Abstract
Given the absence of universal marker genes in the viral kingdom, researchers typically use BLAST (with stringent E-values) for taxonomic classification of viral metagenomic sequences. Since majority of metagenomic sequences originate from hitherto unknown viral groups, using stringent e-values results in most sequences remaining unclassified. Furthermore, using less stringent e-values results in a high number of incorrect taxonomic assignments. The SOrt-ITEMS algorithm provides an approach to address the above issues. Based on alignment parameters, SOrt-ITEMS follows an elaborate work-flow for assigning reads originating from hitherto unknown archaeal/bacterial genomes. In SOrt-ITEMS, alignment parameter thresholds were generated by observing patterns of sequence divergence within and across various taxonomic groups belonging to bacterial and archaeal kingdoms. However, many taxonomic groups within the viral kingdom lack a typical Linnean-like taxonomic hierarchy. In this paper, we present ProViDE (Program for Viral Diversity Estimation), an algorithm that uses a customized set of alignment parameter thresholds, specifically suited for viral metagenomic sequences. These thresholds capture the pattern of sequence divergence and the non-uniform taxonomic hierarchy observed within/across various taxonomic groups of the viral kingdom. Validation results indicate that the percentage of ‘correct’ assignments by ProViDE is around 1.7 to 3 times higher than that by the widely used similarity based method MEGAN. The misclassification rate of ProViDE is around 3 to 19% (as compared to 5 to 42% by MEGAN) indicating significantly better assignment accuracy. ProViDE software and a supplementary file (containing supplementary figures and tables referred to in this article) is available for download from http://metagenomics.atc.tcs.com/binning/ProViDE/
Background
A number of metagenomic studies have been initiated in the past 3-4 years to explore, characterize and compare the taxonomic diversity of viruses present in various environments [1, 2]. Besides cataloguing viral diversity, these studies have identified several hitherto unknown groups of viruses that play a critical role in transferring genes involved in a variety of metabolic functions [1, 3]. Given the absence of universal marker genes (such as 16S rRNA in bacteria / archaea) in the viral kingdom, researchers typically use similarity-based approaches like BLAST (with stringent E-values) for taxonomic classification of viral metagenomic sequences. However, since a majority of sequences in typical metagenomes originate from hitherto unknown viral groups, the use of such stringent thresholds will result in a large fraction of sequences remaining unclassified. Furthermore, using less stringent E-values (observed for BLAST hits with poor alignment quality) will result in a high number of incorrect taxonomic assignments. The recently published SOrt-ITEMS algorithm provides an approach to address the above issues [4]. Based on alignment parameters, an elaborate work-flow is followed by SOrt-ITEMS for assigning reads originating from genomes of hitherto unknown archaeal/bacterial organisms. Alignment parameter thresholds used by SOrt-ITEMS are generated by observing the pattern of sequence divergence within and across various taxonomic groups belonging to bacterial and archaeal kingdoms. However, majority of taxonomic groups within the viral kingdom are characterized by the absence of a typical Linnaean-like taxonomic hierarchy (phylum, class, order, family, genus and species). This motivated us to develop ProViDE (Program for Viral Diversity Estimation), a novel algorithm that uses a customized set of alignment parameter thresholds/ranges, specifically suited for the accurate taxonomic labelling of viral metagenomic sequences. These thresholds take into the account the pattern of sequence divergence and the non-uniform taxonomic hierarchy observed within/across various taxonomic groups of the viral kingdom.
Methodology
Determination of alignment parameter thresholds
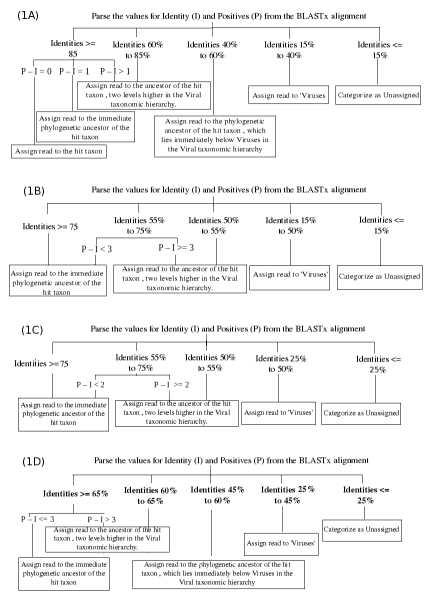
Using MetaSim [5], simulated data sets were generated from 50 diverse viral genomes (Supplementary Table 1). Subsequently the alignment parameter thresholds were determined (Supplementary Figures 1-4, Supplementary Tables 2-5) using a methodology similar to that adopted in SOrt-ITEMS [4]. Based on these, flow charts (Figure 1) were devised (for various query lengths) in order to identify an appropriate taxonomic level of assignment for a given query sequence.
Figure 1.
Flow-charts showing various steps followed to arrive at an appropriate taxonomic level, where the assignment of each read (A) Sanger (˜800 bp length) (B) 454 − Titanium (˜400 bp length) (C) 454- Standard (˜ 250 bp) and (D) 454-GS20 (˜ 100 bp). (I: Identity; P: Positives) is to be restricted. Hit taxon denotes the taxon/organism corresponding to the hit sequence
Steps followed for taxonomic classification of viral metagenomic sequences
Supplementary Figure 5 depicts the various steps followed by ProViDE algorithm. The output of a BLASTx search against the nr database is taken as input for ProViDE. For each hit, ProViDE first parses the values of various alignment parameters. For each read, based on its length, an appropriate taxonomic level of assignment (TL) is subsequently identified (Figure 1). The taxonomic assignment of the read is done using the orthology approach as used in SOrt-ITEMS [4]. The final taxonomic assignment of the read is thus restricted to taxonomic level that lies at or above the TL.
Data-sets and Database variants used for evaluating binning accuracy and specificity
1,40,000 sequences were generated from 35 viral genomes (Supplementary Table 6). These genomes were different from the ones taken for obtaining alignment parameter thresholds. Based on their length, these sequences were divided into four test data-sets, namely Sanger, 454-400, 454-250 and 454-100. To evaluate the performance of ProViDE with respect to sequences originating from unknown viral genomes, sequences in each data-set were queried (using BLASTx) against 2 variants of the nr database, namely, (a) nr database excluding sequences belonging to the query genome ('MINUS SPECIES') and (b) nr database excluding all sequences which fall under the immediate higher level taxonomic group to which the query species belongs (‘MINUS ONE LEVEL UP’). The BLASTx outputs obtained were given as input to ProViDE. The results of ProViDE were also compared with corresponding results generated with a similarity based binning method, MEGAN [6]. Both the programs were run using a min-support value of 2 and a bit score threshold value of 35.
Categorization of taxonomic assignments
The assignments of a read to a taxon that lies in the path between the root and the taxon corresponding to the source organism of the read was categorized as ‘correct’. To quantify the specificity, these ‘Correct assignments’ were subgrouped into two categories. All correct assignments at the level of root or cellular organisms or super-kingdom (Viruses) were considered as ‘non - specific’. Assignments below the level of super-kingdom were considered as ‘specific assignments’. The assignment of a read to a taxon that does not lie in the path between the root and the taxon corresponding to the source organism of the read was categorized as ‘Wrong’. Reads having hits having a bit-score less than 35 and/or an alignment length of less than 25 were categorized as ‘Unassigned’. All reads with no BLAST hits were categorized as ‘No hits’.
Discussion
Table 1 shows evaluation results with respect to the total number of correct assignments, wrong assignments, and the number of sequences categorized as unassigned. As expected, the percentage of total correct assignments is seen to increase with increasing read length. However, it is observed that (for all four test data-sets), the percentage of ‘correct’ assignments by ProViDE is around 1.7 to 3 times higher than that by MEGAN. Since for both methods, most (if not all) correct assignments are at specific levels, the relative specificity obtained with ProViDE is around 1.7 to 3 times higher than that with MEGAN. Furthermore, the percentage of sequences misclassified by ProViDE is in the range of 3-19% (as compared to 5 - 42% by MEGAN) indicating significantly better assignment accuracy. A similar number of sequences are categorized as ‘unassigned’ by both programs indicating that the relatively high levels of accuracy obtained using ProViDE are not at the cost of decreased number of assignments. One of the important aspects of metagenomic sequence analysis is to assign metagenomic sequences to correct taxonomic groups. Given that metagenomic sequence data sets typically contain millions of sequences, majority of which originate from new/hitherto unknown organisms, accurate and specific taxonomic assignment of metagenomic sequences still remains a major computational challenge.
In the current study, we have presented an algorithm (ProViDE) that is specifically customized for taxonomic analysis of viral metagenome data sets. Majority of reads in viral metagenomic data-sets originate from hitherto unknown viral groups, the sequences of which are absent in existing reference databases. Consequently, a majority of these sequences generate poor quality alignments with sequences in reference databases. Assignment of these sequences directly to the taxon corresponding to the best hit (irrespective of alignment quality) is expected to generate a large number of incorrect assignments. Besides, validation results generated in the present study also indicate that the popular binning algorithm, namely MEGAN, which is based on the principle of least common ancestor approach, also has an extremely high misclassification rate (which is as high as 40% for some of the data sets). This high misclassification rate of MEGAN is expected since it uses a single alignment parameter (bit-score) for judging alignment quality (prior to assignment). Consequently, MEGAN ends up misclassifying a majority of reads, especially those having poor quality alignments (with identities as low as 20%). Furthermore, as demonstrated by earlier studies [4], the least common ancestor (LCA) approach used by MEGAN is generally associated with poor binning specificity (especially in metagenomic scenarios wherein majority of reads originate from unknown organisms).
In contrast, multiple alignment parameters like bit-score, identities, positives (thresholds of which were specially identified for viral metagenomic sequences) are used by ProViDE for ascertaining the quality of the alignment. This ensures that assignment of reads at specific levels is done only for those reads that generate high quality alignments with database sequences. As the alignment quality decreases, ProViDE assigns these reads at progressively higher taxonomic levels. Validation results have indicated that employing this approach helps in significantly reducing the number of incorrectly assigned sequences. Validation results also indicate that ProViDE correctly assigns a greater number of sequences at specific levels (as compared to MEGAN). This indicates the overall utility of the ProViDE algorithm for accurate and specific taxonomic assignment of viral metagenomic sequences. A comparative evaluation of binning time indicates that the ProViDE algorithm takes approximately an hour to process the blastx output obtained for a data-set having 100,000 reads. This is marginally higher than the time taken by MEGAN for analysing the same number of reads. Supplementary Figure 6 gives a time comparison analysis plot of this analysis.
Conclusion
Performance evaluation with data-sets or database variants simulating typical metagenomic scenarios indicates that ProViDE has significantly high specificity and accuracy. To the best of our knowledge, ProViDE is the first ever similarity-based binning algorithm that provides an accurate and specific taxonomic label to most of the reads constituting viral metagenomic data sets.
Supplementary material
Footnotes
Citation:Ghosh et al, Bioinformation 6(2): 91-94 (2011)
References
- 1.SJ Williamson, et al. PLoS ONE. 2008;3(1):e1456. doi: 10.1371/journal.pone.0001456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D Lindell, et al. Nature. 2005;438:86. [Google Scholar]
- 3.D Willner, et al. PLoS ONE. 2009;4(10):e7370. doi: 10.1371/journal.pone.0007370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.M Monzoorul Haque, et al. Bioinformatics. 2009;25:1722. doi: 10.1093/bioinformatics/btp317. [DOI] [PubMed] [Google Scholar]
- 5.DC Richter, et al. PLoS ONE. 2008;3(10):e3373. doi: 10.1371/journal.pone.0003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DH Huson, et al. Genome Res. 2007;17:377. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.