Abstract
While many different RNA aptamers have been identified that bind to a plethora of small molecules only very few are capable of acting as engineered riboswitches. Even for aptamers binding the same ligand large differences in their regulatory potential were observed. We address here the molecular basis for these differences by using a set of unrelated neomycin-binding aptamers. UV melting analyses showed that regulating aptamers are thermally stabilized to a significantly higher degree upon ligand binding than inactive ones. Regulating aptamers show high ligand-binding affinity in the low nanomolar range which is necessary but not sufficient for regulation. NMR data showed that a destabilized, open ground state accompanied by extensive structural changes upon ligand binding is important for regulation. In contrast, inactive aptamers are already pre-formed in the absence of the ligand. By a combination of genetic, biochemical and structural analyses, we identified a switching element responsible for destabilizing the ligand free state without compromising the bound form. Our results explain for the first time the molecular mechanism of an engineered riboswitch.
INTRODUCTION
Regulation of gene expression at the level of RNA frequently exploits the conformational flexibility and functional versatility of ribonucleic acids. Riboswitches are genetic regulatory elements which consist solely of RNA. They bind specifically to small molecule metabolites thereby sensing their intracellular concentrations. Metabolite binding induces conformational changes which lead to modulation in gene expression at different levels, such as transcription, translation, splicing, polyadenylation and degradation (1). Riboswitches exploit direct RNA–ligand interaction and fulfill both sensory and regulatory function simultaneously rendering auxiliary protein factors unnecessary. This principle is favorable for the development of conditional gene expression systems.
Approaches to develop engineered riboswitches make use of in vitro selected, small molecule-binding RNA aptamers (2,3). These aptamers bind their respective ligand with high affinity and specificity. Usually, they adopt a unique conformation only upon ligand binding with the ligand becoming an integral part of the complex (4). One approach is to insert aptamers into untranslated regions of eukaryotic mRNAs. Only in complex with its ligand, the aptamer is able to inhibit translation initiation (5–9) (Figure 1A) or pre-mRNA splicing (10). Recently we have shown that aptamer-mediated regulation is efficient enough to conditionally regulate essential genes in yeast (11). The striking advantage of this aptamer-mediated regulation is that aptamers can be selected in vitro against nearly any ligand of choice (12). In addition, most aptamers are small and thus represent only a minor perturbation when introduced into the host mRNA.
Figure 1.
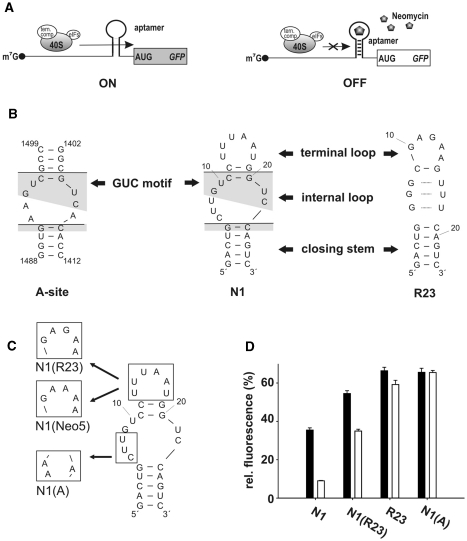
Conditional control by neomycin-binding aptamers. (A) Scheme of aptamer-mediated inhibition of translation initiation. The aptamer is inserted directly upstream of the start codon. Left: without ligand the aptamer does not interfere with ribosomal scanning. Right: the aptamer ligand complex inhibits the small ribosomal subunit and leads to decreased gene expression. (B) Secondary structure of the ribosomal A-site and the neomycin-binding aptamers N1 and R23. The lines dissect the aptamers into the terminal loop, an internal asymmetrical loop and the closing stem. Conserved nucleotides between the A-site and N1 are shaded in gray. (C) Hybrid aptamers. (D) gfp expression in the absence (black bars) and presence (white bars) of 100 µM neomycin. The fluorescence emission of the vector pWHE601 (6) expressing gfp without an aptamer in its 5′UTR was set as 100%. Background level of a vector with no gfp expression was subtracted from all data. Values represent the mean of three independently grown cultures. Measurements were repeated at least twice.
During the last years, it became obvious that only a small fraction of in vitro selected aptamers has the potential to function as riboswitches as described above. Therefore we developed a gfp-based screening system which allowed us to search, within an in vitro selected aptamer pool, for those aptamers which confer neomycin-dependent regulation (13). Screening of aptamers with binding specificity to the drug neomycin (14) resulted in the identification of regulating aptamers (13). Interestingly, these aptamers were highly underrepresented in the original in vitro selected aptamer pool [they were not among 21 different aptamers sequenced after the selection process (14)].
We identified a neomycin-binding aptamer N1 which is with 27 nt the smallest riboswitch reported to date. In the present study we compared N1 with further neomycin-binding aptamers as well as the ribosomal decoding site (A-site), a natural neomycin-binding motif. We performed a comprehensive study to obtain mechanistic insights into the regulation. The combination of genetic, biochemical and structural data lead to the identification of a structural element which is not necessary for ligand binding but for conferring regulation to the aptamer N1 by destabilizing its ground state.
MATERIALS AND METHODS
Plasmid construction
The yeast 2 µ plasmid pWHE601 was used to constitutively express the gfp gene from a pADH1 promoter (6). The vector contains a restriction site for AflII immediately upstream of the start codon with a 5′UTR length of 38 or 44 nt, respectively, depending on the two described transcriptional start sites (15) and a singular site for NheI directly after the start codon. For aptamer insertion, the vector was digested with AflII and NheI. Aptamer pools were generated by PCR using a single-stranded DNA oligonucleotide as template. The PCR products were digested with AflII and NheI prior to ligation. The start codon which was cut out of the vector after AflII/NheI digestion was attached 3′ to the aptamer sequences together with an optimized Kozak sequence for yeast (AAAATG).
For in vitro transcription, aptamers where inserted into the plasmid pSP64 (Promega) digested with NheI and XbaI. Aptamers attached to a T7 promoter sequence were generated using complementary DNA oligonucleotides, designed in a way to comprise compatible 5′ overhangs. Aptamers were transcribed as hammerhead fusion to obtain defined 3′ ends. Stem 3 of the hammerhead ribozyme was mutagenized via PCR to base pair with the 3′ ends of the aptamer sequences. All primer and vector sequences are available upon request.
GFP fluorescence measurement
Saccharomyces cerevisiae strain RS453α (matα ade2-1 trp1-1 can1-100 leu2-3 his3-1 ura3-52) transformed with the respective constructs was grown at 28°C for 48 h in 5 ml of minimal medium [0.2% (w/v) yeast nitrogen base, 0.55% (w/v) ammonium sulfate, 2% (w/v) glucose, 12 µg/ml adenine, MEM amino acids (Gibco BRL)] in the absence or presence of 100 µM neomycin trisulfate (Sigma). Cells were harvested by centrifugation and resuspended in 2 ml phosphate-buffered saline (PBS). For each construct, three independently grown cultures were analyzed. Fluorescence measurements were carried out at 25°C on a Fluorolog FL3-22 (Horiba Jobin Yvon) with the excitation wavelength set to 482 nm and an emission wavelength of 510 nm. Optical density (OD600) was determined to ensure homogeneous cell growth. The vector pVT102-U (16) without gfp+ gene was analyzed in parallel as a blank and its value subtracted from all data. Measurements were repeated at least two times.
In vivo screening
Saccharomyces cerevisiae strain RS453α transformed with the respective plasmid pool was grown at 28°C in minimal medium. Colonies were transferred to 96-well plates containing 200 µl minimal medium and incubated for 24 h. An amount of 20 µl aliquots of each sample were transferred into fresh medium in new plates with and without 100 µM neomycin in a final volume of 200 µl. Fluorescence measurements were performed 48 h after inoculation using an excitation wave length of 484 nm and an emission wave length of 512 nm with a SpectraFluor Plus fluorescence reader (Tecan, Crailsheim). Optical density (OD600) was determined to ensure homogeneous cell growth. Individual candidates were streaked out to single colonies and plasmids were prepared according to Hoffman and Winston (17). After an Escherichia coli passage plasmids were sequenced, retransformed in yeast cells and the fluorescence measurements repeated (see above).
In vitro transcription
The aptamers were transcribed in vitro from an EcoRI-linearized pSP64 plasmid using a T7 promoter. Transcription reactions contained 25 mM magnesium acetate, 200 mM Tris–HCl pH 8.0, 20 mM DTT, 2 mM spermidine, 0.1 mg/ml linearized plasmid, 4 mM of each NTP and 2.5 µl self-made T7 polymerase [prepared according to Davanloo et al. (18)]. After overnight incubation at 37°C precipitated pyrophosphate was pelleted by centrifugation. Residual traces were resolved by adding 20% (v/v) 0.5 M EDTA pH 8.0 to the supernatant. After ethanol/aceton precipitation the RNA products were separated on a 15% denaturing polyacrylamid gel. Aptamer RNA was detected by UV shadowing and eluted from gel slices in 300 mM sodium acetate pH 6.5 overnight at 4°C. The supernatant was filtered using a 0.45 µM filter (Sarstedt) and again ethanol/aceton precipitated. RNA was resuspended in H2O and stored at −20°C. An amount of 3 mM of each 15N-labeled NTP (Silantes) was used for preparation of 15N-labeled N1 RNA.
UV-melting studies
Prior to melting studies 50 µM RNA solutions were heated to 95°C for 5 min, diluted 35- to 40-fold in ice cold water and snap-cooled on ice for 10 min. After folding, buffer was added to a final concentration of 20 mM Na-cacodylate pH 6.8 and 100 mM NaCl. The final concentration of RNA was 1 µM. For ligand-dependent melting studies 10 µM neomycin was added to the sample. Heating and cooling was performed on a UV spectrophotometer V-650 (Jasco) from 15–95°C with a rate of 0.5°C/min. Melting was followed by measuring the OD260 every minute. For each RNA three independent samples were analyzed. Buffer with or without neomycin was analyzed in parallel as a blank and its value was subtracted from all data. Tm values were determined by calculating the first derivative.
ITC measurements
Prior to titration RNA solutions were heated to 95°C for 5 min and snap-cooled on ice for 10 min. After that, folding buffer was added to a final concentration of 20 mM Na-cacodylate, 200 mM NaCl, 10 mM MgCl2, 1 mM spermidine, pH 6.8. The final concentration was 4.5 µM for R23, N1(A) and N1-2/1-CU/A and 4 µM for N1 and N1(R23). Neomycin solution (38 µM) was prepared in the same buffer. ITC experiments were done on a VP-ITC microcalorimeter (MicroCal Inc. Northampton, MA) with the sample cell (1.44 ml) containing RNA and the neomycin solution in the injector syringe. Following thermal equilibration at 37°C, an initial 120 s delay and two initial 1 µl injections, we did 28 serial injections of 10 µl at intervals of 180 s and at a stirring speed of 310 rpm. Raw data were recorded as power (µcal/s) over time (min). The heat associated with each titration peak was integrated and plotted against the respective molar ratio of neomycin and RNA and the resulting experimental binding isotherm was corrected for the effect of titrating neomycin into buffer alone. Thermodynamic parameters were extracted from a curve fit to the corrected data using the one site binding model in the Origin software provided by MicroCal. These are the change in enthalpy ΔH and the dissociation constant KD. As a consequence, the change in Gibbs free energy ΔG and the change in entropy ΔS can be calculated from ΔG = RT ln(KD) and ΔG = ΔH – TΔS, with T being the reaction temperature (in K) and R being the gas constant (1.986 cal/K/mol). Measurements were repeated at least twice.
NMR spectroscopy
Prior to NMR experiments RNA solutions were heated to 95°C for 5 min, diluted 5-fold in ice cold water and snap-cooled on ice for 10 min. After folding, the RNA was concentrated to ∼500 µl using Vivaspin 2 centrifugal concentrators (Sartorius). Concentrated RNA was diluted 4-fold in NMR buffer (25 mM potassium phosphate buffer pH 6.2, 50 mM KCl) and again concentrated to ∼500 µl. Dilution and concentration steps were repeated seven times and RNA concentrated to a final volume of ∼220 µl. The RNA concentration of the NMR samples was ∼150–250 μM for 1D-1H-experiments and titrations with neomycin and 0.6–1 mM when 2D-1H,1H-NOESY experiments were recorded. All spectra were recorded at 283 K in 90% H2O/10% D2O in NMR buffer on Bruker AVANCE 600-MHz or Bruker AVANCE 700-MHz spectrometers equipped with cryogenic TXI-HCN probes and triple axis gradients. Water suppression in the 1D-1H-experiments was achieved using the jump-and-return water suppression scheme (19). In the 2D-1H,1H-NOESY- and the 2D-1H,15N-HSQC-experiments water suppression was achieved by using WATERGATE. For each 1D spectrum, 128–512 transients were accumulated. NOESY-spectra were recorded with 128 transients for each t1 increment and 400 complex points in the F1-dimension. Spectra were processed and analyzed using the Bruker Topspin 2.1 software.
RESULTS
A 3-nt element at the 5′ part of the internal loop of N1 is important for regulation
The secondary structure of the aptamer N1 consists of a 5-bp closing stem, an asymmetric internal loop and a terminal loop separated by two Watson–Crick G:C base pairs. The internal loop contains a 5′-GUC-3′/5′-GUC-3′ sequence (GUC motif) in the upper part and a 5′-CUU-3′ sequence 5′ of the GUC motif (Figure 1B). Mutagenesis studies indicated that both loop regions are important for ligand binding (13). The ribosomal A-site also contains the GUC motif (Figure 1B, shaded in gray) but flanked by three adenine residues, two at the 5′ and one at the 3′ site. The neomycin-binding aptamer, R23, was the most abundant sequence in the in vitro selected aptamer pool [12 out of 21 sequenced clones (14)] but shows no regulation [Figure 1B and D and Table 1; (13)]. It forms a stem loop structure with a neomycin-binding pocket consisting of three consecutive G:U wobble base pairs together with an adenine nucleotide reaching down from the highly structured terminal loop (20).
Table 1.
Activity of neomycin-binding aptamers
| Aptamer | Relative fluorescence (%) 0 µM neomycina | Relative fluorescence (%) 100 µM neomycina | Regulatory Factorb |
|---|---|---|---|
| N1 [N1-3/0-CUU] | 35.5 | 9.0 | 3.9 |
| N1(R23) | 54.5 | 35.0 | 1.6 |
| N1(Neo5) | 56.1 | 28.1 | 2.0 |
| Neo5 | 12.4 | 7.2 | 1.7 |
| R23 | 66.4 | 59.2 | 1.1 |
| N1(A) | 65.6 | 65.4 | 1.0 |
| N1-4/0-CCUU | 44.6 | 11.7 | 3.8 |
| N1-3/0-CUC | 44.2 | 12.7 | 3.5 |
| N1-3/0-CUA | 39.6 | 15.4 | 2.6 |
| N1-3/0-CUG | 15.7 | 9.9 | 1.6 |
| N1-2/0-CU | 26.1 | 7.1 | 3.7 |
| N1-1/0-C | 22.2 | 11.9 | 1.9 |
| N1-1/0-U | 17.8 | 9.7 | 1.8 |
| N1-0/0 | 2.3 | 2.3 | 1.0 |
| N1-2/1-CU/A | 35.3 | 33.2 | 1.1 |
| N1-3/1-CUU/A | 33.5 | 29.2 | 1.1 |
agfp expression in the absence and presence of 100 µM neomycin. The fluorescence emission of the vector pWHE601 expressing gfp without an aptamer in its 5′UTR was set as 100%. Background level of a vector with no gfp expression was subtracted from all data. Values are mean of three independently grown cultures with SD <10%. Measurements were repeated at least twice.
bEfficiency of regulation is given as the ratio of relative fluorescence with and without neomycin.
We mutated the CUU sequence in the lower part of the internal loop of N1 towards the A-site creating the N1-A-site-hybrid N1(A) (Figure 1C). However, this mutation renders N1 inactive [Figure 1D and Table 1; (13)]. We then exchanged the terminal loop of N1 with the loop from R23 and Neo5, respectively, resulting in the hybrids N1(R23) and N1(Neo5) (Figure 1C). Neo5 is also from the in vitro selected pool like R23 and shows marginal regulation [Table 1; (13)]. Both hybrid constructs showed regulation albeit with a reduced dynamic range (Figure 1D and Table 1). This indicates that the CUU sequence of the internal loop is important for regulation whereas the terminal loop has only a modulating effect.
This prompted us to investigate the importance of the CUU sequence by saturating mutagenesis. In a first attempt we cloned four pools with three and two nucleotides, respectively, inserted 5′ of the GUC motif in combination with one or no nucleotide 3′ of the motif. The respective pools were named N1-X/Y with X indicating the number of nucleotides at the 5′ and Y at the 3′ site of the motif (N1-3/0, N1-2/0, N1-3/1 and N1-2/1, Figure 2A). Randomized aptamer sequences were inserted in the 5′UTR of a constitutively expressed gfp reporter gene (6). We obtained complete coverage for the plasmid pools N1-3/0, N1-2/0 and N1-2/1 and 40% coverage for N1-3/1 (Supplementary Table S1). Yeast cells were transformed with these plasmid pools. A total of 192 individual colonies were grown for each pool in 96-well plates and gfp expression was determined in the absence and presence of 100 µM neomycin. Candidates with decreased fluorescence in the presence of neomycin were sequenced and the regulation was verified. The activity of selected candidates is summarized in Figure 2, Table 1 and Supplementary Table S2.
Figure 2.
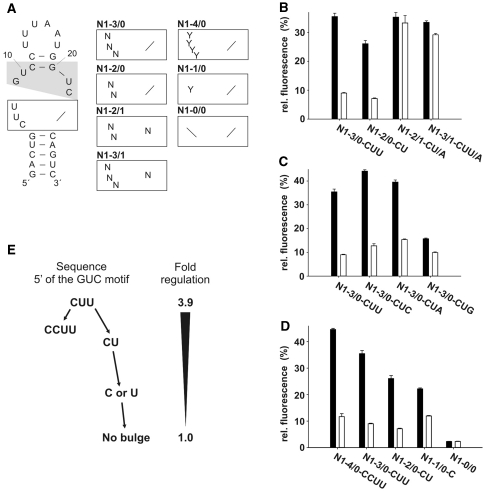
Saturating mutagenesis of the internal loop. (A) Secondary structure of N1. The GUC motif is shaded in gray. The lower part of the internal loop (boxed) was analyzed and randomized. Nucleotides are indicated with N (any nucleotide) or Y (pyrimidines). (B–D) gfp expression in the absence (black bars) and presence (white bars) of 100 µM neomycin. The fluorescence emission of the vector pWHE601 (6) expressing gfp without an aptamer in its 5′UTR was set as 100%. Background level of a vector with no gfp expression was subtracted from all data. Values represent the mean of three independently grown cultures. Measurements were repeated at least twice. (E) Schematic representation of the connection between loop size and regulation.
Interestingly, the wild-type sequence CUU was the most active sequence and no candidates with an increased dynamic range of regulation have been identified. All aptamers with a 3′ nucleotide insertion between the GUC motif and the closing stem (this means all candidates from pool N1-3/1 and N1-2/1) are inactive. In contrast, all aptamers from the N1-3/0 and N1-2/0 pools were active (e.g. Figure 2B). However, the efficiency of regulation is dependent on the sequence (Figure 2B and C, Table 1 and Supplementary Table S2) with a clear preference for pyrimidines over purines. Furthermore, adenines are preferred over guanines which is exemplarily shown for the CUN series from the N1-3/0 pool (Figure 2C). The wild-type sequence CUU is the most active sequence with 3.9-, followed by CUC with 3.5-, CUA with 2.6- and CUG with 1.6-fold regulation.
Next we analyzed the dependence on the length of the inserted sequence at the 5′ site of the GUC motif. In addition to N1-3/0 and N1-2/0 we questioned if four, one or no nucleotide at this position also confer regulation. We cloned the pools N1-4/0, N1-1/0, N1-0/0 but restricted the pools to pyrimidines only. The data are included in Figure 2D, E, Table 1 and Supplementary Table S2. Four nucleotides (e.g. CCUU) result in a similar activity as three (wild-type: CUU) or two nucleotides (e.g. CU). However, one nucleotide at this position (C or U) leads to a significant reduction in regulation (Figure 2D, E and Table 1). The absence of a nucleotide between the GUC motif and the adjacent closing stem results in a complete loss of activity.
These data clearly demonstrate that one to four nucleotides 5′ of the GUC motif are necessary in conferring riboswitch activity to N1. Notably the dynamic range of the regulation is modulated by both the sequence and the length of the insertion.
High ligand-binding affinity and thermodynamic stabilization are important for regulation
Aminoglycosides are known to exert a stabilizing effect upon binding to RNAs (21–23). Therefore, we questioned if an increase in the thermal stability of the aptamer–neomycin complex may be responsible for ligand-dependent translational inhibition.
We have chosen 11 aptamers for melting analyses with varying degrees of regulation including six active [N1, N1-3/0-CUC, N1-2/0-CU, N1-1/0-C, N1-1/0-U, N1(R23)] and five inactive aptamers [R23, N1(A), N1-2/1-CU/A, N1-3/1-CUU/A and N1-0/0]. Melting curves were measured in the absence and presence of 10 µM neomycin and are displayed in Figure 3A and Supplementary Figure S1. All profiles showed monophasic melting. The only exception was N1-2/0-CU with a second melting event at higher temperatures in the absence of neomycin.
Figure 3.
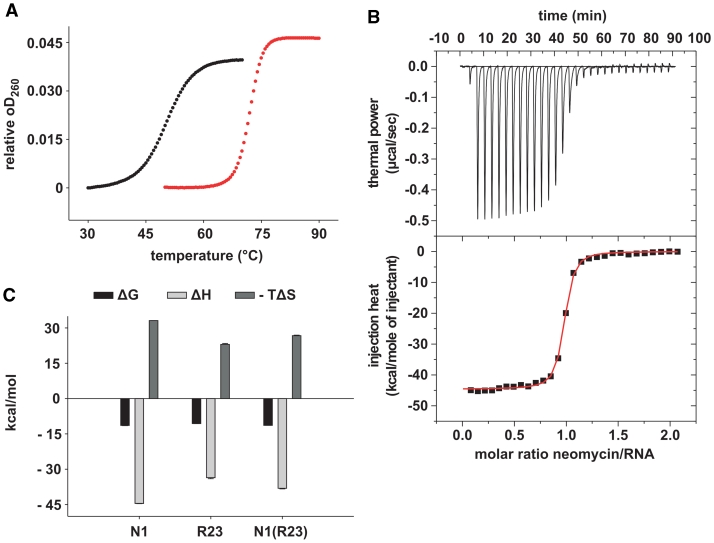
Melting point analyses and determination of the equilibrium dissociation constants (KD) by ITC. (A) Melting curves in the absence (black curves) and presence (red curves) of 10 µM neomycin for 1 µM N1. Melting curves were recorded in triplicates. (B) Upper panel: power required to maintain the temperature (37°C) of the RNA solution (4 µM) recorded over the time of multiple injections (10 µl) of ligand (38 µM neomycin) until saturation was reached (baseline-corrected). Lower panels: integrated heats of interaction per mole of injectant plotted against the molar ratio of ligand over RNA and fitted to a single binding site model. (C) Gibbs free energies (ΔG), enthalpic (ΔH) and entropic (–TΔS) contributions of N1, R23 and N1(R23).
All tested aptamers show a significant increase in thermal stability upon neomycin binding (Figure 3A, Supplementary Figure S1 and Table 2). We observe a correlation between regulation and thermal stabilization. Regulating aptamers are stabilized >20°C compared to 6.3–15.6°C for the inactive variants. Exceptions are the aptamers N1-1/0-C and N1-1/0-U which are only stabilized by 16.1°C. This smaller degree of stabilization seems to result from a more stable ground state compared to the other active aptamers N1, N1-3/0-CUC and N1(R23). Consequently N1-1/0-C and N1-1/0-U markedly interfere with gene expression already in the absence of neomycin (Figure 2D) shifting the regulatory window. We propose that this minor stabilization is sufficient to confer regulation due to the already impaired gfp expression. R23 in contrast which is stabilized to a similar degree (only 0.5°C less) is not active, because of the less impaired basal gene expression (Figure 1D).
Table 2.
Determination of Tm values of different neomycin-binding aptamers
| Aptamer | Tm in °Ca 0 µM neomycin | Tm in °Ca 10 µM neomycin | ΔTm | Regulatory Factorb |
|---|---|---|---|---|
| N1-3/0-CUU | 50.5 ± 0.2 | 71.8 ± 0.5 | 21.3 | 3.9 |
| R23 | 57.5 ± 0.5 | 73.1 ± 0.6 | 15.6 | 1.1 |
| N1(R23) | 51.3 ± 0.5 | 71.5 ± 0.4 | 20.2 | 1.6 |
| N1(A) | 54.6 ± 0.5 | 68.3 ± 0.8 | 13.7 | 1.0 |
| N1-3/0-CUC | 50.5 ± 0.2 | 72.5 ± 0.3 | 22.0 | 3.5 |
| N1-2/0-CU | n.d. | 74.0 ± 0.4 | n.d. | 3.7 |
| N1-1/0-C | 61.0 ± 0.5 | 77.1 ± 0.2 | 16.1 | 1.9 |
| N1-1/0-U | 61.6 ± 0.5 | 77.6 ± 0.7 | 16.1 | 1.8 |
| N1-0/0 | 74.2 ± 0.4 | 80.5 ± 0.5 | 6.3 | 1.0 |
| N1-2/1-CU/A | 60.3 ± 0.3 | 71.4 ± 0.5 | 11.1 | 1.1 |
| N1-3/1-CUU/A | 56.0 ± 0.5 | 69.3 ± 0.8 | 13.3 | 1.1 |
n.d.: not determined. The aptamer N1-2/0-CU shows biphasic melting behavior in the absence of neomycin.
aMelting analysis was performed using 1 µM aptamer RNA in 20 mM Na-cacodylate pH 6.8 and 100 mM NaCl. Melting curves were recorded in triplicates. Buffer with or without neomycin was analyzed in parallel as a blank and substracted from all data.
bEfficiency of regulation is given as the ratio of relative fluorescence with and without neomycin.
We determined the binding constants for two active N1, N1(R23) and three inactive aptamers [R23, N1(A), N1-2/1-CU/A] using isothermal titration calorimetry (Table 3). Both, the active aptamers N1 and N1(R23) bind neomycin in the low nanomolar range, but also, yet somewhat weaker, the inactive R23. In contrast to this N1(A) and N1-2/1-CU/A (both with a nucleotide 3′ of the GUC motif) show very weak binding only (Figure 3B and Supplementary Figure S2). Mutating the terminal loop of N1 towards R23 in N1(R23) does not influence the binding strength but results in reduced regulation (Figure 1D and Supplementary Figure S2). These data show that a high binding affinity is necessary but not sufficient for regulation.
Table 3.
Dissociation binding constants (KD) and thermodynamic parameters for neomycin binding to N1, R23 and N1(R23) at 37°C
| Aptamer | KD (nM) | ΔG (kcal/mol) | ΔH (kcal/mol) | –TΔS (kcal/mol) |
|---|---|---|---|---|
| N1-3/0-CUU | 9.2 ± 1.3 | −11.4 ± 0.1 | −44.5 ± 0.1 | 33.1 ± 0.0 |
| R23 | 34.1 ± 2.1 | −10.6 ± 0.0 | −33.6 ± 0.4 | 23.0 ± 0.4 |
| N1(R23) | 9.5 ± 0.3 | −11.4 ± 0.0 | −38.1 ± 0.3 | 26.7 ± 0.2 |
Parameters extracted from ITC measurements (Figure 3 and Supplementary Figure S2).
All aptamers with high binding affinity show a strongly favorable binding enthalpy ΔH indicating a substantial energy gain from the formation of intra- and intermolecular interactions (Figure 3C and Table 3). The ΔH values correlate with the thermal stabilization, with N1 showing the highest value with −44 kcal/mol. The high enthalpic contribution is counteracted by a significant and unfavorable entropic contribution (Figure 3C and Table 3), indicating a loss of flexibility for both RNA and ligand, and/or water hydration rearrangement upon binding. In line with this, N1 shows the highest –TΔS value and R23 the lowest.
Taken together, genetic and biochemical data reveal that the presence of a sequence element at the 5′ end of the internal loop is important for regulation. The length and sequence of the element show only a modulating effect. In contrast, additional nucleotides 3′ of the GUC motif completely abolish regulation. A high binding affinity as well as a considerable thermal stabilization is necessary for riboswitch activity indicating that major conformational changes occur upon neomycin binding. To characterize the degree and the nature of these structural changes upon ligand binding in more detail we performed NMR spectroscopy.
A largely unstructured ground state is important for regulation
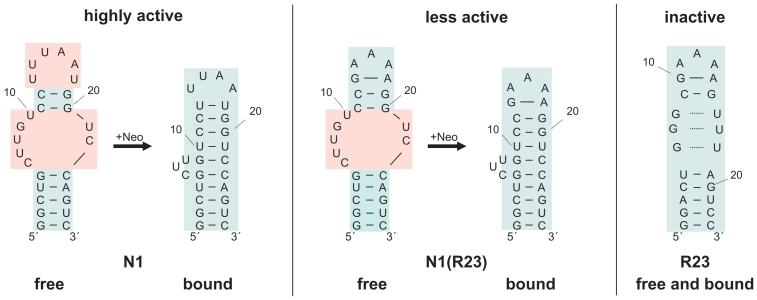
The recently solved structure of the aptamer N1 shows that its ground state is largely unstructured at temperatures >10°C (24). Thus, the free form shows only the 5 bp of the closing stem and two G:C base pairs of the upper helix. All nucleotides within the internal and the terminal loop are apparently not stabilized by hydrogen bonding. However, dramatic conformational changes occur upon neomycin binding (Figure 4). The upper helix is extended by three additional base pairs and stacks on the closing stem. The terminal loop becomes highly structured. The three nucleotides 5′ of the GUC motif are bulged out and exposed to the solvent (Figure 5). They are not directly involved in ligand binding which corresponds to the genetic data. Since the insertion element does not interact with the ligand, its length and the identity of the nucleotides are variable.
Figure 4.
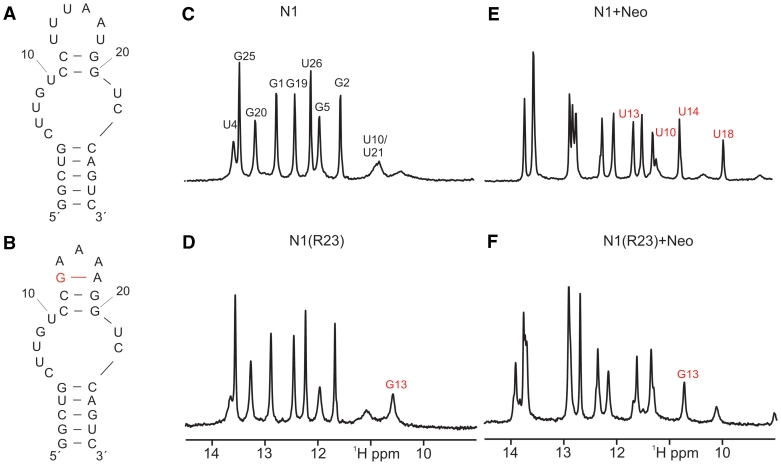
Conformation and neomycin binding of N1 and N1(R23). (A and B) Proposed secondary structure of the free form of N1 and N1(R23) based on the number of observable imino proton signals and NOE-patterns. The additional base pair in the terminal loop of N1(R23) is highlighted in red. (C and D) Comparison of the imino proton region of 1D-1H-spectra of free N1 and free N1(R23). Signal assignments are indicated. The assignment of the signal at ∼10.7 ppm to the imino proton of G13 (marked in red) is based on the comparison with spectra of the original R23 aptamer in its ligand-free state (20). (E and F) Imino proton region of the 1D-1H-spectrum of N1 and N1(R23) in the presence of one equivalent of neomycin. Compared to the spectrum of free N1 and N1(R23) shown in (D and E) novel signals and chemical shift changes are observed as expected due to the formation of stable 1:1 neomycin RNA-complexes in slow exchange on the NMR-time scale.
Figure 5.
Model of proposed secondary structure changes of N1, N1(R23) and R23 upon neomycin binding derived from NMR data. Unstructured elements are shaded in red, structured elements in blue.
In contrast, the inactive R23 is largely pre-structured in its free state with pre-formed non-canonical base pairs and a pre-formed terminal loop [Figure 5; (20)]. The number of NMR signals is similar in the ground and in the ligand-bound form indicating that no new base pairs are formed.
N1(R23) shows decreased regulation compared to N1 and we questioned if this is reflected in the structural pre-formation. The imino proton 1H-spectrum of N1(R23) is very similar to that of N1 in the region typical for imino groups in Watson–Crick base pairs (25) with the same number of signals (Figure 4). Between 10.0 and 12.0 ppm there is one additional signal with a narrow line width. Comparison with the spectrum of R23 (20) indicates that this signal most likely originates from the loop nucleotide G13 in the replaced terminal loop (Figure 4, marked in red). Thus, the free state of N1(R23) is unstructured in the vicinity of the internal bulge region similar to N1, whereas the terminal loop appears to be already structured in the absence of ligand (Figures 4 and 5).
Neomycin binding leads to a change in the chemical shift and a reduction in the line width for the signal between 10.0 and 12.0 ppm as observed for the original R23 aptamer. The neomycin bound form of N1(R23) contains one additional signal in the Watson–Crick region of the spectrum and two additional signals between 10.0 and 12.0 ppm with chemical shifts typical for the imino protons of a U:U base pair (26). Thus, neomycin binding seems to stabilize N1(R23) by extension of the upper helix in a manner very similar to that observed for N1 (Figure 4). In addition, the terminal loop (5′-GAGAA-3′) of N1(R23) functionally replaces the terminal loop of N1 in interacting with the ligand as indicated by the reduced line width and shift change observed for the G13 imino proton signal. Taken together N1(R23) is a hybrid between N1 and R23 with respect to its regulatory abilities and structure which perfectly fits both the genetic and biochemical data.
3′ nucleotide insertion leads to structure stabilization already in the absence of ligand
Insertion of a nucleotide 3′ of the GUC motif completely prevents regulation. We question if this can be explained by the structure of the aptamer in its ligand free state. Comparison of the imino proton 1H- and 2D-1H,1H-NOESY spectra of the free N1(A) with those of N1 reveals the presence of one additional signal with a chemical shift typical for a guanine in a Watson–Crick G:C base pair (Supplementary Figure S3). Another additional signal with a narrow line width is present in the region typical for U:U base pairs (26). NOEs link this uridine signal to two neighboring imino protons from guanines G9 and G20 in G:C base pairs (Supplementary Figure S3). Furthermore, there is a strong intra base pair NOE to one of the broad uridine imino signals in the same region. Thus, the NMR-data demonstrate that the upper helix is extended by a U:U base pair and an additional Watson–Crick G:C base pair in the free state of N1(A) compared to N1 in agreement with the higher melting temperature for free N1(A). Thus, the free forms of N1(A) and N1 differ significantly in terms of structural stability. On the other hand, spectra of the neomycin bound N1(A) strongly resemble those of N1 (compare Supplementary Figure S3G and H). In particular, signals with a chemical shift very similar to those of U10, U13, U14 and U18 in N1 are observed in the 10.0–12.5-ppm region (27). This indicates that neomycin interacts with the terminal loop and the upper helix of N1(A) in a manner very similar to that of N1.
For N1-2/1-CU/A, the imino proton spectrum contains at least one extra signal in the Watson–Crick region when compared to that of N1 (Supplementary Figure S3). Furthermore, for some of the signals in this region (e.g. U4, G5) a significant reduction in line widths with respect to those observed in N1 is obvious which indicates a more stable structure. In addition, four observable signals with chemical shifts ∼10.3 ppm indicate the presence of transiently stable U:U base pairs and consequently a structure where the upper helix is already pre-formed. Thus, the extra adenine residue opposite the bulge region leads to a significant structural stabilization of the free form of the RNA in comparison to N1 (Supplementary Figure S3C and F). In contrast, neomycin binding results in a spectrum that closely resembles N1 with the distinctive signals for U10, U13, U14 and U18 present (Supplementary Figure S3I).
Taken together, NMR data show that aptamers with a nucleotide 3′ of the GUC motif are already pre-formed in the absence of neomycin. In addition, N1 shows contacts of the uppermost G:C base pair of the closing stem to neomycin which enforces coaxial stacking of the upper helix on the closing stem (24). This contact is prevented by insertion of a 3 nucleotide, thereby reducing ligand contacts, probably resulting in the low binding affinity.
DISCUSSION
The initial starting point of our study was the observation that the four neomycin-binding RNA aptamers N1, R23, N1(R23) and N1(A) showed different levels of regulation when employed as riboswitches. Despite obvious similarities with respect to size and secondary structure, only two of them are able to control gene expression. The aim of this study was to investigate differences between active and inactive aptamers and to unravel the determinants for regulation.
Our study shows that differences in the conformation between the ground and ligand bound state is the most important factor in conferring regulation. A sequence element of nucleotides 5′ of the GUC motif keeps the RNA in an open conformation. Ligand binding induces extensive structural changes. The upper helix is extended by three additional base pairs and stacks directly on top of the closing stem, forming a solid continuous 10-bp helix. These conformational changes are accompanied by a dramatic thermal stabilization, also reflected in an extremely high favorable enthalpy for ligand binding. Inactive aptamers, in contrast, are highly pre-formed. R23 is largely pre-structured in its ground state with pre-formed non-canonical base pairs and a pre-formed terminal loop (20). This is reflected by a lower degree of thermal stabilization and a reduced ΔH value. N1(R23) as an intermediate has the terminal loop already pre-structured whereas the internal loop originating from N1 is open and gets stabilized upon ligand binding (Figure 5). The difference in the ground states is also supported by the observation of virtually identical binding affinities for N1 and N1(R23), which, importantly, are composed of different constituting energy terms. This is an example of enthalpy-entropy compensation, which indicates that the same final state is reached, however, on different routes (28,29).
As little as one nucleotide insertion 3′ between the GUC motif and the closing stem results in a complete loss of regulation, independent of the sequence. Structural data show that this nucleotide allows the pre-formation of the GUC motif and interferes with the stacking of the upper helix on the closing stem which abrogates the stabilizing effect of ligand binding. In addition, it dramatically reduces the binding affinity for neomycin.
The asymmetry of the internal loop is clearly the important feature for regulation with the nucleotides 5′ to the GUC motif destabilizing the ground state. The one to four nucleotide small element, which is not implicated in ligand binding, is the functional element rendering the aptamer N1 active. In addition this element allows the tuning of the riboswitch efficiency. Small changes in size and sequence allow the adjustment of the regulatory window of the riboswitch towards different applications.
A flexible unbound state is also present in natural riboswitches. They ensure high selectivity and affinity by interacting with nearly all functional groups provided by their target (30), with the ligand nearly completely enveloped inside the RNA-binding pocket. This suggests that there has to be enough flexibility in the unbound state for the ligand to access the binding pocket. The binding pocket of the purine riboswitches, e.g. is comprised of a three-way junction which is locally disordered in the free state and collapses around the ligand upon binding (31,32). This flexibility in the unbound state is similar for the regulatory aptamer N1, which binds neomycin via conformational selection (24,33).
Riboswitches found in nature are mostly comprised of an aptamer domain and an expression platform, connected by a switching sequence which acts as a communication module. Dependent on the binding status, the switching sequence interacts either with the aptamer domain or the expression platform, thereby favoring one of two mutually exclusive structures and subsequent modulation of gene expression (31). The N1 riboswitch combines aptamer and expression platform in the same domain. Here the ‘switching element’ (the CUU bulge) does not communicate between two otherwise independent domains, but prevents extensive pre-formation of the binding pocket, thereby distinguishing the unbound and bound conformations. This difference in thermal stability between the ‘on-’ and ‘off’-state of an RNA regulator is reminiscent of thermoswitches in prokaryotes. Thermoswitches function in a similar way, just by opening and closing of base pairing interactions with the SD sequence (34).
It will be interesting to analyze if thermal stabilization is a common feature determining riboswitch activity of regulating aptamers. If so, this information is invaluable (i) in predicting the applicability of newly selected aptamers, (ii) in further improving the performance of existing engineered riboswitches or (iii) in creating new riboswitches by rendering regulatory inactive aptamers active. A detailed knowledge of the necessary molecular determinants for riboswitch activity will be of immense value to provide more versatile RNA switches for synthetic biology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Aventis Foundation; the Deutsche Forschungsgemeinschaft (DFG: Cluster of Excellence: Macromolecular Complexes and SU 402/4-1 to B.S., WO 901/2-1 to J.W.); Volkswagenstiftung; Center of Biomolecular Magnetic Resonance (BMRZ); Johann Wolfgang Goethe-University Frankfurt and Heisenberg stipend of the DFG (HA 3459/5) to C.H. Funding for open access charge: DFG.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Ronald Micura, Ulrike Rieder and Renee Schroeder for fruitful discussions. The authors are grateful to Joachim W. Engels and Renee Schroeder for the opportunity to measure melting curves and Olga Frolow and Sabine Stampfl for the kind introduction to the instruments.
REFERENCES
- 1.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suess B, Weigand JE. Engineered riboswitches: overview, problems and trends. RNA Biol. 2008;5:24–29. doi: 10.4161/rna.5.1.5955. [DOI] [PubMed] [Google Scholar]
- 3.Weigand JE, Suess B. Aptamers and riboswitches: perspectives in biotechnology. Appl. Microbiol. Biotechnol. 2009;85:229–236. doi: 10.1007/s00253-009-2194-2. [DOI] [PubMed] [Google Scholar]
- 4.Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 5.Hanson S, Berthelot K, Fink B, McCarthy JE, Suess B. Tetracycline-aptamer-mediated translational regulation in yeast. Mol. Microbiol. 2003;49:1627–1637. doi: 10.1046/j.1365-2958.2003.03656.x. [DOI] [PubMed] [Google Scholar]
- 6.Suess B, Hanson S, Berens C, Fink B, Schroeder R, Hillen W. Conditional gene expression by controlling translation with tetracycline-binding aptamers. Nucleic Acids Res. 2003;31:1853–1858. doi: 10.1093/nar/gkg285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grate D, Wilson C. Inducible regulation of the S. cerevisiae cell cycle mediated by an RNA aptamer-ligand complex. Bioorg. Med. Chem. 2001;9:2565–2570. doi: 10.1016/s0968-0896(01)00031-1. [DOI] [PubMed] [Google Scholar]
- 8.Harvey I, Garneau P, Pelletier J. Inhibition of translation by RNA-small molecule interactions. RNA. 2002;8:452–463. doi: 10.1017/s135583820202633x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Werstuck G, Green MR. Controlling gene expression in living cells through small molecule-RNA interactions. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- 10.Weigand JE, Suess B. Tetracycline aptamer-controlled regulation of pre-mRNA splicing in yeast. Nucleic Acids Res. 2007;35:4179–4185. doi: 10.1093/nar/gkm425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotter P, Weigand JE, Meyer B, Entian KD, Suess B. A fast and efficient translational control system for conditional expression of yeast genes. Nucleic Acids Res. 2009;37:e120. doi: 10.1093/nar/gkp578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sinha J, Reyes SJ, Gallivan JP. Reprogramming bacteria to seek and destroy an herbicide. Nat. Chem. Biol. 2010;6:464–470. doi: 10.1038/nchembio.369. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Weigand JE, Sanchez M, Gunnesch EB, Zeiher S, Schroeder R, Suess B. Screening for engineered neomycin riboswitches that control translation initiation. RNA. 2008;14:89–97. doi: 10.1261/rna.772408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallis MG, von Ahsen U, Schroeder R, Famulok M. A novel RNA motif for neomycin recognition. Chem. Biol. 1995;2:543–552. doi: 10.1016/1074-5521(95)90188-4. [DOI] [PubMed] [Google Scholar]
- 15.Faitar SL, Brodie SA, Ponticelli AS. Promoter-specific shifts in transcription initiation conferred by yeast TFIIB mutations are determined by the sequence in the immediate vicinity of the start sites. Mol. Cell. Biol. 2001;21:4427–4440. doi: 10.1128/MCB.21.14.4427-4440.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vernet T, Dignard D, Thomas DY. A family of yeast expression vectors containing the phage f1 intergenic region. Gene. 1987;52:225–233. doi: 10.1016/0378-1119(87)90049-7. [DOI] [PubMed] [Google Scholar]
- 17.Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 18.Davanloo P, Rosenberg AH, Dunn JJ, Studier FW. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc. Natl Acad. Sci. USA. 1984;81:2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang T, Shaka A. Water suppression that works. Excitation sculpting using arbitrary waveforms and pulsed-field gradients. J. Magn. Reson. 1995;112:275–279. [Google Scholar]
- 20.Jiang L, Majumdar A, Hu W, Jaishree TJ, Xu W, Patel DJ. Saccharide-RNA recognition in a complex formed between neomycin B and an RNA aptamer. Structure. 1999;7:817–827. doi: 10.1016/s0969-2126(99)80105-1. [DOI] [PubMed] [Google Scholar]
- 21.Bernacchi S, Freisz S, Maechling C, Spiess B, Marquet R, Dumas P, Ennifar E. Aminoglycoside binding to the HIV-1 RNA dimerization initiation site: thermodynamics and effect on the kissing-loop to duplex conversion. Nucleic Acids Res. 2007;35:7128–7139. doi: 10.1093/nar/gkm856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pilch DS, Kaul M, Barbieri CM, Kerrigan JE. Thermodynamics of aminoglycoside-rRNA recognition. Biopolymers. 2003;70:58–79. doi: 10.1002/bip.10411. [DOI] [PubMed] [Google Scholar]
- 23.Stampfl S, Lempradl A, Koehler G, Schroeder R. Monovalent ion dependence of neomycin B binding to an RNA aptamer characterized by spectroscopic methods. Chembiochem. 2007;8:1137–1145. doi: 10.1002/cbic.200700030. [DOI] [PubMed] [Google Scholar]
- 24.Duchardt-Ferner E, Weigand JE, Ohlenschlager O, Schmidtke SR, Suess B, Wohnert J. Highly modular structure and ligand binding by conformational capture in a minimalistic riboswitch. Angew. Chem. Int. Ed. Engl. 2010;49:6216–6219. doi: 10.1002/anie.201001339. [DOI] [PubMed] [Google Scholar]
- 25.Furtig B, Richter C, Wohnert J, Schwalbe H. NMR spectroscopy of RNA. Chembiochem. 2003;4:936–962. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- 26.Ohlenschlager O, Wohnert J, Bucci E, Seitz S, Hafner S, Ramachandran R, Zell R, Gorlach M. The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf RNA and its interaction with the proteinase 3C. Structure. 2004;12:237–248. doi: 10.1016/j.str.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 27.Schmidtke SR, Duchardt-Ferner E, Weigand JE, Suess B, Wohnert J. NMR resonance assignments of an engineered neomycin-sensing riboswitch RNA bound to ribostamycin and tobramycin. Biomol. NMR Assign. 2010;4:115–118. doi: 10.1007/s12104-010-9223-z. [DOI] [PubMed] [Google Scholar]
- 28.Cooper A, Johnson CM, Lakey JH, Nollmann M. Heat does not come in different colours: entropy-enthalpy compensation, free energy windows, quantum confinement, pressure perturbation calorimetry, solvation and the multiple causes of heat capacity effects in biomolecular interactions. Biophys. Chem. 2001;93:215–230. doi: 10.1016/s0301-4622(01)00222-8. [DOI] [PubMed] [Google Scholar]
- 29.Moll D, Schweinsberg S, Hammann C, Herberg FW. Comparative thermodynamic analysis of cyclic nucleotide binding to protein kinase A. Biol. Chem. 2007;388:163–172. doi: 10.1515/BC.2007.018. [DOI] [PubMed] [Google Scholar]
- 30.Serganov A. The long and the short of riboswitches. Curr. Opin. Struct. Biol. 2009;19:251–259. doi: 10.1016/j.sbi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu. Rev. Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 32.Schwalbe H, Buck J, Furtig B, Noeske J, Wohnert J. Structures of RNA switches: insight into molecular recognition and tertiary structure. Angew. Chem. Int. Ed. Engl. 2007;46:1212–1219. doi: 10.1002/anie.200604163. [DOI] [PubMed] [Google Scholar]
- 33.Krstic I, Frolow O, Sezer D, Endeward B, Weigand JE, Suess B, Engels JW, Prisner TF. PELDOR spectroscopy reveals preorganization of the neomycin-responsive riboswitch tertiary structure. J. Am. Chem. Soc. 2010;132:1454–1455. doi: 10.1021/ja9077914. [DOI] [PubMed] [Google Scholar]
- 34.Klinkert B, Narberhaus F. Microbial thermosensors. Cell. Mol. Life Sci. 2009;66:2661–2676. doi: 10.1007/s00018-009-0041-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.