Abstract
Food-borne fluke infections/trematodiases are emerging as a major public health problem worldwide with over 40 million people affected and over 10% of world population at risk of infection. The major concentration of these infections is in Southeast Asian and Western Pacific Regions, where the epidemiological factors (including the prevalent socio-cultural food habits) are conducive for transmission of these infections. The preponderance of these infections is usually in food deficit poor communities that lack access to proper sanitary infrastructure. While targeting health for all, especially the poor rural tribal communities, it is imperative to take these infections into account. Bayesian analysis phylogeny of food-borne trematode parasites under study showed that they are closely related phylogenetic groups. To focus the control strategies at the target populations, the aim of the present study was to establish molecular methods for accurate discrimination between common food-borne trematodes parasites Paragonimus (lung fluke), Fasciolopsis (giant intestinal fluke) and Fasciola (liver fluke), the infections of which commonly prevail in NE India. In the first step, we amplified and sequenced the second internal transcribed spacer (ITS2) region of ribosomal DNA, utilizing nucleotide differences between the multiple sequence alignments of the parasites under study. Based upon the differences in nucleotide sequences of conserved regions, we designed species-specific primers that can unequivocally discriminate one species from another. ITS2 sequence motifs allowed an accurate in-silico distinction of the trematodes. The data indicate that ITS2 motifs (≤ 50 bp in size) can be considered promising tool for trematode species identification. Using molecular morphometrics that is based on ITS2 secondary structure homologies, phylogenetic relationships with various isolates of several trematode species have been discussed. The present results suggest that the ITS2 specific primers can be used for epidemiological investigations of the prevalence of trematodiasis.
Keywords: Paragonimus, Fasciolopsis, Fasciola, trematodes, ITS rDNA, species-specific primer
Background
Trematodiases are the zoonoses, caused by trematodes of flukes (Platyhelminthes: Trematoda: Digenea). The flukes are commonly oval or leaf shaped and furnished with two suckers (the anterior oral sucker surrounding the mouth and a posterior ventral sucker or acetabulum for adhering with their hosts). Digenetic trematodes have complicated life cycles that involve one or two intermediate hosts. The cercariae which are shed in water either enter the definitive host directly or encyst on objects or in the bodies of animals (second intermediate hosts), transforming into metacercariae, the infective stage. The definitive host becomes infected by swallowing the metacercariae which develop to sexual maturity in various organs according to the species concerned. The prerequisites for transmission of trematodiases in a community involves presence of water bodies with plenty of water plants to support the large snail populations, consumption of risky foods such as undercooked/pickled crabs, crayfish and fresh water fish or tubers and fruits of water plants and contamination of water bodies with human/animal excreta leading to perpetuation of the foci of human infection of these zoonoses. The preponderance of these infections in usually in food deficit poor communities residing in the interior and the tribal areas due to their food habits (evolved in consonance with the meagre available resources) and unhygienic sanitary habits (practised due to lack of education). Food borne trematodiases are thus a result of environmental, social and economic conditions prevailing in the region. While targeting the health of all, especially the poor tribal communities, it is imperative to take into account. To focus the control strategies at the target populations, knowledge of the distribution and the probable endemic areas of these infections are essential. The lung flukes of the genus Paragonimus have been known as one of the most important zoonotic parasites causing paragonimiasis, also known as endemic haemoptysis, in man. It is estimated that over 20 million people are infected worldwide due to several species of Paragonimus [1]. Over 40 species are known to infect the lung of different mammalian hosts throughout the world [2] and approximately 15 species are known to infect humans. The giant intestinal fluke, Fasciolopsis buski (Trematoda: Fasciolidae), is widely distributed in India and neighboring countries of the continent in South and Southeast Asia [3]. The fluke is the etiological agent of the disease commonly known as fasciolopsosis. The infection occurs by ingestion of raw aquatic vegetation or food plants that are contaminated with the infective encysted larvae, the metacercariae. In endemic zones pigs, dogs and rabbits act as reservoir of infection. In India, the parasite has been reported from different states including those in the Northeast. Variations in the morphology of the fluke have been observed when collected from different geographical regions [4].
The trematode flukes of the genus Fasciola (the sheep liver fluke) are parasites of herbivores and infect humans accidentally causing fascioliasis worldwide. The parasite is very cosmopolitan in distribution being found throughout all regions of the world, both temperate and tropical. F. hepatica is the causative agent of fascioliasis or ‘liver rot’ in ruminants, where it may be an important pathogen. Human infections with F. hepatica are found in areas where sheep and cattle are raised, and where humans consume raw watercress, including Europe, the Middle East and Asia [5]. Infection with F.gigantica on the other hand is found more commonly in tropical regions of the world, areas affected including Africa, Asia, many Pacific islands including Hawaii (where it has been reported in man), the Middle East and Southern Europe and the south of the USA [6]. Both fasciolids follow a similar two-host life cycle, in which freshwater snails of the family Lymnaeidae act as intermediate or vector host and a broad spectrum of mammals including humans, mainly herbivorous large-sized species, act as definitive host [7].
The identification of closely related species based on morphological characters can be difficult. This is particularly the case of soft-bodied animals such as digenean trematodes. However, recent advances in molecular biology, in particular the amplification of specific DNA regions via the polymerase chain reaction (PCR) and improved sequencing techniques, have been employed to resolve taxonomic issues related to various helminth parasites by comparing their DNA. The ribosomal DNA cluster (rDNA), which codes for structural components of ribosomes, is particularly useful for genetic studies because it is highly repeated and contains variable regions flanked by more conserved regions [8]. PCR-based techniques utilizing the rDNA ITS2 sequences, which occur between the 5.8S and 28S coding regions, have proven to be a reliable tool to identify the helminth species and their phylogenetic relationships [9]. The nuclear ribosomal DNA second internal transcribed spacer (ITS2) sequences, which occur between the 5.8S and 28S coding regions, have proven useful for diagnostic purposes at the level of species. Fasciola spp and isolates of Fascioloides magna from different geographical regions were discriminated on the basis of ITS sequences [10]. Studies on phylogeny and/or intraspecific variation in Paragonimus species have also been done using ITS2 region in recent years [11]. The development of objective, reproducible and sensitive diagnostic technique is required for accurate species discrimination and identification of individual flukes. This method could be used for epidemiological investigations of the prevalence of infection and controlling trematodiases. DNA techniques utilizing genetic markers in nuclear ribosomal DNA (rDNA) have been employed to resolve taxonomic issues related to various helminthic parasites. The PCR-based technique has the potential to be used for discrimination and identification of the flukes irrespective of their life cycle stages [12]. In the present study, we focused on the common food-borne trematodes parasites Paragonimus (lung fluke), Fasciolopsis (giant intestinal fluke) and Fasciola (liver fluke), and describe the establishment of species specific markers utilizing the ITS2 sequences, which allows species discrimination and identification of the individual fluke.
Methodology
Parasite material
Naturally infected freshwater edible crabs (Barytelphusa lugubris) were collected from a mountain stream of the suspected focal area Miao, Changlang District in Arunachal Pradesh (Altitude - 213 mASL, Longitude- 96°-15'N and Latitude- 27°-30'E). Paragonimus metacercariae were isolated from the muscles of the crustacean host by digestion technique. Live adult Fasciolopsis buski were obtained from the intestine of freshly slaughtered pig, Sus scrofa domestica at local abattoirs. The worms recovered from these hosts represented the geographical isolates from Assam region of Northeast India. Adult Fasciola were obtained in live form from hepatic biliary ducts of freshly slaughtered cow, Bos indicus. The worms recovered from these hosts represented the geographical isolates from Assam, Northeast India and morphologically resembled Fasciola gigantica (deposition number of paratypes at Zoological Survey of India, Kolkata = W7787/1). Eggs were obtained from mature adult flukes by squeezing between two glass slides.
DNA isolation
The 70% alcohol-fixed metacercariae were further processed for DNA extraction and PCR amplification. For the purpose of extraction, metacercariae recovered from one single host were pooled together, DNA was extracted from metacercariae in FTA card by using Whatman's FTA Purification Reagent and amplified by PCR. For the purpose of DNA extraction, adult flukes collected from different host animals were processed singly; eggs recovered from each of these specimens were also processed separately. The adult flukes were first immersed in digestion extraction buffer (containing 1% SDS, 25 mg Proteinase K) at 37° C for overnight. DNA was then extracted from lysed individual worms by standard ethanol precipitation technique [13] and also extracted on FTA cards using Whatman's FTA Purification Reagent as described elsewhere [14]. DNA from the eggs was extracted only with the FTA card technique.
DNA amplification and sequencing
The rDNA region spanning the ITS2 was amplified from metacercarial DNA by PCR. As primers, we used 3S: 5'-GGTACCGGTGGATCACTC GGCTCGTG-3' (forward) and A28: 5'-GGGATCCTGGTTAGTTTCTT TTCCTCCGC-3' (reverse), which were designed based on the conserved sequences of the 5.8S and 28S genes of Schistosoma species [15]. The PCR amplification was performed following the standard protocol [16] with minor modifications in 100 mM Tris HCL (pH 9.0), 500 mM KCl, 1.5 mM MgCl2, and 0.2 mM deoxynucleotide triphosphates each of dATP, dGTP, dCTP and dTTP, 0.25 mM of each primer and 2.5 units of Taq polymerase (Bangalore Genei Pvt. Ltd., India). DNA was preheated at 94° C for 5 min and added to each PCR reaction. The PCR cocktail (final reaction volume, 25µl) was amplified with the following conditions: 26 cycles of denaturation at 94° C for 30s, annealing at 55° C for 38s and extention at 72° C for 42s followed by a final extension at 72° C for 10 min. The resultant PCR products were separated by electrophoresis through 1.6% (w/v) agarose gels in TAE buffer, stained with ethidium bromide, transilluminated under ultraviolet light and then photographed. The known size fragments of Phi X 174 DNA/ Hae III Digest in agarose gel were used as marker. For DNA sequencing, the PCR products were purified using Genei Quick PCR purification Kit, and sequenced in both directions using PCR primer A28 and 3S on an automated sequencer.
Bayesian phylogenetic analysis
DNA sequences were aligned using ClustalX 2.0.7. The interleaved NEXUS file was edited manually in order for it to be recognized by Mr. Bayes V3.1.2 programme and phylogenetic analysis was carried out using the Bayesian approach with combined datasets and default parameters, wherein each data partition is allowed to have a different evolution rate. The cladogram with the posterior probabilities for each split and a phylogram with mean branch lengths were generated and subsequently read by the tree drawing program Tree view V1.6.6 [17], Motif identification, testing and validation. The ITS sequence motifs were identified from aligned sequences of the data set for the species using PRATT software ( http://genoweb1.irisa.fr/Serveur-GPO/outils_acces.php3?id_syndic=70). The minimum percentage of sequences to match (C%) parameter was adjusted to report pattern matching at 100% of the sequence input. The motifs were expressed using the DNA alphabet (A, T, C, G) in PROSITE language. The validation of the motifs was performed for each species using a “PATTERN MATCHING” Web application ( http://genoweb.univ-rennes1.fr/Serveur-GPO/outils_acces.php3?id_syndic=186).
Predicted ITS2 RNA secondary structures and analyses
Secondary structures of ITS2 sequences of various fasciolid species were reconstructed by aligning their sequences using Bioedit. The acquired structures with restrictions and constrains were submitted in MFOLD. RNA was folded at a fixed temperature of 37° C, and the structure chosen from different output files was the desired 6-helicoidal ring or the one with the highest negative free energy if various similar structures were obtained.
Design of species specific primers and amplification by PCR
To selectively PCR-amplify a DNA fragment, suitable primers need to be designed and synthesized. Primers are short nucleotides often not more than 50 and usually only 18 to 25 base pairs long- containing nucleotides that are complementary to the nucleotides at both ends of the DNA fragment to be amplified. These complementary bases in primer and DNA template facilitate annealing of the primer to the DNA template to which the DNA polymerase can bind and begin with the synthesis of a new DNA strand that is complementary to the DNA template. To establish a more direct PCR procedure for species discrimination and identification, specific reverse primers were designed to target unique regions of the ITS2 sequence of each species. Each specific primer was examined for species-specific amplification with the 3S primer under the PCR conditions described above. The primer set 3S-A28 was used to control for the presence of parasite genomic DNA in each sample.
Results
PCR amplification of ITS region and its analysis
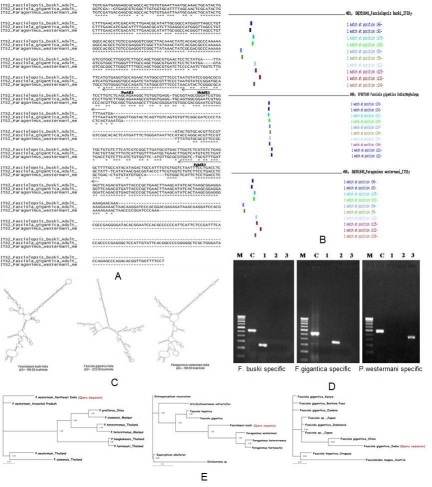
In the first step, we amplified the rDNA region spanning ITS2 from metacercarial DNA using primer set 3S-A28. Agarose gel electrophoresis showed that the generated ITS PCR products were about 500bp in size (including the primer annealing regions) for all the three trematodes. Multiple sequence alignment was done for all the three sequences using ClustalW programme (Figure 1A). Sequence analysis of the ITS2 PCR products of Fasciolopsis buski (DQ351841) Fasciola gigantica (EF027103) and Paragonimus metacercariae (DQ351845) revealed no intra-specific variations in length or composition of the sequences of fasciolid and paragonimid, and the ITS sequences of adult, metacercariae and egg origin were found to be identical in length as well as composition (Figure 1E).We identified hundreds of sequence motifs from the ITS2 areas of the selected sequences. These were then screened and validated against the training set using the pattern matching tool, which allowed a final selection to be made of 10 representative short sequence motifs of sizes inferior or equal to 50 nucleotides and were found to be present in all the sequences under study (Figure 1B).Three predicted RNA secondary structures were reconstructed from the unique sequences with highest negative free energy of Fasciolopsis buski (DQ351845), Fasciola gigantica (EF027103) and Paragonimus westermani (DQ351845) to provide the basic information for phylogenetic analysis (Figure 1C). The ITS2 plus flanking regions of nuclear region ranged from 606bp in F. gigantica India to a minimum length of 481bp in F. buski India. The secondary structural features of ITS2 regions as shown in the figures were analysed based on conserved stems and loops. F. buski and P. westermani isolates from India showed closer similarity in the rRNA folding as revealed by their energy level and had identical secondary structure compared to that of F. gigantica. Moreover, the observed phylogenetic trend was identified with respect to the target accessibility sites for all the three different isolates; the orders of preference being interior loop, bulge loop, multiple branch loop, hairpin loop and exterior loop in all the isolates.The P. westermani-specific, F. buski-specific and F.gigantica-specific primers (PwAR1, FbMR1 and FgMR1 respectively) were designed to target the 3'-terminal position of the ITS2 sequences, and the species specificity of these primers was evaluated by PCR using primer 3S (Figure 1D; see Table 1). As was expected, the primer set 3S-PwAR1 amplified a PCR product only from P. westermani DNA, 3S-FbMR1 amplified a PCR product only from F. buski DNA and 3S-FgMR1 amplified a PCR product only from F. gigantica. Primer set 3S-A28 was used to control for the presence of parasite genomic DNA in each sample. These PCR products were sequenced using the corresponding specific primer and were confirmed to be the ITS2 region of rDNA from the respective species.
Figure 1.
A. Multiple sequence alignments of ITS2 sequence and flanking regions of Fasciolopsis buski, Fasciola gigantica and Paragonimus westermani; B. Pattern matching of ITS motifs of different geographical isolates of fasciolid species: Fasciolopsis buski India (481bp), Paragonimus westermani India (496bp), Fasciola gigantica India (606bp); C. Predicted ITS2 RNA secondary structures and their structure formation enthalpies according to MFOLD; D. Results of PCR amplification of DNA (M= 100-bp DNA ladder marker, C= Control primers 3S-A28); E. Phylogenetic relationships between members of family Paragonimidae and Fasciolidae. The Tree shows hypothetical Bayesian analysis phylogeny based on the secondary structure alignment data of the ITS2 region. The numbers are equivalent to bootstrap percentages.
Discussion
Morphological differences found in adult specimens have been widely used to discriminate between platyhelminth species. However, traditional diagnostic techniques in parasitology are now complemented by a variety of molecular tools to help in resolving the taxonomic issues associated with describing new species or strains on the basis of phenotypic characteristics [18]. The taxonomy of parasitic helminthes has been based mainly on morphological data complemented with ecological, cytological and pathological results as well as clinical manifestations. However, it is impossible to identify the species using adults with damaged internal organs and/or tegument, where the morphological characteristics are insufficient for the unequivocal discrimination of the species. In that case, the PCR-based techniques utilizing the ITS2 sequences can be used as a tool to identify the species. In search for molecular markers for this species, we characterized the ITS region of rDNA.It is possible to distinguish between adult F. hepatica and F. gigantica on the basis of morphology, but much variation exists. Consequently, where both species occur concurrently or in overlapping geographical distribution, it is not possible to be certain as to which species is responsible for the disease. The low number of records of infection with F.gigantica may well be due to the lack of good tools to distinguish this species from F. hepatica [19].We attempted to develop short ITS sequence motifs (each ≤ 50bp) as DNA nucleotide barcode, for an unambiguous and easy identification of parasite species. Three patterns of secondary structures of RNA were constructed, each belonging to a different genus and provided us with additional characteristics that could be used for differentiating the species and their phylogenetic relationships. Secondary structure analysis of the data confirmed the results obtained from the primary sequence data analysis. However, there are difficulties in defining a distance between two related structures with variable topologies [20]. Thus, combined dataset using primary sequence analysis of ITS2 sequences, secondary structure and Bayesian analysis revealed close similarity with members of both Fasciolidae and Paragonimidae, proving that both these families are closely related phylogenetic groups.The PCR-based technique described here allows accurate discrimination between the common food-borne trematodes. To elucidate the applicability of this technique for epidemiological investigations, rigorous testing using a large number of isolates from each species is warranted in order to establish whether intraspecific variations exist in the ITS2 sequence. Thus, the technique described here could present a useful tool for epidemiological investigations of the presence and prevalence of different helminth parasites in the region.
Conclusion
The findings indicate that the different life cycle stages of trematodes do not alter the applicability of the method. The usefulness of the ITS2 region for species discrimination, irrespective of the life cycle stages of the parasite, has also been demonstrated in nematode species. In conclusion, as has already been demonstrated for other parasitic helminthes, ITS can serve as an effective genetic marker for molecular identification. To ascertain intra-specific strain variations, if any, and to determine the population structure, different geographical isolates from the region need to be studied with the use of additional molecular markers.
Supplementary material
Acknowledgments
This study was carried out under the ‘All India Co-ordinated Project on Capacity Building in Taxonomy: Research on Helminths’, sanctioned to VT by Ministry of Environment & Forests, GOI; DBT Project to VT & AC and DSA (UGC-SAP) programme of the University Grants Commission, GOI, in the Department of Zoology and UGC's UPE (Biosciences) programme in the School of Life Sciences at NEHU, Shillong. We thank the Co-ordinator, Bioinformatics Centre, NEHU for allowing access to its facility.
Footnotes
Citation:Prasad et al, Bioinformation 6(2): 64-68 (2011)
References
- 1.C Toscano, et al. Trop Disease Bulletin. 1995;92:1. [Google Scholar]
- 2.D Bunnag, T Harinasuta. Opisthorchiasis, clonorchiasis and paragonimiasis. 1985 [Google Scholar]
- 3.B Roy, V Tandon. International handbook of foodborne pathogens. 2003 [Google Scholar]
- 4.B Roy, V Tandon. Acta Parasitol. 1993;38:72. [Google Scholar]
- 5.MS Mas-Coma, et al. Bull World Health Organ. 1999;77:340. [PMC free article] [PubMed] [Google Scholar]
- 6.JA Hammond, et al. Trans R Soc Trop Med Hyg. 1974;68:253. doi: 10.1016/0035-9203(74)90123-0. [DOI] [PubMed] [Google Scholar]
- 7.S Mas-Coma, et al. Int J Parasitol. 2005;35:1255. doi: 10.1016/j.ijpara.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 8.DM Hillis, MT Dixon. Q Rev Biol. 1991;66:411. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- 9.MJ Nolan, TH Cribb. Adv Parasitol. 2005;60:101. doi: 10.1016/S0065-308X(05)60002-4. [DOI] [PubMed] [Google Scholar]
- 10.RD Adlard, et al. Int J Parasitol. 1993;23:423. doi: 10.1016/0020-7519(93)90022-q. [DOI] [PubMed] [Google Scholar]
- 11.D Blair, et al. Parasitology. 1997;115:411. doi: 10.1017/s0031182097001534. [DOI] [PubMed] [Google Scholar]
- 12.H Sugiyama, et al. Mol Cell Probes. 2002;16:231. doi: 10.1006/mcpr.2002.0417. [DOI] [PubMed] [Google Scholar]
- 13.J Sambrooke, et al. Molecular cloning: a laboratory manual. 1989 [Google Scholar]
- 14.PK Prasad, et al. Parasitol Res. 2007;101:1581. doi: 10.1007/s00436-007-0680-y. [DOI] [PubMed] [Google Scholar]
- 15.J Bowles, et al. Mol Phylogenet Evol. 1995;4:103. doi: 10.1006/mpev.1995.1011. [DOI] [PubMed] [Google Scholar]
- 16.BA White, et al. PCR Protocols current methods and applications. 1993 [Google Scholar]
- 17.JP Huelsenbeck, F Ronquist. Bioinformatics. 2001;17:754. [Google Scholar]
- 18.RC Thompson, et al. Vet Parasitol. 2004;126:15. [Google Scholar]
- 19.SK Semyenova, et al. Acta Parasitologica. 2005;50:3. [Google Scholar]
- 20.BA Shiparo, et al. Comput Appl Biosci. 1998;4:387. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.