Jin B, Jiang X, Xiao Q, Maymani H, Massis LM, Ferreira LC, Newton SM, Klebba PE. J Biol Chem. 2007;282:397–406. doi: 10.1074/jbc.M605333200. Epub 2006 Oct 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh MA, Earhart CF. Biochem Biophys Res Commun. 1976;70:315–322. doi: 10.1016/0006-291x(76)91144-x. [DOI] [PubMed] [Google Scholar]
- Murphy CK, Kalve VI, Klebba PE. J Bacteriol. 1990;172:2736–2746. doi: 10.1128/jb.172.5.2736-2746.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton SM, Igo JD, Scott DC, Klebba PE. Mol Microbiol. 1999;32:1153–1165. doi: 10.1046/j.1365-2958.1999.01424.x. [DOI] [PubMed] [Google Scholar]
- Nikaido H, Rosenberg EY. J Gen Physiol. 1981;77:121–135. doi: 10.1085/jgp.77.2.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H, Vaara M. Microbiol Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novo D, Perlmutter NG, Hunt RH, Shapiro HM. Cytometry. 1999;35:55–63. doi: 10.1002/(sici)1097-0320(19990101)35:1<55::aid-cyto8>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Pawelek PD, Croteau N, Ng-Thow-Hing C, Khursigara CM, Moiseeva N, Allaire M, Coulton JW. Science. 2006;312:1399–1402. doi: 10.1126/science.1128057. [DOI] [PubMed] [Google Scholar]
- Payne MA, Igo JD, Cao Z, Foster SB, Newton SM, Klebba PE. J Biol Chem. 1997;272:21950–21955. doi: 10.1074/jbc.272.35.21950. [DOI] [PubMed] [Google Scholar]
- Postle K. J Bioenerg Biomembr. 1993;25:591–601. doi: 10.1007/BF00770246. [DOI] [PubMed] [Google Scholar]
- Pugsley AP, Reeves P. J Bacteriol. 1976;126:1052–1062. doi: 10.1128/jb.126.3.1052-1062.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley AP, Reeves P. J Bacteriol. 1977;130:26–36. doi: 10.1128/jb.130.1.26-36.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabsch W, Ma L, Wiley G, Najar FZ, Kaserer W, Schuerch DW, Klebba JE, Roe BA, Laverde Gomez JA, Schallmey M, Newton SM, Klebba PE. J Bacteriol. 2007;25:25. doi: 10.1128/JB.00437-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds PR, Mottur GP, Bradbeer C. J Biol Chem. 1980;255:4313–4319. [PubMed] [Google Scholar]
- Saier MH., Jr J Membr Biol. 2000;175:165–180. doi: 10.1007/s00232001065. [DOI] [PubMed] [Google Scholar]
- Scott DC, Newton SM, Klebba PE. J Bacteriol. 2002;184:4906–4911. doi: 10.1128/JB.184.17.4906-4911.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultis DD, Purdy MD, Banchs CN, Wiener MC. Science. 2006;312:1396–1399. doi: 10.1126/science.1127694. [DOI] [PubMed] [Google Scholar]
- Sims PJ, Waggoner AS, Wang CH, Hoffman JF. Biochemistry. 1974;13:3315–3330. doi: 10.1021/bi00713a022. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Wang ZY, Yamakoshi M, Kobayashi M, Nozawa T. Anal Sci. 2003;19:1239–1242. doi: 10.2116/analsci.19.1239. [DOI] [PubMed] [Google Scholar]
- Takeshita S, Sato M, Toba M, Masahashi W, Hashimoto-Gotoh T. Gene. 1987;61:63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- Wang CC, Newton A. J Bacteriol. 1969;98:1142–1150. doi: 10.1128/jb.98.3.1142-1150.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CC, Newton A. J Biol Chem. 1971;246:2147–2151. [PubMed] [Google Scholar]
- Wayne R, Frick K, Neilands JB. J Bacteriol. 1976;126:7–12. doi: 10.1128/jb.126.1.7-12.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita E, Zhalnina MV, Zakharov SD, Sharma O, Cramer WA. Embo J. 2008;27:2171–2180. doi: 10.1038/emboj.2008.137. Epub 2008 Jul 2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharov SD, Sharma O, Zhalnina MV, Cramer WA. Biochemistry. 2008;6:6. doi: 10.1021/bi800865h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.