Abstract
eIF4E binding protein (4E-BP) inhibits translation of capped mRNA by binding to the initiation factor eIF4E and is known to be mostly or completely unstructured in both free and bound states. Using small angle X-ray scattering (SAXS), we report here the analysis of 4E-BP structure in solution, which reveals that while 4E-BP is intrinsically disordered in the free state, it undergoes a dramatic compaction in the bound state. Our results demonstrate that 4E-BP and eIF4E form a ‘fuzzy complex’, challenging current visions of eIF4E/4E-BP complex regulation.
INTRODUCTION
Translation is a complex process that is critical for gene regulation during development, differentiation, nervous system function, aging and disease. In eukaryotes, translation of mRNA begins with specific recognition of the 5′-cap structure by the highly conserved eukaryotic Initiation Factor 4E, eIF4E, a master switch in translation initiation that participates in recruiting mRNAs to the ribosome (1). This cap-dependent initiation process is mainly regulated by the eIF4E binding protein (4E-BP), a small protein of ∼13 kDa, which plays the role of an important repressor of translation.
4E-BP is known to prevent the association of eIF4E with the large scaffolding protein eIF4G that acts as a docking site for several proteins required to bridge the ribosome and the mRNA. 4E-BP shares with eIF4G the eIF4E-binding motif YXXXXLΦ (where X is any amino acid and Φ is a hydrophobic residue) (2) that interacts with the convex dorsal surface of eIF4E. Binding of 4E-BP to eIF4E is classically described as resulting from the interaction between the 4E-BP central consensus motif and eIF4E, which is dependent upon the phosphorylation status of 4E-BP (3,4). Nevertheless, the regulation of the association/dissociation (5) as well as the molecular mechanisms occurring during eIF4E/4E-BP complex formation are not fully understood yet, even if eIF4E and its repressor 4E-BP are to date some of the most characterized chemically and biophysically cap-dependent initiation factors. A new structural study of the eIF4E/4E-BP complex is therefore important to better understand the regulation of cap-dependent initiation.
Previous high-resolution X-ray crystallography revealed that the eIF4E recognition motif contained in the 4E-BP1 peptide (residues 51–67) undergoes a folded transition upon binding to eIF4E forming an L-shaped structure consisting of an extended chain region and a short α-helix (6). Until now, the unfolded nature of 4E-BP outside of the central peptide has been an obstacle for conventional structural methods and consequently no structure of the eIF4E/4E-BP full length complex has been obtained. Nuclear magnetic resonance (NMR) approaches have concluded in a mostly or completely unstructured 4E-BP1 in both free and bound states (7), and have shown in a very indirect manner that the binding footprint of 4E-BP on eIF4E might be larger than the one established by crystallography (8). Nevertheless, it is generally considered that only the central peptide of 4E-BP plays a major role in the eIF4E/4E-BP complex formation, and that consequently, flexible sections that flank the central peptide of 4E-BP do not.
Interestingly, recent data showed that 4E-BP, unlike eIF4G, is still able to interact with eIF4E when the eIF4E-binding site is occupied by the small pharmacologic compound 4EGi-1 (for eIF4E/eIF4G interaction inhibitor) (9). These data are difficult to integrate if 4E-BP remains disordered when associated with eIF4E. Therefore, since this statement is in contradiction with biochemical data (9) and the eIF4E/4E-BP complex formation remains not well understood, we decided to reconsider the folding status of 4E-BP associated with eIF4E. We applied the appropriate and increasingly used method for the structural analysis in solution, namely small angles X-ray scattering (SAXS). SAXS has proven to be a fundamental tool for the study of biological molecules in solution (10–13). The originality of this method lies in the fact that it can provide structural information on molecules exhibiting some intrinsic disorder, flexibility or heterogeneity, which usually constitutes a major obstacle for the other classical structural methods. Applying this technique to study the eIF4E/4E-BP complex, we observed for the first time the entire structure at low resolution of the complex eIF4E/full-length 4E-BP and show a dramatic compaction of 4E-BP while it interacts with eIF4E. The combination of our results to other previous structural analysis, allowed us to build a new model of interaction, involving a much larger binding footprint of 4E-BP on eIF4E.
MATERIALS AND METHODS
Proteins expression and purification
The construction of the plasmid pAr encoding eIF4E1 from mus musculus has already been described (14) and the protocol to produce and purify eIF4E1 tagged FLAG was performed as described herein. The cloning of Strongylocentrotus purpuratus Sp4E-BP was performed using a S. purpuratus cDNA library. The 342-bp fragment corresponding to the Sp4E-BP coding region was amplified by PCR and inserted into the pFO4 vector digested with BamH1 and EcoR1. 4E-BP, with an Histidine tag, was overexpressed in the BL21 (DE3) strain Escherichia coli (Novagen). Cellular pellet from bacteria were resuspended in 5 volumes buffer A (50 mM sodium phosphate, 300 mM NaCl, 80 mM imidazole, pH 8). Cells were lysed using French Press with an amplitude of 1.6. The lysate was then incubated 30 min at 4°C with 1% Triton (Sigma) and clarified by centrifugation at 10 000g for 30 min. The clarified supernatant was then injected on an affinity column, packed with a chelating sepharose Fast Flow Resin preloaded with Ni2+ ions (Amersham Pharmacia Biotech), previously equilibrated with buffer A. The column was washed with buffer A and the protein was eluted with a gradient of imidazole from 80 to 500 mM. Eluates were analyzed by SDS–PAGE for the presence of the desired product. The fractions containing the recombinant product were combined, and concentrated using Amicon® Ultra-4 Centrifugal Filter Units (10 kDa). The protein was then loaded onto a Superdex 75 column (Amersham Pharmacia Biotech) and eluted in buffer C (50 mM Phosphate, 300 mM NaCl, pH 8). The proteins were concentrated with Amicon® Ultra-4 Centrifugal Filter Units (10 kDa) and the concentration of the purified proteins was estimated with a Nanodrop ND-1000.
After production and purification, 1.5 mg of eIF4E1 was incubated for 1 h with 500 µl of m7GTP sepharose beads (Amersham Pharmacia Biolabs) in buffer CAP (50 mM HEPES, 150 mM KCl, 1 mM EDTA, 5% glycerol, pH 7.7). After washing, the beads were incubated for 1 h in buffer CAP containing 1 mg/ml of BSA, 0.5% Igepal with 4 mg of 4E-BP. After extensive washing, the complex was eluted from the affinity column with 200 µM of free m7GTP in buffer CAP. After concentration with Amicon® Ultra-4 Centrifugal Filter Units (10 kDa), the complex was then loaded onto a Superdex 75 column (Amersham Pharmacia Biotech) and eluted in buffer CAP. The peak containing the complex was concentrated with an Amicon® Ultra-4 Centrifugal Filter Units (10 kDa). The quality of the purified proteins was analyzed by SDS–PAGE (Supplementary Figure S1). As expected we observed two bands with the sample containing the complex of eIF4E and 4E-BP. Quantification with the image analysis software ImageQuant reveals a ratio of 2.1 between the two bands corresponding, respectively, to eIF4E and 4E-BP, consistent with a stoichiometry ratio of 1:1 since 4E-BP is two times smaller than eIF4E.
SAXS experiments
SAXS experiments were carried out twice on the beamline ID14-3 at the European Synchrotron facility (ESRF), Grenoble, France and on the beamline X33 at the DORIS-III storage ring at DESY (Deutsches Elektronen-Synchrotron), Hamburg, Germany. Data have been treated on both beamlines, showing essentially the same results. However, the higher brilliance of the beam at the ESRF consequently was more suitable for analyzing our protein concentrations, which is why the data shown here only come from the beamline ID14-3. On ID14-3, the wavelength was 0.931 Å and the sample-to-detector distance was 1.67 m, leading to scattering vectors q ranging from 0.1 to 5 nm−1. Due to the low concentration of some of our samples (∼1 mg/ml), the data beyond 0.25 Å−1 were statistically not exploitable and were therefore truncated in order to enhance the analysis quality. The scattering vector is defined as q = 4π/λ sin θ, where 2θ is the scattering angle. The detector used was an X-ray detector Pilatus 1 M from Dectris coupled to a CCD camera and 10 successive frames of 10 s exposure time were recorded for each sample. Each frame was carefully checked for possible bubble formation or radiation-induced aggregation. If such effects were not observed, the individual frames were averaged. A 4.8 mg/ml solution of BSA was measured as a reference and for calibration procedures. Up to five different concentrations of each protein sample were measured to test for consistency and eventually detect and eliminate concentration dependent effects. Background scattering was quantified before and after each protein measurement by measuring the corresponding buffer, which is then subtracted from the protein patterns using the Primus suite (15). All experiments were performed at 16°C.
Data evaluation
For high concentrations of 4E-BP, the experimental SAXS data were not linear in the Guinier plot of the low q region, indicating that the proteins undergo aggregation. We only used the scattering profile for concentrations below 6 mg/ml. The radius of gyration, Rg, was derived by the Guinier approximation: I(q) = I(0)exp(−q2Rg2/3) up to qRg < 1.0. The radii of gyration, Rg, calculated for different 4E-BP concentrations display concentration dependence at high concentration, indicating the presence of attractive interactions of the particles in solution. The Rg was approximated for a theoretical concentration of 0 mg/ml. The Rg calculated for different complex eIF4E/4E-BP concentrations display no concentration dependence and indicate the absence of interaction in solution. The program GNOM (16) was used to compute the pair-distance distribution functions P(r). This approach gives the maximum dimension of the macromolecule Dmax and offers an alternative calculation of Rg, which is based on the entire scattering curve. Data evaluation for free eIF4E is reported in the Supplementary Data.
Ab Initio modeling of the overall shapes
The overall shapes of all assemblies were restored from the experimental data by three independent programs (detailed in Supplementary Data): DAMMIN, GASBOR and DAMMIF (17). The scattering profiles were used up to qmax = 0.25 Å−1. Nine low-resolution models obtained from different runs were compared using the program DAMAVER (18) to give an estimate of the reproducibility of the results inferred from the ab initio shape calculation and to construct the average model representing the general structural features of all the reconstruction filtered by DAMFILT.
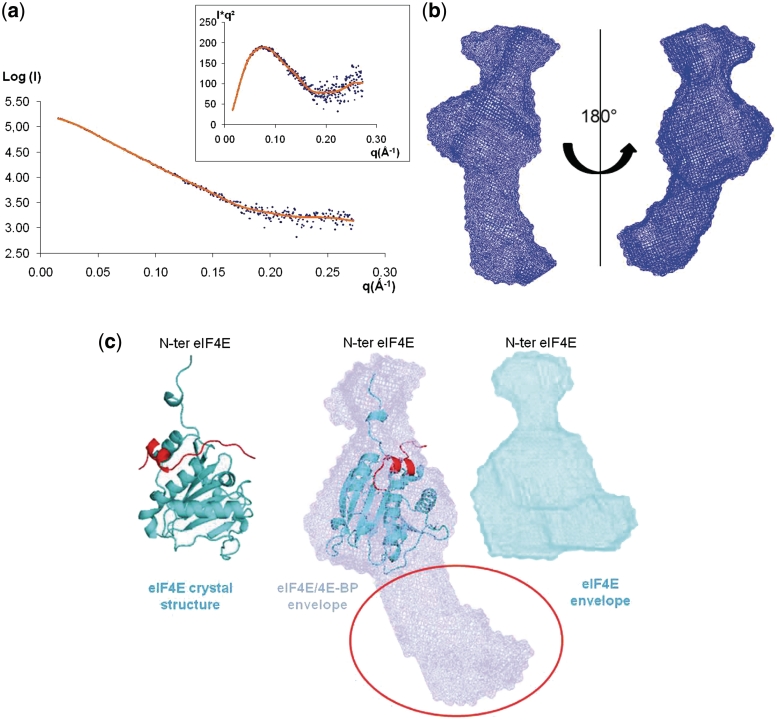
The ‘drag’ option of the program Pymol (The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC) was then used to position manually the crystal structure of eIF4E (19) into the low-resolution envelope of eIF4E obtained by SAXS measurements (Supplementary Figure S2). The same method was applied to position the crystal structure of the complex eIF4E/ 4E-BP peptide (20) into the low-resolution envelope of the complex eIF4E/4E-BP, respectively (Figure 2c). To establish this crystal structure, Tomoo et al. (20) used a 4E-BP peptide containing the amino acids from 36 to 70 but only the residues 47–66 are finally present in the structure because they were the only ones visible on the electron density. The Protein Data Bank accession codes for eIF4E and eIF4E/4E-BP peptide crystal structures are respectively 1L8B and 1WKW.
Figure 2.
Structure of 4E-BP bound to eIF4E in solution. (a) Experimental SAXS profile of the complex 4E-BP/eIF4E (blue dots) fitted by the scattering curves calculated with the program DAMMIF (brown). Inset: Kratky plot [I(q)q2 versus q] showing a typical profil of a partially unfolded protein. (b) Example of the most typical shape of the complex 4E-BP/eIF4E, as obtained by DAMMIF, shown in two orientations rotated by 180°. (c) Example of the most typical shape of the complex 4E-BP/eIF4E displayed in a blue transparent surface representation, superimposed with the crystal structure of eIF4E lacking the 30 amino acids on N-terminal (blue) and complexed with the 4E-BP peptide (red) (PDB number: 1WKW) (20). The position of the N-terminal extremity has been determined in the Supplementary Figure S2. The red circle indicates the position of the extended extremities of 4E-BP full-length. In cyan blue is displayed the average low resolution model for the free eIF4E.
These structural superimpositions of the partial crystal structures and the low resolution solution structures obtained by SAXS measurements demonstrate that the overall shape of the eIF4E/4E-BP complex does not coincide with the shape of eIF4E free in solution but clearly results from the association of 4E-BP and eIF4E. The goodness of fit for all atomic models, as well as the low resolution models, was determined using the χ2 discrepancy (15) reported in the experimental data (Table 1).
Table 1.
Experimental and model SAXS parameters
| Proteins | Exp. I(0)a | Conc. (mg/ml)b | Estimated Mr (kDa)c | Mrd (kDa) | Number of amino acids | Rg (GNOM)e | Rg (Guinier)e | Dmaxf (nm) | Χ(over) | Χ(crysol) | Damaver NSDg |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 4E-BP | 4883 | 1.86 | 20 | 12.8 | 117 | 4.86 ± 0.22 | 4.88 ± 0.17 | 16.0 ± 0.8 | 0.430 | NA | 2.50 ± 0.14 |
| eIF4E | 4256 | 1.26 | 26.6 | 25.05 | 217 | 2.34 ± 0.11 | 2.23 ± 0.44 | 6.8 ± 0.6 | 0.433 | 1364 | ND |
| eIF4E/4E-BP Complex | 5922 | 1.11 | 41.9 | 37.058 | 334 | 3.60 ± 0.03 | 3.52 ± 0.04 | 12.1 ± 0.6 | 1.360 | NA | 1.40 ± 0.08 |
| BSA | 40247 | 4.8 | 66.3 | 582 | ND | 3.1 ± 0.2 | 9.0 ± 0.4 | ND | ND | ND |
aValues for I(0) have been extrapolated by the Guinier approximation from the experimental scattering profiles.
bConcentration of the protein used for the calculation of the estimated Mr.
cRelative molecular mass estimated from I(0) and the concentration of the protein through BSA calibration.
dRelative molecular mass predicted from the sequence.
eRg (Guinier), Rg (GNOM), radius of gyration given by the Guinier approximation, and calculated by the program GNOM, respectively, given in nm.
fMaximum dimension of the macromolecules.
Χ(over) Discrepancy between the SAXS profile and its fit by the overall shapes-models calculated by DAMMIF, and Χ(crysol) the average discrepancy of the best atomic models estimated with the program CRYSOL (Supplementary Data). ND, not determined; NA, not appropriate.
gAverage of the normalized spatial discrepancies (NSD) obtained by Damaver for the different ab initio models generated by DAMMIF, DAMMIN and GASBOR.
Construction of the structural model of 4E-BP embracing eIF4E—construction of the structural model of 4E-BP embracing eIF4E
The residues of yeast eIF4E that were affected by the binding of 4E-BP have been identified by NMR titration measurements in ref. (8). We have transferred these residues onto eIF4E1 crystal structure from mouse by structural superimposition of the respective three-dimensional (3D) structures. Since no specific secondary or tertiary structuration of 4E-BP is assumed within the fuzzy complex, the most extended residue conformation (i.e. β-strand) was chosen to mimic the intrinsically unstructured ‘arms’ of 4E-BP. Consequently, starting from the crystal structure of eIF4E in complex with the 4E-BP1 peptide (20), the according N- and C-terminal residues were sequentially added to the central 4E-BP peptide with standard extended β-strand conformation following the residues on eIF4E that were affected by 4E-BP binding, using the interactive graphical software TURBO (21). Residues were added, covering the external surface of eIF4E, until ‘free dangling’ ends of 4E-BP were produced. The ending residues were chosen arbitrarily to be situated five residues beyond the surface of eIF4E and no molecular contact closer than 4.0 Å with eIF4E was designed. Glycine residues are well known to be involved in hinge regions. Considering that the central binding peptide of 4E-BP is flanked by two sequential glycines (G38 and G39) on the N-terminal side and by repeated glycines at positions 79, 86 and 88 on the C-terminal side, these glycine residues were chosen to perform rotational adjustments for the extended arms of 4E-BP to follow the external surface of eIF4E without producing sterical clashes. Sterical clashes were evaluated by distance–any atom of a residue was checked to be no closer than 3.0 Å.
RESULTS
In our study, as a representative translational repressor, we used the 4E-BP protein from the sea urchin (S. purpuratus), a sister group to chordates (22), that has only one 4E-BP. In vertebrates, the ancestral gene for 4E-BP underwent duplication giving rise to three 4E-BPs (4E-BP1, 4E-BP2 and 4E-BP3) in mammals. Both 4E-binding motif and phosphorylation sites of 4E-BPs are strongly conserved in distantly-related species (22) as well as the entire sequence of the protein (Supplementary Figure S5). Moreover, we previously conducted an extensive functional analysis showing that variants mimicking hyperphosphorylation of Sp 4E-BP and its mammalian counterpart displayed similar properties (5). The wild-type version of these proteins represses translation to a similar extent when added in a mammalian reticulocyte lysate (Supplementary Figure S4), indicating that Sp 4E-BP is functional to inhibit mammalian eIF4E.
Disorder predicted from the primary sequence
Intrinsically disordered protein prediction is a necessary prerequisite for the complete understanding of protein folding. Initially, we analyzed the primary sequence of S. purpuratus 4E-BP (Sp 4E-BP, Supplementary Figure S3) using various algorithms that predict disorder, in order to localize consensus disordered regions as described in ref. (23). This approach showed that 4E-BP is a mostly intrinsically disordered protein. Only a short central domain, containing the eIF4E-binding motif (YDRNFLLM) that displayed a hydrophobic cluster, is predicted to be structured. We observed the same pattern for the human 4E-BP1 (data not shown). The central domain predicted to be structured in our analysis corresponds to the one that undergoes a folded transition in an α-helix upon binding to eIF4E in the crystal structure (6,20).
Solution structure of the free 4E-BP
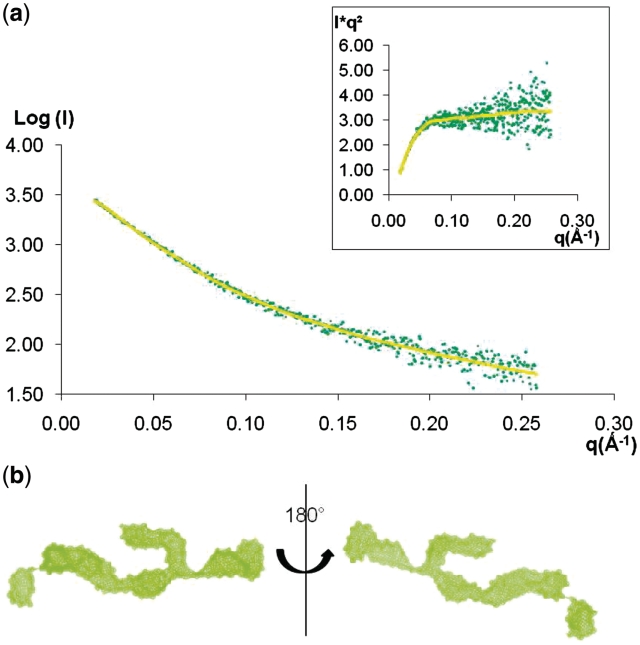
To determine the solution structure of 4E-BP, we used SAXS analysis. The 4E-BP scattering profile (Figure 1a) was obtained as described in Methods. The Krakty profile (Figure 1a, inset), derived from the scattering profile, had a plateau-shaped appearance typical of unfolded proteins or extended molecules (24). Analysis of the scattering curve for low q gave a radius of gyration (Rg) of 4.88 nm determined by the Guinier approximation, suggesting that the protein possesses a very extended state with a maximum distance of 16.0 nm (Table 1). In comparison, a typical globular protein of same molecular mass, such as for example Neocarzinostatin (25), has a Rg value of 1.4 nm. The overall envelope of 4E-BP was calculated ab initio from its scattering profile using DAMMIF and the most typical shape is provided Figure 1b. The program DAMAVER allowed us to align the ab initio models and gave us an average envelope of 4E-BP free in solution displaying a normalized spatial discrepancy (NSD) of 2.50 (Table 1). The numerous independent runs yielded recurrent features consistent with an extended three-branched shape. Conceptually, a structure with three branches can only be formed if there is a folding in the central region of the disordered protein, consistent with the prediction of a central structured region. This would imply that, in solution, the central region of 4E-BP is to some extent pre-structured, facilitating the interaction with eIF4E by protecting the hydrophobic region containing the eIF4E-binding site.
Figure 1.
Structure of 4E-BP free in solution. (a) Experimental SAXS profile of 4E-BP (green dots) fitted by the scattering curves calculated with the program DAMMIF (yellow). Inset: Kratky plot [I(q)q2 versus q] with a plateau illustrating specifically the unfolded nature of 4E-BP. (b) Example of the most typical shape of 4E-BP, as obtained by DAMMIF, shown in two orientations rotated by 180°.
4E-BP compacts upon binding to eIF4E
This successful SAXS analysis of 4E-BP then led us to determine the eIF4E/4E-BP complex structure using full length recombinant 4E-BP. SAXS analysis of the complex produced the scattering curve (Figure 2a), with the Kratky profile in the inset, consisting of an approximate bell-shaped curve typical of a largely folded conformation (24). The binding affinities of homologous proteins (non-phosphorylated 4E-BPs for human eIF4E) are high (estimated KD values of 10−8–10−9) (26), and suggest that, after gel filtration of the formed complex, we are not in presence of an equilibrium between the free and the complexed proteins in solution. Moreover, the calculated molecular mass of the scattering particles of the complex, derived from the experimental forward scattering intensity I(0) that was measured (Table 1), is in total agreement with the expected molecular mass, validating the fact that only the complex is present in solution. Analysis of the scattering curve in the low q region with the Guinier approximation gave an Rg of 3.52 nm with a maximal distance (Dmax) of 12.1 nm (Table 1). We first controlled that the Rg and Dmax of the complex were superior to the ones we obtained for eIF4E (Rg Guinier= 2.23 nm; Dmax = 6.8 nm) (Table 1). We then compared these values to the ones obtained from the scattering curve of 4E-BP free in solution. Rg and Dmax of the complex were clearly inferior, thus demonstrating that the complex eIF4E/4E-BP is much less extended than 4E-BP itself. These results show that the 4E-BP in complex with eIF4E is more compact than free in solution and undergoes partial refolding upon interaction with eIF4E. The overall envelopes of the complex were calculated ab initio from its scattering profile using DAMMIF and subsequently compared with the program DAMAVER that computes the NSD value for the obtained shapes. The various calculations led to similar forms with an average of the NSD values (1.40) significantly lower than the average found for 4E-BP free state (2.50) (Table 1), providing additional evidence that 4E-BP undergoes a compaction in the bound state. The recurrent shape of the complex was composed of three modules (Figure 2b), one central that always displays the same shape, and two others with slightly different orientations relative to the central shape in the different runs, characteristic of disordered regions. The most typical shape of the complex obtained with DAMMIF is illustrated in Figure 2b. We superimposed the crystal structure of eIF4E lacking the N-terminal 30 residues fragment, complexed with a 4E-BP peptide (20) (Figure 2c), by rotation and translation to fit the envelope. The atomic structure of eIF4E superimposed adequately and unambiguously with the central module of the low-resolution model. The N-terminal 30 residues fragment of eIF4E has never been observed in the eIF4E crystal structure due to its high flexibility. Consequently, we determined that these N-terminal residues constitute the adjacent module, using SAXS data collected for recombinant full length eIF4E free in solution (Supplementary Figure S2). The third module (red circle, Figure 2c) therefore could not be attributed to the crystal structure of eIF4E, and obviously belonged to 4E-BP.
DISCUSSION
The intrinsically disordered nature of 4E-BP in solution has hampered its full-length structure determination in the past (6,7). We have applied an original structural method which is receiving a growing interest, that gives access to low-resolution structures of biological molecules in solution (10). By comparing the experimental SAXS scattering curves of eIF4E and 4E-BP alone with the one for the purified eIF4E/4E-BP complex and by deriving ab initio solution structures, we have gained new structural insight into the determinant interactions for complex formation.
The SAXS envelope obtained for the complex is constituted by three modules, in which we localized eIF4E. Given that the central peptide of 4E-BP is known to interact with eIF4E at the dorsal molecular surface of the eIF4E cap-binding pocket (6,20), we conclude that the parts adjoining each side of the central motif also interact along eIF4E and that the extremities of 4E-BP form a disordered portion corresponding to the third module of the SAXS envelope (Figure 2c). Therefore, the interaction of eIF4E with the central peptide and its adjoining sides causes the partial folding of 4E-BP and supports the fact that 4E-BP reorganizes and compacts when it interacts with eIF4E. In this picture, 4E-BP, as an intrinsically disordered protein, has the advantage to ‘catch’ more easily eIF4E with its extended arm-like structures. Using its extremities to embrace eIF4E, 4E-BP would then strengthen the interaction while forming a fuzzy complex (27): a ‘fuzzy complex’ is constituted of at least one intrinsically unstructured/disordered protein. The parts of the complex that contribute productively to binding and function are structurally ill-defined, and cannot be described by a classical single conformational state. Comparing with complexes of ordered and globular proteins, intrinsically disordered proteins employ distinct molecular principles of protein−protein recognition and the interfaces are more hydrophobic and often cover a much larger part of the surfaces of the protein (28). Taking all together, our data strongly suggest that the interaction between eIF4E and 4E-BP not only relies on the central peptide, as currently thought, but that the binding footprint of 4E-BP appears to be larger and should involve contacts outside of this sequence.
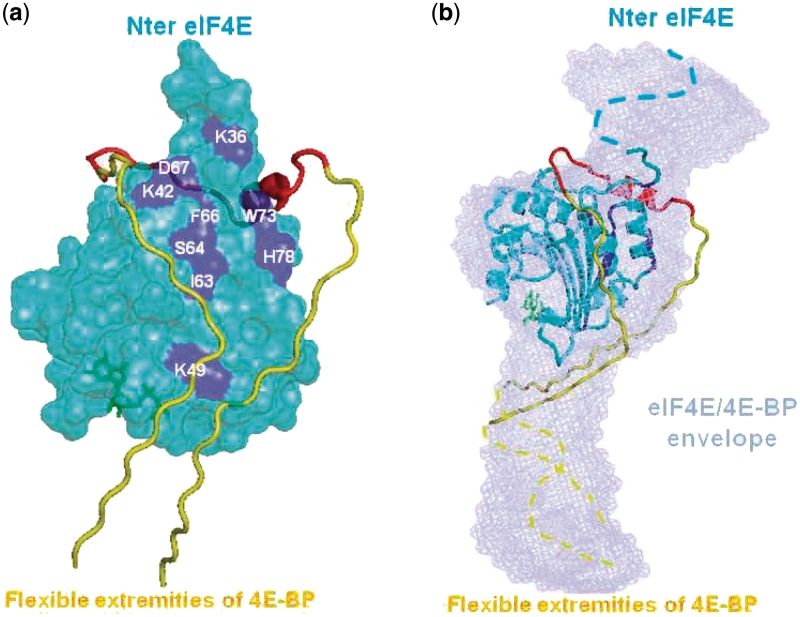
A previous NMR study (8) has indirectly determined the potential sites on eIF4E that are affected when 4E-BP binds eIF4E. From these data, we designed a model of interaction between eIF4E and 4E-BP taking into account the compaction of 4E-BP upon binding to eIF4E (Figure 3). Following the potential 4E-BP binding sites on eIF4E identified by NMR, we were able to add the N- and C- terminal stretches of 4E-BP onto the crystal structure of the peptide complex, designing residues until 4E-BP extremities reached the eIF4E boundaries (roughly from residues 25 to 88, Figure 3a). We further observe that this model fits perfectly with the SAXS complex surface (Figure 3b).
Figure 3.
Structural model of 4E-BP embracing eIF4E. (a) eIF4E crystal surface (lacking the 30 amino acids on N-terminal) is represented in cyan blue, on which the 4E-BP binding sites established by NMR are indicated in dark blue (labeled residues are the one visible from this orientation). The 4E-BP1 peptide (residues 47−66) (20) (red helix) has been prolonged in N- and C-terminal with standard extended β-strand conformation in yellow until five residues beyond the surface of eIF4E to visualize the entrance in the third module of the complex SAXS envelope. The cap (m7GTP) is represented in green. (b) The structural model built in (a) is superimposed with the SAXS envelope of the eIF4E/4E-BP full-length complex. The eIF4E crystal structure is displayed in cartoons instead of surface in (a) and rotated by 90°. The blue dotted-line represents the disordered module of the 30 amino acids on the N-terminal of eIF4E, missing on the crystallographic structure. The yellow dotted-line symbolized 4E-BP extremities that constitute the disordered section corresponding to the third module of the complex SAXS envelope.
In addition, multiple sequence alignments of 4E-BP sequences from distantly-related species (Supplementary Figure S5) reveal that the amino acids from 30 to 86 are highly conserved with >90% identity between the species. The apparent evolutionary pressure towards conservation of primary structure within these regions supports the notion that not only the L-shaped central domain [residues 47−66, (20)] but also its adjoining sides have a functional significance. Thus, these flanking parts of the central motif (residues 30−46 and 67−86) may provide a physical link through dynamic contacts with eIF4E. In the C-terminal extremity, only one or two residues are conserved (105−107) apart from the mTOR-signaling (TOS) (29) motif at the very end of the sequence. In contrast, residues of the N-terminal region, plus the RAIP motif (named after its sequence) (29), are mostly conserved suggesting that the N-terminal extremity would be more interesting for investigating potential avidity hot spots involved in the interaction with eIF4E, as suggested by Abiko et al. (30).
This new model of interaction helps to reinterpret data that could not be explained by binding of the 4E-BP central peptide alone to eIF4E. For instance, it could explain why in presence of 4EGi-1 (9), 4E-BP still binds to eIF4E while eIF4G cannot bind. Indeed, a NMR study (31) showed that eIF4G establishes extensive contacts with the normally disordered N-terminal extremity of eIF4E, in addition to the binding of the consensus motif. In contrast, our model of full-length 4E-BP with eIF4E rather covers the structured part of eIF4E and does not appear to interact with the disordered N-terminal part. This is in agreement with the observation that there is an apparent overlapping but non-identical binding site of eIF4G and 4E-BPs (31).
The vision of a larger footprint brings also new insights for the understanding of the association/dissociation of the complex controlled by 4E-BP phosphorylation that is not fully understood yet (5). Indeed, it was shown by Oulhen et al. (5) that the phosphorylation of the following sites, namely T37, T46, S65, T70 on 4E-BP, was necessary but not sufficient to prevent the association between eIF4E and 4E-BP, contrary to what was currently thought. Our model gives thus new leads to investigate potential modifications, well outside of the central motif, that would participate and would be essential in the release of 4E-BP from eIF4E.
As a crucial translational repressor, the demonstration that unstructured free 4E-BP reorganizes subsequent to eIF4E binding changes the current vision of the regulation of the eIF4E/4E-BP complex formation and stabilization, and gives new perspectives for the understanding of translational control.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
UPMC Univ Paris 06 grants (BQR); Ligue Nationale contre le Cancer (Délegations Départementales Finistère, Morbihan, Vendée et Côte d’Armor to B.C.); Conseil Régional de Bretagne and Conseil Général du Finistère. Funding for open access charge: Institutional funding (CNRS/UPMC).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank N. Sonenberg, M. Bidinosti and G. Wessel for critical reading of the article. We also thank the staff members of the beamline ID14-3, particularly A. Round, at the European Synchrotron Radiation Facilities, Grenoble, France, for beam time and technical assistance. We acknowledge A. Kikhney, A. Zozulya and D. Svergun for the use of and assistance with the X33 SAXS beamline of the European Molecualr Biology Laboratory outstation, Deutsches Electronen Synchrotron, Hamburg.
REFERENCES
- 1.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation-factor Eif-4e binds to a common motif shared by the translation factor Eif-4-gamma and the translational repressors 4e-binding proteins. Mol. Cell. Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 4.Gingras AC, Kennedy SG, O'Leary MA, Sonenberg N, Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev. 1998;12:502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oulhen N, Boulben S, Bidinosti M, Morales J, Cormier P, Cosson B. A variant mimicking hyperphosphorylated 4E-BP inhibits protein synthesis in a sea urchin cell-free, cap-dependent translation system. PLoS ONE. 2009;4:e5070. doi: 10.1371/journal.pone.0005070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell. 1999;3:707–716. doi: 10.1016/s1097-2765(01)80003-4. [DOI] [PubMed] [Google Scholar]
- 7.Fletcher CM, Wagner G. The interaction of eIF4E with 4E-BP1 is an induced fit to a completely disordered protein. Protein Sci. 1998;7:1639–1642. doi: 10.1002/pro.5560070720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsuo H, Li H, McGuire AM, Fletcher CM, Gingras AC, Sonenberg N, Wagner G. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat. Struct. Biol. 1997;4:717–724. doi: 10.1038/nsb0997-717. [DOI] [PubMed] [Google Scholar]
- 9.Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, Gross JD, Degterev A, Yuan J, Chorev M. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 10.Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, 2nd, Tsutakawa SE, Jenney FE, Jr, Classen S, Frankel KA, Hopkins RC, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat. Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Longhi S, Receveur-Brechot V, Karlin D, Johansson K, Darbon H, Bhella D, Yeo R, Finet S, Canard B. The C-terminal domain of the measles virus nucleoprotein is intrinsically disordered and folds upon binding to the C-terminal moiety of the phosphoprotein. J. Biol. Chem. 2003;278:18638–18648. doi: 10.1074/jbc.M300518200. [DOI] [PubMed] [Google Scholar]
- 12.Pang SS, Berry R, Chen Z, Kjer-Nielsen L, Perugini MA, King GF, Wang C, Chew SH, La Gruta NL, Williams NK, et al. The structural basis for autonomous dimerization of the pre-T-cell antigen receptor. Nature. 2010;467:844–848. doi: 10.1038/nature09448. [DOI] [PubMed] [Google Scholar]
- 13.Rosenberg OS, Deindl S, Sung RJ, Nairn AC, Kuriyan J. Structure of the autoinhibited kinase domain of CaMKII and SAXS analysis of the holoenzyme. Cell. 2005;123:849–860. doi: 10.1016/j.cell.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 14.Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 1999;18:270–279. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 2003;36:1277–1282. [Google Scholar]
- 16.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 17.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niedzwiecka A, Marcotrigiano J, Stepinski J, Jankowska-Anyszka M, Wyslouch-Cieszynska A, Dadlez M, Gingras AC, Mak P, Darzynkiewicz E, Sonenberg N, et al. Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol. 2002;319:615–635. doi: 10.1016/S0022-2836(02)00328-5. [DOI] [PubMed] [Google Scholar]
- 20.Tomoo K, Matsushita Y, Fujisaki H, Abiko F, Shen X, Taniguchi T, Miyagawa H, Kitamura K, Miura K, Ishida T. Structural basis for mRNA Cap-binding regulation of eukaryotic initiation factor 4E by 4E-binding protein, studied by spectroscopic, X-ray crystal structural, and molecular dynamics simulation methods. Biochim. Biophys. Acta. 2005;1753:191–208. doi: 10.1016/j.bbapap.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 21.Roussel A, Cambillau C. Silicon Graphics Geometry Partners Directory. CA: Silicon Graphics, Mountain View; 1991. TURBO-FRODO; p. 89. [Google Scholar]
- 22.Morales J, Mulner-Lorillon O, Cosson B, Morin E, Belle R, Bradham CA, Beane WS, Cormier P. Translational control genes in the sea urchin genome. Dev. Biol. 2006;300:293–307. doi: 10.1016/j.ydbio.2006.07.036. [DOI] [PubMed] [Google Scholar]
- 23.Leyrat C, Gerard FC, de Almeida Ribeiro E, Jr, Ivanov I, Ruigrok RW, Jamin M. Structural disorder in proteins of the Rhabdoviridae replication complex. Protein Pept. Lett. 2010;17:979–987. doi: 10.2174/092986610791498939. [DOI] [PubMed] [Google Scholar]
- 24.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 25.Perez J, Vachette P, Russo D, Desmadril M, Durand D. Heat-induced unfolding of neocarzinostatin, a small all-beta protein investigated by small-angle X-ray scattering. J. Mol. Biol. 2001;308:721–743. doi: 10.1006/jmbi.2001.4611. [DOI] [PubMed] [Google Scholar]
- 26.Youtani T, Tomoo K, Ishida T, Miyoshi H, Miura K. Regulation of human eIF4E by 4E-BP1: binding analysis using surface plasmon resonance. IUBMB Life. 2000;49:27–31. doi: 10.1080/713803582. [DOI] [PubMed] [Google Scholar]
- 27.Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem. Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 28.Meszaros B, Tompa P, Simon I, Dosztanyi Z. Molecular principles of the interactions of disordered proteins. J. Mol. Biol. 2007;372:549–561. doi: 10.1016/j.jmb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 29.Beugnet A, Wang X, Proud CG. Target of rapamycin (TOR)-signaling and RAIP motifs play distinct roles in the mammalian TOR-dependent phosphorylation of initiation factor 4E-binding protein 1. J. Biol. Chem. 2003;278:40717–40722. doi: 10.1074/jbc.M308573200. [DOI] [PubMed] [Google Scholar]
- 30.Abiko F, Tomoo K, Mizuno A, Morino S, Imataka H, Ishida T. Binding preference of eIF4E for 4E-binding protein isoform and function of eIF4E N-terminal flexible region for interaction, studied by SPR analysis. Biochem. Biophys. Res. Commun. 2007;355:667–672. doi: 10.1016/j.bbrc.2007.01.198. [DOI] [PubMed] [Google Scholar]
- 31.Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JE, Wagner G. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell. 2003;115:739–750. doi: 10.1016/s0092-8674(03)00975-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.