Abstract
In most eukaryotes, telomeric DNA consists of repeats of a short motif that includes consecutive guanines and may hence fold into G-quadruplexes. Budding yeasts have telomeres composed of longer repeats and show variation in the degree of repeat homogeneity. Although telomeric sequences from several organisms have been shown to fold into G-quadruplexes in vitro, surprisingly, no study has been dedicated to the comparison of G-quadruplex folding and stability of known telomeric sequences. Furthermore, to our knowledge, folding of yeast telomeric sequences into intramolecular G-quadruplexes has never been investigated. Using biophysical and biochemical methods, we studied sequences mimicking about four repetitions of telomeric motifs from a variety of organisms, including yeasts, with the aim of comparing the G-quadruplex folding potential of telomeric sequences among eukaryotes. G-quadruplex folding did not appear to be a conserved feature among yeast telomeric sequences. By contrast, all known telomeric sequences from eukaryotes other than yeasts folded into G-quadruplexes. Nevertheless, while G3T1-4A repeats (found in a variety of organisms) and G4T2,4 repeats (found in ciliates) folded into stable G-quadruplexes, G-quadruplexes formed by repetitions of G2T2A and G2CT2A motifs (found in many insects and in nematodes, respectively) appeared to be in equilibrium with non-G-quadruplex structures (likely hairpin-duplexes).
INTRODUCTION
Telomeres are specialized nucleoprotein complexes that cap and protect the extremities of linear eukaryotic chromosomes. In most eukaryotes telomeric DNA consists of a tandem array of a short motif of 5–8 nt, that includes two, three or four consecutive guanines, with an impressive prevalence of the G3T2A motif (1).
The G3T2A motif is found in many phylogenetically distant organisms, including vertebrates (2,3), Trypanosoma brucei (4,5), several fungi (6), slime moulds (7,8) and several aquatic invertebrates (9). Variants of this motif are found in many other organisms: G3TA in the parasite Giardia intestinalis (10); G3T3A in a variety of plants (11), such as Arabidopsis thaliana (12); G3T4A in the unicellular green alga Chlamydomonas reinhardtii (13); G2T2A in the silkworm Bombyx mori and in many other insects (14); G2CT2A in the nematodes Ascaris lumbricoides (15), Caenorhabditis elegans and in other nematodes (16,17). Telomeric motifs bearing four consecutive guanines are found in ciliates: G4T2 in holotrichous ciliates such as Tetrahymena (18) and G4T4 in hypotrichous ciliates such as Oxytricha, Euplotes and Stylonychia (19). Degenerated telomeric motifs have been reported for several organisms: GGGTTTA and GGGTTCA in Plasmodium species (20,21); GGGTTTA and GGGTTAA in Lycopersicon esculentum (tomato plant) (22); GGGGTT and GGGTTT in Paramecium species (23,24), where a single telomerase RNA, encoding G4T2 repeats, has been identified (25).
Unlike in most eukaryotes, telomeres have rapidly evolved among budding yeasts; these organisms possess the most variable set of telomeric repeats (26,27). A number of budding yeast species have telomeres composed of long repeats (8–26 bp) and show variations in the degree of repeat homogeneity (28,29). The budding yeast Saccharomyces cerevisiae has short heterogeneous telomeric repeats of the general form TG2–3(TG)1–6 (30,31), encoded by a single telomerase RNA (32); the most abundant type of repeat is TG3TG (29). G3T2A repeats are found in the immediate subtelomeric region of S. cerevisiae, suggesting that G3T2A-like telomeres were present prior to yeast divergence (26). Despite length and sequence diversity, a TG-rich motif (a potential binding site for the repressor-activator protein 1 RAP1) appears to be a conserved feature of telomeric motifs among a variety of budding yeasts (28,29). Heterogeneous telomeric repeats, encoded by a single telomerase RNA (33,34), are also observed in the fission yeast Schizosaccharomyces pombe; G2T2AC and G2T2ACA are by far the most frequently found motifs (33–36).
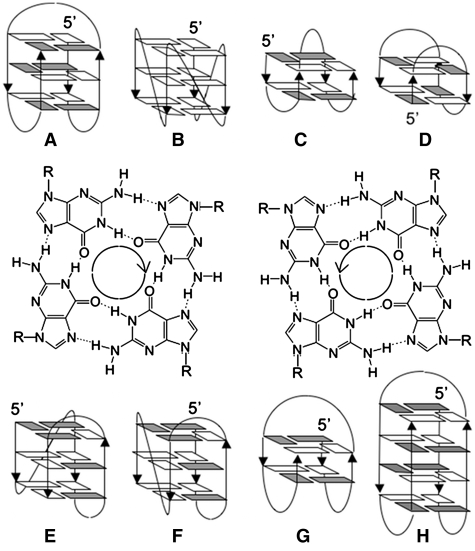
Bearing runs of consecutive guanines, 5′ to 3′ telomeric strands from most eukaryotes may potentially fold into G-quadruplexes. G-quadruplexes are a family of nucleic acid secondary structures based on the formation and stacking of quartets of coplanar guanines (G-quartets) (Figure 1); they are stabilized by several cations, in particular potassium and sodium (37). G-quadruplexes can form either by the folding back of a single strand (intramolecular) or by the association of two (bimolecular) or four (tetramolecular) different strands (38–40). Ciliate telomeric repeats were the first to be studied for their G-quadruplex folding potential in vitro (38,39,41,42), and ciliates are the first organisms for which direct involvement of G-quadruplexes in telomere biology has been unambiguously demonstrated (43–45). Evidence for G-quadruplex formation by vertebrate telomeric repeats has been gathered in vitro, but evidence for G-quadruplex formation at telomeres in vivo is mostly indirect. The G-quadruplex folding potential of telomeric motifs has been emphasized since early studies (46,47). Several G-quadruplex structures formed by telomeric repeats from different organisms have been resolved, notably the intramolecular G-quadruplexes formed by Oxytricha and Tetrahymena telomeric sequences in sodium (48–50), by human (vertebrate) telomeric sequences in sodium and in potassium (51–58) and by Bombyx mori and Giardia modified telomeric sequences in potassium (59,60) (Figure 1).
Figure 1.
Schematic structures of intramolecular G-quadruplexes formed by about four repetitions of telomeric motifs from (A) vertebrates in Na+ (51), (B, E–G) vertebrates in K+ (52–57), (B and C) Giardia in K+ (60), (D) Bombyx in K+ (59), (F) Tetrahymena in Na+ (50) and (H) Oxytricha in Na+ (49). Anti and syn guanines are in white and grey, respectively. Structures of Giardia and Bombyx G-quadruplexes were obtained with modified sequences. The two G-quartets illustrate the possible donor-to-acceptor hydrogen-bond orientations (left: clockwise, right: anticlockwise).
Although several telomeric sequences have been shown to fold into G-quadruplexes, data have been acquired with different techniques and under different conditions, hence do not allow a straightforward comparison of G-quadruplex folding and stability of telomeric sequences among eukaryotes. Furthermore, to our knowledge, folding of yeast telomeric sequences into G-quadruplexes has never been proved, with the exception of sequences mimicking two or three repetitions of S. cerevisiae heterogeneous telomeric motifs (46,61). Using biophysical and biochemical methods, we undertook an investigation of sequences consisting of about four repetitions of telomeric motifs from a variety of organisms (the minimal length required to allow formation of intramolecular G-quadruplexes); the aim was to (i) probe their propensity to fold into G-quadruplexes, (ii) provide some information about their folding and (iii) study/compare their stability. The studied sequences are listed in Table 1. Among yeasts, we chose to limit our study to S. cerevisiae and S. pombe (which are model organisms in the study of telomeres), to Candida glabrata and Candida guillermondii. Among the diverse telomeric sequences of budding yeasts (other than species of the genus Saccharomyces), C. glabrata and C. guillermondii telomeric sequences present the shortest loops (6 nt) connecting consecutive guanines (28,29); they should hence be the most prone to fold into G-quadruplexes. Actually, several studies on model sequences indicate that increasing loop length leads to a decrease in G-quadruplex stability (62–64). S. cerevisiae and S. pombe have heterogeneous telomeric sequences; we chose to study repetitions of the most frequently found motifs. In a previous study, we provided evidence of folding into stable G-quadruplexes by repetitions of Plasmodium degenerated telomeric motif (GGGTTYA, where Y is T or C) under the same conditions used here (65); melting temperatures and thermodynamic data from this work were included in Table 1.
Table 1.
Sequences, melting temperatures and thermodynamic parameters for G-quadruplex folding of the studied oligonucleotides
| Sequence name (species) | Sequence (from 5′ to 3′) | NaCl |
KCl |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tm (°C) | ΔH° (kcal mol−1) | ΔS° (cal K−1 mol−1) | ΔG°(310 K) (kcal mol−1) | Tm (°C) | ΔH° (kcal mol−1) | ΔS° (cal K−1 mol−1) | ΔG°(310 K) (kcal mol−1) | |||
| G2 motifs | ||||||||||
| Bom17 | (Bombyx) | (GGTTA)3GG | a | a | ||||||
| Asc20 | (Ascaris) | (GGCTTA)3GG | a | a | ||||||
| Spom20 | (S. pombe) | (GGTTAC)3GG | no G4b | ndc | ||||||
| Spom23 | (S. pombe) | (GGTTACA)3GG | no G4b | no G4b | ||||||
| Cgi26 | (C. guillermondii) | (GGTGTACT)3GG | no G4 | no G4 | ||||||
| G3 motifs | ||||||||||
| Gia18 | (Giardia) | (GGGTA)3GGG | 60 | −53 ± 7 | −157 ± 19 | −3.7 ± 0.6 | 68 | −63 ± 4f | −185 ± 12f | −5.7 ± 0.4f |
| Hum21 | (H. sapiens) | (GGGTTA)3GGG | 59 | −55 ± 4 | −167 ± 12 | −3.7 ± 0.4 | 65 | −60 ± 3 | −177 ± 9 | −5.0 ± 0.3 |
| Par21 | (Parameciumd) | (GGGTTT)3GGG | 50 | −53 ± 8 | −162 ± 23 | −2.2 ± 0.5 | 64 | −64 ± 10f | −189 ± 29f | −5.2 ± 0.9f |
| Scer21 | (S. cerevisiae) | (GGGTGT)3GGG | 50 | −48 ± 5 | −148 ± 15 | −1.9 ± 0.2 | 68 | −68 ± 8f | −200 ± 25f | −6.2 ± 0.9f |
| Ara24 | (Arabidopsis) | (GGGTTTA)3GGG | 57 | −60 ± 5 | −179 ± 14 | −3.6 ± 0.4 | 64 | −73 ± 10 | −216 ± 28 | −5.7 ± 0.9 |
| Plasmodiume | (GGGTTYA)3GGG, Y=T or C | 55 ± 2 | −53 ± 4 | −158 ± 11 | −2.8 ± 0.4 | 64 ± 1 | −63 ± 5 | −186 ± 14 | −4.9 ± 0.6 | |
| Tom24 | (L. esculentumd) | (GGGTTAA)3GGG | 51 | −54 ± 4 | −168 ± 13 | −2.3 ± 0.2 | 64 | −77 ± 5 | −228 ± 13 | −6.1 ± 0.4 |
| Chla27 | (Chlamydomonas) | (GGGTTTTA)3GGG | 46 | −56 ± 4 | −176 ± 13 | −1.6 ± 0.3 | 57.5 | −68 ± 8 | −207 ± 24 | −4.2 ± 0.6 |
| G4 motifs | ||||||||||
| Tet22 | (Tetrahymena) | (GGGGTT)3GGGG | 64 | −54 ± 8 | −161 ± 24 | −4.4 ± 0.7 | >80 | |||
| Oxy28 | (Oxytricha) | (GGGGTTTT)3GGGG | 66 | −82 ± 13 | −242 ± 38 | −6.9 ± 1.3 | >80 | |||
| G3/G4 motif | ||||||||||
| Gla26 | (C. glabrata) | G4T(CTG3TGCTGTG4T)CTG3 | 48 | −53 ± 12 | −164 ± 36 | −1.8 ± 0.5 | 63 | −70 ± 12 | −206 ± 34 | −5.4 ± 0.8 |
The number in the sequence name denotes the sequence length (in nucleotides); telomeric motifs are in italic font.
Melting temperatures (Tm) reported in this table depended neither on wavelength nor on oligonucleotide strand concentration (3, 10 and 30 µM).
Melting curves at 295 nm were analysed according a two-state equilibrium model and assuming linear low- and high-temperature absorbance baselines. Standard enthalpy and entropy changes (ΔH° and ΔS°) for folding were determined by linear fitting lnK versus 1/T, where K is the equilibrium constant between the folded and the unfolded state; standard Gibbs free energy changes (ΔG°) were extrapolated at 310 K, from the relation ΔG°(T) = ΔS° – TΔS°. ΔH°, ΔS° and ΔG° reported in this table are the mean values obtained from analysis of melting curves at 3 and 30 µM strand concentration upon varying linear low- and high-temperature absorbance baselines ± maximum deviation.
aThe presence of a G-quadruplex and of a non-G-quadruplex competing structure made Tm determination not straightforward.
bA very minor fraction of oligonucleotide may be folded into G-quadruplexes at low temperatures.
cNot determined: Tm of Spom20 in KCl could not be determined accurately because of the incertitude in low-temperature absorbance baseline.
dParamecium and L. esculentum (tomato plant) have degenerated telomeric motifs: GGGKTT in Paramecium (K=T or G) and GGGTTWA (W=A or T) in tomato plant; for W=T, the tomato telomeric motif comes back to the Arabidopsis one; for K=G, the Paramecium telomeric motif comes back to the Tetrahymena one.
eData from a previous study (65) (mean values from analysis of the eight possible variant sequences); for Y=T, the Plasmodium telomeric sequence comes back to the Arabidopsis one.
fFor Gia18, Par21 and Scer21 in KCl at 30 µM strand concentration a two-state model is not appropriate, as revealed by non-denaturing PAGE; nevertheless linear van’t Hoff graphs were obtained.
MATERIAL AND METHODS
Oligonucleotides
Reverse-Phase Cartridge•Gold™ (RPC) purified oligonucleotides were purchased from Eurogentec (Seraing, Belgium), dissolved in bi-distilled water at a concentration of 200 µM and stored at −20°C. Concentrations were determined by ultraviolet (UV) absorption using the extinction coefficients provided by the manufacturer. For spectroscopic measurements, RPC-purified oligonucleotides were used without further purification; for non-denaturing PAGE experiments, oligonucleotides were purified by PAGE according to standard protocols. For Asc20, Gia18 and Tet22, spectroscopic measurements were carried out on two different batches of RPC purified oligonucleotides; for Gia18, spectroscopic measurements were also carried out on a third batch, PAGE purified by the manufacturer: data reproducibility was excellent. The quality of Gia18 oligonucleotides (RPC and PAGE purified) was checked by denaturing PAGE.
Thermal difference spectra, UV-melting curves and circular dichroism
UV-absorbance measurements were acquired on an Uvikon 940 (Kontron) spectrophotometer or, for tomato plant and Paramecium variant sequences, on an Uvikon XL (Secomam). For melting experiments, samples were heated at 92°C for a few minutes, cooled from 92 to 2°C at a rate of 0.2°C min−1, kept at 2°C for 30 min and heated from 2 to 92°C at a rate of 0.2°C min−1. The temperature was varied with a circulating water bath; evaporation at high temperatures and condensation at low temperatures were prevented by a layer of mineral oil and by a dry air flow in the sample compartment, respectively. Temperature was measured with an inert glass sensor immersed into a water-filled quartz cell or, for tomato plant and Paramecium variant sequences, with a sensor embedded in a solid cuvet. The absorbance was monitored at 245, 260, 273, 295 and 405 nm; the absorbance at 405 nm was subtracted from each wavelength. For each melting curve, the melting temperature (Tm) was graphically determined as the intercept between the melting curve and the median line between low-temperature and high-temperature absorbance linear baselines (66). For each sample, the thermal difference spectrum (TDS) was obtained by subtracting the absorbance spectrum at 2°C from the one at 92°C (67). The spectrum at 92°C was recorded after heating the sample at 92°C for a few minutes, whereas the spectrum at 2°C was recorded after annealing from 92 to 2°C at a rate of 0.2°C min−1. Circular dichroism (CD) spectra were recorded on a Jasco J-810 spectropolarimeter or, for tomato plant and Paramecium variant sequences, on a Jasco J-815, at 4°C, after annealing from 92°C at a rate of 0.2°C min−1. Each spectrum was obtained by averaging three scans at a speed of 500 nm min-1 and by subtracting the contributions of the solvent and quartz cell. All experiments (melting, TDS and CD) were carried out at 3, 10 and 30 µM oligonucleotide strand concentration (±5%), in a cacodylic acid buffer (10 mM) at pH 7.2 (adjusted with LiOH), containing KCl (100 mM) or NaCl (100 mM), using 1 cm (for 3 and 10 µM strand concentration) or 0.2 cm (for 30 µM strand concentration) path-length quartz cells. Stock solutions (200 µM) were heated at 90°C for a few minutes immediately before sample preparation. TDS, UV-melting and CD experiments were carried out at least twice.
Non-denaturing PAGE
Stock solutions (200 µM) of PAGE-purified oligonucleotides were heated at 90°C for a few minutes immediately before sample preparation. Oligonucleotides (30 µM strand concentration) were annealed from 92 to 2°C at a rate of 1°C min−1 in the same solvents used for spectroscopic measurements (10 mM cacodylic acid buffer, pH 7.2 adjusted with LiOH, 100 mM NaCl or 100 mM KCl). Samples, supplemented with sucrose (11.5%), were then loaded (10 µl) into a 16.5 cm × 14.5 cm × 0.1 cm 12% polyacrylamide gel (acrylamide:bisacrylamide 19:1, prepared in a TBE buffer supplemented with 20 mM NaCl or KCl). Electrophoresis was run in a cold room at 4°C, for 2 h and 30 min, at 3 W per gel, in a TBE buffer supplemented with NaCl (20 mM) or KCl (20 mM); the temperature attained in the gels was about 15°C. DNA was detected by UV-shadow at 254 nm with a G:BOX (Syngene). An oligothymidylate marker (dT21) and two double-stranded markers of 9 and 12 bp (dx9: 5′GCGTATCGG3′ + 5′CCGATACGC3′; dx12: 5′GCGTGACTTCGG3′ + 5′CCGAAGTCACGC3′) were loaded into the gels to provide internal migration markers. We point out that dTn oligonucleotides are not appropriate markers for single-strand DNA migration in PAGE (68).
Fluorescence resonance energy transfer melting curves
The melting of the two double-dye labelled oligonucleotides FBom17T and FAsc20T was followed by fluorescence resonance energy transfer (FRET) on a FluoroMax-3 spectrofluorimeter (FBom17T: FAM-5′GGTTAGGTTAGGTTAGG3′-TAMRA, FAsc20T: FAM-5′GGCTTAGGCTTAGGCTTAGG3′-TAMRA, FAM: 6-carboxyfluorescein, TAMRA: carboxytetramethylrhodamine, purchased from Eurogentec, Seraing, Belgium). The temperature was raised from 5 to 90°C with a circulating water bath; FAM emission (excitation at 470 nm, emission at 520 nm) was recorded at intervals of 1°C, after an equilibration time of 30 s. FAM emission versus temperature plots were normalized between 0 and 1 and the temperature of half-dissociation (T1/2), corresponding to an emission value of 0.5, was determined. FRET-melting experiments were carried out at 0.2 µM oligonucleotide strand concentration, in a cacodylic acid buffer (10 mM) at pH 7.2 (adjusted with LiOH), containing NaCl (100 mM) or KCl (100 mM).
RESULTS
Thermal difference spectra
A simple method to have an insight into the structure adopted by a nucleic acid sequence is to record its absorbance spectra at high temperature (unfolded form) and at low temperature (folded form). The difference between these spectra, called thermal difference spectrum (TDS), provides a spectroscopic signature for nucleic acid structures (67,69).
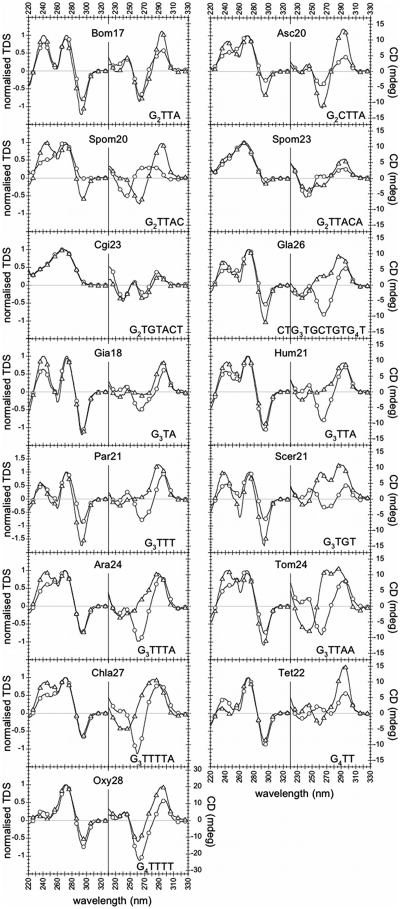
TDS spectra are shown in Figure 2. With the exception of the C. guillermondii (Cgi26) and of the S. pombe (Spom20 and Spom23) sequences, all the studied telomeric sequences displayed TDS signatures characteristic of G-quadruplex structures, both in NaCl and in KCl: two major positive peaks around 240 and 270 nm and a negative peak around 295 nm.
Figure 2.
CD spectra at 4°C (right panels) and normalized TDS (left panels) of all the sequences listed in Table 1, in NaCl (circles) and KCl (triangles), at 3 µM oligonucleotide strand concentration. The CD axis scale is the same for all sequences (–17/+17 mdeg), with the exception of Oxy28 (–32/+32 mdeg). TDS and CD spectra did not depend on strand concentration (3, 10 and 30 µM), with the exception of Gia18, Tet22 and Scer21 CD spectra in KCl.
The C. guillermondii sequence (Cgi26) neither in NaCl nor in KCl displayed a G-quadruplex-type TDS signature. Concerning the S. pombe sequences (Spom20 and Spom23), an unambiguous TDS G-quadruplex signature was obtained only for Spom20 in KCl. TDS of Spom20 in NaCl and Spom23 in NaCl and KCl exhibited a very small negative peak at 295 nm (autoscaled TDS are shown in Figure S1).
CD spectra
CD spectra were recorded at 4°C, a temperature at which the equilibrium was strongly shifted towards the folded forms, as shown by thermal melting profiles (Figures S2 and S3). CD spectra at 3 µM oligonucleotide strand concentration are shown in Figure 2. The Bombyx (Bom17), Ascaris (Asc20) and Oxytricha (Oxy28) sequences exhibited a negative band around 260 nm and a positive band around 290 nm (a ‘Type II’ signature), both in NaCl and in KCl. All the other G-quadruplex folding sequences (i.e. sequences displaying G-quadruplex TDS) exhibited distinct CD signatures in NaCl and in KCl: a Type II signature in NaCl (with the exception of Tet22), and a positive peak around 290 nm and a positive peak or shoulder in the 250–270-nm region in KCl; this supports structural differences in NaCl and in KCl.
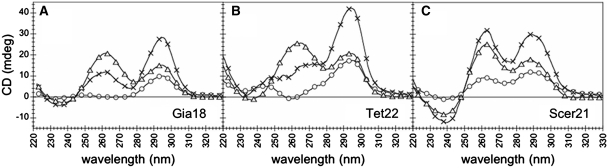
With the exception of the Giardia (Gia18), Tetrahymena (Tet22) and S. cerevisiae (Scer21) sequences in KCl, the CD spectra of all the other sequences were independent from oligonucleotide strand concentration (3, 10 or 30 µM), both in NaCl and in KCl. Conversely, the CD spectra of Giardia (Gia18), Tetrahymena (Tet22) and S. cerevisiae (Scer21) sequences in KCl strongly depended on oligonucleotide strand concentration (Figure 3). For Gia18 and Tet22 increasing the oligonucleotide concentration from 3 to 30 µM led to the appearance of a CD positive peak at about 260 nm (Figure 3A and B). For Scer21, increasing the concentration led to a change in the ratio r between the CD peak at about 260 nm and the one at about 290 nm: (r < 1 for 3 µM, r ≈ 1 for 10 µM and r > 1 for 30 µM) (Figure 3C). The dependence of Gia18, Tet22 and Scer21 CD spectra on strand concentration in KCl supports the presence of multi-stranded structures.
Figure 3.
Circular dichroism spectra at 4°C of (A) Gia18, (B) Tet22 and (C) Scer21 in KCl at 3, 10 and 30 µM oligonucleotide strand concentration (circles, crosses and triangles, respectively).
Concerning C. guillermondii (Cgi26) and S. pombe (Spom20 and Spom23) sequences, only the S. pombe sequence Spom20 in KCl displayed an unambiguous Type II G-quadruplex CD signature, consistently with the results obtained by TDS.
Hence, TDS provided evidence of G-quadruplex folding for most of the sequences listed in Table 1; CD allowed (i) highlighting structural differences in NaCl and in KCl for most of the sequences (at least for Gia18, Hum21, Par21, Scer21, Ara24, Tom24, Chla27, Tet22 and Gla26) and (ii) providing evidence of the presence of multi-stranded structures for a few sequences (Gia18, Tet22 and Scer21 in KCl). Individual CD spectra will not be discussed in detail for the following reason. Empirically, two types of CD spectra are considered indicative of DNA G-quadruplexes: spectra with a negative band around 240 nm and a dominant positive band around 260 nm (‘Type I’ spectra), and spectra with a negative band around 260 nm and a positive band around 290 nm (‘Type II’ spectra). Theoretical calculations give an insight into the origin of Type I and Type II signals: stacking of two G-quartets with the same hydrogen-bond orientation originates a Type I signal, stacking of two G-quartets with opposite hydrogen-bond orientation originates a Type II signal (70,71) (G-quartet hydrogen-bond orientation is illustrated in Figure 1). Nevertheless, G-quadruplexes are highly polymorphic and these empirical and theoretical evidences do not allow an unambiguous interpretation of CD spectra of potential G-quadruplex folding sequences.
UV melting experiments
TDS allowed verifying G-quadruplex folding at low temperature. To study stability, we recorded melting profiles at wavelengths of absorbance maximum variations upon folding, determined by TDS, i.e. 245, 273 and 295 nm.
All melting profiles were reversible (melting curves at 295 nm are reported in Supplementary Figures S2 and S3). Melting transitions of all G-quadruplex folding sequences presented a single transition (a single first derivative peak) and occurred (with the exception of Spom20 in KCl) in a relatively narrow range of temperature (about 30°C for Bom17 and Asc20 and about 20°C for the other sequences), thereby the graphically determined melting temperatures (Tms) did not significantly depend on the choice of the high- and low-temperature absorbance linear baselines; Tms could then be determined with a precision of ± 1°C or better.
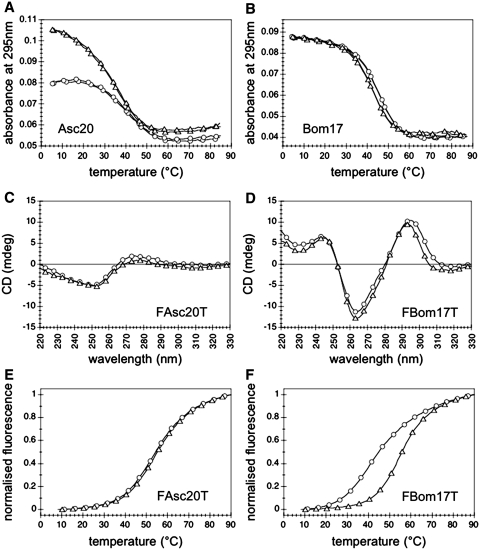
Tms of sequences bearing G3 and G4 motifs (included Gla26) were higher in potassium than in sodium: Tms ranged from 46°C (Chla27) to 64–66°C (Tet22 and Oxy28) in NaCl and from 57.5°C (Chal27) to >80°C (Tet22 and Oxy28) in KCl; they did not depend on wavelength (245, 273 and 295 nm) and are reported in Table 1. The flatness of the S. pombe Spom20 melting profile in KCl at 295 nm (Supplementary Figures S2 and S3) prevented us from determining an accurate Tm (incertitude in the determination of a low-temperature absorbance baseline); its Tm was roughly estimated to be around 27°C. Bombyx (Bom17) and Ascaris (Asc20) differed from all the other G-quadruplex folding sequences for several features. (i) Surprisingly, the graphically determined Tms at 295 nm were higher in sodium than in potassium: 46°C in NaCl and 44°C in KCl for Bom17, 41°C in NaCl and 39°C in KCl for Asc20. The maxima of the first derivatives of their melting curves (Tmax) showed the same trend than melting temperatures determined by graphical analysis: Tmax(NaCl) > Tmax(KCl). (ii) Asc20 Tms depended on wavelength (Tms at 245 nm were around 45°C both in sodium and potassium); this feature suggests the presence of a competing structure. (iii) The amplitude of Asc20 melting transition at 295 nm in NaCl was about half of the amplitude in KCl (Figure 4A); Bom17 melting transitions at 295 nm in NaCl and in KCl had nearly equal amplitudes (Figure 4B). Conversely, for all G-quadruplexes with Tm(NaCl) < Tm(KCl), the amplitudes of melting transitions at 295 nm in NaCl were larger than in KCl (Figures S2 and S3) (this feature appears to be characteristic of many other G-quadruplexes).
Figure 4.
(A and B) Thermal melting (cooling and heating) followed by absorbance at 295 nm of (A) Asc20 and (B) Bom17 in NaCl (circles) and KCl (triangles), at 3 µM oligonucleotide strand concentration. (C and D) CD spectra at 4°C of (C) FAsc20T and (D) FBom17T in NaCl (circles) and KCl (triangles), at 3 µM oligonucleotide strand concentration. (E and F) Thermal melting (heating) followed by FRET (excitation at 470 nm, emission at 520 nm) of (E) FAsc20T and (F) FBom17T in NaCl (circles) and KCl (triangles), at 0.2 µM oligonucleotide strand concentration.
Finally, for all the sequences, Tm did not depend on oligonucleotide strand concentration (3, 10 and 30 µM), not even for those sequences displaying concentration-dependent CD spectra, i.e. Gia18 and Scer21 in KCl (Tet22 Tm in KCl was too high to be determined). In particular, we verified that Tm of Gia18 in KCl did not vary upon increasing oligonucleotide strand concentration up to 100 µM (data not shown). The independence of Tm from oligonucleotide strand concentration suggests formation of intramolecular structures.
FRET melting experiments
The ‘non-classical’ behaviour of Bombyx (Bom17) and Ascaris (Asc20) prompted us to a deeper investigation. We studied the double-dye labelled sequences FAsc20T (FAM-5′(GGCTTA)3GG3′-TAMRA) and FBom17T (FAM-5′ (GGTTA)3GG3′-TAMRA). FAsc20T neither in NaCl nor in KCl folded into G-quadruplexes, as revealed by CD spectroscopy (Figure 4C); it folded into a structure where the two dyes were close to each other, the stability of which (T1/2 = 54°C) did not depend on the cation (very likely a hairpin-duplex), as revealed by thermal melting followed by FRET (Figure 4E). Conversely, FBom17T folded into G-quadruplexes, as revealed by CD spectroscopy (Figure 4D), more stable in potassium (T1/2 = 55°C) than in sodium (T1/2 = 45°C) (Figure 4F).
Gel electrophoresis
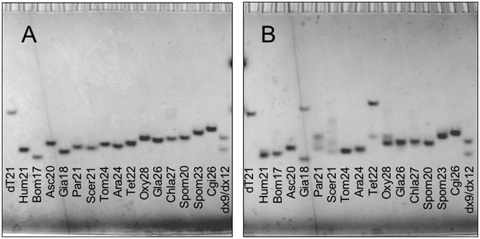
Non-denaturing PAGE was carried out at 30 µM oligonucleotide strand concentration, i.e. the highest concentration used for spectroscopic measurements. DNA was detected by UV-shadow (Figure 5). The temperature of the gels was about 15°C; at this temperature, all the G-quadruplex folding sequences (i.e. all the sequences except Spom20, Spom23 and Cgi26 in NaCl and Spom23 and Cgi26 in KCl) were folded both in KCl and in NaCl, as could be inferred by the inspection of melting profiles (Supplementary Figures S2 and S3). We used an oligothymidylate marker (dT21) and 9- and 12-bp duplexes (dx9 and dx12, respectively) as internal migration standards.
Figure 5.
Non-denaturing PAGE in (A) NaCl and (B) KCl at 30 µM oligonucleotide strand concentration. dT21 is an oligothymidylate marker; dx9 and dx12 are two double-stranded markers of 9 and 12 bp, respectively. Oligonucleotides were detected by UV-shadow.
In NaCl, a single band was detected for all the telomeric sequences (Figure 5A). All the oligonucleotides displayed a higher mobility than dT21; most of them had a mobility intermediate between the ones of dx9 and dx12. In NaCl, the mobility of the sequences displaying a TDS G-quadruplex signature (i.e. all the sequences except Spom20, Spom23 and Cgi26) slightly decreased with the length of the sequence, with the exception of Asc20 and Tet22. The yeast Spom20, Spom23 and Cgi26 sequences, which did not fold into G-quadruplexes in NaCl, as revealed by TDS, were the slowest migrating species.
In KCl, the migration patterns were more complex (Figure 5B). Most of the sequences (Hum21, Bom17, Asc20, Tom24, Ara24, Gla26, Chla27, Spom20, Spom23 and Cgi26) migrated as single bands and faster than dT21. Among these single-band migrating sequences, Spom23 and Cgi26 had the lowest electrophoretic mobility; according to spectroscopic measurements in KCl, these two sequences are not folded into G-quadruplexes. The Giardia (Gia18) and Tetrahymena (Tet22) sequences displayed a fast- and a slow-migrating band, the latter migrating slower than the internal migration standard dT21. For the Paramecium (Par21) and the Oxytricha (Oxy28) sequences, two bands were detected, both fast migrating as compared to dT21. In particular, the two bands of Oxy28 (a principal and a secondary one) were very close to each other. Finally, three bands were detected for the S. cerevisiae sequence (Scer21): two fast migrating bands (between dx9 and dx12) and a slow migrating band (between dx12 and dT21).
The migration pattern obtained in KCl for Gia18, Tet22, Scer21, Par21 and Oxy28 provides evidence of the presence of at least two (or three, for Scer21) different structures at the tested oligonucleotide strand concentration (30 µM). For Gia18, Tet22 and Scer21, the slow migrating band detected in KCl may be ascribed to the presence of multi-stranded species; this hypothesis is supported by changes in their CD spectra upon increasing oligonucleotide strand concentration from 3 to 30 µM (Figure 3).
DISCUSSION
In the present work we studied telomeric sequences from different organisms, with the aim of probing their propensity to fold into G-quadruplex structures, providing some information about their folding and studying their stability.
G-quadruplex folding
Thermal difference spectra provided evidence of the formation of G-quadruplex structures, both in sodium and in potassium, for all the studied telomeric sequences, with the exception of the C. guillermondii (Cgi26) and S. pombe (Spom20 and Spom23) sequences. By contrast, TDS and CD in sodium and in potassium did not support formation of G-quadruplexes by Cgi26. Spom20 in potassium displayed unambiguous TDS and CD G-quadruplex signatures. Finally, the very small negative peak at 295 nm in TDS of Spom20 in sodium and Spom23 in both sodium and potassium suggests that a very minor fraction of these sequences folds into G-quadruplexes; this fraction may contribute to the observed CD spectra.
Molecularity
For the telomeric sequences displaying a TDS G-quadruplex signature, three lines of evidence supported folding into intramolecular structures in sodium: the independence of (i) melting temperatures and (ii) CD spectra from oligonucleotide strand concentration; (iii) the presence of a single and fast migrating band (as compared to dT21) in a non-denaturing PAGE experiment. The same conclusion could be drawn for most of the sequences in potassium (Bom17, Asc20, Hum21, Ara24, Tom24, Gla26, Chla27 and Spom20).
For the Paramecium (Par21) and Oxytricha (Oxy28) sequences in potassium, two bands were detected by PAGE; the mobility of these bands (as compared to the internal markers) and the independence of Par21 Tm and Par21–Oxy28 CD spectra from oligonucleotide strand concentration suggest folding into intramolecular structures of different electrophoretic mobilities (Oxy28 Tm in potassium were too high to be determined).
The PAGE migration pattern of Giardia (Gia18), Tetrahymena (Tet22) and S. cerevisiae (Scer21) sequences in potassium provided evidence of the presence of dramatically different structures (at least two for Gia18 and Tet22 and at least three for Scer21). For these three sequences, the slow migrating band may be ascribed to multi-stranded structures. This hypothesis is supported by the change in Gia18, Tet22 and Scer21 CD spectra upon increasing oligonucleotide strand concentration. Furthermore, the PAGE migration pattern of Gia18 in KCl was shown to be concentration-dependent: the slow migrating band was not detected at 100-fold lower oligonucleotide strand concentration (300 nM) (gel detected by Syber Gold staining, data not shown). This demonstrates that the slow migrating band of Gia18 corresponds to multi-stranded species.
On the other hand, the independence of Gia18 and Scer21 Tms from oligonucleotide strand concentration (up to 100 µM for Gia18) does not support folding into intermolecular structures, such as bimolecular or tetramolecular G-quadruplexes (Tet22 Tm in potassium were too high to be determined). This puzzling behaviour might be explained by folding into intramolecular G-quadruplexes (monomers) and formation of stacked monomers upon increasing oligonucleotide strand concentration from 3 to 30 µM. The appearance of a positive CD band around 260 nm might arise from stacking of terminal G-quartets with the same hydrogen-bond orientation (70,71). The hypothesis of stacked intramolecular G-quadruplexes is not appropriate (or is only partially appropriate) for Scer21, which present a more complex non-denaturing PAGE migration pattern.
Structural differences in sodium and in potassium
Besides providing evidence of a concentration-dependent behaviour of Gia18, Tet22 and Scer21 in potassium, CD allowed highlighting structural differences in sodium and potassium.
The CD spectra of most of the telomeric sequences (Gia18, Hum21, Ara24, Chla27, Par21, Scer21, Tom24 and Tet22) strongly depended on the nature of the cation present in solution (sodium or potassium), suggesting different G-quadruplex conformations or a different equilibrium among multiple possible G-quadruplex conformations according to the nature of the cation. Only the Bombyx (Bom17), Ascaris (Asc20) and Oxytricha (Oxy28) sequences displayed a similar (although not strictly identical) CD signature in sodium and in potassium.
It would be unwarranted to speculate about CD signatures and G-quadruplex folding topologies, for several reasons. (i) The simple empirical rule often used to interpret CD spectra (Type I spectrum = parallel folding, Type II spectrum = antiparallel folding) is based on a few resolved structures and is not of general validity (71). (ii) Interpretation of CD spectra in terms of relative strand orientation is not formally correct (70,71). (iii) Interpretation of CD spectra in terms of folding topologies relies on the implicit assumption that a sequence folds into a single G-quadruplex structure. A single sequence may fold into multiple G-quadruplex structures in the same solution; this is the case, for example, of human and Giardia telomeric sequences in potassium (Figure 1). Structure determination requires high-resolution structural studies, such as nuclear magnetic resonance (NMR) or X-ray crystallography, and is beyond the scope of this work.
Stability of G3 and G4 motifs
All sequences bearing three or more consecutive guanines folded into G-quadruplexes more stable in potassium than in sodium (higher Tms), as expected for G-quadruplexes. Sequences bearing three consecutive guanines (including Gla26, for a total of eight motifs) had Tms ranging from 46 to 60°C in sodium and from 57.5 to 68°C in potassium. Notably, for the G3TnA motif sequences, Tm slightly decreased with n for n = 1, 2 and 3 (Gia18, Hum21 and Ara24, respectively); increasing n to 4 (Chla27) led to a dramatic decrease in Tm (about 10°C). The Tetrahymena (Tet22) and Oxytricha (Oxy28) sequences, bearing four consecutive guanines, were the most stable; they had roughly the same Tms in sodium (64 and 66°C, respectively) and Tms higher than 80°C in potassium.
We carried out a thermodynamic analysis of melting transitions (Table 1). A two-state equilibrium model, assuming linear low- and high-temperature absorbance baselines, resulted in linear van’t Hoff graphs. Non-denaturing PAGE experiments revealed that such a model is not appropriate for Gia18, Par21 and Scer21 in KCl, nevertheless linear van’t Hoff graphs could be obtained even for these sequences in KCl. Thermodynamic values did not depend on oligonucleotide strand concentration (3 and 30 µM). For G3 and G3/G4 motifs, ΔH° of folding varied between −60 and −48 kcal mol−1 in NaCl and between −77 and −60 kcal mol−1 in KCl; ΔG° of folding, extrapolated at 37°C, varied between −3.7 and −1.6 kcal mol−1 in NaCl and between −6.2 and −4.2 kcal mol−1 in KCl. The analysis of G4 motifs was possible only in NaCl: standard enthalpy change of Oxy28 folding (ΔH° = −82 ± 13 kcal mol−1) was more negative than standard enthalpy changes of Tet22 folding (ΔH° = −54 ± 8 kcal mol−1) and of G3 and G3/G4 motifs. For Tet22, G3 and G3/G4 motifs in NaCl, Tm and ΔG° were linearly correlated (linear regression coefficient R = 0.9877). In KCl, Tm and ΔG° correlation was poorer; this might be ascribed to a greater error in ΔG° determination in KCl than in NaCl (due to a greater uncertainty in high-temperature baseline determination and to extrapolation of ΔG° at a temperature far below the temperature ranges where melting transitions occurred).
The influence of G-tract length, loop length and loop sequence on G-quadruplex structure and stability has been explored on model sequences (62–64,72–77); nevertheless, no general rules has emerged (78). In particular, the number of consecutive guanines does not necessarily correspond to the number of G-quartets (50,57,60). NMR studies of the Oxytricha 5′(G4T4)3G43′ and of the Tetrahymena 5′(T2G4)43′ telomeric sequences in sodium revealed four stacked G-quartets in the Oxytricha G-quadruplex (48,49) (Figure 1H) and only three stacked G-quartets in the Tetrahymena G-quadruplex, connected by a GTTG lateral loop, a central TTG lateral loop and a TT double-chain-reversal loop (50) (Figure 1F). Our data do not allow drawing conclusions about the number of stacked G-quartets in the Tetrahymena telomeric sequence Tet22, which differed from the one used in the NMR study by two nucleotides (TT). Nevertheless, in sodium, the amplitude in absorbance of the 295 nm melting transition of Tet22 (0.047 variation in absorbance recorded at 3 µM oligonucleotide strand concentration and 1 cm optical path length) was very similar to the amplitudes of the sequences bearing three consecutive guanines (0.049 ± 0.008 on average) and significantly smaller than the amplitude of Oxy28 (0.072) (Figure S2). This feature, along with ΔH° values discussed above, might argue stacking of three G-quartets in Tet22 and of four G-quartets in Oxy28.
For a comprehensive analysis of tomato plant and Paramecium degenerated telomeric sequences, we studied the sixteen following variant sequences: 5′(GGGTTWA)3GGG3′ (where W is A or T) (tomato) and 5′(GGGKTT)3GGG3′ (where K is T or G) (Paramecium) (Table 2). Not surprisingly, all these sequences folded into stable G-quadruplexes (TDS are shown in Supplementary Figures S4 and S5). Tm did not strongly vary among tomato variant sequences: it ranged from 55 to 58°C in NaCl and from 64 to 69°C in KCl (Table 2). Tms of Paramecium variant sequences spanned a broader range: from 52 to 60°C in NaCl and from 68 to 76°C in KCl (Table 2). Two correlations between sequence and stability and between sequence and CD signature emerged. (i) Among Paramecium variant sequences, stability in NaCl correlated with the sequence of the third repeated unit (more precisely of nucleotides 16–18): G-quadruplexes formed by 5′(G3KTT)2G3TTTG33′ sequences were less stable (Tms of 52–54°C) than the ones formed by 5′(G3KTT)2G3GTTG33′ sequences (Tms of 58–60°C). (ii) Among tomato plant variant sequences, the sequence of the first loop connecting runs of guanines (nucleotides 4–7) correlated with CD spectra in KCl: 5′G3TTAA(G3TTWA)2G33′ sequences had Tom24-like CD spectra, while 5′G3TTTA(G3TTWA)2G33′ sequences had Ara24-like CD spectra (Figure S4).
Table 2.
Tomato plant and Paramecium variant telomeric sequences and Tms determined by analysis of melting curves recorded at 295 nm, at 3 µM oligonucleotide strand concentration
| Sequence name | Sequence (from 5′ to 3′) | Tm (°C) in NaCl | Tm (°C) in KCl |
|---|---|---|---|
| Tomato plant | |||
| Tom24 | G3TTAAG3TTAAG3TTAAG3 | 55 | 67 |
| TAA | G3TTTAG3TTAAG3TTAAG3 | 58 | 65 |
| ATA | G3TTAAG3TTTAG3TTAAG3 | 57 | 69 |
| AAT | G3TTAAG3TTAAG3TTTAG3 | 58 | 67 |
| TTA | G3TTTAG3TTTAG3TTAAG3 | 58 | 66 |
| TAT | G3TTTAG3TTAAG3TTTAG3 | 57 | 64 |
| ATT | G3TTAAG3TTTAG3TTTAG3 | 56 | 67 |
| Ara24 | G3TTTAG3TTTAG3TTTAG3 | 58 | 66 |
| Paramecium | |||
| Par21 | G3TTTG3TTTG3TTTG3 | 52 | 68 |
| GTT | G3GTTG3TTTG3TTTG3 | 54 | 72 |
| TGT | G3TTTG3GTTG3TTTG3 | 52 | 68 |
| TTG | G3TTTG3TTTG3GTTG3 | 59 | 72 |
| GGT | G3GTTG3GTTG3TTTG3 | 54 | 71 |
| GTG | G3GTTG3TTTG3GTTG3 | 58 | 74 |
| TGG | G3TTTG3GTTG3GTTG3 | 58 | 73 |
| GGG | G3GTTG3GTTG3GTTG3 | 60 | 76 |
Tms reported in this table were obtained on a different spectrophotometer from those reported in Table 1.
Temperature was measured with a sensor in a sealed cuvet provided by the manufacturer; this may explain the slightly higher Tms reported in this table as compared to Tms reported in Table 1 for Tom24, Ara24 and Par21.
TDS and CD spectra are reported in Supplementary Figures S4 and S5.
Stability of G2 motifs
Among the telomeric sequences bearing two consecutive guanines, only the Bombyx (Bom17) and the Ascaris (Asc20) sequences folded into G-quadruplexes both in sodium and in potassium. Permuting a cytosine from the first position (Asc20) to the last position (Spom20) of the G-tract connecting sequences abolished G-quadruplex folding in sodium. The analysis of Bom17 and Asc20 melting transitions at 295 nm revealed higher Tms in sodium than in potassium (about 2°C). This behaviour is unique to Bom17 and Asc20 sequences, since the model sequences 5′(G2Tn)3G23′ with n = 3 and n = 4 folded into G-quadruplexes with higher Tms in potassium than in sodium (data not shown). An apparent higher Tm at 295 nm in sodium than in potassium can result from the equilibrium between a G-quadruplex and a non-G-quadruplex structure. Asc20 and Bom17 can in principle fold into (hairpin-) duplexes with, respectively, 2 out of 3 and 1 out of 3 nucleotides engaged in Watson–Crick base pairs. The presence of a competing structure is suggested by results obtained in LiCl and results obtained with fluorescent conjugates. Folding of Asc20 and Bom17 into non-G-quadruplex structures, melting at about 38 and 25°C, respectively, was indeed observed in LiCl (Figure S6); folding into these structures did not result into absorbance variation at 295 nm. Results obtained with the double-dye-labelled sequences suggest than both Asc20 and Bom17 fold into two structures (a G-quadruplex and a non-G-quadruplex structure) and that the presence of the dyes completely shift the equilibrium of the Ascaris sequence toward the non-G-quadruplex structure and the equilibrium of the Bombyx sequence toward the G-quadruplex (a ‘classical’ G-quadruplex, i.e. more stable in potassium than in sodium). If a competing structure is present, then the Tm at 295 nm is not a ‘true’ G-quadruplex Tm, even if the competing structure does not contribute to melting profiles at 295 nm. More precisely, if f is the G-quadruplex folded fraction at low temperatures, then the graphically determined Tm at 295 nm is the temperature at which a fraction f/2 is folded into a G-quadruplex; this temperature, which depend on f (i.e. on the equilibrium between the G-quadruplex and the competing structure) cannot be used as a parameter for G-quadruplex stability. The presence of a non-G-quadruplex competing structure may also explain the small amplitude of Asc20 melting transition at 295 nm in sodium compared to the one in potassium (Figure 4A). This feature may result from a smaller G-quadruplex folded fraction in sodium than in potassium at low temperatures. This would mean that G-quadruplexes formed by Asc20 are less stable in sodium than in potassium (under the reasonable hypothesis that the stability of the competitive structure does not depend on the nature of the cation). This hypothesis is not in contradiction with higher Tms at 295 nm in sodium than in potassium, as discussed above. Finally, the independence of Asc20 and Bom17 melting temperatures from oligonucleotide strand concentration (3, 10 and 30 µM) supports the intramolecular nature of both the G-quadruplex and the non-G-quadruplex structures. The PAGE migration patterns of Asc20 and Bom17 are not inconsistent with the presence of G-quadruplexes and hairpin-duplexes, when compared to the internal migration standard dx9 and dx12.
The inability of the Cgi26 and Spom23 sequences to fold into G-quadruplexes or stable G-quadruplexes does not result just from the length of the loops separating the runs of guanines (6 and 5 nt, respectively). Actually the model sequence 5′(G2T5)3G23′folded into G-quadruplexes both in sodium and in potassium (with a Tm of 16 and 25°C, respectively), whereas the model sequence 5′(G2T6)3G23′ folded into stable G-quadruplexes in potassium only (with a Tm of 18°C) (data not shown). Hence, the behaviour of the telomeric sequences bearing just two consecutive guanines (from Bom17 to Cgi26) arises from their peculiar loop sequences.
In summary, with regard to the ability of folding into intramolecular G-quadruplexes, our study highlights at least two different sets of telomeric motifs: (i) yeast telomeric motifs and (ii) telomeric motifs from other eukaryotes. On the one hand, the ability to fold into G-quadruplex structures does not appear to be a conserved feature among the exceptionally diverse yeast telomeric sequences: unlike C. glabrata, C. guillermondii telomeric sequence did not fold into G-quadruplexes; unlike repetitions of the TG3TG motif (frequently occurring in S. cerevisiae telomeres), repetitions of G2TTAC(A) motifs (frequently occurring in S. pombe telomeres) were not prone to fold into stable G-quadruplexes. On the other hand, evolution of telomeric sequences in eukaryotes other than yeasts (repetitions of G3T1-4A in a variety of organisms, G4T2,4 in ciliates, G2T2A in many insects and G2CT2A in nematodes) appears to have preserved the G-quadruplex folding potential, even if these structures pose a potential problem to telomeric G-strand replication (79,80). Notably, telomeric sequences with three or more consecutive guanines folded into G-quadruplexes with melting temperatures above temperatures where organisms known to have these telomeric sequences thrive. Curiously, no telomeric motif with an adenine following the run of guanines has been found, with the exception of the putative G6AAT telomeric motif of the hot-spring unicellular red alga Cyanidioschyzon merolae (81). Intriguingly, an adenine as the first base in the loops connecting runs of two or three consecutive guanines is detrimental to G-quadruplex folding (47,77). Cyanidioschyzon merolae is an extremophilic organism, thriving in acid hot springs (pH 1.5, 45°C). Its telomeric sequence 5′(G6AAT)3G63′ folded into very stable G-quadruplexes, melting at 70°C in NaCl and stable at temperatures as high as 90°C in KCl (Supplementary Figure S7). Unlikely, telomeric motifs of other thermophilic eukaryotes (such as fungi able to grown at 55–60°C, hot-spring algae other than C. merola, the worms Paralvinella sulfincola and Alvinella pompejana, tardigrades) are not known, to our knowledge.
In conclusion, the present study provides an insight into intramolecular G-quadruplex folding potential of telomeric sequences from a variety of organisms. (i) All the telomeric sequences bearing three or more consecutive guanines (in particular G3T1-4A motifs and G4T2,4 ciliate motifs) folded into stable G-quadruplexes both in potassium and in sodium. (ii) The G2TTA telomeric motif sequence (found in many insects, such as Bombyx mori) and the G2CTTA telomeric motif sequence (found in the nematodes Ascaris lumbricoides and Caenorhabditis elegans) folded into G-quadruplexes both in potassium and in sodium. Our results strongly suggest that, for these two sequences, the G-quadruplex structures are in equilibrium with non-G-quadruplex structures (likely hairpin-duplexes). (iii) G-quadruplex folding does not appear to be a characteristic feature of budding yeast telomeres, despite a widespread conserved TG-rich motif of about six base pairs (28). Actually, unlike repetitions of the S. cerevisiae frequent occuring motif TG3TG and of the Candida glabrata CTGTG4TCTG3TG motif, the Candida guillermondii telomeric sequence (TACTG2TG motif) was unable to fold into G-quadruplexes. (iv) The most frequent telomeric motifs in the fission yeast Schizosaccharomyces pombe are G2TTAC and G2TTACA. Only repetitions of the GGTTAC motif folded into G-quadruplexes and just in potassium. In order to verify if the S. pombe expanded consensus telomeric motif ggggggGGTTACAc (where lower case letters denote rare insertions) (34,35) is prone to fold into G-quadruplex structures a deeper investigation of combinations of possible S. pombe telomeric units is required. (v) For most of the telomeric sequences folding into G-quadruplexes, structural differences between the sodium and the potassium forms were revealed by CD. (vi) Finally, circular dichroism and PAGE supported folding of the Giardia intestinalis GGGTA, of the Tetrahymena GGGGTT and of the S.cerevisiae TGGGTG motif sequences into intramolecular and multi-stranded structures in potassium; the independence of their Tms from oligonucleotide strand concentration suggests that these multi-stranded structures may be stacked intramolecular G-quadruplexes.
Besides allowing comparison of G-quadruplex folding and stability of eukaryotic telomeric sequences, the present study enriches the dataset necessary to build and refine algorithms predicting G-quadruplex stability (82).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Institut National de la Santé et de la Recherche Médicale (INSERM), Centre National de la Recherche Scientifique (CNRS), Muséum National d'Histoire Naturelle (MNHN); Fondation pour la Recherche Médicale (FRM); University of Bordeaux 2; the Agence Nationale de la Recherche (G4-Toolbox ANR-09-BLAN-0355); Région Aquitaine grants. Funding for open access charge: Institut National de la Santé et de la Recherche Médicale (INSERM).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Guédin (Pessac) for her help in a few experiments. We thank N. Smith (Pessac) for her careful reading of the manuscript. We thank L. Lacroix (Paris), and A. Bourdoncle (Pessac) for helpful discussions.
REFERENCES
- 1.Wellinger RJ, Sen D. The DNA structures at the ends of eukaryotic chromosomes. Eur. J. Cancer. 1997;33:735–749. doi: 10.1016/S0959-8049(97)00067-1. [DOI] [PubMed] [Google Scholar]
- 2.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl Acad. Sci. USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyne J, Ratliff RL, Moyzis RK. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc. Natl Acad. Sci. USA. 1989;86:7049–7053. doi: 10.1073/pnas.86.18.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackburn EH, Challoner PB. Identification of a telomeric DNA sequence in Trypanosoma brucei. Cell. 1984;36:447–457. doi: 10.1016/0092-8674(84)90238-1. [DOI] [PubMed] [Google Scholar]
- 5.Van der Ploeg LH, Liu AY, Borst P. Structure of the growing telomeres of Trypanosomes. Cell. 1984;36:459–468. doi: 10.1016/0092-8674(84)90239-3. [DOI] [PubMed] [Google Scholar]
- 6.Schechtman MG. Characterization of telomere DNA from Neurospora crassa. Gene. 1990;88:159–165. doi: 10.1016/0378-1119(90)90027-o. [DOI] [PubMed] [Google Scholar]
- 7.Emery HS, Weiner AM. An irregular satellite sequence is found at the termini of the linear extrachromosomal rDNA in Dictyostelium discoideum. Cell. 1981;26:411–419. doi: 10.1016/0092-8674(81)90210-5. [DOI] [PubMed] [Google Scholar]
- 8.Forney J, Henderson ER, Blackburn EH. Identification of the telomeric sequence of the acellular slime molds Didymium iridis and Physarum polycephalum. Nucleic Acids Res. 1987;15:9143–9152. doi: 10.1093/nar/15.22.9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinclair CS, Richmond RH, Ostrander GK. Characterization of the telomere regions of scleractinian coral, Acropora surculosa. Genetica. 2007;129:227–233. doi: 10.1007/s10709-006-0001-x. [DOI] [PubMed] [Google Scholar]
- 10.Le Blancq SM, Kase RS, Van der Ploeg LH. Analysis of a Giardia lamblia rRNA encoding telomere with [TAGGG]n as the telomere repeat. Nucleic Acids Res. 1991;19:5790. doi: 10.1093/nar/19.20.5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuchs J, Brandes A, Schubert I. Telomere sequence localisation and karyotype evolution in higher plants. Plant Syst. Evol. 1995;196:227–241. [Google Scholar]
- 12.Richards EJ, Ausubel FM. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell. 1988;53:127–136. doi: 10.1016/0092-8674(88)90494-1. [DOI] [PubMed] [Google Scholar]
- 13.Petracek ME, Lefebvre PA, Silflow CD, Berman J. Chlamydomonas telomere sequences are A+T-rich but contain three consecutive G-C base pairs. Proc. Natl Acad. Sci. USA. 1990;87:8222–8226. doi: 10.1073/pnas.87.21.8222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okazaki S, Tsuchida K, Maekawa H, Ishikawa H, Fujiwara H. Identification of a pentanucleotide telomeric sequence, (TTAGG)n, in the silkworm Bombyx mori and in other insects. Mol. Cell. Biol. 1993;13:1424–1432. doi: 10.1128/mcb.13.3.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muller F, Wicky C, Spicher A, Tobler H. New telomere formation after developmentally regulated chromosomal breakage during the process of chromatin diminution in Ascaris lumbricoides. Cell. 1991;67:815–822. doi: 10.1016/0092-8674(91)90076-b. [DOI] [PubMed] [Google Scholar]
- 16.Cangiano G, La Volpe A. Repetitive DNA sequences located in the terminal portion of the Caenorhabditis elegans chromosomes. Nucleic Acids Res. 1993;21:1133–1139. doi: 10.1093/nar/21.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wicky C, Villeneuve AM, Lauper N, Codourey L, Tobler H, Muller F. Telomeric repeats (TTAGGC)n are sufficient for chromosome capping function in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA. 1996;93:8983–8988. doi: 10.1073/pnas.93.17.8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blackburn EH, Gall JG. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J. Mol. Biol. 1978;120:33–53. doi: 10.1016/0022-2836(78)90294-2. [DOI] [PubMed] [Google Scholar]
- 19.Klobutcher LA, Swanton MT, Donini P, Prescott DM. All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc. Natl Acad. Sci. USA. 1981;78:3015–3019. doi: 10.1073/pnas.78.5.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dore E, Pace T, Ponzi M, Scotti R, Frontali C. Homologous telomeric sequences are present in different species of the genus Plasmodium. Mol. Biochem. Parasitol. 1986;21:121–127. doi: 10.1016/0166-6851(86)90015-0. [DOI] [PubMed] [Google Scholar]
- 21.Bottius E, Bakhsis N, Scherf A. Plasmodium falciparum telomerase: de novo telomere addition to telomeric and nontelomeric sequences and role in chromosome healing. Mol. Cell. Biol. 1998;18:919–925. doi: 10.1128/mcb.18.2.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganal MW, Lapitan NL, Tanksley SD. Macrostructure of the tomato telomeres. Plant Cell. 1991;3:87–94. doi: 10.1105/tpc.3.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baroin A, Prat A, Caron F. Telomeric site position heterogeneity in macronuclear DNA of Paramecium primaurelia. Nucleic Acids Res. 1987;15:1717–1728. doi: 10.1093/nar/15.4.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forney JD, Blackburn EH. Developmentally controlled telomere addition in wild-type and mutant paramecia. Mol. Cell. Biol. 1988;8:251–258. doi: 10.1128/mcb.8.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCormick-Graham M, Romero DP. A single telomerase RNA is sufficient for the synthesis of variable telomeric DNA repeats in ciliates of the genus Paramecium. Mol. Cell. Biol. 1996;16:1871–1879. doi: 10.1128/mcb.16.4.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teixeira MT, Gilson E. Telomere maintenance, function and evolution: the yeast paradigm. Chromosome Res. 2005;13:535–548. doi: 10.1007/s10577-005-0999-0. [DOI] [PubMed] [Google Scholar]
- 27.Lue NF. Plasticity of telomere maintenance mechanisms in yeast. Trends Biochem. Sci. 35:8–17. doi: 10.1016/j.tibs.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McEachern MJ, Blackburn EH. A conserved sequence motif within the exceptionally diverse telomeric sequences of budding yeasts. Proc. Natl Acad. Sci. USA. 1994;91:3453–3457. doi: 10.1073/pnas.91.8.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohn M, McEachern MJ, Blackburn EH. Telomeric sequence diversity within the genus Saccharomyces. Curr. Genet. 1998;33:83–91. doi: 10.1007/s002940050312. [DOI] [PubMed] [Google Scholar]
- 30.Shampay J, Szostak JW, Blackburn EH. DNA sequences of telomeres maintained in yeast. Nature. 1984;310:154–157. doi: 10.1038/310154a0. [DOI] [PubMed] [Google Scholar]
- 31.Wang SS, Zakian VA. Sequencing of Saccharomyces telomeres cloned using T4 DNA polymerase reveals two domains. Mol. Cell. Biol. 1990;10:4415–4419. doi: 10.1128/mcb.10.8.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 33.Leonardi J, Box JA, Bunch JT, Baumann P. TER1, the RNA subunit of fission yeast telomerase. Nat. Struct. Mol. Biol. 2008;15:26–33. doi: 10.1038/nsmb1343. [DOI] [PubMed] [Google Scholar]
- 34.Webb CJ, Zakian VA. Identification and characterization of the Schizosaccharomyces pombe TER1 telomerase RNA. Nat. Struct. Mol. Biol. 2008;15:34–42. doi: 10.1038/nsmb1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hiraoka Y, Henderson E, Blackburn EH. Not so peculiar: fission yeast telomere repeats. Trends Biochem. Sci. 1998;23:126. doi: 10.1016/s0968-0004(98)01176-1. [DOI] [PubMed] [Google Scholar]
- 36.Trujillo KM, Bunch JT, Baumann P. Extended DNA binding site in Pot1 broadens sequence specificity to allow recognition of heterogeneous fission yeast telomeres. J. Biol. Chem. 2005;280:9119–9128. doi: 10.1074/jbc.M414511200. [DOI] [PubMed] [Google Scholar]
- 37.Neidle S, Balasubramanian S. Quadruplex Nucleic Acids. Cambridge: RSC Biomolecular Sciences; 2006. [Google Scholar]
- 38.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 39.Sundquist WI, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 40.Sen D, Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 41.Henderson E, Hardin CC, Walk SK, Tinoco I, Jr, Blackburn EH. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell. 1987;51:899–908. doi: 10.1016/0092-8674(87)90577-0. [DOI] [PubMed] [Google Scholar]
- 42.Oka Y, Thomas CA., Jr The cohering telomeres of Oxytricha. Nucleic Acids Res. 1987;15:8877–8898. doi: 10.1093/nar/15.21.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paeschke K, Juranek S, Simonsson T, Hempel A, Rhodes D, Lipps HJ. Telomerase recruitment by the telomere end binding protein-beta facilitates G-quadruplex DNA unfolding in ciliates. Nat. Struct. Mol. Biol. 2008;15:598–604. doi: 10.1038/nsmb.1422. [DOI] [PubMed] [Google Scholar]
- 45.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps H. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 46.Petracek ME, Berman J. Chlamydomonas reinhardtii telomere repeats form unstable structures involving guanine-guanine base pairs. Nucleic Acids Res. 1992;20:89–95. doi: 10.1093/nar/20.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murchie AI, Lilley DM. Tetraplex folding of telomere sequences and the inclusion of adenine bases. EMBO J. 1994;13:993–1001. doi: 10.1002/j.1460-2075.1994.tb06344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith FW, Feigon J. Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature. 1992;356:164–168. doi: 10.1038/356164a0. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, Patel DJ. Solution structure of the Oxytricha telomeric repeat d[G4(T4G4)3] G-tetraplex. J. Mol. Biol. 1995;251:76–94. doi: 10.1006/jmbi.1995.0417. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Patel DJ. Solution structure of the Tetrahymena telomeric repeat d(T2G4)4 G-tetraplex. Structure. 1994;2:1141–1156. doi: 10.1016/s0969-2126(94)00117-0. [DOI] [PubMed] [Google Scholar]
- 51.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 52.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 53.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Phan AT, Luu KN, Patel DJ. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006;34:5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dai J, Carver M, Punchihewa C, Jones RA, Yang D. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35:4927–4940. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lim KW, Amrane S, Bouaziz S, Xu W, Mu Y, Patel DJ, Luu KN, Phan AT. Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009;131:4301–4309. doi: 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Phan AT, Kuryavyi V, Luu KN, Patel DJ. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007;35:6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amrane S, Ang RW, Tan ZM, Li C, Lim JK, Lim JM, Lim KW, Phan AT. A novel chair-type G-quadruplex formed by a Bombyx mori telomeric sequence. Nucleic Acids Res. 2009;37:931–938. doi: 10.1093/nar/gkn990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu L, Lim KW, Bouaziz S, Phan AT. Giardia telomeric sequence d(TAGGG)4 forms two intramolecular G-quadruplexes in K+ solution: effect of loop length and sequence on the folding topology. J. Am. Chem. Soc. 2009;131:16824–16831. doi: 10.1021/ja905611c. [DOI] [PubMed] [Google Scholar]
- 61.Venczel EA, Sen D. Parallel and antiparallel G-DNA structures from a complex telomeric sequence. Biochemistry. 1993;32:6220–6228. doi: 10.1021/bi00075a015. [DOI] [PubMed] [Google Scholar]
- 62.Hazel P, Huppert J, Balasubramanian S, Neidle S. Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 63.Guédin A, De Cian A, Gros J, Lacroix L, Mergny JL. Sequence effects in single-base loops for quadruplexes. Biochimie. 2008;90:686–696. doi: 10.1016/j.biochi.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 64.Guedin A, Gros J, Alberti P, Mergny JL. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010;38:7858–7868. doi: 10.1093/nar/gkq639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De Cian A, Grellier P, Mouray E, Depoix D, Bertrand H, Monchaud D, Teulade-Fichou MP, Mergny JL, Alberti P. Plasmodium telomeric sequences: structure, stability and quadruplex targeting by small compounds. ChemBiochem. 2008;9:2730–2739. doi: 10.1002/cbic.200800330. [DOI] [PubMed] [Google Scholar]
- 66.Mergny JL, Lacroix L. UV melting of G-quadruplexes. Curr. Protoc. Nucleic Acids Chem. 2009:17.1.1–17.1.15. doi: 10.1002/0471142700.nc1701s37. [DOI] [PubMed] [Google Scholar]
- 67.Mergny JL, Li J, Lacroix L, Amrane S, Chaires JB. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005;33:e138. doi: 10.1093/nar/gni134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kejnovska I, Kypr J, Vorlickova M. Oligo(dT) is not a correct native PAGE marker for single-stranded DNA. Biochem. Biophys. Res. Commun. 2007;353:776–779. doi: 10.1016/j.bbrc.2006.12.093. [DOI] [PubMed] [Google Scholar]
- 69.Mergny JL, Phan AT, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 70.Gray DM, Wen JD, Gray CW, Repges R, Repges C, Raabe G, Fleischhauer J. Measured and calculated CD spectra of G-quartets stacked with the same or opposite polarities. Chirality. 2008;20:431–440. doi: 10.1002/chir.20455. [DOI] [PubMed] [Google Scholar]
- 71.Masiero S, Trotta R, Pieraccini S, De Tito S, Perone R, Randazzo A, Spada GP. A non-empirical chromophoric interpretation of CD spectra of DNA G-quadruplex structures. Org. Biomol. Chem. 2010;8:2683–2692. doi: 10.1039/c003428b. [DOI] [PubMed] [Google Scholar]
- 72.Risitano A, Fox KR. Stability of intramolecular DNA quadruplexes: comparison with DNA duplexes. Biochemistry. 2003;42:6507–6513. doi: 10.1021/bi026997v. [DOI] [PubMed] [Google Scholar]
- 73.Rachwal PA, Findlow IS, Werner JM, Brown T, Fox KR. Intramolecular DNA quadruplexes with different arrangements of short and long loops. Nucleic Acids Res. 2007;35:4214–4222. doi: 10.1093/nar/gkm316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rachwal PA, Brown T, Fox KR. Effect of G-tract length on the topology and stability of intramolecular DNA quadruplexes. Biochemistry. 2007;46:3036–3044. doi: 10.1021/bi062118j. [DOI] [PubMed] [Google Scholar]
- 75.Bugaut A, Balasubramanian S. A sequence-independent study of the influence of short loop lengths on the stability and topology of intramolecular DNA G-quadruplexes. Biochemistry. 2008;47:689–697. doi: 10.1021/bi701873c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smargiasso N, Rosu F, Hsia W, Colson P, Baker ES, Bowers MT, De Pauw E, Gabelica V. G-quadruplex DNA assemblies: loop length, cation identity, and multimer formation. J. Am. Chem. Soc. 2008;130:10208–10216. doi: 10.1021/ja801535e. [DOI] [PubMed] [Google Scholar]
- 77.Guedin A, Alberti P, Mergny JL. Stability of intramolecular quadruplexes: sequence effects in the central loop. Nucleic Acids Res. 2009;37:5559–5567. doi: 10.1093/nar/gkp563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kumar N, Maiti S. A thermodynamic overview of naturally occurring intramolecular DNA quadruplexes. Nucleic Acids Res. 2008;36:5610–5622. doi: 10.1093/nar/gkn543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 80.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nozaki H, Takano H, Misumi O, Terasawa K, Matsuzaki M, Maruyama S, Nishida K, Yagisawa F, Yoshida Y, Fujiwara T, et al. A 100%-complete sequence reveals unusually simple genomic features in the hot-spring red alga Cyanidioschyzon merolae. BMC Biol. 2007;5:28. doi: 10.1186/1741-7007-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stegle O, Payet L, Mergny JL, MacKay DJ, Huppert JL. Predicting and understanding the stability of G-quadruplexes. Bioinformatics. 2009;25:i374–i382. doi: 10.1093/bioinformatics/btp210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.