Abstract
Informatics for Integrating Biology and the Bedside (i2b2) is a database system to facilitate sharing and reuse of clinical patients' data collected in individual hospitals. The i2b2 provides an ontology based object-oriented database system with highly simple and flexible database schema which enables us to integrate clinical patients' data from different laboratories and different hospitals. 392 patients' data including carcinoma and non-carcinoma specimens from cancer patients are transported from the Integrated Clinical Omics Database (iCOD) to the i2b2 database for a feasibility study to check applicability of i2b2 ontology and database schema on Japanese clinical patients’ data. No modification is required for the i2b2 data model to deal with Japanese characters. Some modification of ontology is required to integrate biomedical information extracted from the cancer patients’ data. We believe that the i2b2 system will be practical infrastructure to integrate Japanese clinical databases if appropriate disease ontology for Japanese patients is provided.
Background
Patients' records are rich in biomedical information to be analyzed, investigated and interpreted for application to clinical use in translational informatics. Translational informatics has a recently emerging research domain targeting computationally analysis of genomic information on individual patients in association with the clinical patients' data extracted from electronic medical records (EMRs) in hospitals. Translational informatics shall discover a clue to prediction and prevention of diseases by the use of personal genomic information, to provide better medication optimized person by person and healthy life for an individual patient.To facilitate translational informatics, there is a need of collection of clinical patients' data formalized ready for computational analysis. However extraction of the biomedical information from electronic medical records often requires expert knowledge and manual handling. Most of the biomedical information is buried in texts of doctors' comments (‘clinical findings’ in technical terms in medicine) in medical records. Much of the information leaves behind extracted and integrated into a database to be available for further scientific analysis.The computational infrastructure called 'Informatics for Integrating Biology and the Bedside' (i2b2) [1, 2] has been developed in US to facilitate the use of the clinical patients' data in the translational informatics. It is an open-source database system on the basis of object-oriented database schema and ontology-based knowledge definition. The system also provides statistical data analysis facilities and Natural Language Processing (NLP) tools for term extraction from free texts in EMRs. The i2b2 project has been funded by NIH in five years from the fall of 2004 by 20 million dollars in total [3]. Masses of clinical patients' data have already been accumulated by hospitals participating in this project which medicate and study multifactor diseases such as asthma, hypertension, type 2 diabetes, Huntington's disease, depressive disorder, rheumatoid arthritis, and obesity.
In this study, we constructed the i2b2 database with 392 clinical patients' data collected in a university hospital of Tokyo Medical and Dental University (TMDU). The patients' data includes biomedical information extracted from carcinoma and non-carcinoma specimens of cancer patients recorded in ‘Integrated Clinical Omics Database’ (iCOD) [4]. The clinical patients' data includes 8,580 English and 54,579 Japanese descriptions mostly uncategorized. In order to deal with the uncategorized description, we developed our original ontology to represent a conceptual structure of the TMDU data and also extraction of disease names from doctors' comments in Japanese. Such modification was necessary to fill the gap between the iCOD data and the original i2b2 ontology which is composed of ICD (International Statistical Classification of Diseases and Related Health Problems) [5] and LOINC (Logical Observation Identifiers Names and Codes) [6].
Methods and results
Installation of i2b2 database server and clients
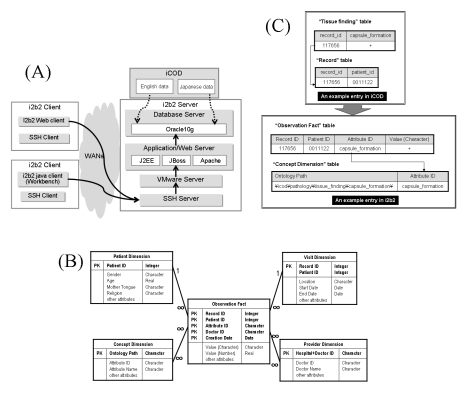
Our i2b2 database server is constructed following the instruction provided at i2b2 web site [7] (Figure 1A). Oracle 10g is used for the backend database and built our application server on JBoss, Tomcat, Apache and Eclipse on the base of VMware and CentOS 5.
Figure 1.
System configuration of the i2b2, the star schema, and an example of data conversion. A: System configuration of the i2b2 we installed in Japanese environment. Solid lines indicate transporting routs of a user's query to database. Dot lines indicate transferring routes of clinical patients' data from the iCOD to the i2b2. Because our gateway server allows only SSH protocol in communication from the Internet for security reasons, we used SSH port forwarding to access the server. Modified IP address of the i2b2 web server is used for the feasibility study so as to meet the SSH port forwarding protocol. Global IPs must be needed to make the i2b2 database publicly accessible. B: Star schema. This figure shows the entity relationship diagram of five tables composing the star schema. PK stands for ‘Primary Key’. C: An example of data conversion from the iCOD to the i2b2. We take an entry in Tissue_finding table of the iCOD as an example. A direct relation between a primary key (i.e. record_id) and an attribute (i.e. capsule_formation) was converted into a quadruplet relation among patient_id (i.e. 0011122), record_id (i.e. 117656), attribute (i.e capsule_formation), and attribute value (i.e. (+)), so as to be transferred into Observation Fact table. The attribute (i.e. capsule_formation) was assigned a semantic location in ontology and transferred into Concept Dimension table with its ontology path (i.e. \icod\pathology\tissue_finding\capsule_formation\).
Analysis of the i2b2 database schema
The data model provided in the i2b2 database is called ‘star schema’ where tables are connected as a star. The star schema plays an important role bridging object-oriented ontological data model and physical data representation on a relational database. Figure 1B shows the star schema consists of Observation Fact surrounded by Patient Dimension, Visit Dimension, Concept Dimension, and Provider Dimension. In this schema, the Observation Fact table represents a patient object and other four dimensions represent its attributes such as who (patient information), when (dates), what (ontology for clinical patients' data) and where (hospital and doctor names), respectively.Generally, sets of polynomial relations called ‘tables’ represent a data model of a target domain in an ordinary relational database system. In the i2b2, however, ontology represents a data model of a target domain. Various biomedical attributes of individual patient data can be retrieved indirectly by tracking linkages to ontology that systematizes all the attributes in the i2b2. Ontology is stored in Concept Dimension table, which contains a symbolic name of an individual attribute (Attribute ID) and a path from a root of ontology to an individual attribute (Ontology Path). Every attribute is allowed to have only one conceptual path, so that ontology in the i2b2 includes no multiple inheritance.
i2b2 with Japanese character set
The i2b2's availability with Japanese characters is crucial when it is applied to collect Japanese patients' data in Japanese medical environment. Practically speaking, it is almost impossible to write all patients' records in English or any language other than Japanese at ordinal Japanese hospitals. Therefore, we investigated the availability of Japanese character set in the i2b2 system.According to our investigation on eight application modules on the i2b2 system, the i2b2 web client can handle Japanese characters in all the modules but the i2b2 workbench fails in half of the modules in default (Table 1 see Table 1). This means that careful modification of the i2b2 source codes is necessary to handle Japanese characters in the i2b2 database.
Clinical patients' data collected in TMDU university hospital
Tanaka's group of TMDU had developed the iCOD [4] for integration of clinical patients' data and genome-wide molecular data measured in carcinoma and non-carcinoma specimens from cancer patients. The iCOD provides not only biomedical information on patients but also information on patients' lifestyle in detail such as dietary, smoking, and toiletry habits. All the clinical patients' data were collected manually by experts having licenses of hospital nurses. The iCOD currently includes clinical patients' data on hepatic carcinoma, intestinal caner, and oral cancer. It is planned to collect patients' data from all the kinds of carcinomas in the future. It currently contains 392 patients in Japanese description, of which 130 patients have their descriptions also in English. It consists of five schemas and 52 tables, of which 34 tables contain clinical patients' data.
Data transfer from the iCOD to the i2b2
As the first step of our data transfer from the iCOD to the i2b2 system, 25 tables out of 34 tables are transferred. We eliminate nine tables mainly because they include only Japanese texts and have nothing to do with our feasibility study. The tables containing only English characters and numeric values can be transferred automatically. However, care must be taken for the tables which contain Japanese free text such as Past_history_of_disease field of Medical history table in the iCOD which contains very informative information to identify patients' diseases in detail. In order to deal with this issue, we conducted to use NLP technologies to extract disease names from the medical history texts automatically.The example of data conversion is demonstrated in Figure 1C. The differences of the iCOD and the i2b2 data are the following: Direct linking in table property in the iCOD is converted to an indirect link represented by Attribute ID (e.g. ‘capsule_formation’) in the i2b2. It is also regarded as a conversion from the third norm form to the fourth norm form in the relational database theory. Note that the semantics of Attribute ID can be represented by an ontology path which locates the position of the term in the i2b2 categorical hierarchy. According to our implementation, 8,580 indirect links are generated from the 25 iCOD tables. In the 8,580 links, 7,973 links were generated from manual translation from Japanese words to English words, and 607 links were extracted and translated by our NLP operations. 684 Attribution IDs are used for representing the 8,580 links in the i2b2.
Development of an ontology for the iCOD data
In order to handle 684 attributes newly generated from iCOD database, iCOD ontology must be added to the i2b2 ontology. In order to solve this issue systematically as much as possible, we adopt the strategy to use the iCOD table names as categories of the iCOD ontology in the i2b2. The table names were then summarized into upper concepts and 684 attributes were systematized under the upper concepts by refereeing to ICD10 if applicable. The final iCOD ontology consists of eight levels of depth is available at Supplementary File 1(available in http://bio-omix.tmd.ac.jp/disease/i2b2/). The top two levels are shown in Table 1 (available in http://bio-omix.tmd.ac.jp/disease/i2b2/). The comparison of the iCOD ontology and other standard ontologies in clinical medicine is the following. More than half of the iCOD categories are consistent with either ICD10 or LOINC. However, several categories such as Prognosis, Pathology, Molecular information, and Occupation diet lifestyle are the iCOD original. We must say that our ontology is not formally systematized in the sense of explicit specification conceptualization [8]. In the i2b2 system, ontology primarily plays a role of representing a data model so as to navigate a user to individual entries. The primarily role makes it difficult for us to build ontology being consistent with established ontology like OBO Foundry in the biomedical domain [9] based on SNAP and SPAN model.
Term extraction by NLP from doctors' comments
NLP is one of plausible information technologies to extract technical terms useful for translational research from doctors' comments in text form. In case of the iCOD, 10 tables contain plain texts in Japanese. There are two approaches to extract English technical terms from Japanese texts. One is to translate Japanese technical terms extracted from Japanese texts with Japanese NLP tools. The other one is to apply English NLP tools such as GATE to English texts translated from Japanese text. GATE is an open-source NLP framework including tokenizing, Part-of-Speech tagging, and noun phrases parsing [10]. Extracting technical terms from doctors' comments in EMRs has been reported with customized GATE [11]. However, we gave up this approach due to the technological difficulty to translate Japanese doctors' comments to English ones correctly.Our proposed method to analyze Japanese texts of doctors' comments consists of the computational pipeline of part-of-speech tagging, morphological analysis and dictionary search of technical terms. MeCaB [12] was adopted for morphological analysis with Japanese clinical dictionary (MEDIS) of ‘Standard Disease Names in Japan corresponding to ICD10 for electronic medical record‘ [13]. MEDIS provides 22,439 of relations between ICD10 codes and corresponding Japanese names covering all over ICD10 domain.
In our feasibility study, 225 records of the doctors' comments on past histories of diseases were computationally processed. The 225 records had been manually collected from EMRs by experts having licenses of hospital nurses. Our NLP operation results in 66% recall (421 terms / 641 terms) and 73% precision (421 terms / 578 terms) in Japanese term extraction from Japanese texts. According to the careful investigation of 220 terms of true negatives, the low recall rate resulted from the following four reasons: A) Use of synonyms for standard disease names defined in MEDIS dictionary (177 terms), B) Mismatch of 1byte and 2byte characters in alphabets (six terms), C) Typing mistakes in iCOD entries (three terms), and D) Use of clinical terms not included in ICD10 (34 terms). We further investigated cases of A) and classified them into three sub-categories: A-1) Use of common names instead of scientific names, A-2) Use of obsolete names, and A-3) Alphabetical variations in Japanese words (Hiragana, Katakana, and Kanji notations for the same word).According to our post processing investigation, the call rate was dramatically improved (from 66% to 95%) by just expanding term dictionary and pre-processing of the doctors' comments with regard to 2 bytes characters in alphabets and typographic errors. However, care must be taken for the handling of clinical terms. We found that 34 terms are still failed in automatic term extraction after tuning due to the mismatch with ICD10. These terms seem to either disease names of ambiguity or not disease names but symptoms. We shall further investigate the terms in our next study.
As for the precision rate, 157 terms of false positives resulted from the following two reasons: (1) Duplicated extraction of disease names from one compound noun (156 terms) and (2) Failure in tokenizing with regard to the two character word ‘shou-jou’ in Japanese which means ‘symptom’ in English. Typical example of duplicated extraction is ‘abdominal aneurysm’ and ‘aneurysm’. This problem can be easily removed by checking if a compound noun including a clinical term and the clinical term is extracted from the same sentence.The latter case is rather special and specific to Japanese sentences. The two character word ‘shou-jou’ consists of two Kanji characters ‘shou’ and ‘jou’. The Kanji character ‘shou’ is often used as the last character of disease names. Therefore, wrong disease name may be extracted if tokenizer separates ‘shou’ and ‘jou’ instead of one word ‘shou-jou’ in parsing of Japanese sentences, in precisely, in wakachi-gaki analysis. One of the ad-hoc ways to solve this problem is to check the occurrence of ‘jou’ after the detected disease name in Japanese texts in post-processing.
Finally, with the extended dictionary (Supplementary File 2 available in http://bio-omix.tmd.ac.jp/disease/i2b2/) and the post-processing program, we have succeeded to extract 607 terms out of 641 terms (recall 95%) without false positive (precision 100%) for 225 records of the doctors' comments on past histories of diseases (Supplementary Table 2 available in http://bioomix.tmd.ac.jp/disease/i2b2/). This indicates that automatic extraction of English clinical term from Japanese texts would be possible if appropriate dictionary and translation rules are provided. We shall further study so as to extract all the clinical terms from doctors' comments contained in the iCOD.The extracted 607 Japanese terms are translated into English terms using ICD10 codes with MEDIS dictionary. A pair of English and Japanese terms with the same ICD codes can be considered as the same terms in the medical domain. Applying this rule, we automatically obtained 607 English terms from the extracted Japanese terms. The conceptual hierarchy is available at http://root/Patient_background/Medical_history/Past_history_of_disease/ in our i2b2 ontology.
Application of the patients' clinical data contained in the i2b2
We have not yet established a security policy that meets the requirements of protection rules of personal privacy in accepting public access to our i2b2 data. Our i2b2 is currently used only by the authors of this paper. This paper, instead, shows an instance of how to apply our data to translational research (Supplementary Figure 1 available in http://bio-omix.tmd.ac.jp/disease/ i2b2/). A research paper reported on a combined effect of metformin (a drug for diabetes) and doxorubicin (a drug for breast cancer) on mice [14]. The combined treatment caused a significant decrease of breast cancer stem cells. This paper indicates a possibility that this combination of the drugs can be effective also for patients of breast cancer. We then investigated the comparable incidents contained in our i2b2 database. Such incidents shall help us in better designing clinical tests for the combined therapy, because we can utilize information on patients who had chances to take both the drugs at the same time. Such patients' information enables us to make clinical trials efficient, less expensive, and short-term successful.We retrieved data of ‘patients diagnosed as cancer and have medical histories of diabetes’ from our i2b2 database (Figure 1). Since our i2b2 database contains no data for breast cancer patients, we retrieved data for other cancers as a test case. 50 patients were retrieved (Supplementary Figure 1 available in http://bioomix.tmd.ac.jp/disease/i2b2/). The i2b2 accepts queries of any combination of attributes consisting ontology, so that you can investigate a variety of combined conditions in clinical patients' properties.
Discussion
We discussed with medical doctors before our construction of the i2b2 in Japanese environment. The issues were 1) linguistic problem and 2) security problem.If the i2b2 can process both English and Japanese, which language we should use in constructing the i2b2 database in Japanese environment. Our conclusion was Japanese. We use Japanese in hospitals everyday. Giving consideration to the fact that medical doctors occupy themselves in heavy duties in hospitals, clinical patients' data should be collected in the same language as the medical doctors use everyday. That is Japanese in Japan. On the other hand, there is a need to develop the database in English so as to share and reuse Japanese patients' data with people overseas. We shall be able to solve this linguistic problem by a strategy as we tried in this study: building an i2b2 database with Japanese data first, and then translating the data into English. We will apply NLP technologies to translate the Japanese data into English.
Security is always a big issue whenever developing a database containing personal information. At present, it is impossible to open clinical patients' data in public at any countries in the world because of protection of personal rights. Actually, data in the i2b2 databases developed in US are shared only by the i2b2 project members of hospitals and research institutes in a closed network. SHRINE is a secured network technology developed in the i2b2 project to establish a secured data retrieving environment for the membership hospitals [15]. The ‘avatar patient’ is another idea to tackle the security issue. The ‘avatar’ means imaginary patients' data created by random sampling from a prior distribution of a certain patient's attribute. There can be no problem to open the imaginary patients' data to the public regardless of protection of personal rights. Despite such an attractive idea, there should be many studies conducted on how to find appropriate prior distributions to certain clinical patients' attributes.
In our next step, we shall develop a pipeline that automats all the operations of transporting data to the i2b2. We should integrate processes of term extraction from doctors' comments, translation of Japanese terms into English, making associations with patients' IDs, conversion of data formats, and uploading the data to the i2b2 server. We shall also further investigate what combinations of attributes have significant effects on prognosis of cancer patients using statistical tests by R software, after our preliminary analysis using the i2b2 data. We have already studied a significant correlation between intake a drug for atherosclerosis and recurrence of colon cancer (manuscript is under preparation).
Supplementary material
Acknowledgments
We thank Dr. P. Tonellato, Dr. S. Murphy, Dr. P. Kos, Dr. V. Fusaro, Dr. D. MacFadden, Dr. M. Palchuk, Dr. P. Emerson, and Dr. E. Rubin for giving us much useful information on the i2b2. We thank Dr. Y. Natsumeda, Dr. Y. Yamaguchi, Dr. A. Nemoto, Dr. S. Inoue, and Dr. M. Yamada for useful discussion on how to practically apply the i2b2 in Japanese hospitals. Educational Program for Bio-Medical OMICS Information was supported by Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
Citation:Takai - Igarashi et al, Bioinformation 6(2): 86-90 (2011)
References
- 1.S Murphy, et al. Genome Res. 2009;19:1675. [Google Scholar]
- 2. https://www.i2b2.org/
- 3. http://www.boston.com/news/education/higher/articles/2005/07/03/harvar_project_to_scan_millions_of_medical_files/
- 4. http://omics.tmd.ac.jp/icod_pub_eng/.
- 5. http://apps.who.int/classifications/apps/icd/icd10online/
- 6. http://loinc.org/
- 7. https://www.i2b2.org/software/
- 8. http://www-ksl.stanford.edu/knowledge-sharing/papers/onto-design.rtf.
- 9.B Smith, et al. Nat Biotechnol. 2007;25:1251. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. http://gate.ac.uk/
- 11.QT Zeng, et al. BMC Med Inform Decis Mak. 2006;6:30. doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. http://mecab.sourceforge.net/
- 13. http://www2.medis.or.jp/stdcd/byomei/
- 14.HA Hirsch, et al. Cancer Res. 2009;69:7507. doi: 10.1158/0008-5472.CAN-09-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.GM Weber, et al. J Am Med Inform Assoc. 2009;16:624. doi: 10.1197/jamia.M3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.