Abstract
The relationship between virion protein maturation and genomic RNA dimerization of human immunodeficiency virus type 1 (HIV-1) remains incompletely understood. We have constructed HIV-1 Gag cleavage site mutants to enable the steady state observation of virion maturation steps, and precisely study Gag processing, RNA dimerization, virion morphology and infectivity. Within the virion maturation process, the RNA dimer stabilization begins during the primary cleavage (p2-NC) of Pr55 Gag. However, the primary cleavage alone is not sufficient, and the ensuing cleavages are required for the completion of dimerization. From our observations, the increase of cleavage products may not put a threshold on the transition from fragile to stable dimeric RNA. Most of the RNA dimerization process did not require viral core formation, and particle morphology dynamics during viral maturation did not completely synchronize with the transition of dimeric RNA status. Although the endogenous virion RT activity was fully acquired at the initial step of maturation, the following process was necessary for viral DNA production in infected cell, suggesting the maturation of viral RNA/protein plays critical role for viral infectivity other than RT process.
INTRODUCTION
The genome of retrovirus such as human immunodeficiency virus type 1 (HIV-1) is a single-stranded, positive-sense RNA. The viral genome always occurs as a dimer in virus particles, and the interaction is non-covalent since heating easily dissociates purified dimeric genomes into monomers. Genomic RNA dimerization is believed to be a crucial step for the life cycle of retroviruses. Template strand switching between two genomes during reverse transcription is often observed in the retroviral lifecycle (1). It is likely that the presence of two genomes in one virion helps the virus survive by providing genetic variety for their progeny (2). However, this may not fully explain why the virion is required to carry two identical RNAs in spite of severe space limitation, since retroviruses with little sequence variety such as HTLV-1 (3) are also dimeric. Identification of cis-acting signals for retrovirus genome dimerization, called the dimer linkage structure (DLS), was initially attempted in an in vitro assay (4–8). The proposed DLS regions of HIV-1 is located within the untranslated region between LTR and the gag gene (4,9). Although the DLS on viral RNA is suggested to be involved in dimer formation and its close relationship to the packaging signal has been studied (2), there remains incompletely understood issues about the overall mechanisms and the precise nature of retroviral genome dimerization.
The retrovirus dynamically converts the morphology of its particle interior during particle release, termed ‘maturation’. Maturation changes virion morphology from the immature particle, called donut-shaped particle, to the mature virion; a particle lined with viral matrix proteins containing a condensed core composed of a viral capsid shell caging ribonucleoprotein (RNP) complex, comprised of viral RNA, nucleocapsid and enzymes (10). Maturation prepares the virus for infection of adjacent hosts and is inevitably essential for particle infectivity. Although many aspects about how the process of virion maturation contributes to achieving infectivity remain unclear, it is a well-accepted idea that viral RNA within the virion forms a stable and uniform dimer only after complete virion maturation. Obviously, viral protease (PR) activity to process Gag precursor protein (Pr55) is required for stable genomic RNA dimerization in the virion. It has been suggested that Gag precursor, as well as viral NC protein, have RNA chaperone activity and are required for the proper formation of dimeric RNA in the virion (11,12). A defect in its capability to stably dimerize genomic RNA was found in a ΔPR virus (13,14), which led to a hypothesis that one or more Gag cleavage products help form or stabilize genomic RNA dimers. There are five cleavage sites in the HIV-1 Gag protein and the sequential processing of Gag by PR has been discussed so far (15). Some preceding studies suggested that specific Gag cleavage sites or protein regions contribute to viral genome dimerization (16–20). In light of these findings, we constructed two sets of Gag mutants which could represent cleavage intermediates, effectively snapshooting the process of virion maturation in this study. To systematically clarify the dynamic correlation between viral RNA and protein maturation in viral life cycle, virion protein, genomic RNA, virion morphology and infectivity of these mutants were examined comprehensively. We found that NC maturation is critical for the accomplishment of RNA dimerization and viral infectivity, but unnecessary for proper reverse transcription of viral RNA and virion maturation. The mutual relationship between viral protein and RNA maturation was discussed for a further understanding of the retroviral life cycle.
MATERIALS AND METHODS
Constructs
The plasmid pNLNh (21), which contains a 4-base insertion mutation to abrogate Env protein expression on the env gene of HIV-1 proviral clone pNL4-3 (22), was used as the wild-type (WT) and as the progenitor for the mutant constructs described. The amino acid sequence of the Gag cleavage sites and the constitution of the mutants are summarized in Figure 1. The Gag cleavage site mutants MA/CA, CA/p2, p2/NC, NC/p1 and p1/p6, were constructed according to the previous reports (16,23) using PCR stitch mutagenesis (24). The double cleavage site mutant Step2, containing CA/p2 and NC/p1 mutations, was constructed by introducing N432T amino acid substitution to the CA/p2 mutant. The triple cleavage site mutant Step1.1, containing MA/CA, CA/p2 and NC/p1 mutations, was constructed by introducing Y132I amino acid substitution to the Step2 mutant. The quadruple cleavage site mutant Step1, containing all four cleavage site mutations except p2/NC, was constructed by introducing F448S amino acid substitution to the Step1.1 mutant. The quintuple cleavage site mutant Step0, containing all Gag cleavage sites mutations, was constructed by introducing M377V amino acid substitution to the Step1 mutant. The HIV-1 proviral mutant MS172, which lacks PR activity, was previously described (25).
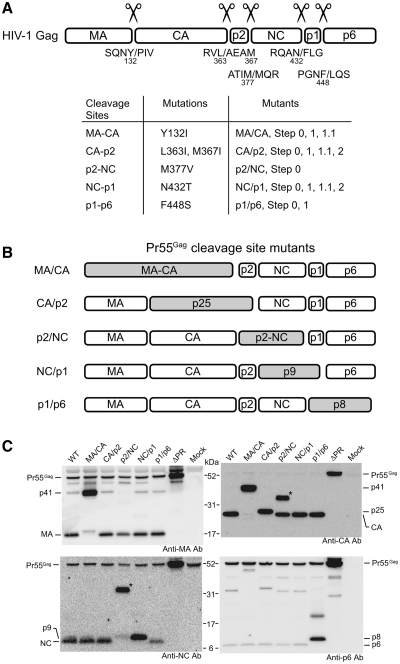
Figure 1.
Gag-cleavage site mutations of HIV-1. (A) Summary of the mutations used in this study. Upper figure represents HIV-1 Gag and its cleavage site sequences. The slashes represent the position of cleavage and the numbers beneath the letters designate amino acid positions in Pr55. The correlation between the mutation and the mutants constructed are summarized in the table. Details on the mutations Y132I, M377V, N432T and F448S have been reported (23), as well as the L363I and M367I mutations (16). The M367I mutation was introduced to block an HIV-1 PR-mediated cryptic cleavage site within the p2 protein because this cryptic cleavage has been reported to occur when proteolytic cleavage between CA and p2 was inhibited by L363I mutation. (B) Schematic representations of the single Gag-cleavage site mutants. Shaded boxes represent fusion proteins which resulted from the mutations. (C) Detection of HIV-1 protein produced in pelleted virions by western blotting with various anti-Gag antibodies. Positions of Gag proteins and precursors were indicated. The asterisks indicate aberrant proteins (‘Results’ section).
Cell culture and transfection
293T cells (26) were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, penicillin and streptomycin. The cells were transfected with the calcium phosphate method (27). To recover virus particles, 5 µg of plasmid DNA was used to transfect the cells in a 100 mm dish. Production of particles containing a mixture of quadruple and quintuple Gag cleavage sites mutants was achieved by co-transfection of Step1 and 0 plasmids in various ratios.
Virus purification
Supernatants of transfected cells were collected 72 h post-transfection and treated with DNase I for elimination of plasmid (28). Then they were centrifuged for 20 min at 4°C and 1570 g to remove cellular debris. The clarified supernatants were concentrated by ultracentrifugation through a 20% sucrose cushion in PBS by using an Optima L-100XP ultracentrifuge and a SW41Ti rotor (Beckman Coulter Inc., Brea, CA, USA) at 151 000 g for 45 min at 4°C.
Western blotting analysis
Equal amounts of viral lysate from each sample were resuspended in NuPAGE LDS Sample Buffer (Invitrogen, Carlsbad, CA, USA). Proteins were resolved by SDS–polyacrylamide gels (4–12%) and transferred to nitrocellulose membranes by using iBlot (Invitrogen). The membranes were treated with mouse anti-MA monoclonal antibody (29), mouse anti-CA monoclonal antibody (Advanced Biotechnologies Inc., Columbia, MD, USA), rabbit anti-NC polyclonal antibody, and rabbit anti-p6 polyclonal antibody (30). ECL Western blotting detection reagents (Nacalai Tesque, Kyoto, Japan) were used for signal detection with horseradish peroxidase conjugated anti-mouse and anti-rabbit IgG (Vector Laboratories, Burlingame, CA, USA). Chemiluminescence was visualized by LAS-1000 (Fujifilm, Tokyo, Japan) according to the manufacturer’s protocols.
Isolation of viral RNA
Purified virions were resuspended in 150 µl of lysis buffer (50 mM Tris–HCl pH 7.5, 100 mM NaCl, 10 mM EDTA, 1% SDS). Viral lysate was treated with 0.2 µg/µl proteinase K (Invitrogen) at room temperature for 60 min, followed by Tris–EDTA-saturated phenol–chloroform extraction, chloroform extraction and ethanol precipitation to collect purified viral RNA.
Native northern blotting analysis
Pelleted RNA was resuspended in T-buffer (31), and the thermostability of dimeric viral RNA was determined by incubating RNA aliquots for 10 min at 25, 35, 40, 44, 48 and 52°C (Figure 2), or 25, 35, 40, 45 and 50°C (Figure 3). One quarter of the total amount of purified viral RNA was applied for each sample. The proportions of the dimers and monomers were measured by electrophoresis at room temperature on a non-denaturing 0.7% agarose gel in 0.5X Tris–borate–EDTA buffer. Field inversion gel electrophoresis was performed for better separation of large RNA molecules. The conditions for field inversion gel electrophoresis were as follows: forward, 5 V/cm and 0.6 s; reverse, 5 V/cm and 0.1 s. The agarose gel was then treated with 10% formaldehyde at 65°C for 30 min before being washed with H2O three times, and the RNA was blotted electrically onto a Hybond-N+ nylon membrane (GE healthcare, Chalfont St Giles, UK).
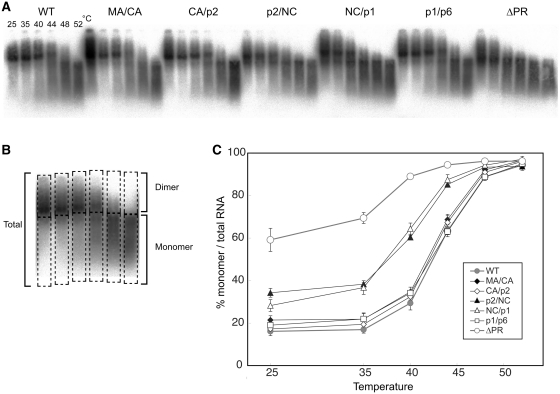
Figure 2.
Thermal stability of Viral RNA dimers of single Gag-cleavage site mutants. (A) Virion RNA profiles detected by northern blotting in a native agarose gel. Viruses were prepared by transfection of 293T cells with the wild-type (WT; pNLNh) virus or its derivative mutants. Aliquots of RNA extracted from virions were resuspended in T-buffer and incubated for 10 min in parallel reactions. The temperatures in which aliquots were incubated are indicated for each lane. (B) A schematic figure for dimer/monomer calculation. Each lane of the blot was separated into two parts, dimer or larger and monomer or smaller molecules as shown in boxes with dashed lines. The relative amounts of monomeric and total RNAs in each lane were quantitated with ImageGauge, and the percentage of the total represented by monomeric RNA was calculated for each RNA sample. (C) Thermal dissociation kinetics of RNA dimers. Results are the average of three independent experiments. Error bars represent SEM.
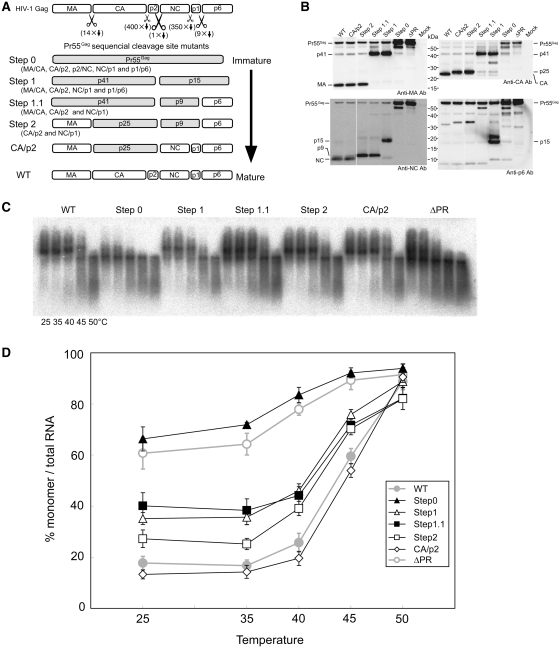
Figure 3.
The mutants to mimic sequential processing of HIV-1 Gag. (A) Schematic representations of the mutants. CA/p2 and NC/p1 mutants were added for reference. Shaded boxes represent fusion proteins which resulted from the mutations. (B) Detection of HIV-1 protein produced in pelleted virions by western blotting with various anti-Gag antibodies. Positions of Gag proteins and precursors are indicated. (C) Raw virion RNA profiles detected by northern blotting in a native agarose gel as in Figure 2. (D) Thermal dissociation kinetics of RNA dimers. The experiments were similarly performed as described in Figure 2. Results are the average of three to five independent experiments. Error bars represent SEM.
RNA northern hybridization analysis was performed as described previously (31). The plasmid pT7pol (31) and T7 RNA polymerase (Takara Bio Inc., Shiga, Japan) were used to synthesize the riboprobe, which specifically detects unspliced viral RNAs, for northern hybridization. Hybridization was carried out in the presence of Rapid-Hyb buffer (GE healthcare). Membranes were washed extensively with 0.1× SSC (1× SSC was comprised of 15.0 mM NaCl and 15 mM sodium citrate [pH 7.0]) 0.1% SDS at 70°C. In experiments designed to assess the conversion of dimers to monomers, the relative amounts of both RNA species were quantitated with a FLA5000 image analyzer and BAS imaging plate (Fujifilm). To characterize the profile of genomic RNA, a lane on the blot was scanned by ImageGauge software (Fujifilm) and the signal intensity and distance migrated from each well were measured.
Electron microscopy
293T cells were transfected with appropriate plasmids. Supernatants were collected 36 h post-transfection and ultracentrifuged for 30 min at 4°C and 49 000 g through a 20% sucrose cushion. Pellets were washed twice with 0.15 M phosphate buffered saline (PBS, pH 7.2) and fixed with 2% glutaraldehyde in PBS at 4°C for 120 min. The fixed pellets were washed five times with PBS. The fixed pellets were further fixed in 1% osmium tetroxide in the same buffer at 4°C for 60 min, washed in the buffer, dehydrated in a graded ethanol series and embedded in epoxy resin (Plain Resin; Nisshin-EM, Tokyo, Japan). Ultrathin sections were cut using a Porter Blum ultramicrotome (Sorvall MT-5000; Du Pont, Newton, CT, USA) and mounted on a copper grid (300 mesh) supported by a carbon-coated collodion film. Finally, the sections were double stained with uranyl acetate and lead citrate. All the sections were observed under a transmission electron microscope (H-7650; Hitachi, Tokyo, Japan) with an accelerating voltage of 80 kV. Electron micrographs were obtained at 10 000-fold magnification.
Endogenous RT assay
Purified virions were suspended with PBS(−) and endogenous RT assay was performed essentially as previously described (32). Aliquots of virus suspensions were incubated in 30 µl of endogenous reverse transcription reaction mixture (0.01% Triton X-100, 50 mM NaCl, 50 mM Tris–HCl [pH 8.0], 10 mM dithiothreitol, 5 mM MgCl2, 100 µM each dATP, dCTP, dGTP and dTTP). As a negative control, reactions without dTTP were performed in parallel. After 2 h of incubation at 37°C, the reaction was terminated by addition of 120 µl of stop mix (50 µg of proteinase K per ml, 20 µg of tRNA per ml, 1.5 mM EDTA [pH 8.0]). After incubation at 60°C for 1 h, the proteinase K was heat-inactivated by incubation at 95°C for 15 min. 4 µl aliquots of the reaction mixtures were subjected to real-time PCR analysis. A portion of virion suspension with PBS (−) was dispensed and viral RNA was purified directly by High Pure Viral RNA Kit (Roche Applied Science, Mannheim, Germany). Viral RNA was applied for quantitation with real-time PCR.
Infection assay
The infection of env-pseudotyped virus into human T-cell lines were performed essentially as previously described (25). M8166/H1Luc cells (33), containing HIV-1 LTR-luciferase vector, were used for single-round infection experiments to determine overall infectivity of virus by Tat expression. Luciferase activity of cells was determined by the Bright-Glo luciferase assay system (Promega, Madison, WI, USA). TRIM5-KD/Luc-KD Jurkat cells (kind gifts from Dr Jeremy Luban) and MT-4 cells (34) were used for viral infection and viral DNA quantitation by real-time PCR assay. 1 × 106 cells per sample were infected with purified virus and total cellular DNA was extracted with GenElute mammalian genomic DNA miniprep kit (Sigma-Aldrich, St Louis, MO, USA) at 18 h post infection. Equal amount of total DNA was applied for real-time PCR analysis.
Real-time PCR
Real-time PCR analysis was performed essentially as previously described (35). For detection of viral RNA from purified virion, Gag primers (TM-GagF: GCAGCCATGCAAATGTTAAAAGAG, and TM-GagR: TCCCCTTGGTTCTCTCATCTGG) were applied with One Step SYBR PrimeScript PLUS RT-PCR kit (Takara Bio Inc.). For detection of viral DNA from MT-4 cells, primers and Taqman probes were selected according to criteria described elsewhere (36). For detection of viral DNA from TRIM5-KD/Luc-KD Jurkat cells, Gag primers and a probe (NL1359F: AGTGGGGGGACATCAAGCAGCCATGCAAAT, NL1500R: TGCTATGTCAGTTCCCCTTGGTTCTCT, and NLgag1400TqmnFB: FAM-ATCAATGAGGAAGCTGCAGAATGGGA-BHQ-1) were applied
RESULTS
Maturation of NC is important for the completion of genome dimerization
To investigate which cleavage site(s) of HIV-1 Gag protein is responsible for genomic RNA dimerization, five single Gag-cleavage site mutants, MA/CA, CA/p2, p2/NC, NC/p1 and p1/p6 (Figure 1B) were constructed. Protein profiles of the mutants released from 293T cells were assessed by Western blotting, using antibodies against MA, CA, NC and p6 (Figure 1C). The mutant MA/CA yielded an MA–CA fusion product (p41) and the mutant CA/p2 yielded a CA-p2 fusion product (p25), as expected. Only a faint band corresponding to the p2-NC fusion product (p9) was observed in the p2/NC mutant and a strong signal was observed as the p32 or p33 intermediate, which was consistent with a previous report (17). This can be considered as an aberrant product comprised by CA-p2-NC fusion protein and may be due to the block of the primary cleavage at p2/NC of Gag, resulting in an aberrant exhibition of other cleavage sites to PR leading to insufficient processing. However, there was also a clear CA protein signal on anti-CA blot in the p2/NC mutant (Figure 1C). As the CA/p2 junction must be cleaved for CA release, it is reasonable to assume that a certain amount of p2-NC protein existed in the mutant. The faint signal from the p9 on the anti-NC blot would suggest that the antibody recognition epitope of NC is somehow masked on the p2-NC fusion protein. The Asn to Thr substitution (amino acid #432) in the mutant NC/p1 was located just proximal to the heptameric slippery sequence, UUUUUUA, where the programmed ribosomal frame shifting for Gag-Pol precursor translation occurs (37). Nonetheless the heptameric sequence itself was conserved completely, the mutation introduced was expected to have little effect on frame shifting. The mutant NC/p1 actually yielded p9 properly. The mutant p1/p6 yielded a p1–p6 fusion product (p8) and the substitution did not affect the peptide sequence of Pol protein. Taken together, all the mutations expressed as expected.
To evaluate the genome RNA dimer stability of these single cleavage site mutants, genomic RNA isolated from mutant virions were incubated at various temperatures and their thermostability was examined with native northern blot analysis (Figure 2A). Genomic RNA was mainly observed as a dimer at 25°C in all mutant virions except for the ΔPR mutant (Figure 2C). The amounts of monomeric RNA for the p2/NC and NC/p1 mutants at 25°C were slightly higher than WT (NLNh) (Figure 2C and D). On the other hand, the amounts of monomers for the other mutants (MA/CA, CA/p2 and p1/p6) were similar to that of WT from 25°C to 44°C. These results indicated that both cleavage sites at the NC termini are important for genomic RNA dimerization. It is also possible that insufficient Gag-cleavage of the p2/NC mutant might contribute to the relatively low stability of its dimeric genome.
The mutants to mimic cleavage intermediates of Pr55
It was previously demonstrated that the processing rates of the five Pr55 cleavage sites were not equal (38). The processing rate of the cleavage site between p2 and NC (p2/NC) is the fastest, while p1/p6 and MA/CA are second and the third, respectively. The processing rates of the two remaining sites are much slower than those of the three sites mentioned above. Although the results from single cleavage site mutants (p2/NC and NC/p1) showed the importance of NC in RNA dimerization (Figure 2), such composition of intermediate proteins would hardly appear during native virion maturation. As virion maturation is suggested to occur during or immediately after virion budding (39,40), it is very difficult to observe this maturation process in a time series. Therefore, four sequential HIV-1 Gag mutants were constructed (Figure 3A) in an attempt to generate a sequential series of Gag maturation intermediates and to help study the RNA dimer transition from fragile to stable, which occurs in virions during maturation. Step0 carries all cleavage site mutations and its Pr55 would not be processed by PR. The mutants Step1, 1.1 and 2 carried four, three and two cleavage site mutations, respectively. Thus, these sequential mutants were expected to mimic the maturation process of Gag in a step-wise manner. All mutations were confirmed to yield expected fusion proteins, respectively by western blotting analysis (Figure 3B). Step0 mostly expressed unprocessed Pr55. Step1 contained p41 and p15 fusion proteins, Step1.1 contained p41 and p9 fusion proteins, and Step2 contained p25 and p9 fusion proteins.
When genomic RNA isolated from the sequential mutants was examined by native northern blotting, the sequential mutants contained a higher rate of monomeric genomes than the WT (Figure 3C and D). The genomic RNA from Step0 was exclusively monomeric, similar to the ΔPR. The monomer rate of Step1, where Gag was cleaved only at p2/NC, was dramatically reduced (<40%) in comparison to that of Step0, and was comparable to the rate of Step1.1. Viral genomes from immature particles were observed as monomers on a native gel, and were proposed to be unstable and fragile dimers (13). These results suggested that the initial cleavage of Gag, which processes p2/NC, was critical for genomic RNA dimer stabilization in HIV-1. The monomer rate of the Step2 was gradually reduced to ∼25%, but still higher than that of WT. Finally, the CA/p2 mutant showed a similar level of monomer content to WT. Taken together, viral genome dimerization showed triphasic states during virion protein maturation.
Consecutive Gag processing is required for genome dimer maturation
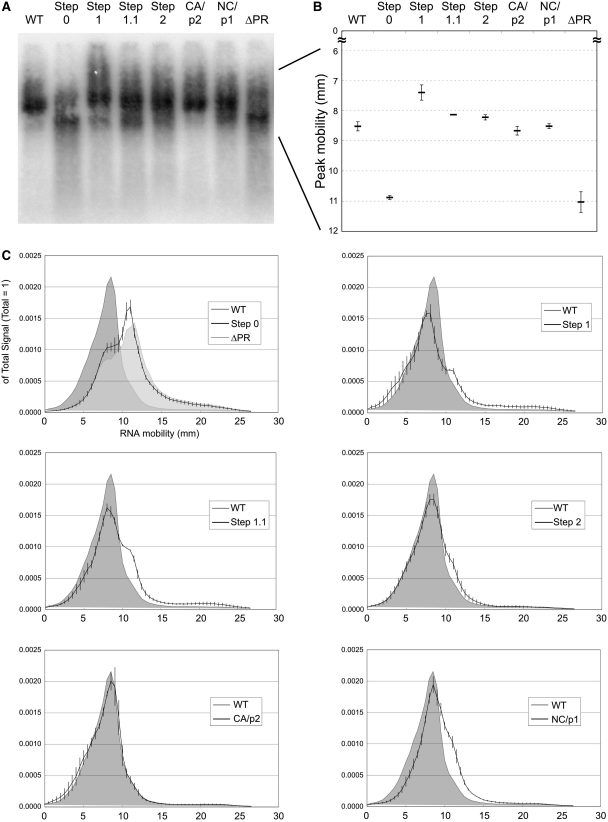
As shown in Figure 3, our data indicated that the proportions of genomic RNA dimers in the Step1 and 1.1 mutants were similar, but we did notice that their RNA profiles were not completely identical. To compare the RNA profiles of these samples more precisely, we scanned the RNA signals on the blot and densitometrically analyzed their distribution (Figure 4). We first focused on the peak position of RNA dimer in each mutant (Figure 4B). The mobility of the dimer in Step1 was considerably lower than that of WT, suggesting its apparent molecular mass to be relatively high. The peak mobility of Step1.1 and 2 were slightly lower than the mobility of WT, while the dimer mobility of CA/p2 and NC/p1 mutants were very similar to WT. We then examined the RNA signal distribution pattern of the mutants on the blots (Figure 4C) When genomic RNA were analyzed on the gel under native conditions (35°C), an intense signal of monomeric RNA from ΔPR mutant formed a sharp peak, although it was preceded by a smaller peak at the dimer position. The RNA signal of Step0 formed the highest peak at the same position as ΔPR and overall signal distributions looked very similar. Interestingly, Step1 formed a broader RNA signal range in comparison to WT. Although the peak signal was detected at the dimer position, the mobility was shorter and signal intensity was relatively weak. Additional signals were observed at larger molecule areas than the normal dimer and at a monomer position. The RNA signal of Step1.1 formed the highest peak at the dimer position, with a modest intensity at the monomer area, which was also observed for Step1. The signal pattern of Step2 was similar to that of Step1.1, but with reduced signals at the monomer area. The signal of CA/p2 mutant was almost the same shape as that of WT, whereas that of NC/p1 was more aberrant and analogous to that of Step1, but the peak signal of dimer was more concentrated. These results suggested that p2/NC processing alone was not sufficient to complete the dimerization of genomic RNA at WT levels and the ensuing cleavages were also required to change the genome into uniform dimers.
Figure 4.
Dissection of RNA profiles of the mutants in native condition. Phosphorimage of RNA incubated at 35°C was taken, its signal intensity distribution analyzed and represented by the line chart. (A) Raw RNA profiles from northern blot. (B) Mobility of peak signals of monomeric RNA (Step0 and ΔPR) or dimeric RNA (others). (C) The RNA profile transition from immature (ΔPR or Step0) to mature (WT) conditions. The profile of WT was drawn as shaded charts in each graph. Signal intensities were measured at every 0.01 mm and total value of each signal was set to one. The plotted data are the value at every 0.5 mm. The data are the average of three separate experiments, and the error bars represent SEM.
Stoichiometric analysis of Gag processing and RNA dimerization
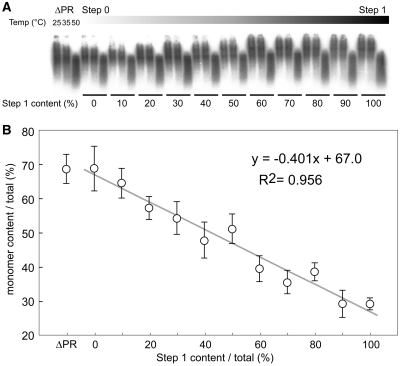
The results presented in Figure 3 revealed that p2/NC processing was the most important step for stable dimeric RNA formation. This suggested that the progression of the initial Pr55 cleavage would stimulate the genomic RNA maturation. To investigate the extent of the required processing, and whether a threshold for the RNA dimerization switch from fragile to stable exists, we attempted to snapshot the initial virion maturation process of HIV-1 through co-transfection. The ratios of Step1 to Step0 were changed from 10:0 to 0:10 successively by mixing two plasmids for co-transfection and the series of resultant viral particles were harvested. The monomeric viral genome content from various particles were visualized and analyzed by northern blot (Figure 5A). The graph represents the relationship between monomeric genome content and proportion of processed Gag (Step1) within the virion (Figure 5B), and the data were fit to a linear regression model (R2 ≈ 0.956). These results indicated that the increase of cleavage products may not have a threshold for the transition from monomeric to dimeric RNA genomes.
Figure 5.
Stoichiometric analysis of Gag processing and RNA dimerization. ΔPR mutant plasmid and the mixtures of Step0 and 1 plasmids were applied for transfection, and a series of resultant viral particles were harvested. The ratios of Step 1 to Step 0 was changed successively from 10:0 to 0:10. (A) Virion RNA profiles were detected by northern blotting in a native agarose gel as in Figure 2. (B) The monomer content of each RNA in native condition was calculated and represented by a scatter diagram. The data are the average of three independent experiments and the error bars represent SEM. The formula and R-squared value of the straight-line approximation are shown.
Virion morphology and genome dimerization
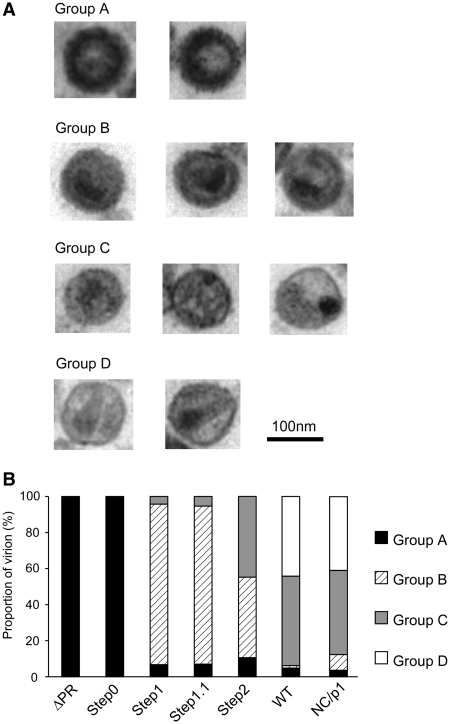
It is well-known that HIV-1 virion morphology dramatically changes during Pr55 processing. To investigate whether the morphological changes of the virion are correlated with genomic RNA dimerization, virion morphological features of the four sequential step mutants and NC/p1 mutant were observed with electron microscopy (Figure 6). Virion morphologies were classified into four main groups (Figure 6A) and morphological classifications were based on viral core and membrane conditions (Table 1). The morphology of all ΔPR virions were typical immature particles (Group A) as demonstrated by many previous reports [e.g. (41)]. As expected, virions from the Step0 mutant were very similar to those of ΔPR, containing a thick electron-dense ring and devoid of a conical-shaped core, termed a doughnut-shaped morphology. The morphologies of the WT, as well as the NC/p1 virions, were classified into two major groups. One was the typical mature particle (Group D), with a conical core surrounded by an envelope (Figure 6A). The second was an enveloped particle containing some amorphous structures rather than an obvious core (Group C). Most of the virions from Step1 and 1.1 were immature (Group B), but did not have an apparent electron-dense ring like Group A particles, with envelopes that were thicker than those of Group C. The virions from Step2 contained both Group B and C particles. We classified the virions from each step into groups and drew a chart to represent the morphological transition of the HIV-1 virion during maturation (Figure 6B). These results indicated that dimer stabilization of the HIV-1 genome, occurring at the timing of p2-NC cleavage, was consistent with the start of virion morphological change from Group A to B, but not with viral core formation.
Figure 6.
Virion morphology of the mutants. (A) Classification of virion morphology judged by electron microscopy analysis. The criterion for judgment was summarized in Table 1. (B) The distribution chart of virion morphology in each mutant. A minimum of one hundred particles were surveyed and classified for each mutant.
Table 1.
Classification of virion morphology based on electron-microscopic observation
| Group | Core | Envelope |
|---|---|---|
| A | Ring-shaped | Thick |
| B | Amorphous | Thick |
| C | Amorphous or non-conical | Thin |
| D | Conical | Thin |
Endogenous RT activity, infectivity and maturation process
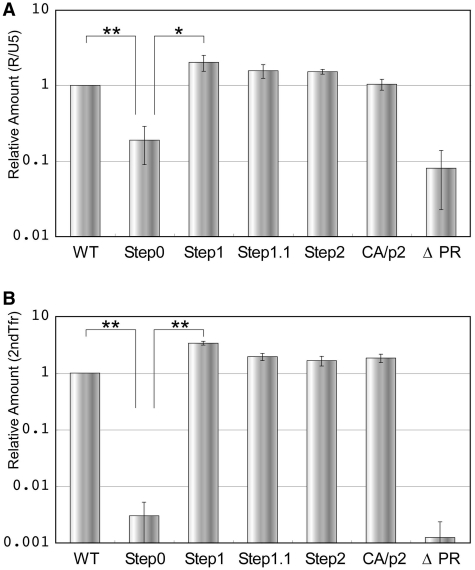
Although the maturation process is required for virion infectivity, the sequential steps to acquire infectivity, remains unclear. To identify the maturation step which prepares viral RNA ready for reverse transcription, the endogenous RT activity of the mutant virions were assessed. The virions were purified from transfectant supernatants, re-suspended in PBS (−) then incubated with detergent and dNTPs. The amount of viral DNA was determined by real-time PCR with two sets of primers, R/U5 (early products) and 2ndTfr (late products) (Figure 7). The RT product of Step0 was reduced by one order of magnitude when compared to WT for the early products, and was attenuated by more than two orders for the late products. Step1 RT products were increased by 2- to 3-fold in both at early and late RT stages. A gradual reduction in the amount of viral DNA was observed, which eventually reached WT levels by the fourth cleavage event (CA/p2 mutant). These data indicated that the initial Pr55 cleavage prepares the viral RNA for RT within virion. The enzymatic activity of viral reverse transcriptase was confirmed by exogenous RT assay. All mutants, with the exception of ΔPR, showed similar RT activity as the WT (data not shown).
Figure 7.
The endogenous RT activity in the virions of mutants. The original values are calculated by dividing the viral DNA amount by the viral RNA amount in virions. Relative amount to WT of early (R-U5: A) and late (2nd Tfer :B) products of RT are shown. The data are the average of at least three independent experiments, and the error bars represent SEM. The statistical significance was evaluated by a Student’s t-test. *P < 0.05; **P < 0.01.
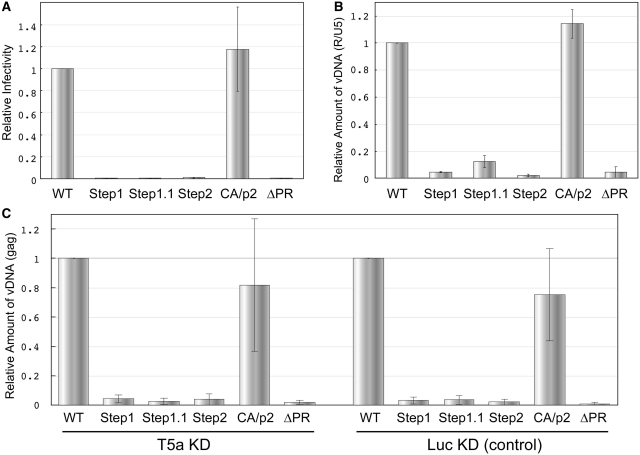
We finally compared the infectivity of the mutants in T-cell lines. Single-round virus infectivity assays using marker cells (M8166/H1Luc) revealed that only the CA/p2 mutant showed similar infectivity to WT (Figure 8A). We performed real-time PCR analysis of viral DNA within infected cells (MT-4) to determine when the defect occurred during virion maturation (Step1, 1.1 and 2), and located the defect at a step before and/or early-RT (Figure 8B). The TRIM5alpha protein (T5a) restricts retrovirus infection during the uncoating process prior to the initiation of RT (42). Although human T5a is suggested to have little anti-HIV-1 effect, immature virions, may be affected by it. To determine the effect of human T5a on the mutants, T5a knock-down (KD) and control cells were applied for infection assay (Figure 8C). The viral DNA production in both cells was very similar, suggesting no participation of T5a restriction with immature virions.
Figure 8.
The infectivity of the mutants into T-cell lines. (A) Overall infectivity into M8166/H1Luc cells. (B) Viral DNA production measured by real-time PCR assay within infected cells (MT-4) 18 h post-infection. (C) Viral DNA production within TRIM5alpha+/− Jurkat cells. In each figure, the value of WT is set as one. The data are the average of at least three independent experiments, and the error bars represent SEM.
DISCUSSION
We have described a sequential mutational analysis series of the HIV-1 Gag cleavage sites to clarify the relationship between virion maturation and genome RNA dimerization, adding further mechanical insight into previously reported observations.
The results from the five single cleavage site mutants in Figures 1 and 2 showed that NC/p1 site as well as p2/NC is involved in genomic RNA dimerization, which is consistent with previous studies (17,18). NC maturation, indicated by the release of NC from p2 and p1, would be a critical factor for genomic RNA dimerization. NC has been reported to convert fragile RNA dimers into stable dimers in a cell-free system, suggesting that NC is an important contributor to RNA dimer formation (43). The partially processed NCs, p2-NC and NC-p1, may have attenuated RNA dimerization properties, indicated by the failure to form normal RNA dimers and leaving them in an unstable state. The dissociation temperature of the RNA dimer in our experiments, ∼45°C, was relatively low in comparison to those in other studies (18,44,45). This might be due to the subtle differences in assay conditions, such as the presence of formamide in RNA buffer.
The examination of genomic RNAs derived from four sequential mutants showed many interesting results (Figure 3). The RNA from Step0 mutant was very similar to that from ΔPR. This result clearly shows that Pr55 cleavage, and not PR activity itself, is required for RNA maturation. Although ΔPR and Step0 mutant show immature RNA phenotype, the blots change with increasing temperature. The RNA in the ΔPR have been suggested to form fragile dimers (13). In addition, the retroviral RNA in the virion contains many nicks regardless of PR cleavage (46). Thus we think that ΔPR/Step0 RNA in native condition partially forms fragile dimers, and it gradually dissociate and is dissected in fragments to be smeared signals as the temperature rises.
The blot also indicated that the main transition of the RNA from a monomer to a dimer state occurred between the Step0 and Step1 mutants (Figure 3). Pr55 Gag is initially cleaved by PR into intermediate proteins p41 (MA-CA-p2) and p15 (NC-p1-p6). Previous reports showed that p15, as well as NC of HIV-1, interacted with in vitro synthesized viral RNA, and GST-fused recombinant p15 protein efficiently induced dimerization of viral RNA fragments (47,48). Our results showed that p15 also promoted genomic RNA dimerization in a nascent virion. A recent report showed the nucleotide binding ability of Pr55 Gag was ∼10-fold higher than NC proteins (p15, p9 and NC), while its destabilizing effect on double-stranded nucleotides was much lower than the NC proteins (49). Taken together with our results, we assumed that the primary processing of Pr55, which is anchored to the viral inner membrane during virion assembly, would release p15 within the virion. Then the tight, non-specific binding between nucleotide and Pr55 is relieved, enabling the NC region of p15 to interact with the proper sites on genomic RNAs to initiate dimer maturation.
The melting temperature (Tm) of RNA from Step2 seems similar to those of Step1/1.1, although the monomer contents are different. We regard that the thermal stability of ‘already dimerized’ RNA in the mutants are similar regardless of dimer formation efficiency. Thus the thermal dissociation curve of the mutants would be converged to present similar Tm.
The molecular mass of dimerized genomic RNA isolated from the Step1 mutant included larger and smaller components than WT (Figure 4). We propose two hypotheses on this observation. First, non-viral RNAs may interact with the detected genomic RNA dimer. Previous reports have suggested that in addition to viral genomes, non-selective RNAs derived from the host cell are packaged in virions (50,51). It is possible that p15 from the Step1 mutant is somewhat different from p9 or WT NC in its ability to recognize its proper viral RNA target. Thus, the slower-migrating complex detected in the Step1 mutant may be composed of genomic RNAs and other non-viral RNAs. Second, genomic RNA dimers in the Step1 mutant may be in a relaxed conformation. It has been often observed that heated genomic RNA dimers have slower mobility in a native gel (e.g. see 44°C lanes of NLNh in Figure 2A), which is considered to be the reason why RNA dimers are relaxed by thermal energy. In either case, ensuing Gag processing and resultant NC maturation would lead to the proper dimerization of genomic RNA and acquire infectivity. As the viral RNA from Step1.1 no longer contained larger components, p15 processing into p9/p6 might play a role(s) to form tight dimers. These results suggested the complexities of dimer maturation and were consistent with the recent report describing a three-step dimerization process of the HIV-1 genome (52).
Some unexpected results were obtained from the stoichiometrical analysis of Gag processing and RNA dimerization (Figure 5). We first considered the uniformity of the ‘chimeric’ virion. There could be an argument whether what we are observing is a changing proportion of two distinct populations of homo-multimeric particles or whether the particles are becoming individual, chimeric virions. Thus we showed the model of virus particle composition here. To simplify the model, we first imposed precondition: that one particle includes 1000 molecules of Gag and the two species of Gag (Step0 and Step1) distributes evenly within the cell. The particle assembly driven by the Gag proteins follows a binomial distribution manner. Under these conditions, the calculations suggested that ∼99.9% of the particle includes 450–550 Gag of each species if the ratios of two species are 1:1. Similar results are obtained in various ratios. This outcome strongly suggests that most of the particle composition reflects the mixed ratio of the plasmids applied for transfection. Thus, we believe the data in Figure 5 represents the RNA status of a chimeric virion.
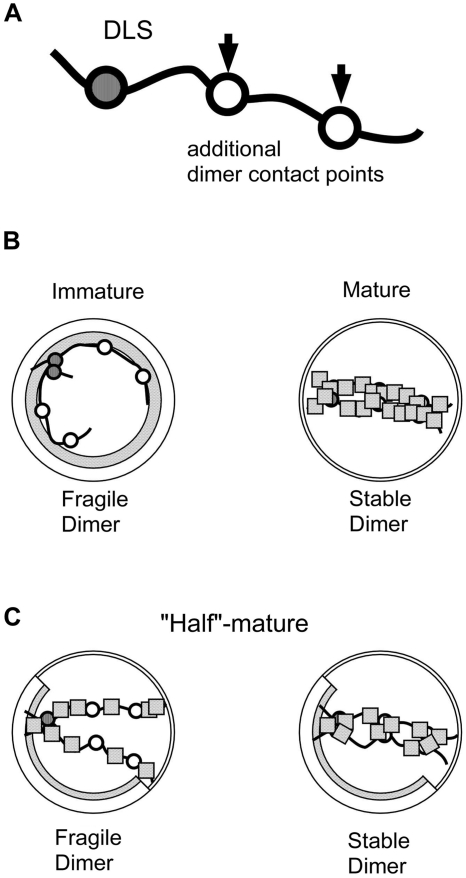
It has been suggested that NC protein has RNA chaperone activity and one NC molecule/20 nt would be sufficient for promoting viral RNA dimerization (43). Thus, we assumed that there should be a p15 molecular concentration threshold imposed on the dimer transition from fragile-to-stable during Pr55 processing. It is reasonable to consider that a certain accumulation of processed Gag would trigger dimer stabilization. However, as in Figure 5, ‘intermediately-processed’ virions packaged stable and unstable dimers about equally. This means that even within the same virion protein composition, a population undergoing genome dimer stabilization encountered a stochastic decision process. To explain this observation, we present one working theory in Figure 9. Since NC displays both non-specific and specific RNA binding, it has been proposed that NC first interacts with the packaging signal and then additional NC domains coat the viral RNA in a sequence-independent manner. The binding ability of NC to RNA has been suggested to be generally in the range of 1 protein molecule per 5–7 nucleotides. (53–55). Thus around 2500–4000 NCs would be required to coat the entire dimeric retroviral genome, which is approximately 9000 nucleotides per strand. Because a retroviral virion is suggested to be composed of about 2500–5000 Gag molecules according to the latest reports (56,57), the quantity of NC proteins in a virion is not in excess, but in rather adequate numbers for a dimerized genome. As some reports have suggested (31,58,59), there is a possibility that additional dimer contact points on viral RNA other than the DLS, which mediate mature ribonucleoprotein complex formation within virion, may exist. Here, we assume that there are several such points on each genome (Figure 9A) and NC proteins binding to these points were simply non-specific and random, such as binding to normal nucleotides. In the fully-processed virions (Step1; Figure 9B, right), all the points are coated with sufficient NC and produced the stable dimer formation. Conversely, if no points are bound with NC, the dimeric genome would remain fragile in immature particles, like particles from the Step0 or ΔPR mutants (Figure 9B, left). In the case of ‘intermediately-processed’ particles, like the virions produced from the Step0–Step1 mixtures, the dimers could be both fragile and stable. As NC (p15) numbers are not sufficient in such particles, the genomes would be only partially covered by NC. If NCs covered the contact points on the genomes and properly mediated the RNA–RNA interaction, the dimer would be stable (Figure 9C, right). On the other hand, if NCs failed to cover the points correctly, the dimer formation would remain fragile (Figure 9C, left).
Figure 9.
Schematic hypothetical models of virion and genome dimerization in various maturation conditions. (A) A diagram of a viral genome carrying one DLS and putative ‘dimer contact points (DCP)’ (B) Images of matured (right) and immature (left) virions. (C) Images of ‘intermediately-matured’ virions. Although both genomes were only partially covered with NC, the particle on left fails to form a stable genome dimer, whereas the particle on the right forms a stable dimer. The dimerization efficiency depends on the extent of coverage of DCP by NC. Solid lines represent viral RNA, Shaded circles represent DLS on the RNA, Open circles represent hypothetical DCP, and dotted squares represent NC (p15).
Electron microscopy analysis of the sequential cleavage site mutants showed successive morphological changes during Pr55 Gag maturation (Figure 6). Virions classified into Group A were <10% from Step0 to Step1, and most of the remaining virions changed to Group B, another immature particle classification. As the change from Group A to B was characterized by disappearance of an electron dense ring inside the viral membrane, the observed ring could be composed by the p15 region within Pr55. After the cleavage at p2-NC position, the ordered array of the p15 region would be lost and p15 would be released from perimembrane. This transition was synchronous with initial dimeric RNA stabilization, suggesting the release of genome RNA from a perimembrane localization to a space inside the virion where p15 would trigger dimer maturation. The cleavage at p1–p6 (from Step 1 to 1.1) did not affect virion morphology, although this process contributed to a uniformly dimerized genome (Figure 4). The thin-membraned virions (Group C) became abundant after the third proteolytic processing at the MA/CA region, represented by the Step2 mutant. A certain portion of Group C particles could easily contain the conical cores of Group D, which are meant to be matured virions, but cut transversely the morphology can appear different. Nonetheless half of Step2 mutant particles were classified into Group B and thus showing unique morphology. Viral membranes of Group B may appear to be thick because of Pr55 CA region fusion to MA, which lines the inner membrane. It has been speculated that viral dimeric RNA could mature in the absence of the viral core (44). Our study not only confirmed this hypothesis, but additionally revealed that RNA maturation would begin and approach completion before core formation in the virion. In addition, although morphological proportion of the virions produced from the NC/p1 mutant was almost similar to WT (Figure 6B), genome dimerization in the NC/p1 mutant was less efficient than in WT (Figure 2). This suggests that proper core formation alone may be insufficient to fully accomplish RNA maturation. Taken together, particle morphology dynamics during viral maturation did not clearly synchronize with the transition of dimeric RNA status.
The processing speed of the NC-p1 cleavage site in vitro has been suggested to be very low and nearly equal to that of CA-p2 (38). This observation leads to the idea that an intermediary such as the NC/p1 mutant could be generated as much as the CA/p2 mutant during the final phase of virion maturation. However, the data from viral RNA profiles (Figures 2 and 4) indicated that NC/p1 mutant contained aberrant RNA and could not support infection. In contrast, the RNA profiles or infectivity of CA/p2 mutant were very similar to WT. In addition, CA/p2 mutant has been reported to contain tightly condensed internal RNP cores with a size and morphology similar to those of WT particles (16). These observations imply that CA-p2 cleavage, but not NC-p1 cleavage, would occur during the final phase of virion maturation. This insight might be explained by the findings of the recent paper (60) which presented the stimulation of p15 (NC-p1-p6) cleavage by PR in the presence of ssRNA/DNA. Due to a simple biochemical assay which ignored the existence of ssRNA genome interacting with most of Pr55 molecules, the processing rate of NC-p1 site within the virions may be underestimated.
The infectivity assay of the mutants unveiled very intriguing results. The endogenous RT activity of viral RNA in the virion was fully detected following the initial cleavage of Pr55, whereas the production of viral DNA in the infected cell was not observed until completion of NC maturation. These results suggested that although viral RNA was fully active as the template for viral reverse transcriptase just after initial cleavage of the precursor, much more virion maturation processes were required for the RT of viral RNA within cells. We form two possibilities to explain these results. One is the existence of cellular restriction system to eliminate immature viral RNA just after the penetration into the cytoplasm. As the restriction activity was not mediated by human T5a (Figure 8C), we assume it could be cytoplasmic RNases or some other enzymes to corrupt RNP of virus. The second possibility is a defect in penetration into the host cell by the mutant. As the most of the mutants failed to form proper virion structure, there may be some defects in the release of RNP into the cells somewhere during viral fusion and uncoating. However, when we examined the infectivity of NC/p1 mutant, we found that NC/p1 produced only <10% early viral DNA in infected cell in comparison to WT (data not shown). As virion morphology of the NC/p1 mutant is nearly similar to the WT (Figure 6B), the uncoating defects of the mutants alone may not fully explain this difference.
In the in vitro assay, p9 or p15 proteins, which are immature NC intermediates, were suggested to obtain insufficient RNA chaperone activity required for strand transfer during reverse transcription (49). Although our results of the endogenous RT assay were not consistent with the study, different experimental conditions can offer partial explanation to this discrepancy. As the samples applied for the endogenous RT assay are purified virions, many viral/host factors are present within the assay conditions and would participate in the reaction.
In summary, we suggested here that mature NC or the intermediate proteins containing NC region are involved in genomic RNA dimerization within the HIV-1 particle. Dimer stabilization begins during the primary cleavage (p2-NC) of Pr55 Gag. However, the primary cleavage alone is not sufficient for completion of dimerization and ensuing cleavages are required to generate a stable and uniform dimer. The majority of this process does not require a mature viral core. In contrast, the primary cleavage is more than enough to initiate endogenous RT activity of viral RNA, whereas overall viral infectivity, as well as intracellular viral DNA production, required the near-completion of viral maturation.
Other retroviruses also have NC and dimeric RNA genomes, suggesting that RNA dimerization could occur in the same manner as HIV-1. Future studies with HIV-1 or other retroviruses will contribute to clarify the general and specific roles and mechanisms of retroviral RNA dimerization, which continues to be an age-old problem.
FUNDING
Funding for open access charge: Ministry of Education, Culture, Sports, Science and Technology; the Ministry of Health, Labour and Welfare; the Health Science Foundation, Japan.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Mr Yoshihiko Fujioka of the Department of Microbiology and Infection Control, Osaka Medical College, for his technical help in Electron Microscopy study and Dr Seiga Ohmine of the Mayo Clinic, for critically reading the article.
REFERENCES
- 1.Hu WS, Temin HM. Retroviral recombination and reverse transcription. Science. 1990;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 2.Paillart JC, Shehu-Xhilaga M, Marquet R, Mak J. Dimerization of retroviral RNA genomes: an inseparable pair. Nat. Rev. Microbiol. 2004;2:461–472. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 3.Greatorex JS, Laisse V, Dockhelar MC, Lever AM. Sequences involved in the dimerisation of human T cell leukaemia virus type-1 RNA. Nucleic Acids Res. 1996;24:2919–2923. doi: 10.1093/nar/24.15.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darlix JL, Gabus C, Nugeyre MT, Clavel F, Barre-Sinoussi F. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J. Mol. Biol. 1990;216:689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- 5.Prats AC, Roy C, Wang PA, Erard M, Housset V, Gabus C, Paoletti C, Darlix JL. cis elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. J. Virol. 1990;64:774–783. doi: 10.1128/jvi.64.2.774-783.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy C, Tounekti N, Mougel M, Darlix JL, Paoletti C, Ehresmann C, Ehresmann B, Paoletti J. An analytical study of the dimerization of in vitro generated RNA of Moloney murine leukemia virus MoMuLV. Nucleic Acids Res. 1990;18:7287–7292. doi: 10.1093/nar/18.24.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marquet R, Baudin F, Gabus C, Darlix JL, Mougel M, Ehresmann C, Ehresmann B. Dimerization of human immunodeficiency virus (type 1) RNA: stimulation by cations and possible mechanism. Nucleic Acids Res. 1991;19:2349–2357. doi: 10.1093/nar/19.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darlix JL, Gabus C, Allain B. Analytical study of avian reticuloendotheliosis virus dimeric RNA generated in vivo and in vitro. J. Virol. 1992;66:7245–7252. doi: 10.1128/jvi.66.12.7245-7252.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marquet R, Paillart JC, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of human immunodeficiency virus type 1 RNA involves sequences located upstream of the splice donor site. Nucleic Acids Res. 1994;22:145–151. doi: 10.1093/nar/22.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adamson CS, Salzwedel K, Freed EO. Virus maturation as a new HIV-1 therapeutic target. Expert Opin. Ther. Targets. 2009;13:895–908. doi: 10.1517/14728220903039714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng YX, Campbell S, Harvin D, Ehresmann B, Ehresmann C, Rein A. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J. Virol. 1999;73:4251–4256. doi: 10.1128/jvi.73.5.4251-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng YX, Copeland TD, Henderson LE, Gorelick RJ, Bosche WJ, Levin JG, Rein A. HIV-1 nucleocapsid protein induces "maturation" of dimeric retroviral RNA in vitro. Proc. Natl Acad. Sci. USA. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu W, Gorelick RJ, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu W, Rein A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 1993;67:5443–5449. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pettit SC, Sheng N, Tritch R, Erickson-Viitanen S, Swanstrom R. The regulation of sequential processing of HIV-1 Gag by the viral protease. Adv. Exp. Med. Biol. 1998;436:15–25. doi: 10.1007/978-1-4615-5373-1_2. [DOI] [PubMed] [Google Scholar]
- 16.Wiegers K, Rutter G, Kottler H, Tessmer U, Hohenberg H, Krausslich HG. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 1998;72:2846–2854. doi: 10.1128/jvi.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shehu-Xhilaga M, Kraeusslich HG, Pettit S, Swanstrom R, Lee JY, Marshall JA, Crowe SM, Mak J. Proteolytic processing of the p2/nucleocapsid cleavage site is critical for human immunodeficiency virus type 1 RNA dimer maturation. J. Virol. 2001;75:9156–9164. doi: 10.1128/JVI.75.19.9156-9164.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kafaie J, Dolatshahi M, Ajamian L, Song R, Mouland AJ, Rouiller I, Laughrea M. Role of capsid sequence and immature nucleocapsid proteins p9 and p15 in Human Immunodeficiency Virus type 1 genomic RNA dimerization. Virology. 2009;385:233–244. doi: 10.1016/j.virol.2008.11.028. [DOI] [PubMed] [Google Scholar]
- 19.Fu W, Dang Q, Nagashima K, Freed EO, Pathak VK, Hu WS. Effects of Gag mutation and processing on retroviral dimeric RNA maturation. J. Virol. 2006;80:1242–1249. doi: 10.1128/JVI.80.3.1242-1249.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jalalirad M, Laughrea M. Formation of immature and mature genomic RNA dimers in wild-type and protease-inactive HIV-1: differential roles of the Gag polyprotein, nucleocapsid proteins NCp15, NCp9, NCp7, and the dimerization initiation site. Virology. 2010;407:225–236. doi: 10.1016/j.virol.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 21.Sakuragi J, Ueda S, Iwamoto A, Shioda T. Possible role of dimerization in human immunodeficiency virus type 1 genome RNA packaging. J. Virol. 2003;77:4060–4069. doi: 10.1128/JVI.77.7.4060-4069.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pettit SC, Henderson GJ, Schiffer CA, Swanstrom R. Replacement of the P1 amino acid of human immunodeficiency virus type 1 Gag processing sites can inhibit or enhance the rate of cleavage by the viral protease. J. Virol. 2002;76:10226–10233. doi: 10.1128/JVI.76.20.10226-10233.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohishi M, Shioda T, Sakuragi Ji. Retro-transduction by virus pseudotyped with glycoprotein of vesicular stomatitis virus. Virology. 2007;362:131–138. doi: 10.1016/j.virol.2006.12.030. [DOI] [PubMed] [Google Scholar]
- 25.Sakuragi J, Iwamoto A, Shioda T. Dissociation of genome dimerization from packaging functions and virion maturation of human immunodeficiency virus type 1. J. Virol. 2002;76:959–967. doi: 10.1128/JVI.76.3.959-967.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DuBridge RB, Tang P, Hsia HC, Leong PM, Miller JH, Calos MP. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aldovini A, Walker BD. Techniques in HIV Research. New York: Stockton Press; 1990. [Google Scholar]
- 28.Koh KB, Fujita M, Adachi A. Elimination of HIV-1 plasmid DNA from virus samples obtained from transfection by calcium-phosphate co-precipitation. J. Virol. Methods. 2000;90:99–102. doi: 10.1016/s0166-0934(00)00224-x. [DOI] [PubMed] [Google Scholar]
- 29.Ota A, Liu X, Fujio H, Sakato N, Ueda S. Random expression of human immunodeficiency virus-1 (HIV-1) p17 (epitopes) on the surface of the HIV-1-infected cell. Hybridoma. 1998;17:73–75. doi: 10.1089/hyb.1998.17.73. [DOI] [PubMed] [Google Scholar]
- 30.Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakuragi JI, Panganiban AT. Human immunodeficiency virus type 1 RNA outside the primary encapsidation and dimer linkage region affects RNA dimer stability in vivo. J. Virol. 1997;71:3250–3254. doi: 10.1128/jvi.71.4.3250-3254.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masuda T, Planelles V, Krogstad P, Chen IS. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J. Virol. 1995;69:6687–6696. doi: 10.1128/jvi.69.11.6687-6696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagao T, Yoshida A, Sakurai A, Piroozmand A, Jere A, Fujita M, Uchiyama T, Adachi A. Determination of HIV-1 infectivity by lymphocytic cell lines with integrated luciferase gene. Int. J. Mol. Med. 2004;14:1073–1076. [PubMed] [Google Scholar]
- 34.Harada S, Koyanagi Y, Yamamoto N. Infection of HTLV-III/LAV in HTLV-I-carrying cells MT-2 and MT-4 and application in a plaque assay. Science. 1985;229:563–566. doi: 10.1126/science.2992081. [DOI] [PubMed] [Google Scholar]
- 35.Sakuragi J, Sakuragi S, Shioda T. Minimal region sufficient for genome dimerization in the human immunodeficiency virus type 1 virion and its potential roles in the early stages of viral replication. J. Virol. 2007;81:7985–7992. doi: 10.1128/JVI.00429-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Julias JG, Ferris AL, Boyer PL, Hughes SH. Replication of phenotypically mixed human immunodeficiency virus type 1 virions containing catalytically active and catalytically inactive reverse transcriptase. J. Virol. 2001;75:6537–6546. doi: 10.1128/JVI.75.14.6537-6546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dulude D, Theberge-Julien G, Brakier-Gingras L, Heveker N. Selection of peptides interfering with a ribosomal frameshift in the human immunodeficiency virus type 1. RNA. 2008;14:981–991. doi: 10.1261/rna.887008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettit SC, Moody MD, Wehbie RS, Kaplan AH, Nantermet PV, Klein CA, Swanstrom R. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaplan AH, Manchester M, Swanstrom R. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J. Virol. 1994;68:6782–6786. doi: 10.1128/jvi.68.10.6782-6786.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bukrinskaya AG. HIV-1 assembly and maturation. Arch. Virol. 2004;149:1067–1082. doi: 10.1007/s00705-003-0281-8. [DOI] [PubMed] [Google Scholar]
- 41.Peng C, Ho BK, Chang TW, Chang NT. Role of human immunodeficiency virus type 1-specific protease in core protein maturation and viral infectivity. J. Virol. 1989;63:2550–2556. doi: 10.1128/jvi.63.6.2550-2556.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakayama EE, Shioda T. Anti-retroviral activity of TRIM5 alpha. Rev. Med. Virol. 2010;20:77–92. doi: 10.1002/rmv.637. [DOI] [PubMed] [Google Scholar]
- 43.Muriaux D, De Rocquigny H, Roques BP, Paoletti J. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop–loop complex into a stable dimer. J. Biol. Chem. 1996;271:33686–33692. doi: 10.1074/jbc.271.52.33686. [DOI] [PubMed] [Google Scholar]
- 44.Moore MD, Fu W, Soheilian F, Nagashima K, Ptak RG, Pathak VK, Hu WS. Suboptimal inhibition of protease activity in human immunodeficiency virus type 1: effects on virion morphogenesis and RNA maturation. Virology. 2008;379:152–160. doi: 10.1016/j.virol.2008.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paillart JC, Dettenhofer M, Yu XF, Ehresmann C, Ehresmann B, Marquet R. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J. Biol. Chem. 2004;279:48397–48403. doi: 10.1074/jbc.M408294200. [DOI] [PubMed] [Google Scholar]
- 46.Johnson SF, Telesnitsky A. Retroviral RNA dimerization and packaging: the what, how, when, where, and why. PLoS Pathog. 2010;6:e1001007. doi: 10.1371/journal.ppat.1001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weiss S, Konig B, Morikawa Y, Jones I. Recombinant HIV-1 nucleocapsid protein p15 produced as a fusion protein with glutathione S-transferase in Escherichia coli mediates dimerization and enhances reverse transcription of retroviral RNA. Gene. 1992;121:203–212. doi: 10.1016/0378-1119(92)90123-7. [DOI] [PubMed] [Google Scholar]
- 48.Barat C, Lullien V, Schatz O, Keith G, Nugeyre MT, Gruninger-Leitch F, Barre-Sinoussi F, LeGrice SF, Darlix JL. HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J. 1989;8:3279–3285. doi: 10.1002/j.1460-2075.1989.tb08488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cruceanu M, Urbaneja MA, Hixson CV, Johnson DG, Datta SA, Fivash MJ, Stephen AG, Fisher RJ, Gorelick RJ, Casas-Finet JR, et al. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 2006;34:593–605. doi: 10.1093/nar/gkj458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muriaux D, Mirro J, Harvin D, Rein A. RNA is a structural element in retrovirus particles. Proc. Natl Acad. Sci. USA. 2001;98:5246–5251. doi: 10.1073/pnas.091000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rulli SJ, Jr, Hibbert CS, Mirro J, Pederson T, Biswal S, Rein A. Selective and non-selective packaging of cellular RNAs in retrovirus particles. J. Virol. 2007;81:6623–6631. doi: 10.1128/JVI.02833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song R, Kafaie J, Yang L, Laughrea M. HIV-1 viral RNA is selected in the form of monomers that dimerize in a three-step protease-dependent process; the DIS of stem-loop 1 initiates viral RNA dimerization. J. Mol. Biol. 2007;371:1084–1098. doi: 10.1016/j.jmb.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 53.Rein A, Henderson LE, Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 54.Freed EO. HIV-1 replication. Somat Cell Mol. Genet. 2001;26:13–33. doi: 10.1023/a:1021070512287. [DOI] [PubMed] [Google Scholar]
- 55.Shvadchak VV, Klymchenko AS, de Rocquigny H, Mely Y. Sensing peptide–oligonucleotide interactions by a two-color fluorescence label: application to the HIV-1 nucleocapsid protein. Nucleic Acids Res. 2009;37:e25. doi: 10.1093/nar/gkn1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Briggs JA, Johnson MC, Simon MN, Fuller SD, Vogt VM. Cryo-electron microscopy reveals conserved and divergent features of gag packing in immature particles of Rous sarcoma virus and human immunodeficiency virus. J. Mol. Biol. 2006;355:157–168. doi: 10.1016/j.jmb.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 57.Briggs JA, Simon MN, Gross I, Krausslich HG, Fuller SD, Vogt VM, Johnson MC. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 2004;11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 58.Tchénio T, Heidmann T. The dimerization/packaging sequence is dispensable for both the formation of high-molecular-weight RNA complexes within retroviral particles and the synthesis of proviruses of normal structure. J. Virol. 1995;69:1079–1084. doi: 10.1128/jvi.69.2.1079-1084.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ortiz-Conde BA, Hughes SH. Studies of the genomic RNA of leukosis viruses: implications for RNA dimerization. J. Virol. 1999;73:7165–7174. doi: 10.1128/jvi.73.9.7165-7174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mirambeau G, Lyonnais S, Coulaud D, Hameau L, Lafosse S, Jeusset J, Borde I, Reboud-Ravaux M, Restle T, Gorelick RJ, et al. HIV-1 protease and reverse transcriptase control the architecture of their nucleocapsid partner. PLoS One. 2007;2:e669. doi: 10.1371/journal.pone.0000669. [DOI] [PMC free article] [PubMed] [Google Scholar]