Abstract
In eukaryotes, the transcription of tRNA genes is initiated by the concerted action of transcription factors IIIC (TFIIIC) and IIIB (TFIIIB) which direct the recruitment of polymerase III. While TFIIIC recognizes highly conserved, intragenic promoter elements, TFIIIB binds to the non-coding 5′-upstream regions of the tRNA genes. Using a systematic bioinformatic analysis of 11 multicellular eukaryotic genomes we identified a highly conserved TATA motif followed by a CAA-motif in the tRNA upstream regions of all plant genomes. Strikingly, the 5′-flanking tRNA regions of the animal genomes are highly heterogeneous and lack a common conserved sequence signature. Interestingly, in the animal genomes the tRNA species that read the same codon share conserved motifs in their upstream regions. Deep-sequencing analysis of 16 human tissues revealed multiple splicing variants of two of the TFIIIB subunits, Bdp1 and Brf1, with tissue-specific expression patterns. These multiple forms most likely modulate the TFIIIB–DNA interactions and explain the lack of a uniform signature motif in the tRNA upstream regions of animal genomes. The anticodon-dependent 5′-flanking motifs provide a possible mechanism for independent regulation of the tRNA transcription in various human tissues.
INTRODUCTION
The genetic code is degenerate and the full set of the 20 naturally occurring amino acids (except for Met and Trp) are decoded by multiple codons. Theoretically, a set of tRNA species that decode each codon individually can translate the entire genetic information from all mRNAs in a cell. Considering the wobbling effect by which one tRNA species can recognize more than one codon, the minimal tRNA set required for translation would be approximately 30 species. In bacterial genomes, the number of tRNA genes ranges from 29 to 120 genes. The low number of tRNA genes in these organisms has probably resulted from genome minimization favoring rapid replication and relatively simple regulation (1). The number of the tRNA genes increases parallel with the increase of the complexity of the organisms. The genomes of higher eukaryotes encode several distinct tRNA species (isoacceptors) to read the 61 sense codons, which also differ in their copy number among the genomes (2). A significant fraction of the tRNAs represents also species that bear the same anticodon but differ in the sequence elsewhere in the tRNA body and are defined as isodecoders (3). According to the recent version of the genomic tRNA database (November 2010) (4,5), 506 tRNAs genes in human, 605 in Caenorhabditis elegans, 430 in mouse and 630 in Arabidopsis thaliana decode the standard 20 amino acids. Systematic analysis of the tRNA composition in human tissues has revealed that the concentration of tRNA isoacceptors and isodecoders varies by as much as 10-fold among different tissues and stages of differentiations (2,3,6–8) and is therefore unlikely to be driven by neutral genetic drift. The mature tRNAs are very stable, total tRNA levels likely reflect the tRNA transcription rates (9–11). The tRNA composition differs so that it mirrors the expression needs of the cell: some tRNA species can be upregulated to meet the larger demand for some specific tRNAs by overproduction of proteins with overrepresented codons or by enhanced cell proliferation in cancer cells (6–8). Clearly, the eukaryotic genomes encode a complex tRNA pool; however, the expression of the tRNA species is not random and is subject to tight regulation. Transcriptional control of the tRNA genes might play a role in the development and cell differentiation of tissues of higher eukaryotes (12,13). The information embedded in the genome to maintain the differential regulation and transcription of the tRNA genes remains elusive.
In eukaryotes, the tRNA genes are transcribed by RNA polymerase III (pol III) [reviewed in (14–16)] and transcription is primarily dependent on intragenic promoter elements. The highly conserved A and B boxes of tRNA genes (coding for the D- and TΨC-stem loops) serve as internal promoters to which the transcription factor IIIC (TFIIIC) binds (17). TFIIIC-binding is followed by a recruitment of a second transcription factor, TFIIIB, which recognizes upstream region of the tRNA genes and directs the recruitment of pol III (14). TFIIIB is a multisubunit complex consisting of pol III-specific B-double prime 1 (Bdp1), TFIIB-related factor 1 (Brf1) and TATA-binding protein (TBP) (18). In vertebrates, TFIIIB exists in two isoforms: TFIIIBα (composed of Bdp1, Brf2 and TBP), which is specialized to transcribe genes with promoter elements upstream of the initiation site, e.g. U6-type RNA pol III promoters (19), and TFIIIBβ (comprising Bdp1, Brf1 and TBP), which is required for transcription of genes with internal promoter elements, e.g. tRNA and 5S RNA pol III promoters (20,21). TBP is a shared element with the polymerase II (pol II) and recognizes pol II and pol III-specific transcription factors through different regions of its surface (22). TBP occupies a key position in the TFIIIB: analogous to its function in the pol II promoters, TBP contacts the TATA-like elements in the promoter sequences and through distortion of these segments strengthens the binding of the transcription factor complex (23) Brf1 plays an integral role for the functional stability of the TFIIIB complex by interacting with both, TBP and Bdp1 [interactions are reviewed in (14)]. The third component, Bdp1, determines the physical stabilities of the TFIIIB complex by modulating the DNA deformation in sequence-independent fashion (24). A cDNA library derived from human breast cancer cells reveals a presence of several active Brf1 isoforms which are generated by alternative splicing and are suggested to serve as molecular adaptors that link the TFIIIB to the DNA-recognition complexes (25). Similarly, various Bdp1 forms with different molecular mass and TFIIIC activity have been isolated from human embryonic kidney cells (26) which are most likely a result of an alternative splicing (27).
TATA-box-like elements and TA-rich sequences are present in the upstream regions of some tRNA genes usually between positions −34 and −29 (numbered from the 5′-end of the mature tRNA) (28,29). TFIIIB can directly be recruited to a typical TATA box even in the absence of TFIIIC and independently initiate transcription by pol III recruitment in yeast (30,31). In silkworms, TFIIIB binds stably to the promoter sequence, but is transcriptionally incompetent in the absence of TFIIIC (32). Importantly, TA-enriched regions located immediately upstream of the tRNA coding sequence (within the first 50 nt) enhance transcription, whereas multiple copies of such sequences in the far upstream regions reduce the tRNA transcription levels; sequestration of the TFIIIB complex at multiple binding sites decreases its effective concentration thus reducing the formation of the initiation complex with pol III (32). In addition, the regions upstream of the 5′-initial nucleotide of the mature tRNAs in plant and yeast (usually at positions −7 to −3) bear another conservative CAA motif which together with the TATA boxes cooperatively enhances transcription (28,33). Not all active tRNA genes, however, have this promoter structure. In general, the average proportion of AT-nucleotides in the promoter region is very high in plants (72.5%) and yeast (69.4%) compared to mammals (52.5%) (28). In a set of human (302 species) and mouse (267 species) tRNAs, the upstream region lacks TA-rich motifs (34). Similarly, the analysis of 267 species of Drosophila melanogaster did not reveal the typical TATA box which is most likely due to the replacement of TBP with the transcription factor TRF in this organism (35). Taken together, these data imply that sequences outside the tRNA coding regions are essential for transcription and suggest that they might play a crucial role in regulating the tRNA transcription.
Taking advantage of the large number of sequenced and annotated genomes, we have undertaken a systematic analysis of the upstream promoter regions of tRNA genes in several multicellular eukaryotic genomes. Our data revealed a clear difference in the conserved motifs located upstream of the transcription initiation site of plant versus animal tRNAs. Using deep-sequencing data of the transcriptome of 16 human tissues, we identified multiple splicing variants of Bdp1 and Brf1 with a tissue-specific expression pattern. We explain the lack of a common conserved motif in the upstream promoter region of animal tRNAs by the presence of different splice forms of Bdp1 and Brf1, which through their structural variations may differently modulate the TFIIIB–DNA interactions. This finding suggests that distinct mechanisms in the regulation of the tRNA transcription exist in plant and animal genomes.
MATERIALS AND METHODS
tRNA information and genome sequences
The tRNA genes encoded in the nuclear genomes were extracted from the genomic tRNA database (gtRNAdb, http://gtrnadb.ucsc.edu/) (4,5). Pseudo tRNA genes were excluded from the analysis. Note that the tRNA genes encoded in mitochondrial and chloroplast genomes were not included in the search. The following genomes (release version and date are indicated) were used: Homo sapiens (human; hg19, NCBI build 37.1, February 2009), Mus musculus (mouse; mm9, July 2007), Rattus norvegicus (rat; rn4, November 2004), Gallus gallus (galGal3, May 2006), D. melanogaster (release 5, April 2006), Anopheles gambiae (anoGam1, February 2003); C. elegans (ce4, January 2007), A. thaliana (February 2004); Medicago truncatula (March 2009), Populus trichocarpa (version 2.0, January 2010), Glycine max (soybean; glyma 1.0, December 2008).
Statistical analysis of the upstream sequences of the tRNA genes
Motif ‘word’ search
The upstream sequences of the tRNA genes (nucleotide positions −100 to −1 nt; numbered from the first 5′-nt of the mature tRNA) were retrieved from the genomic tRNA data base and a simple ‘word’ search to extract a conserved sequence using the algorithm originally proposed by Waterman and Jones (36) and implicated for the search of consensus motifs in the upstream regions of yeast tRNAs (34). The 100 nt-long upstream sequences were screened for the frequency of occurrence of 6-nt ‘word’ comprising all possible combinations of 6 nt, thereby allowing none or one mismatch within a 9-nt sliding window. The ‘word’ length was set to 6 nt which is the common length of transcription factor binding sites in eukaryotes (37–39) and has been used in sequence analysis of transcription factor binding motifs elsewhere (34,40). The ‘word’ with the highest frequency is identified as the best frequency ‘word’ for each genome. To avoid a bias toward the highly homologous sequences of mature tRNAs, the word search was terminated at position −9. To sort out significant motifs (peaks), we set a threshold which is defined as already described (34). Briefly, the individual sequences of the upstream region for each organism were randomized and the mean, minimum and maximum values occurring within this randomized data set were calculated to determine the random noise range.
Elicitation of conserved motifs via MEME
To identify conserved motifs in the upstream sequences of tRNA genes, we used the MEME version 4.4.0 (available online at http://meme.sdsc.edu/meme4_4_0/cgi-bin/meme.cgi) (41). The parameters were set as motif minimum width of 6, maximum width of 10, max number of motifs of 3 or 5 and the option ‘zero or one per sequence’.
Deep-sequencing data sets
The deep-sequencing data sets of poly(A)+-selected mRNAs of 16 human tissues and organs were analyzed as a part of the ‘Human Body Map 2.0′ project and the sequencing was performed using an Illumina HiSeq 2000 platform with an error rate of 0.2–0.6%. The sequencing reads with a length of 75 nt were kindly provided by Gary Schroth and his group at Illumina Inc.
RESULTS
Conserved motifs are present in the upstream sequences of plant tRNA genes but are lacking in the animal genomes
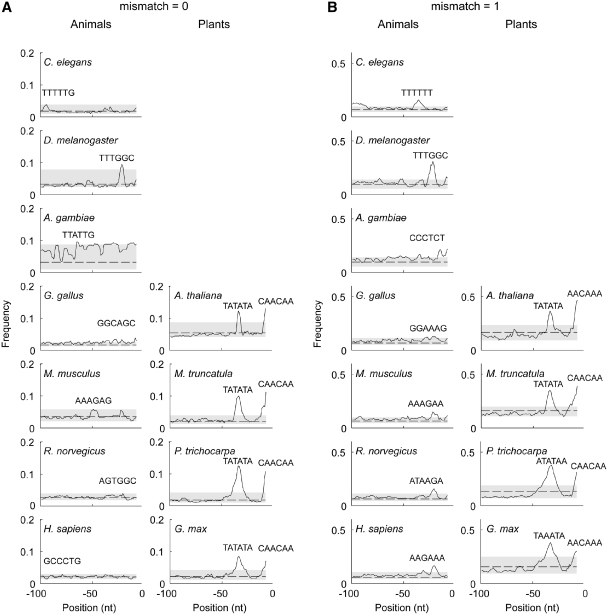
TFIIIB recognizes conserved TATA-box-like elements in the upstream regions of the tRNA genes in plants and yeast through its TBP subunit (28,34,42). To address whether a conserved sequence motif is a characteristic feature of the upstream sequences of all eukaryotic tRNAs we performed a ‘word’ search within the 100 upstream nucleotides of the tRNA genes and compared seven animal with four plant genomes (Figure 1). A significant peak around position −35 with a TATA motif was detected in all plant genomes as reported already for A. thaliana and S. cerevisiae (28,34,42,43). In contrast, the upstream tRNA regions of the seven animal genomes did not reveal any uniform and conserved motif (Figure 1); motifs with rather individual signature for each genome were identified and the sequence and position of the motifs was dependent on the search settings (Figure 1; compare the outcome of the search with none or one nucleotide mismatch). Allowing 1 nt mismatch in the search yielded slightly more significant peaks positioned at ∼−40 to −10 nt upstream of the 5′-initial nucleotide of the mature tRNA in all seven animal genomes (Figure 1B). Extending the search up to 2000 nt upstream did not reveal any further conserved sequences in the animal genomes (Supplementary Figure S1). The height of the identified highest peaks in animal genomes is one-third of those specified in the plant genomes. An exception among the animal genomes is D. melanogaster: independent of the search parameters (none or one mismatch) a clear 6-nt motif (TTTGGC) at position −22 was detected.
Figure 1.
Conserved patterns are only present in the upstream sequences of plant tRNA genes. The best frequency ‘word’ search was performed in the 5′-region (positions −100 to −9 nt) upstream of the initial nucleotide of the mature nuclear-encoded tRNAs of different organisms allowing none (A) or one mismatch (B) (for details see ‘Materials and Methods’ section). The occurrence of the most frequent six-letter ‘word’ at each position (x-axis) is plotted on the y-axis. Position 0 corresponds by the first nucleotide of the mature tRNA. The six-letter ‘word’ with the highest frequency (highest peak) is indicated above the corresponding peak. The CAA-motifs immediately upstream of the 5′-initial nucleotide of the mature tRNAs are also depicted. The dashed line represents the average frequency value obtained from randomized sequences. The shadowed area indicates the area between the maximum and minimum frequency values derived from the randomized data set. nt, nucleotide.
Furthermore, the ‘word’ search revealed an interesting feature in the plant genomes: a significant CAA motif was detected at positions −7 to −3 upstream of the 5′-initial nucleotide of the mature tRNAs (Figure 1). The signal is completely absent in all analyzed animal genomes. The distance between the processed 5′-end of the mature tRNA and the transcription initiation site is usually 9–12 bases [G. Kassavetis, a supplement article to (44)] and conserved motifs in these segments play a central role in transcriptional initiation (33), particularly in plants (28,43). Interestingly, the same motif was identified in the unicellular yeast S. cerevisiae while it was absent in Schizosaccharomyces pombe (Supplementary Figure S2). The best six-letter ‘word’ identified in S. cerevisiae was the TATA motif which resembled the sequence identified in the non-coding upstream regions of the plant tRNA genes (Supplementary Figure S2). Notably, in the two yeast species, the motifs with the highest frequencies centered at positions identical to those identified for the plant tRNA upstream regions (Figure 1).
To further verify the presence of a conserved motif signature within the upstream sequences of the tRNA genes, we used the algorithm introduced by Bailey and Elkan (41). Similar to the ‘word’ search, this algorithm validated the presence of the TA-rich motif in the upstream sequences of plant tRNA genes, whereas more variable GC-rich sequences were extracted from the animal genomes (Supplementary Figure S3). Taken together, these analyses suggest that distinct patterns of regulation of tRNA transcription in animal and plant genomes may exist.
Animal tRNA genes share common motifs in the upstream regulatory regions in an anticodon-dependent manner
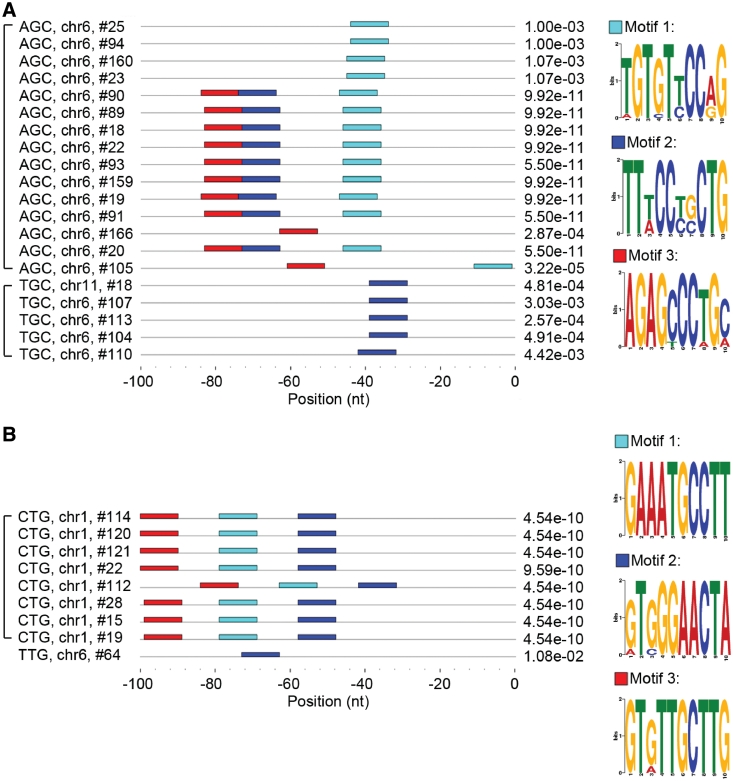
Since we could not identify a common, conserved motif in the upstream segments of animal tRNA genes, we next analyzed separately the isoacceptor and isodecoder tRNAs that read codons encoding the same amino acid. Importantly, this search extracted anticodon-dependent motifs in the upstream segments of the majority of the tRNA families (Figure 2 and Supplementary Figure S4) in all tested animal genomes (Table 1 and Supplementary Table S1); however, no such dependence was observed for any of the plant genomes. In general, this trend is slightly more prominent in the genomes of higher vertebrates than those of lower, invertebrate animals. Comparison of the tRNA species pairing to the codons for alanine (AGC, TGC and CGC are the corresponding anticodons) revealed a unique TGTGTtCCaG motif for Ala-tRNAs with the AGC anticodon and a characteristic TTtCCtgCTG motif for the Ala-tRNAs with the TGC anticodon (Figure 2A). The TTtCCtgCTG motif centers at position −40 to −30 upstream of the 5′-site of the mature Ala-tRNAs with the TGC anticodon; it is also present within the 5′-upstream flanking region of the Ala-tRNAs that contain the AGC anticodon albeit at a different position (Figure 2A). In contrast, the upstream regions of the third type of Ala-tRNAs bearing the CGC anticodon do not have any motif in common. Another example, i.e. the genes for Gln-tRNA, showed a more complex pattern: tRNAs bearing the CTG anticodon share three different motifs GAAATGCCTT, gTgGGAACTA, GTgTTGCTTG, whereas Gln-tRNAs with the TTG anticodon possess only the gTgGGAACTA motif which is also present in Gln-tRNAs with the CTG anticodon (Figure 2B). The number of the common motifs varies among the tRNA species harboring the same anticodon; common motifs are also shared with other tRNAs bearing the same amino acid but pairing to another, alternative codon (Supplementary Figure S4A). Next to Trp and Met which are encoded by a single codon, due to the wobbling some amino acids, albeit encoded by different codons, are decoded by tRNA species bearing one and the same anticodon. Interestingly, these tRNA families also share one to three common motifs (Supplementary Figure S4B). Different combinations among the common motifs divide the tRNA families into subgroups suggesting also a possibility for differential regulation of the expression even among the tRNA families bearing the same anticodon. The variations in the significance value (P-value) imply that the homologous motifs observed within a tRNA family bearing the same anticodon do not originate from simple duplications of the tRNA genes (Figure 2 and Supplementary Figure S4).
Figure 2.
Animal tRNA genes with the same anticodon bear conserved motifs in their upstream regions. Examples of typical anticodon-dependent motifs in the upstream sequences of the human tRNAs encoding Ala (A) and Gln (B) identified with MEME 4.4. Each tRNA is specified by its anticodon, chromosome location and serial chromosome number (data were retrieved from the genomic tRNA database). The square brackets on the left group the tRNAs with the same anticodon. Each horizontal line represents the 100-nt-long region upstream of the 5′-end of the mature tRNA genes; position 0 determines the first nucleotide of the mature tRNA. The positions of the three most significant motifs are colored (cyan, blue and red) and the corresponding sequence logos are presented on the right side of each group. The numbers on the right side represent the combined P-value and are the product of the P-values of all motifs detected within the upstream sequence; they are inversely proportional to the significance of the motifs. Motifs with P < 0.05 were considered as significant.
Table 1.
Statistical analysis of anticodon-dependent motifs of animal genomes
| Genomea | Number of amino acids with anticodon- dependent upstream tRNA motif | Number of amino acids without anticodon- dependent upstream tRNA motif |
|---|---|---|
| Homo sapiens (human) | 10 | 4 |
| Rattus norvegicus (rat) | 8 | 5 |
| Mus musculus (mouse) | 8 | 4 |
| Gallus gallus (hen) | 9 | 5 |
| Drosophila melanogaster | 5 | 7 |
| Anopheles gambiae | 5 | 7 |
| Caenorhabditis elegans | 8 | 5 |
aA detailed list of the motifs within the tRNA upstream sequences dependent on the anticodon is given in Supplementary Table S1. Note that amino acids decoded by tRNAs with one and the same anticodon are not included in the table.
Tissue-specific expression patterns of TFIIIB isoforms may explain the anticodon-specific regulation of tRNA expression
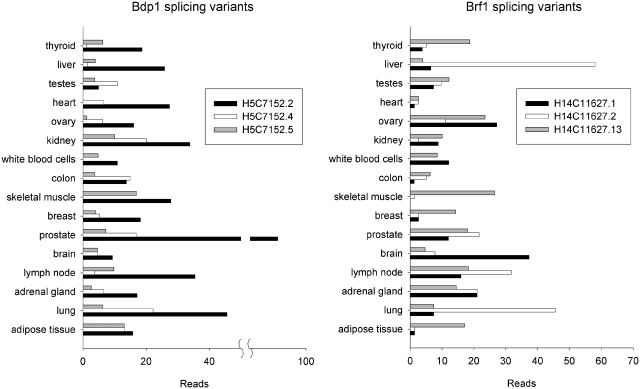
TFIIIB is the essential factor that recognizes specific motifs in the upstream sequences of tRNA genes [preferably TA-rich regions in plants (28,43) and yeast cells (42)] and its binding stability to the cognate motif determines the expression level within one family of isodecoders (32). In human cells, expression of the genes encoding the three subunits of the TFIIIB complex, Brf1, Bdp1 and TBP, is either coordinated (45) or differentially modulated (46,47). The human BRF1 and BDP1 genes encode various isoforms of considerably different molecular masses [UniProt database (http://www.uniprot.org/)] that result from alternative splicing [http://genome.ewha.ac.kr/ECgene/ (48)] (Supplementary Table S2). We hypothesized that the anticodon-dependent motifs detected in the tRNA families of the animal genomes might bear the information for differential regulation of the expression of the tRNA sets for each codon in different tissues. This, in turn, would require a differential expression of the TFIIIB isoforms in various tissues and cell types. We tested this hypothesis using high-throughput deep-sequencing data of total mRNA isolated from various human tissues. To search for specific splice variants of Brf1 and Bdp1, we aligned all deep-sequencing reads (75 nt) to the specific junction of the target splicing variants. Only reads that aligned to at least 10 nt upstream and downstream of the splice junction (allowing thereby a maximum of one mismatch) were recorded as hits. The read hits were then normalized to the total read count of the data sets to calculate the relative concentrations of the different isoforms (Figure 3 and Supplementary Figure S5). Intriguingly, various Bdp1 and Brf1 isoforms are differentially enriched in different tissues so that a clear tissue-specific pattern emerges. For example, the H5C7152.4 isoform of Bdp1 dominates in the thyroid, whereas the isoform H5C7152.2 is the main species in the liver and prostate gland (Figure 3 and Supplementary Figure S5). Liver and lung tissues are enriched in the H14C11627.2 isoform of Brf1 and H14C11627.13 is the dominating isoform in the skeletal muscle (Figure 3 and Supplementary Figure S5). Variations in the expression of the isoforms in each tissue suggest that they may modulate the TFIIIB-binding affinity to various promoter elements and thus may serve as a regulatory tool to modulate the tRNA expression levels of the isodecoders or isoacceptors in each tissue.
Figure 3.
Various RNA splice variants of BDP1 and BRF1 genes are expressed in different human tissues. Normalized read hits for specific splicing junctions of three Bdp1 and Brf1 isoforms in 16 human tissues. All read hits were normalized to 100 million total sequence reads. Numbering of the splice variants is according to the ECgene alternative splicing database (Supplementary Table S2). The expression of other isoforms is presented in Supplementary Figure S5.
DISCUSSION
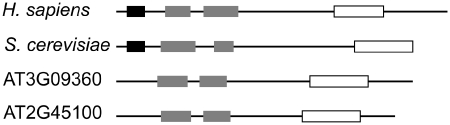
We have compared on genome-wide scale the conserved signatures within the upstream regions of nuclear tRNAs from 11 different multicellular genomes. The upstream segments of plant tRNAs showed a conserved TA-enriched motif, followed by a CAA motif immediately upstream of the 5′-initial nucleotide of the mature tRNA transcript. Similar motifs were identified in the upstream regions of yeast tRNAs [Supplementary Figure S2 and (29)]. In contrast, the 5′-flanking regions of the tRNAs from animal genomes are highly heterogeneous and vary significantly among the different species. This observation was surprising to us as TBP is present as a subunit of both, plant and animal TFIIIB complexes. TBP recognizes TATA-like elements in the promoter regions; however, the entry of Brf1 and Bdp1 into the TFIIIB complex alters the dynamics of TBP–DNA interactions (24). For example, the sequence preference of the assembled TFIIIB complex with a mutant TBP differs significantly from the mutant TBP alone (49). The crystal structure of the yeast TFIIIB complex shows that the convex surface of the TBP is extensively engaged in exceptionally extended contacts with the Brf1 subunit which are three to five times greater than the contacts observed in the pol II complexes: a total of 31 Brf1 amino acid residues interact with 29 residues of TBP through hydrogen bonds and van der Waals contacts (22). Based on sequence conservation between human and yeast Brf1, it can be predicted that 28 of the 31 hydrogen bonds observed in the yeast complex TBP-Brf1 are retained (22) suggesting also a large interaction interface between Brf1 and TBP in the human TFIIIB complex. In addition, Brf1 is structurally flexible and undergoes large unfolding transitions by complex formation with TBP (22). The primary sequence variations in the human Brf1 and Bdp1 isoforms may introduce some structural variations among them, which consequently may differently modulate the TFIIIB–DNA interactions and consequently the interactions with TFIIIC. Consistent with this interpretation, isolated forms of Bdp1 with molecular masses of 220 and 250 kDa show higher TFIIIC activity compared to the smaller Bdp1 forms with apparent masses of 150, 100 and 90 kDa (26). In the animal genomes, the presence of multiple isoforms of Bdp1 and Brf1 correlates with the lack of a uniform signature in the upstream regions of the tRNA genes suggesting that different Brf1 and Bdp1 splice forms may differently modulate the specificity and dynamics of TFIIIB-promoter interactions. A genome-wide search in the A. thaliana genome revealed a single, uncharacterized gene (AT4G39160) which possesses limited overall sequence homology to human and yeast Bdp1 (below 9% amino acid similarity). A fragment of it, compassing the phylogenetically most conserved region of Bdp1, the SANT domain (14), showed higher amino acid similarity to the human and yeast Bdp1 proteins (55–63% amino acid similarity). Currently, however, there is no evidence for a contribution of this protein to the tRNA transcription initiation in plants. Two genes, which might be homologous to the Brf1 were identified in Arabidopsis (Figure 4). Note, that so far no splice variants have been identified for Brf1-like transcripts in plants.
Figure 4.
Putative plant homologues of Brf1 share some conservative elements with other eukaryotic Brf1 proteins. Homology searches with the S. cerevisiae and H. sapiens Brf1 splice variant H14C11627.9 (Supplementary Table S2) identified two putative homologues, AT3G09360 and AT2G45100 from A. thaliana. The conserved motifs derived from the sequence alignment (Supplementary Figure S6) are depicted in boxes: gray—cyclin fold boxes, white—Brf1-like TBP-binding domain. Both Arabidopsis proteins do not possess the conserved TFIIB zinc-binding domain (black box). Note that each conserved segment is scaled to the position within the corresponding nucleotide sequence (black line).
The genomic sequence database is updated frequently based on the higher accuracy of the new sequencing technologies and improved genome annotations. Consequently, the automated annotation of tRNA genes increases rapidly (5). For example, 446 tRNA genes were identified in the human genome in 2006 (2); this number increased to 506 species and additional 110 pseudogenes in 2009 (hg19, NCBI Build 37.1, February 2009). The differential expression of tRNA genes in a tissue- and developmental stage-specific manner (6,8) requires a sophisticated mode to control the transcription. Degenerate regulatory information embedded in the upstream non-coding sequences of the tRNA genes would reduce the complexity of regulation, which may result in lower metabolic costs and thus provide an evolutionary advantage. This may explain the presence of conserved signature motifs in the flanking 5′-upstream regions of tRNA genes bearing the same anticodon. The presence of more than one motif for the majority of the tRNAs with the same anticodon suggests a complex regulation mechanism of transcription. Importantly, even within these groups of tRNAs, the number of the conserved motifs varies (Figure 2A, Supplementary Figure S4) implying a fine-tuning function of some of the motifs. The different motifs might have evolved to bind various TFIIIB isoforms coexisting in some tissues; however, in different concentration (Figure 3 and Supplementary Figure S5), so that differential expression of a tRNA species bearing one and the same codon can be achieved in a tissue-specific manner. The new information, we present here on the distinct modes of regulation of tRNA transcription in plants and animals provides an excellent cross-validation with the wealth of experimental data in plants and yeast and presents new twist to understand variations in the tRNA regulation in multicellular eukaryotic organisms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: The Deutsche Forschungsgemeinschaft (grant IG 73/10-1 to Z.I.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
We are grateful to Gary Schroth and his group at Illumina for providing the deep-sequencing data sets from 16 the human tissues and organs.
REFERENCES
- 1.Sharp PM, Bailes E, Grocock RJ, Peden JF, Sockett RE. Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res. 2005;33:1141–1153. doi: 10.1093/nar/gki242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodenbour JM, Pan T. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res. 2006;34:6137–6146. doi: 10.1093/nar/gkl725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geslain R, Pan T. Functional analysis of human tRNA isodecoders. J. Mol. Biol. 2010;396:821–831. doi: 10.1016/j.jmb.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hentzen D, Chevallier A, Garel JP. Differential usage of iso-accepting tRNASer species in silk glands of Bombyx mori. Nature. 1981;290:267–269. doi: 10.1038/290267a0. [DOI] [PubMed] [Google Scholar]
- 8.Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37:7268–7280. doi: 10.1093/nar/gkp787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Czech A, Fedyunin I, Zhang G, Ignatova Z. Silent mutations in sight: co-variations in tRNA abundance as a key to unravel consequences of silent mutations. Mol. Biosyst. 2010;6:1767–1772. doi: 10.1039/c004796c. [DOI] [PubMed] [Google Scholar]
- 10.Komar AA. A pause for thought along the co-translational folding pathway. Trends Biochem. Sci. 2009;34:16–24. doi: 10.1016/j.tibs.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Zhang G, Fedyunin I, Miekley O, Valleriani A, Moura A, Ignatova Z. Global and local depletion of ternary complex limits translational elongation. Nucleic Acids Res. 2010;38:4778–4787. doi: 10.1093/nar/gkq196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouyang C, Martinez MJ, Young LS, Sprague KU. TATA-Binding protein-TATA interaction is a key determinant of differential transcription of silkworm constitutive and silk gland-specific tRNA(Ala) genes. Mol. Cell. Biol. 2000;20:1329–1343. doi: 10.1128/mcb.20.4.1329-1343.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reynolds WF. Developmental stage-specific regulation of Xenopus tRNA genes by an upstream promoter element. J. Biol. Chem. 1995;270:10703–10710. doi: 10.1074/jbc.270.18.10703. [DOI] [PubMed] [Google Scholar]
- 14.Geiduschek EP, Kassavetis GA. The RNA polymerase III transcription apparatus. J. Mol. Biol. 2001;310:1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- 15.Huang Y, Maraia RJ. Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human. Nucleic Acids Res. 2001;29:2675–2690. doi: 10.1093/nar/29.13.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paule MR, White RJ. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galli G, Hofstetter H, Birnstiel ML. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981;294:626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- 18.Kassavetis GA, Braun BR, Nguyen LH, Geiduschek EP. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- 19.Teichmann M, Seifart KH. Physical separation of two different forms of human TFIIIB active in the transcription of the U6 or the VAI gene in vitro. EMBO J. 1995;14:5974–5983. doi: 10.1002/j.1460-2075.1995.tb00286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schramm L, Pendergrast PS, Sun Y, Hernandez N. Different human TFIIIB activities direct RNA polymerase III transcription from TATA-containing and TATA-less promoters. Genes Dev. 2000;14:2650–2663. doi: 10.1101/gad.836400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teichmann M, Wang Z, Roeder RG. A stable complex of a novel transcription factor IIB- related factor, human TFIIIB50, and associated proteins mediate selective transcription by RNA polymerase III of genes with upstream promoter elements. Proc. Natl Acad. Sci. USA. 2000;97:14200–14205. doi: 10.1073/pnas.97.26.14200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juo ZS, Kassavetis GA, Wang J, Geiduschek EP, Sigler PB. Crystal structure of a transcription factor IIIB core interface ternary complex. Nature. 2003;422:534–539. doi: 10.1038/nature01534. [DOI] [PubMed] [Google Scholar]
- 23.Patikoglou G, Burley SK. Eukaryotic transcription factor-DNA complexes. Annu. Rev. Biophys. Biomol. Struct. 1997;26:289–325. doi: 10.1146/annurev.biophys.26.1.289. [DOI] [PubMed] [Google Scholar]
- 24.Grove A, Kassavetis GA, Johnson TE, Geiduschek EP. The RNA polymerase III-recruiting factor TFIIIB induces a DNA bend between the TATA box and the transcriptional start site. J. Mol. Biol. 1999;285:1429–1440. doi: 10.1006/jmbi.1998.2347. [DOI] [PubMed] [Google Scholar]
- 25.McCulloch V, Hardin P, Peng W, Ruppert JM, Lobo-Ruppert SM. Alternatively spliced hBRF variants function at different RNA polymerase III promoters. EMBO J. 2000;19:4134–4143. doi: 10.1093/emboj/19.15.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weser S, Gruber C, Hafner HM, Teichmann M, Roeder RG, Seifart KH, Meissner W. Transcription factor (TF)-like nuclear regulator, the 250-kDa form of Homo sapiens TFIIIB”, is an essential component of human TFIIIC1 activity. J. Biol. Chem. 2004;279:27022–27029. doi: 10.1074/jbc.M312790200. [DOI] [PubMed] [Google Scholar]
- 27.Kelter AR, Herchenbach J, Wirth B. The transcription factor-like nuclear regulator (TFNR) contains a novel 55-amino-acid motif repeated nine times and maps closely to SMN1. Genomics. 2000;70:315–326. doi: 10.1006/geno.2000.6396. [DOI] [PubMed] [Google Scholar]
- 28.Yukawa Y, Sugita M, Choisne N, Small I, Sugiura M. The TATA motif, the CAA motif and the poly(T) transcription termination motif are all important for transcription re-initiation on plant tRNA genes. Plant J. 2000;22:439–447. doi: 10.1046/j.1365-313x.2000.00752.x. [DOI] [PubMed] [Google Scholar]
- 29.Dieci G, Yukawa Y, Alzapiedi M, Guffanti E, Ferrari R, Sugiura M, Ottonello S. Distinct modes of TATA box utilization by the RNA polymerase III transcription machineries from budding yeast and higher plants. Gene. 2006;379:12–25. doi: 10.1016/j.gene.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 30.Gerlach VL, Whitehall SK, Geiduschek EP, Brow DA. TFIIIB placement on a yeast U6 RNA gene in vivo is directed primarily by TFIIIC rather than by sequence-specific DNA contacts. Mol. Cell. Biol. 1995;15:1455–1466. doi: 10.1128/mcb.15.3.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kassavetis GA, Bardeleben C, Kumar A, Ramirez E, Geiduschek EP. Domains of the Brf component of RNA polymerase III transcription factor IIIB (TFIIIB): functions in assembly of TFIIIB-DNA complexes and recruitment of RNA polymerase to the promoter. Mol. Cell. Biol. 1997;17:5299–5306. doi: 10.1128/mcb.17.9.5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parthasarthy A, Gopinathan KP. Transcription of individual tRNA1Gly genes from within a multigene family is regulated by transcription factor TFIIIB. FEBS J. 2005;272:5191–5205. doi: 10.1111/j.1742-4658.2005.04877.x. [DOI] [PubMed] [Google Scholar]
- 33.Geiduschek EP, Tocchini-Valentini GP. Transcription by RNA polymerase III. Annu. Rev. Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- 34.Giuliodori S, Percudani R, Braglia P, Ferrari R, Guffanti E, Ottonello S, Dieci G. A composite upstream sequence motif potentiates tRNA gene transcription in yeast. J. Mol. Biol. 2003;333:1–20. doi: 10.1016/j.jmb.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 35.Takada S, Lis JT, Zhou S, Tjian R. A TRF1:BRF complex directs Drosophila RNA polymerase III transcription. Cell. 2000;101:459–469. doi: 10.1016/s0092-8674(00)80857-0. [DOI] [PubMed] [Google Scholar]
- 36.Waterman MS, Jones R. Consensus methods for DNA and protein sequence alignment. Methods Enzymol. 1990;183:221–237. doi: 10.1016/0076-6879(90)83016-3. [DOI] [PubMed] [Google Scholar]
- 37.Loots GG, Ovcharenko I, Pachter L, Dubchak I, Rubin EM. rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 2002;12:832–839. doi: 10.1101/gr.225502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bilu Y, Barkai N. The design of transcription-factor binding sites is affected by combinatorial regulation. Genome Biol. 2005;6:R103. doi: 10.1186/gb-2005-6-12-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Husbands A, Bell EM, Shuai B, Smith HM, Springer PS. Lateral organ boundaries defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 2007;35:6663–6671. doi: 10.1093/nar/gkm775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang G, Wang X, Wang Y, Yang JY, Li L, Nephew KP, Edenberg HJ, Zhou FC, Liu Y. Identification of transcription factor and microRNA binding sites in responsible to fetal alcohol syndrome. BMC Genomics. 2008;9(Suppl. 1):S19. doi: 10.1186/1471-2164-9-S1-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 42.Joazeiro CA, Kassavetis GA, Geiduschek EP. Alternative outcomes in assembly of promoter complexes: the roles of TBP and a flexible linker in placing TFIIIB on tRNA genes. Genes Dev. 1996;10:725–739. doi: 10.1101/gad.10.6.725. [DOI] [PubMed] [Google Scholar]
- 43.Yukawa Y, Mizutani T, Akama K, Sugiura M. A survey of expressed tRNA genes in the chromosome I of Arabidopsis using an RNA polymerase III-dependent in vitro transcription system. Gene. 2007;392:7–13. doi: 10.1016/j.gene.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 44.Bachman N, Eby Y, Boeke JD. Local definition of Ty1 target preference by long terminal repeats and clustered tRNA genes. Genome Res. 2004;14:1232–1247. doi: 10.1101/gr.2052904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong S, Johnson DL. The JNKs differentially regulate RNA polymerase III transcription by coordinately modulating the expression of all TFIIIB subunits. Proc. Natl Acad. Sci. USA. 2009;106:12682–12687. doi: 10.1073/pnas.0904843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cabarcas S, Jacob J, Veras I, Schramm L. Differential expression of the TFIIIB subunits Brf1 and Brf2 in cancer cells. BMC Mol. Biol. 2008;9:74. doi: 10.1186/1471-2199-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Athineos D, Marshall L, White RJ. Regulation of TFIIIB during F9 cell differentiation. BMC Mol. Biol. 2010;11:21. doi: 10.1186/1471-2199-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim P, Kim N, Lee Y, Kim B, Shin Y, Lee S. ECgene: genome annotation for alternative splicing. Nucleic Acids Res. 2005;33:D75–D79. doi: 10.1093/nar/gki118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsihlis ND, Grove A. The Saccharomyces cerevisiae RNA polymerase III recruitment factor subunits Brf1 and Bdp1 impose a strict sequence preference for the downstream half of the TATA box. Nucleic Acids Res. 2006;34:5585–5593. doi: 10.1093/nar/gkl534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.