Abstract
Recently, many small non-coding RNAs (sRNAs) with important regulatory roles have been identified in bacteria. As their eukaryotic counterparts, a major class of bacterial trans-encoded sRNAs acts by basepairing with target mRNAs, resulting in changes in translation and stability of the mRNA. RNA interference (RNAi) has become a powerful gene silencing tool in eukaryotes. However, such an effective RNA silencing tool remains to be developed for prokaryotes. In this study, we described first the use of artificial trans-encoded sRNAs (atsRNAs) for specific gene silencing in bacteria. Based on the common structural characteristics of natural sRNAs in Gram-negative bacteria, we developed the designing principle of atsRNA. Most of the atsRNAs effectively suppressed the expression of exogenous EGFP gene and endogenous uidA gene in Escherichia coli. Further studies demonstrated that the mRNA base pairing region and AU rich Hfq binding site were crucial for the activity of atsRNA. The atsRNA-mediated gene silencing was Hfq dependent. The atsRNAs led to gene silencing and RNase E dependent degradation of target mRNA. We also designed a series of atsRNAs which targeted the toxic genes in Staphyloccocus aureus, but found no significant interfering effect. We established an effective method for specific gene silencing in Gram-negative bacteria.
INTRODUCTION
The small non-coding RNAs (sRNAs) have attracted great interest as ubiquitous regulators in all kingdoms of life. The eukaryotic microRNA (miRNA) and short interfering RNA (siRNA) have been making a splash during the past few years. Recently, with the development of the experimental and computational approaches, hundreds of sRNAs have been identified in bacteria, especially in Escherichia coli [reviewed in ref. (1,2)]. The bacterial sRNAs constitute a structurally diverse class of molecules that are typically of 50–250 nt in length and do not contain expressed open reading frames (ORFs). They have been shown to be involved in many cellular processes in prokaryotes, including translational quality control, protein inhibition, iron homeostasis, outer membrane protein biogenesis, sugar metabolism, quorum sensing, survival in stationary phase and virulence regulation of pathogens (3–8). According to the researches in Gram-negative bacteria, those sRNAs whose functions have been characterized can be sorted into three general categories: sRNAs that have intrinsic catalytic activity or are components of ribonucleoproteins, sRNAs that affect protein activity by structurally mimicking other nucleic acids and sRNAs that regulate gene expression by basepairing with target mRNAs, changing the translation and stability of the mRNA. sRNAs in the latter category appear to be the best-characterized and most abundant in Gram-negative bacteria. Most of these identified sRNAs are trans-encoded, located at chromosomal positions different from their targets. The majority of these trans-encoded sRNAs, such as DicF, MicF, OxyS, Spot42, RyhB and GcvB, repress the translation of target mRNAs. Compared to the Gram-negative bacterium, far fewer sRNAs were identified in Gram-positive pathogens, and there exist many differences in length, structure and function mode between the sRNAs from these two groups of bacteria (4,9–16).
In Gram-negative bacteria, a large class of trans-encoded sRNAs silences their target mRNAs by binding tightly to Hfq, a highly abundant RNA chaperone protein that also binds the target mRNA in a number of cases studied (17–19). Base pairing between these sRNAs and their target mRNAs requires Hfq which is proposed to enhance the stability of sRNAs in vivo, by protecting them from degradation. Hfq has also been shown to interact with proteins such as poly(A) polymerase I (PAP I), polynucleotide phosphorylase (PNP) and RNase E which are involved in mRNA decay and it is shown to form fibres in vitro, the physiological significance of which is unknown (20–24). It is necessary to mention that Ribonuclease (RNase) E is the major endoribonuclease participating in RNA turnover in E. coli, which initiates the decay of RNAs by cutting them internally near their 5′-end. Additionally, the binding of Hfq and E. coli sRNAs could protect sRNAs from endonucleolytic attack. More interestingly, some endogenous sRNAs in E. coli have been shown to form a specific ribonucleoprotein complex with RNase E and Hfq, resulting in translation inhibition and RNase E dependent degradation of target mRNAs (25–27). In contrast to this, Hfq had been supposed to be dispensable for sRNA-mediated riboregulation in the Gram-positive species for a long time. But a recent study provided the first evidence for Hfq-dependent antisense regulation in Gram-positive bacteria Listeria monocytogenes (28,29).
The small RNAs in eukaryotes, such as miRNA and siRNA, have become a powerful experimental technique for silencing gene expression both in cultured cells and living organisms (30,31). However, such an effective RNA silencing tool remains to be developed for prokaryotes in which the main methods of deciphering gene function are still homologous recombination and transposon mutagenesis. Additionally, the application of traditional antisense RNA (asRNA) is also limited due to its low efficacy (32,33). Therefore, effective and convenient RNA silencing methods are urgently needed for prokaryotic organisms. Recently, the emergence of wide spread trans-encoded sRNAs that act as regulators of gene expression by basepairing with their targets in bacteria provide us a promising perspective for gene function investigation.
In the present study, we described first the use of artificial trans-encoded sRNAs (atsRNAs) for specific gene silencing in bacteria. A series of atsRNAs targeting specific genes were designed and synthesized, and some of them markedly suppressed the expression of target genes in an Hfq-dependent manner. Our results demonstrated that atsRNA was an efficient approach for specific gene silencing in E. coli, giving rise to a new research method for deciphering gene functions in bacteria.
MATERIALS AND METHODS
Strains and plasmids
Escherichia coli MC4100 (F- araD139 Δ(argF-lac) U169 rpsL150 relA1 deoC1 rbsR fthD5301 fruA25 λ-) (a gift from R. Gary Sawers) and CSH26 (F- ara Δ(lac-pro) thi) (a gift from Masaaki Wachi) were used as the parent wild-type strains. The hfq mutant strain GSO81 (MC4100 hfq-1::Ω) was a gift from Gisela Storz; the temperature-sensitive rne-1 mutant strain HAT103 (CSH26 zce-726::Tn10 rne-1) was a gift from Masaaki Wachi; the Staphyloccocus aureus RN4220 strain was conserved by our lab. EGFP gene was inserted into pET-24a (+) (Novagen, USA), the yielding recombinant plasmid pET24a-EGFP. A constitutive promoter P1 was cloned from plasmid pBR322 and inserted into pET24a-EGFP to drive the transcription of EGFP gene. The fragment of P1 promoter, EGFP gene and T7 transcriptional terminator was amplified and inserted into pACYC177 carrying a P15A origin of replication and kanamycin resistance gene (New England Biolabs, USA), creating recombinant plasmid pACYC-P1-EGFP. The high-copy E. coli–S. aureus shuttle plasmid pSB2035 was a gift from Hill.
Constructs for atsRNA biosynthesis
The mRNA base pairing regions of atsRNAs were designed according to the 5′-UTR of target exogenous EGFP gene, uidA gene (a non-essential gene for E. coli), hla gene and SEA gene. (U00096). The other two modules (Hfq binding site and Rho-independent terminator) were extracted from well-known natural Hfq-dependent trans-encoded sRNAs. The three elements were selected by simple random sampling techniques and assembled into a series of atsRNA candidates from which atsRNA was chosen based on the predicted secondary structure by MFOLD (34). Then two complementary DNA oligonucleotides for atsRNA biosynthesis were annealing into double strands. For EGFP and uidA series atsRNAs, the DNA was double digested with EcoRI/HindIII and inserted into the same sites of pRI (a gift from Gisela Storz), for hla and SEA series atsRNAs, the double stranded DNA was double digested with NdeI/SalI and inserted into the same sites of pSB2035 (a gift from Philip Hill). pRI was a high copy vector compatible with pACYC-P1-EGFP and carrying a pBR322 origin of replication and a bla ampicillin resistance gene. The transcription of atsRNA gene would precisely start at the putative +1 site of atsRNA under the control of tac promoter (35). pSB2035 was also a relatively high-copy shuttle vector carrying inducible P3 promoter, which would endure the effective expression of atsRNAs in S. aureus. It also contained chloramphenicol resistance gene which would facilitate the screening of positive clones (36). The sequences of synthesized atsRNA genes were listed in Supplementary Tables S1, S2, S6 and S7.
Fluorescence measurement
Escherichia coli strains carrying the plasmid pACYC-P1-EGFP were cultured at 30°C and the isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM to induce the expression of atsRNA genes when OD600 of the culture reached 0.5. To measure the fluorescence, after induction for 30 min, bacterial cells were collected by centrifugation at 5000 g and 4°C for 5 min, washed and re-suspended in 500 μl of phosphate-buffered saline (50 mM, pH 7.0). Cells were disrupted with Vcx750 watt ultrasonic processor (Sonics and Materials Inc., USA) at 0°C and 9 kHz for 2 min, and then the cell free extract was obtained by centrifugation at 15 000 g and 4°C for 15 min. The protein concentration was assayed with BCA Protein Assay kit (Pierce, USA). As much as 300 mg of proteins were transferred to a 96-well microtitre plate (BD Falcon, USA) with the final volume adjusted with phosphate-buffered saline to 200 μl. Fluorescence was measured using SpectraMax M5 (Molecular devices, USA) with excitation filter at 474 nm and emission filter at 515 nm. Three independent sets of experiments were performed.
Activity assay of β-glucuronidase
The activity of β-glucuronidase was assayed using the 4-methyl-umbelliferyl-β-d-glucuronide (4-MUG) as substrate. Bacterial strain was cultured at 30°C with IPTG added to a final concentration of 1 mM when OD600 reached 0.5, and conduct the assay described by Wickes and Edman (37,38).
Activity assay of α-hemolysin
Bacteria were cultured in LB to the post-exponential growth phase (OD600 of 2.5). α-hemolysin levels in bacterial culture supernatants were determined by using S. aureus α-hemolysin ELISA Kit (Shanghai Jingtian, China) as instructed.
Activity assay of SEA
Bacteria were cultured in LB to the post-exponential growth phase (OD600 of 2.5). SE levels in bacterial culture supernatants were determined by using S. aureus SE ELISA Kit (Shanghai Jingtian, China) as instructed. To further test the SEA level, we conducted western blot analysis under the conditions described by Towbin (39).
Northern blot analysis
Bacterial total RNA was isolated using Trizol reagent (Invitrogen, USA) according to the manufacturer’s instructions. Digoxigenin-labeled RNA probes against atsRNAs, uidA mRNA, 5 S rRNA and 16 S rRNA were obtained from the specific polymerase chain reaction (PCR) products containing a T7 promoter by in vitro transcription using the DIG Labeling kit (Roche Diagnostics, Germany). The primer sequences used for northern blotting were as follows: forward primer of atsRNA genes, 5′-GTTGACAATTAATCATCGGCT-3′, reverse primer of atsRNA genes, 5′-CTAATACGACTCACTATAGGGAGCTCTCATCCGCCAAAACAG-3′; forward primer of uidA gene, 5′-TATACGCCATTTGAAGCC-3′, reverse primer of uidA gene, 5′-CTAATACGACTCACTATAGGGAGAAGCCAGTAAAGTAGAACG-3′; forward primer of 5 S rRNA gene, 5′-GAATTTGCCTGGCGGCAGTAGCGCG-3′, reverse primer of 5 S rRNA gene, 5′-CTAATACGACTCACTATAGGGAGATGCCTGGCAGTTCCCTACTCTCGC-3′; forward primer of 16 S rRNA gene, 5′-TGGCGGACGGGTGAGTAATG-3′, reverse primer of 16 S rRNA gene, 5′-CTAATACGACTCACTATAGGGAGGGCTGCTGGCACGGAGTTAG-3′. Northern blot was conducted as described by Storz (12). Probe hybridization and detection were carried out according to the supplier’s instruction (DIG RNA Labeling kit, Roche Diagnostics, Germany). For stability experiments, transcription initiation was inhibited by adding rifampicin to growing cells to a final concentration of 500 µg/ml (time 0). Northern blots were performed as above except that rifampicin was added.
Electrophoretic mobility shift assays
The hfq gene without stop codon was amplified from MC4100 and inserted into the BamHI/XhoI sites of pET-24a, creating recombinant plasmid pET24a-hfq. The His-tagged Hfq protein was overproduced from BL21 (DE3) strain and purified by Ni2+-NTA column (Qiagen, Germany). Digoxigenin-labeled atsRNAs were obtained from specific PCR products containing a T7 promoter by in vitro transcription using the DIG RNA Labeling kit (Roche Diagnostics). The primer sequences used were as follows: forward primer of atsRNA genes, 5′-CTAATACGACTCACTATAGGGAGGTTGACAATTAATCATCGGCT-3′, reverse primer of atsRNA genes, 5′-CTCTCATCCGCCAAAACAG-3′. Electrophoretic mobility shift assay (EMSA) was carried out as described by Storz (12). Then the Dig-labeled RNA-protein complexes were detected according to the supplier’s instruction (Roche Diagnostics, Germany).
Quantitative real-time polymerase chain reaction
Bacterial total RNA was isolated using RNeasy Midi Kit (Qiagen, Germany) and cDNA was synthesized using High Capacity cDNA Archive Kit (Applied Biosystems, USA) following manufacturer’s protocols, respectively. The primer sequences used for quantitative real-time polymerase chain reaction (QPCR) were as follows: forward primer of EGFP gene, 5′-CGGCATGGACGAGCTGTAC-3′, reverse primer of EGFP gene, 5′-GCTTCCTTTCGGGCTTTGT-3′; forward primer of uidA gene, 5′-TGAGCGTCGCAGAACATTACA-3′, reverse primer of uidA gene, 5′-GCCACTGGCGGAAGCAA-3′; forward primer of 16 S rRNA gene, 5′-TGACGCTCAGGTGCGAAAG-3′, reverse primer of 16 S rRNA gene, 5′-CAAGGGCACAACCTCCAAGT-3′. 16 S rRNA gene was used as the normalizer. Reactions were run in a 7500 real-time PCR system (Applied Biosystems, USA) under standard reaction conditions. Three independent sets of experiments were performed.
RESULTS
Designing principle of atsRNA
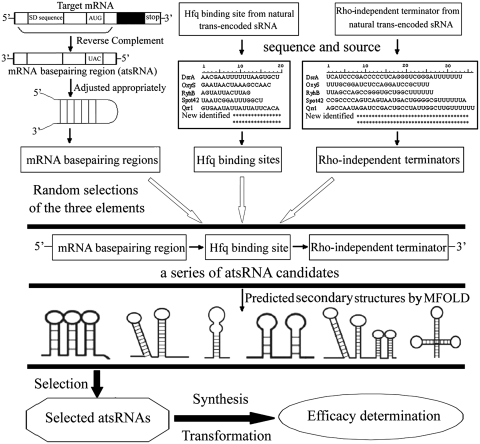
We first analyzed the sequences of well-known bacterial trans-encoded sRNAs in E. coli, such as Spot42 (4,15), RyhB (5,20), MicF (10,40), OxyS (12,17,41), DsrA (42,43) and Qrr1 (7,44), and divided them into three modules that were referred to as mRNA base pairing region, Hfq binding site and Rho-independent terminator in this study (Supplementary Figure S1). The mRNA base pairing region of the sRNA was imperfectly complementary with the 5′-untranslated region (5′-UTR) of the target mRNA, especially the Shine-Dalgarno (SD) sequences. Although the Hfq binding sites of natural sRNAs shared no homology, the common characteristic of these sequences was a high A+U-content. The Rho-independent terminators typically shared common structural features which consisted of a GC rich inverted repeat followed by a run of U residues. Next, the secondary structures of these well-known natural sRNAs were predicted by MFOLD program (34), and then their common characteristics were analyzed. These sRNAs were highly structured, forming at least two stem loops by mRNA base pairing region and Rho-independent terminator, respectively. In general, the Hfq binding site located at the AU rich area between these two stem loops in the predicted secondary structure.
Based on the common structural characteristics of natural bacterial trans-encoded sRNAs in E. coli, we developed the principle and process for atsRNA design (Figure 1). (i) atsRNA should be a modular structure consisting of three elements: mRNA base pairing region, Hfq binding site and Rho-independent terminator. (ii) The mRNA base pairing region (20 to 30 nt in length) should be designed according to the 5′-UTR (or part of the coding regions) of target mRNA, especially the SD sequence, and then the sequence composition should be adjusted appropriately to make it form a stem loop structure. (iii) The Hfq binding sites and Rho-independent terminators were extracted from the well-studied endogenous bacterial trans-encoded sRNAs, such as RyhB, Spot42, DsrA and OxyS. The Hfq binding sites generally corresponded to a 12–19 nt AU-rich region. (iv) The three component parts were selected randomly and then assembled into a series of atsRNA candidates according to the combination order listed in Figure 1. Then their secondary structures were predicted by MFOLD program. (v) atsRNAs should be selected from atsRNA candidates whose predicted secondary structures meet the following criteria: atsRNAs should form two to four stem loops, including one formed by mRNA base pairing region and one formed by Rho-independent terminator. The Hfq binding site should locate between two stem loops. (vi) The start codon AUG should be avoided in atsRNA in order to prevent the translation. (vii) The selected atsRNAs were synthesized, cloned into the expression vector and transformed into host bacteria. Then the efficacies of these atsRNAs were determined.
Figure 1.
Schematic representation of the designing process for atsRNA.
Designing and characterization of atsRNAs targeting against specific genes
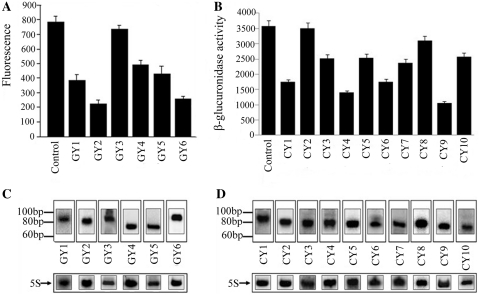
To evaluate the feasibility of this method in E. coli, the exogenous EGFP gene (enhanced green fluorescent protein) was selected as the target. According to the above designing principle, six atsRNAs (GY1 to GY6) targeting against EGFP gene were designed and synthesized (Table 1 and Supplementary Table S1), then inserted into the expression vector pRI (35), The pACYC-P1-EGFP vector and pRI vector carrying atsRNA genes were co-transformed into E. coli MC4100. As shown in Figure 2A, GY2 and GY6 efficiently suppressed the expression of EGFP gene; and GY1, GY4 and GY5 did so slightly; while GY3 almost had no repressing effect. To further test the application of this method, 10 atsRNAs (CY1 to CY10) targeting against endogenous uidA gene that encodes β-glucuronidase were designed and synthesized (Table 1 and Supplementary Table S2). Plasmid pRI carrying these atsRNA genes were transformed into E. coli MC4100, respectively and the expression level of uidA gene was measured by detecting the activity of β-glucuronidase. As shown in Figure 2B, CY1, CY4, CY6 and CY9 suppressed the expression of uidA gene obviously; and CY3, CY5, CY7 and CY10 did so slightly; while CY2 and CY8 almost had no effect. The most effective atsRNA CY9 triggered a 71% reduction in expression of target genes. To determine whether the repression of target genes was indeed due to the expression of atsRNAs, we then analyzed the transcripts of atsRNAs by northern blot. As shown in Figure 2C and D, atsRNAs were transcribed with the predicted length, indicating that the reduction on target gene was due to the expression of atsRNA. And the QPCR analysis indicated that atsRNAs generally expressed at 1–2 orders of magnitude greater levels than that of some native sRNAs in E. coli such as RyhB, DicF and DsrA (data not shown). Furthermore, to confirm whether the expression of atsRNAs interfere with the physiology of host bacteria, we first detected the growth curve of host bacteria after the induction of atsRNA. As shown in Supplementary Figure S2, the expression of atsRNAs had no effect on growth rate of host bacteria. Then we swapped atsRNA expression plasmids (CY series for GY series and vice versa) as a test of specificity. As shown in Supplementary Figure S3, the expression level of EGFP and uidA gene had no reduction after the induction of atsRNA. These results demonstrated that atsRNA was effective in suppressing the expression of specific bacterial genes.
Table 1.
Summary of atsRNAs in E. coli
| Name | Sequence information |
dG(kcal/ mol)e | Target | Inhibitory efficiencyf (%) | |||
|---|---|---|---|---|---|---|---|
| Lengtha (nt) | mRNA base pairing region |
Hfq binding sited (nt) | |||||
| Lengthb (nt) | Complementary sequencesc (nt) | ||||||
| GY1 | 82 | 38 | 21 | 19 | −24.4 | EGFP | 50.7 |
| GY2 | 81 | 31 | 16 | 19 | −24.7 | EGFP | 71.1 |
| GY3 | 83 | 32 | 22 | 17 | −35.9 | EGFP | 6.1 |
| GY4 | 76 | 31 | 16 | 12 | −25.2 | EGFP | 37.6 |
| GY5 | 73 | 32 | 19 | 15 | −26.2 | EGFP | 45.1 |
| GY6 | 87 | 38 | 21 | 18 | −26.8 | EGFP | 65.6 |
| CY1 | 86 | 31 | 19 | 19 | −16.9 | uidAg | 51.2 |
| CY2 | 81 | 31 | 20 | 17 | −22.7 | uidA | Null |
| CY3 | 81 | 31 | 20 | 19 | −28.1 | uidA | 29.6 |
| CY4 | 82 | 32 | 24 | 19 | −25.1 | uidA | 63.9 |
| CY5 | 82 | 32 | 24 | 17 | −24.5 | uidA | 28.9 |
| CY6 | 79 | 32 | 24 | 19 | −21.5 | uidA | 50.8 |
| CY7 | 77 | 32 | 24 | 19 | −18 | uidA | 33.5 |
| CY8 | 79 | 28 | 23 | 17 | −26.89 | uidA | 13.4 |
| CY9 | 73 | 28 | 23 | 12 | −21.8 | uidA | 70.5 |
| CY10 | 71 | 28 | 23 | 18 | −13.1 | uidA | 27.5 |
aThe full length of atsRNA.
bThe length of mRNA base pairing region of atsRNA.
cThe length of predicted complementary sequences between atsRNA and target mRNA.
dThe length of Hfq binding site of atsRNA.
eThe minimum free energy of atsRNA predicted at 37°C by MFOLD program.
fThe repression efficiency was calculated after the expression of each atsRNA for 30 min and bacterial cells containing empty vector pRI was used as the control.
gThe uidA gene is an endogenous gene that encodes β-glucuronidase in E. coli. The detailed sequences of these atsRNAs were listed in Supplementary Tables S1 and S2.
Figure 2.
Effect of atsRNAs on gene silencing in E. coli. (A) The effect of atsRNAs on target EGFP gene. (B) The effect of atsRNAs on target uidA gene. (C) Northern blot analysis of atsRNA transcripts targeting EGFP gene. (D) Northern blot analysis of atsRNA transcripts targeting uidA gene. In (A) and (B), the expression of target gene was measured after the induction of atsRNA with 1 mM IPTG for 30 min. Bacterial cells containing the empty vector pRI were used as the control. Average values were from triplicate experiments with std values indicated. In (C) and (D), total RNAs were purified from cells after the expression of atsRNAs were induced with 1 mM IPTG for 30 min and analyzed using probes specific for atsRNAs respectively. The positions of RNA size standards are indicated on the left. The 5S rRNA was used as the loading control.
Structural and functional relationship of atsRNA
To gain more information for designing atsRNAs, the sequences and predicted structures of those effective and non-effective atsRNAs were compared. From the primary sequences of atsRNAs, there were no obvious differences in full length of the effective and non-effective atsRNAs (Table 1, Supplementary Tables S1 and S2). The mRNA base pairing regions of those atsRNAs imperfectly paired with the 5′-UTR of the target mRNAs according to the predicted results by LALIGN (Supplementary Figure S4, Tables S1 and S2). This result indicted that atsRNA acted by basepairing with target mRNA. The complementary sequences between atsRNAs and their targets ranged mainly from 19 to 23 nt in length, and there were no obvious differences in sequence composition between them (Supplementary Figure S4, Tables S1 and S2). Since the putative interacting sequences were maintained in all atsRNAs (effective and non-effective), we presumed that the different effect of these atsRNAs was due to the secondary structure or stability or the other two component parts of atsRNA.
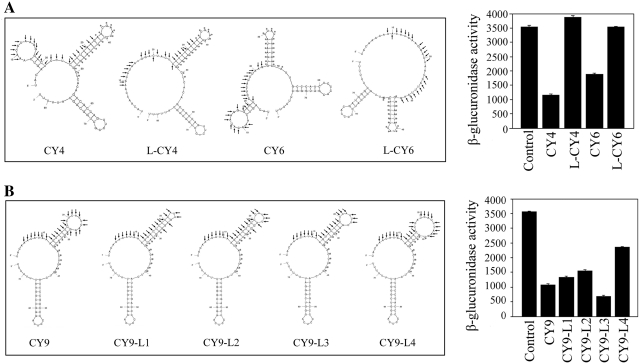
In the predicted secondary structure, all the effective atsRNAs showed two or three stem loop structures and putative AU rich Hfq binding sites locating between two stem loops (Supplementary Figure S5). Their mRNA base pairing regions formed a stem–loop structure, while two atsRNA candidates without stem loop structure in mRNA base pairing region were non-effective. To confirm the role of stem loop structure, the mRNA base pairing regions of CY4 and CY6 were adjusted appropriately to make them lost the stem loop in predicted secondary structure, yielding atsRNA mutants L-CY4 and L-CY6, respectively (Figure 3A). The complementary sequences of L-CY4 and L-CY6 were similar with that of CY4 and CY6 (Supplementary Figure S6) and the Hfq binding site and Rho-independent terminators remained constant in the mutant construct, respectively (Supplementary Table S3). As shown in Figure 3A, L-CY4 and L-CY6 lost repressing effect on the target gene (P < 0.01). Northern blot results showed that the amounts of L-CY4 and L-CY6 decreased compared to that of the corresponding atsRNAs (Supplementary Figure S7). However, the reduced amounts of atsRNA mutants could not account for the loss of repressing effect on target gene. After normalization of the interference efficiency of each atsRNAs to relative RNA amounts, the relative interference efficiency of L-CY4 and L-CY6 decreased dramatically (Supplementary Figure S7). This result suggested that the lost activity of mutants L-CY4 and L-CY6 was due to the loss of stem loop structure but not the reduced stability. These results demonstrated that the stem loop structure formed by mRNA base pairing region was necessary for the activity of atsRNA.
Figure 3.
Secondary structure and function of atsRNA. (A) The predicted secondary structures and effect of atsRNA mutants L-CY4 and L-CY6 designed based on CY4 and CY6, respectively. Compared with the corresponding atsRNAs, L-CY4 and L-CY6 lost the stem loop of mRNA base pairing region in secondary structure. (B) The predicted secondary structures and effect of atsRNA mutants designed based on CY9. CY9-L1, CY9-L2, CY9-L3 and CY9-L4 were CY9 mutants with different number of unpaired nucleotides in the loop structure of mRNA base pairing region. The sequences of predicted pairing region (indicated with arrows) and Hfq binding site (indicated with asterisks) of atsRNA are marked. The secondary structure of atsRNA is predicted at 37°C by MFOLD program. Bacterial cells containing empty vector pRI were used as the control.
The initial interaction between endogenous sRNA and mRNA was a stretch of unpaired nucleotides in a loop structure of sRNA in some cases (15,17,20,45). The length of the unpaired nucleotides in the loop formed by mRNA base pairing region of CY9 was 9 nt. To determine the optimal length of unpaired nucleotides in the loop, the number of unpaired nucleotides of CY9 was changed appropriately, yielding atsRNA mutants CY9-L1 (3 nt in the loop), CY9-L2 (5 nt in the loop), CY9-L3 (7 nt in the loop) and CY9-L4 (13 nt in the loop). All of the mutants were maintained similar secondary structure with CY9 (Figure 3B). In addition, the complementary sequences of these mutants were similar to that of CY9 (Supplementary Figure S8) and the Hfq binding site and Rho-independent terminators remained constant in these mutant constructs (Supplementary Table S3). As shown in Figure 3B, the interference efficiency of CY9-L3 achieved 73%, while the interference efficiency of CY9-L1, CY9-L2 and CY9-L4 was lower than that of CY9 (P < 0.01). Furthermore, the relative interference efficiency of CY9-L3 was higher than that of other atsRNAs (Supplementary Figure S9). This result demonstrated that the length of unpaired nucleotides in the loop structure formed by mRNA base pairing region was important for the function of atsRNA and the optimal length of unpaired nucleotides was seven for CY9.
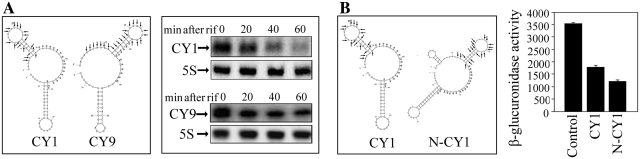
The secondary structure of CY1 was similar to that of CY9 (Figure 4A), however, its interference efficiency was lower than that of CY9 (Figure 2B). The minimum free energy of CY1 (−16.9 kcal/mol) was higher than that of CY9 (−21.8 kcal/mol), suggesting that it was easier to be degraded. To verify the prediction, the stability of CY1 and CY9 was determined by northern blot. As shown in Figure 4A, the half-life of CY9 was much longer than that of CY1. Furthermore, to increase the stability of CY1, atsRNA mutant N-CY1 was constructed by adding a 5′-stem–loop structure without pairing sequence with target mRNA (Figure 4B). The mutant N-CY1 had the same mRNA base pairing region, Hfq binding site and Rho-independent terminator with that of CY1 (Supplementary Table S3). As expected, the predicted minimum free energy of N-CY1 was reduced to −20.7 kcal/mol and the stability of N-CY1 was improved according to the northern blot results (Supplementary Figure S10). The interference efficiency was significantly increased compared with that of CY1 (P < 0.05) (Figure 4B). Additionally, the relative interference efficiency of N-CY1 was also slightly higher than that of CY1 (Supplementary Figure S10). These results demonstrated that the lower interference efficiency of CY1 was due to its unstable structure and the stability of atsRNA was crucial for its activity.
Figure 4.
Stability and function of atsRNA. (A) The predicted secondary structure and stability of atsRNAs CY1 and CY9. The expression of atsRNAs was induced with 1 mM IPTG for 30 min, and then rifampicin was added. The incubation was continued further and cells were harvested at the indicated time points for RNA preparation. Total RNA was analyzed using probes specific for CY1, CY9 and 5S rRNA respectively. (B) The predicted secondary structure and repressing effect of CY1 and N-CY1. N-CY1 was constructed by adding a 5′-stem–loop structure without pairing sequence with target mRNA. It held same mRNA base pairing region, Hfq binding site and Rho-independent terminator with CY1. The activity of β-glucuronidase was assayed after the expression of atsRNA was induced with 1 mM IPTG for 30 min. Bacterial cells containing empty vector pRI were used as the control. Average values were from triplicate experiments with std values indicated. In (A) and (B), the sequences of predicted pairing region (indicated with arrows) and Hfq binding site (indicated with asterisks) of atsRNA are marked.
The mRNA base pairing region and Hfq binding site are crucial for the activity of atsRNA
To confirm the effect of each component of atsRNA, a series of atsRNA mutants were designed. Since the secondary structure and stability played important roles in the functioning of atsRNA, the original structure and predicted minimum free energy of these mutants should be maintained to the most extent.
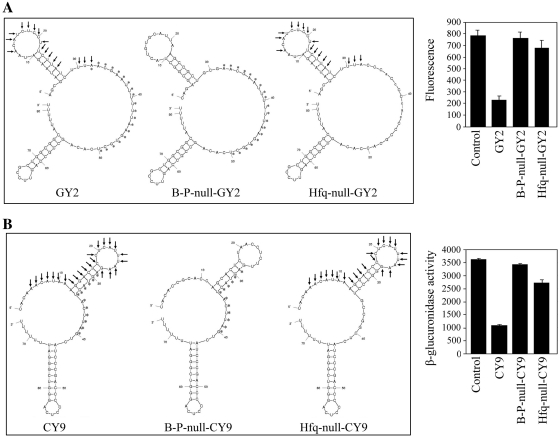
First, the mRNA base pairing regions of GY2 and CY9 were replaced by non-pairing sequences with target mRNA, respectively, yielding B-P-null-GY2 and B-P-null-CY9 (Figure 5A and B). The two mutants held similar secondary structure and minimum free energy with the corresponding atsRNAs. And the Hfq binding site and Rho-independent terminator remained constant during these mutations (Supplementary Table S4). Northern blot results showed that these mutations did not alter the stability of the two corresponding atsRNAs (Supplementary Figure S11). As shown in Figure 5A and B, the two mutants lost the repressing effect on target genes, demonstrating that mRNA base pairing region was necessary for the functioning of atsRNA. This result also suggested that atsRNA acted by pairing with target mRNA. Second, the AU rich Hfq binding sites of GY2 and CY9 were replaced by GC rich sequences, respectively, yielding Hfq-null-GY2 and Hfq-null-CY9 with similar secondary structure and minimum free energy to the corresponding atsRNAs (Figure 5A and B). The mRNA base pairing region and Rho-independent terminator remained constant during these mutations (Supplementary Table S4). It was found that the interference efficiency of Hfq-null-GY2 and Hfq-null-CY9 was attenuated dramatically in comparison with that of GY2 and CY9. Then, the stability of mutants Hfq-null-GY2 and Hfq-null-CY9 were determined by northern blot analysis (Supplementary Figure S12). Interestingly, the half-lives of the two mutants were obviously shorter than that of the corresponding atsRNAs. The reduced stability of mutants was presumed to be due to the reduced ability to bind Hfq. To confirm whether the decreased repressing effect was due to the reduced RNA amounts or not, we normalized the interference efficiency of each atsRNAs to relative RNA amounts. As shown in Supplementary Figure S12, the relative interference efficiency of the two mutants decreased certainly compared with that of the corresponding atsRNAs. This result indicated that the decreased interference efficiency was due to the lost Hfq binding site but not the reduced RNA amounts in vivo. It demonstrated that Hfq binding site was crucial for the stability and functioning of atsRNA.
Figure 5.
Effect of mRNA base pairing region and Hfq binding site on the functioning of atsRNA. (A) The predicted secondary structures and effect of atsRNA mutants B-P-null-GY2 and Hfq-null-GY2 designed based on GY2. (B) The predicted secondary structures and effect of atsRNA mutants B-P-null-CY9 and Hfq-null-CY9 designed based on CY9. In (A) and (B), the sequences of predicted pairing region (indicated with arrows) and Hfq binding site (indicated with asterisks) of atsRNA are marked. The secondary structure was predicted at 37°C by MFOLD program. Compared with the corresponding atsRNAs, the mRNA base pairing regions of B-P-null-GY2 and B-P-null-CY9 were non-pairing sequences with target mRNAs and the Hfq binding sites of Hfq-null-GY2 and Hfq-null-CY9 were GC rich sequences. Furthermore, the two other modules of atsRNA remained constant during this mutation. The expression of target gene was measured after the induction of atsRNA with 1 mM IPTG for 30 min. Bacterial cells containing empty vector pRI were used as the control. Average values were from triplicate experiments with std values indicated.
The atsRNA-mediated gene silencing is Hfq dependent
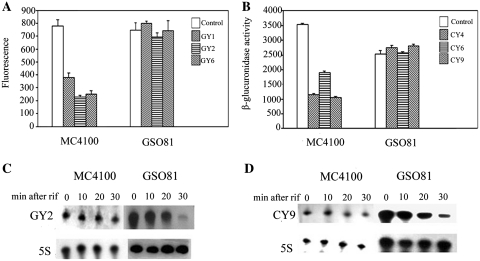
To further confirm the role of Hfq, the EGFP expression vector pACYC-P1-EGFP and effective atsRNAs GY1, GY2 and GY6 were first co-transformed into E. coli MC4100 and its hfq mutant strain GSO81 (17), respectively. These atsRNAs significantly suppressed the expression of EGFP gene in MC4100; however, they lost the repressing effects in GSO81 (Figure 6A). In addition, atsRNAs CY4, CY6 and CY9 were able to suppress the expression of target gene uidA in MC4100 but not in GSO81 (Figure 6B). Furthermore, the interference efficiencies of these atsRNAs were recovered after a multicopy plasmid containing hfq gene was transformed into the mutant strain GSO81 (Supplementary Figure S13). It is worthwhile noting that the expression of target gene uidA decreased dramatically in GSO81 compared to the wild-type strain MC4100 in the absence of atsRNAs (Figure 6B). The importance of Hfq in cellular physiology has been acknowledged when the broadly pleiotropic phenotypes of an E. coli hfq mutant were characterized (46). Deletion of hfq could lead to the increase on oxidation of carbon sources. In this study, the target gene uidA encodes β-glucuronidase which is involved in the degradation of carbon compounds. Hfq was hypothesized to have effect on utilization of carbon sources and therefore affect the expression of uidA gene. Nonetheless, these findings demonstrated that the atsRNA-mediated gene silencing was Hfq dependent.
Figure 6.
Effect of Hfq on atsRNA-mediated gene silencing. (A) The effect of atsRNA GY1, GY2 and GY6 on EGFP gene in strains MC4100 and its hfq mutant GSO81. (B) The effect of atsRNA CY4, CY6 and CY9 on uidA gene in strains MC4100 and its hfq mutant GSO81. (C) The stability of atsRNA GY2 in MC4100 and its hfq mutant GSO81. (D) The stability of atsRNA CY9 in MC4100 and its hfq mutant GSO81. In (A) and (B), bacteria cells containing empty vector pRI were used as the control. The expression of target gene was measured after the induction of atsRNA with 1 mM IPTG for 30 min. Average values were from triplicate experiments with standard values indicated. In (C) and (D), the expression of atsRNAs was induced with 1 mM IPTG for 30 min, and then rifampicin was added. The incubation was continued further and cells were harvested at the indicated time points for RNA preparation. Total RNA was analyzed using probes specific for GY2, CY9 and 5S rRNA, respectively.
Given that Hfq is important for the stability of natural trans-encoded sRNAs (14,44), we then determined the stability of atsRNAs GY2 and CY9 in isogenic hfq+ MC4100 and its hfq mutant strain GSO81. As shown in Figure 6C and D, the atsRNAs was very stable in MC4100, whereas it was rapidly degraded in the hfq mutant. This result demonstrated that Hfq was required for atsRNA stability. The effect of Hfq on atsRNA stability and function suggested that Hfq is binding to atsRNAs. To validate this conclusion, we determined the binding affinity of Hfq for atsRNAs GY2, CY9 and their corresponding Hfq binding site null mutants by electromobility shift assay (EMSA) in vitro (Supplementary Figure S14). Hfq was found to bind to atsRNA GY2 and CY9 with high affinity, while it was defective in binding to Hfq-null-GY2 and Hfq-null-CY9. This result demonstrated that Hfq does bind with atsRNA and the Hfq binding site of atsRNA is crucial for the binding affinity. Then we detected whether Hfq could facilitate the interaction between atsRNA and target mRNA in vitro. As shown in Supplementary Figure S15, Hfq strongly enhanced the complex formation between atsRNA and target mRNA, indicating that Hfq could also facilitate the binding between atsRNA and target.
atsRNA leads to gene silencing and RNase E dependent degradation of target mRNA
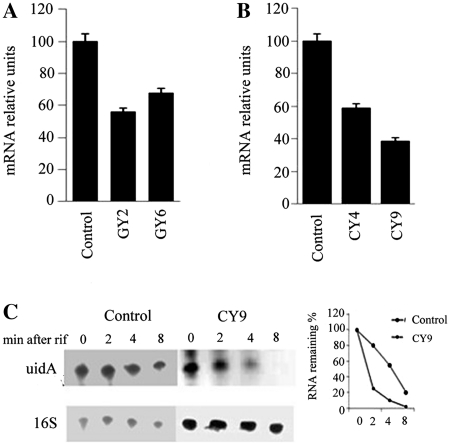
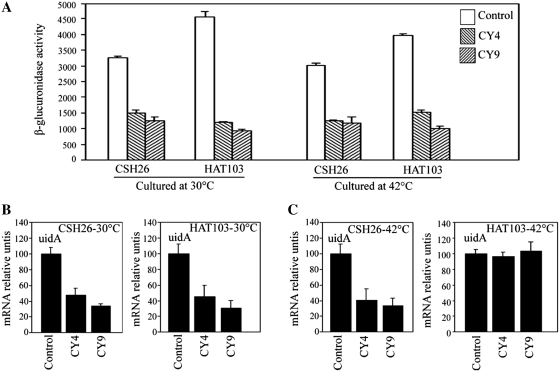
As described above, atsRNAs led to gene silencing of target gene (Figures 2 and 5). Here, we tested whether atsRNA caused the degradation of target mRNAs. As shown in Figure 7A and B, the amount of target mRNAs decreased obviously after the induction of atsRNA by QPCR. Furthermore, northern blot result showed that the half-life of uidA gene was reduced dramatically after the induction of CY9 (Figure 7C). These results demonstrated that atsRNAs led to the destabilization of target mRNA. To further elucidate the mechanism of atsRNA-mediated gene silencing, atsRNAs CY4 and CY9 was transformed into E. coli K12 strain CSH26 and its temperature-sensitive rne-1 mutant strain HAT103 (47), respectively. As shown in Figure 8A, CY4 and CY9 effectively suppressed the expression of uidA gene in both CSH26 and HAT103 strains at 30°C. The two atsRNA still repressed the expression of target gene in HAT103 at 42°C, indicating that the atsRNA-mediated gene silencing was RNase E independent (Figure 8A). The amount of uidA mRNA decreased obviously after the induction of atsRNAs in both CSH26 and its rne-1 mutant strain HAT103 at 30°C (Figure 8B), whereas the amount of uidA mRNA had no changes in HAT103 at 42°C (Figure 8C). Furthermore, the two atsRNAs could lead to degradation of target uidA mRNA in HAT103 even when it was cultured at 42°C after a multicopy plasmid containing rne gene was transformed into the mutant strain (Supplementary Figure S16). These results demonstrated atsRNA led to RNase E dependent degradation of target mRNA. Taken together, atsRNA down-regulated target mRNA expression primarily by inhibiting translation, and that the RNase E dependent degradation of target mRNA was not necessary for gene silencing.
Figure 7.
atsRNA leads to the degradation of target mRNA. (A) Quantitative real-time PCR analysis of EGFP mRNA in MC4100. (B) Quantitative real-time PCR analysis of uidA mRNA in MC4100. In (A) and (B), the mRNA level was detected after the expression of atsRNA was induced with 1 mM IPTG for 30 min. Bacterial cells containing empty vector pRI were used as the control. The value was normalized to the level of 16 S rRNA in each sample. (C) Northern blot analysis of stability of uidA mRNA in cells containing empty vector pRI and CY9. The expression of atsRNAs was induced with 1 mM IPTG for 30 min, and then rifampicin was added. The incubation was continued further and cells were harvested at the indicated time points for RNA preparation. Total RNA was analyzed using probes specific for uidA mRNA and 16 S rRNA respectively. Bacterial cells containing empty vector pRI were used as the control.
Figure 8.
Effect of RNase E on the functioning of atsRNAs and degradation of target mRNA. (A) The effect of atsRNA CY4 and CY9 on uidA gene in strains CSH26 and its temperature-sensitive rne-1 mutant HAT103. Overnight cultures grown at 30°C were diluted 1:100 into LB medium. Then the bacterial strains were grown at 30°C or 42°C, respectively. The activity of β-glucuronidase was assayed after the expression of atsRNA was induced with 1 mM IPTG for 30 min. Bacterial cells containing empty vector pRI were used as the control and average values were from triplicate experiments with std values indicated. (B) Quantitative real-time PCR analysis of uidA mRNA in strains grown at 30°C. (C) Quantitative real-time PCR analysis of uidA mRNA in strains grown at 42°C. In (B) and (C), the mRNA level was detected after the expression of atsRNA was induced with 1 mM IPTG for 30 min. Bacterial cells containing empty vector pRI were used as the control. The value was normalized to the level of 16 S rRNA in each sample.
Characterization of atsRNA targeting against bacterial essential genes
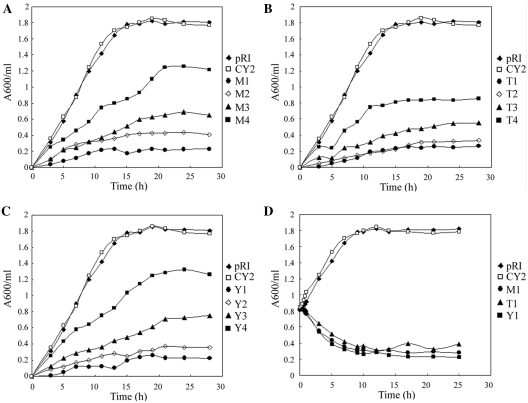
To determine whether atsRNA-mediated gene silencing are suitable for study of bacterial essential gene, a series of atsRNAs targeting against identified essential genes were designed according to the above designing principle. The essential genes murA (encoding UDP-N-acetylglucosamine enolpyruvoyl transferase), trmA (encoding tRNA (m5-U54) methyltransferase) and ygjD (encoding a putative O-sialoglycoprotein endopeptidase) were chosen as the targets (48–50). Twelve atsRNAs were designed and synthesized (M1–M4 for murA, T1–T4 for trmA and Y1–Y4 for ygjD). The sequence and secondary structure of these atsRNAs were listed in Supplementary Figure S17 and Table S5. Plasmid pRI carrying atsRNA genes were transformed into E. coli K12 strain MC4100, respectively. The efficacies of these atsRNAs were first determined by QPCR analysis (Supplementary Figure S18). According to the QPCR results, atsRNA M1, T1, Y1 and Y2 reached a reduction >80% on the expression of target gene and most of the other atsRNAs (except for M4 and Y4) inhibited the expression of target gene obviously (>50%). As the three target genes have been demonstrated to be crucial for bacterial growth rate, we then characterized the effect of atsRNA on cell growth rate by detecting the growth curve of host bacteria. As shown in Figure 9, all the atsRNAs suppressed the growth rate of host bacteria obviously, indicating that atsRNA repressed the expression of target gene efficiently. To further confirm the effect of atsRNAs M1, T1 and Y1, IPTG was added to a final concentration of 0.1 mM when OD600 reached 0.8 and then the growth curve was detected. As shown in Figure 9D, the bacterial growth rate decreased dramatically after the induction of atsRNA, suggesting that atsRNAs were effective in repressing the expression of target gene. This result demonstrated that the atsRNA-mediated silencing method would be a valuable tool for study of bacterial essential gene in which knockouts are notoriously tedious to perform.
Figure 9.
Effect of atsRNAs targeting against essential genes on growth rate of host bacteria. Growth curves of MC4100 strains after the expression of atsRNAs targeting against murA (A), trmA (B) and ygjD (C). Overnight cultures were diluted 1:100 into LB medium and bacterial strains were grown with 0.1 mM IPTG at time 0 at 30°C. (D) Growth curve of MC4100 strain after the expression of atsRNAs M1, T1 and Y1. Bacterial cells containing empty vector pRI and atsRNA CY2 were used as the control. Turbidity of the cultures was monitored at 600 nm.
atsRNA can not efficiently downregulate its targets in S. aureus
To test whether atsRNA-mediated gene silencing are suitable for study of Gram-positive bacteria genes, two series of atsRNAs targeting against identified S. aureus genes were designed according to the above designing principle. The toxic genes hla (encoding α-hemolysin) and SEA (encoding Enterotoxin A) were chosen as the targets. Fourteen atsRNAs were designed and synthesized (H1–H8 for hla and S1–S6 for SEA). The sequence and other detailed information of these atsRNAs were listed in Supplementary Tables S6, S7 and S8. Plasmid pSB2035 carrying atsRNA genes were transformed into S. aureus RN4220.
The efficacies of H series atsRNAs were determined by using α-hemolysin activity assay (Supplementary Figure S19). H1, H2, H3 and H4 shared the same Hfq binding site and Rho-independent terminator with CY9, and had 22, 16, 11 and 6 base pairings with mRNA, respectively. The results indicated that all the four atsRNAs only slightly suppressed the expression of α-hemolysin, and the silencing abilities were positively related with mRNA base pairing numbers. Although H5 had no Hfq binding site compared with H1, it also triggered the same interference efficiency with H1. H6 also had no Hfq binding site, and contained fewer mRNA base pairings compaired with H5. The suppression to gene expression was weaker than that of H5. Compared with H1, H7 and H8 had the same mRNA base pairing regions but both were likely to be easily degraded (the minimum free energy of them is −19.6 and −16.7 kcal/mol, respectively), accordingly, they lost repressing effect on the target gene as H1. The efficacies of S series atsRNAs were determined by using SE ELISA Kit and Western blot analysis (Supplementary Figure S20). It also showed the same tendencies with that of H series atsRNAs. As shown in Supplementary Figure S21, the expression of atsRNAs had no effect on growth rate of host bacteria.
These results demonstrated that atsRNAs designed following the principle mentioned above may not lead to effective inhibitions to the expression of target genes in S. aureus. However, it was likely that these atsRNAs to trigger a slight antisense mechanism, which related with mRNA base pairing numbers and its own stabilities. And Hfq was dispensable in this process.
DISCUSSION
In this study, we have developed an effective and convenient method to downregulate the expression of specific gene using atsRNAs in E. coli. atsRNA was designed to be a modular structure composed of mRNA base pairing region, Hfq binding site and typical Rho-independent terminator. Using this method, we successfully repressed the expression of exogenous EGFP gene and endogenous uidA gene in E. coli. atsRNA acted by base pairing with the 5′-UTR of target mRNA and the atsRNA-mediated gene silencing was Hfq dependent. atsRNA led to translation inhibition and RNase E dependent degradation of target mRNA and the translation inhibition was the primary event for gene silencing.
The atsRNA-mediated gene silencing holds many advantages over conventional methods. It is an easy and fast way of down regulating the expression of specific genes, and more importantly, it is an inducible system. The existing methods for functional decipherment of genes in prokaryotes, such as homologous recombination and transposon mutagenesis, significantly limit the study of bacterial essential genes. The atsRNA-mediated silencing is promised to be an alternative RNA tool of deciphering gene function in bacteria whereas gene knockout was time consuming. In addition, atsRNA acting at post-transcriptional level provides a simple way to impose an over-arching level of regulation on a group of genes or operons that may be regulated in many different ways at the level of transcription. Due to its low cost and easy designing, atsRNA makes the automatic and high-throughput functional decipherment of prokaryotic genes possible.
Bacterial sRNAs that act by base pairing can be divided into two classes: cis-encoded and trans-encoded antisense sRNAs. The former are located in the same DNA region and are, therefore, fully complementary to their targets over a long sequence stretch, whereas the latter are located in another chromosomal location, and are only partially complementary to their target mRNAs. The traditional antisense RNA (asRNA) strategy has been developed based on the cis-encoded antisense RNAs. Thus, asRNAs were fully complementary to the target genes over a long region (usually >300 nt). Although the asRNA technology worked well in some cases (51), it usually led to a 40–60% reduction in the expression of target mRNA (52,53). In this study, the atsRNA strategy was developed based on the trans-encoded antisense RNAs. They were complementary to the target mRNAs over a stretch of ∼20 nt (Supplementary Figure S4, Tables S1 and S2), and even within this stretch, the complementarity is not complete. The full length of atsRNAs was generally <100 nt. In this study, the interference efficiency of most effective atsRNA could reach ∼80%, which is higher than that of asRNAs. Stability was important for the efficacy of both asRNA and atsRNA. The half-life of atsRNA as well as the natural trans-encoded sRNAs was usually >10 min (15,54,55), while the half-life of asRNA was usually >5 min (53,56). This may partially explain the improved efficacy of atsRNA.
The RNA chaperone protein Hfq is important for the functioning of sRNA; it facilitates the interaction between some of well-characterized sRNA and their targets (54,57). Hfq has been shown to exert a stabilizing effect on endogenous sRNAs by protecting them from endonucleolytic cleavage (25,58). Our results also demonstrated that Hfq was required for the stability of atsRNAs. The interaction between Hfq and atsRNA was presumed to increase the stability and then enhance the efficacy of atsRNA. Hfq appears to bind preferentially to unstructured AU-rich regions, which were frequently located between structured loop regions of sRNA (15,17,43). And the AU rich Hfq binding site of atsRNA has been demonstrated to be crucial for the binding affinity to Hfq (Supplementary Figure S14). In this study, atsRNAs with long putative Hfq binding site in secondary structure, such as GY2, GY6, CY4 and CY9, had higher interference efficiency than those with short putative Hfq binding site (e.g. GY3, GY5 and CY6). Therefore, the length of putative Hfq binding site was proposed to be crucial for the interaction between atsRNA and Hfq. In order to enhance the efficacy of atsRNA, the putative Hfq binding site could be appropriately elongated during the designing process. However, longer putative Hfq binding site is not always better. For instance, increase of the length of putative Hfq binding site of CY9 did not result in an increase of interference efficiency. According to our results, a length of 10 nt of putative Hfq binding site was believed to be enough for the functioning of atsRNA.
The efficacy of atsRNA depends on several factors, including the structure and stability itself, the binding kinetics (e.g. binding rate and binding strength) between atsRNA and target mRNA and the Hfq binding affinity. The lower inhibitory efficiency of CY1 and CY10 was presumed to be due to the unstable structure according to the predicted minimum free energy. Since the short putative Hfq binding sites, the less effective efficiency of GY3, GY5, CY3 and CY6 was hypothesized to be due to the low binding affinity with Hfq. The secondary structure of atsRNA is an important factor for inhibitory efficiency of atsRNA. Our results indicated the stem–loop structure within the mRNA base pairing region is necessary for atsRNA regulation. Especially, the size of the loop structure is crucial for the activity of atsRNA (Figure 3). However, the optimal number of unpaired nucleotides in the loop is not always the same for all atsRNAs. For instance, the optimal number of loop nucleotides for atsRNA GY2, GY6 and CY4 was 9, 11 and 8 nt, respectively (data not shown). We presume that the optimal size of the loop depends on the secondary structure of both atsRNA and target mRNA, as well as the nucleotide composition in the loop. In addition, according to the calculated thermodynamic pairing energy value of atsRNA-target duplex formation, there is no direct correlation between the inhibitory efficiency and the stability of the hybrids (Supplementary Figure S4). It is worth noting that the inhibitory efficiency of classical cis-acting antisense RNAs has proven to be primarily controlled by the binding rate constant rather than the binding affinity between the sense–antisense complexes (59). However, the relationship between inhibitory efficiency and binding kinetics of atsRNA requires further studies. Since the participation of Hfq in atsRNA regulation, the binding kinetic features of atsRNA would be more complex than that of cis-acted antisense RNA. In this study, the number of hybrid base pairings of atsRNA ranged mainly from 19 to 23 nt in length. Some effective asRNA cassettes with a small number of base pairings (12 nt) have been reported, indicating that relatively short antisense sequences can also suffice for efficient binding and regulation (53). However, in the absence of knowledge about target accessibility, we still favor large number of base pairing sequences during atsRNA design process, which are more likely to contain structures that efficiently seed hybridization. Furthermore, a large number of antisense nucleotides would be helpful for the inhibitory specificity of atsRNA.
In this study, the effective atsRNA could result in a 4- to 7-fold repression of target genes, and this is enough for functional investigation of bacterial genes. However, over-expression of natural trans-encoded sRNAs could generally repress the target genes from 7- to hundreds-fold (12,60–63). Therefore, we will further optimize the above-discussed design parameters to increase the efficacy of atsRNA. More importantly, research progress on natural trans-encoded sRNA will be contributive to the improvement of atsRNA technology. Additionally, taking into account of the structural accessibility of target mRNA during the design process would be helpful for the efficacy of atsRNA. Since the binding between many natural sRNAs and their corresponding targets initiates through a loop–loop structure (45,64–66), the target site for loop nucleotides of atsRNA should also be located in an accessible loop structure.
In E. coli, the endogenous sRNAs, SgrS and RyhB have been shown clearly to form a specific ribonucleoprotein complex with RNase E and Hfq, resulting in translation inhibition and RNase E dependent degradation of target mRNAs (26,27). Translation inhibition is the primary event for gene silencing rather than the target mRNA degradation (67). In this study, atsRNA also led to gene silencing and RNase E dependent degradation of target mRNA and gene silencing occurred in the absence of target mRNA degradation. This is quite similar with the endogenous sRNAs RyhB and SgrS. According to the mechanism similarity between endogenous trans-encoded sRNA and atsRNA, we conclude that atsRNA functions also by associating with the RNase E/Hfq complex. The atsRNA-Hfq-RNase E complex acts on the SD sequences of target mRNA through base pairing sequences, resulting in translation inhibition and RNase E dependent degradation of target mRNA. For both the two endogenous sRNA and atsRNA, the decay of target mRNAs is just a consequence of translational inhibition. However, the physiological relevance of the RNase E-dependent degradation of target mRNAs would be necessary to make gene silencing irreversible.
Compared to Gram-negative bacteria, much fewer sRNAs have been identified in Gram-positive species, and there exists many differences in length, structure and function mode between the sRNAs from the two bacterial groups (9,16). In our study, atsRNAs targeting hla and SEA genes designed by the above principle were transformed into S. aureus strain RN4220 in which Hfq is produced. Unfortunately, the results indicated that both H and S series of atsRNAs failed to achieve significant repression effect on the respective target mRNA. It is necessary to mention that the synthesis of virulence factors in S. aureus is controlled by a regulatory RNA molecule, RNAIII, encoded by the agr locus. Transcription of genes coding for secreted toxins and enzymes is stimulated by RNAIII. More importantly, the ribosome binding site of the hla transcript is blocked by intramolecular base pairing. Hybridization with RNAIII prevents this intramolecular base pairing and makes the hla mRNA accessible for translation initiation (68). So initially we doubted that the inhibition effect of H series atsRNAs might be counteracted by the competitive binding of RNAIII and hla transcript. But later sequence alignments showed that H series atsRNAs were mainly complementary to nucleotides 305–345 of the hla transcript including the start codon and SD region, whereas RNAIII forms base pairing with the anti-SD region (nucleotides 179–186) of the hla mRNA. So it seemed unlikely that S. aureus RNAIII, which is heavily expressed at the stationary phase of growth, would compete with the atsRNAs involved in this study. Moreover, the S series atsRNAs also triggered weak downregulation of SEA gene. Taken together, these results indicated that the designing principle was not applicable for gene silencing in S. aureus. Therefore we speculated that these atsRNAs probably affected gene expression only by a slight antisense mechanism.
In low GC Gram-positive bacteria, the function of Hfq is unclear. Although Hfq has been shown to interact with Bacillus subtilis SR1 and SR2 as well as S. aureus RNAIII, it neither affects their stability nor facilitates the interactions between the antisense RNAs and their target mRNAs. A study from S. aureus failed to identify significant phenotype changes for an Hfq mutant strain. So it has been supposed that Hfq is dispensable for riboregulation by sRNAs in Gram-positive bacteria (69–73). But some notions may question this view. First, Hfq homologs are present in one or more copies in several Gram-positive species, and many of the key amino acids involved in RNA binding are conserved (57,74). Second, Hfq in L. monocytogenes contributes to stress tolerance and pathogenesis in mice and interacts with the sRNAs, indicating a role for Hfq in sRNA-mediated riboregulation in this Gram-positive pathogen (16,75). Third, although Hfq protein of E. coli has a longer C terminal-tail than that of S. aureus, the tail appears to be dispensable in the RNA binding function of Hfq (76). So the possibility of S. aureus Hfq functioning as a RNA chaperon should not be excluded. Just as expected, a recent work suggested that the Hfq protein in S. aureus could also act as an RNA binding protein such as in E. coli (77). However, the mechanism how the relevant genes can be regulated by Hfq remains to be uncovered. Last but not least, some studies indicated that hfq gene was differently expressed in several S. aureus strains. Even in strains such as RN6390, Hfq can not be detected at all (73,77). So it is supposed that while the protein-mediated riboregulation is likely to exist in Gram-positive bacterium, Hfq should not be regarded as the unique RNA chaperone, in other word, there may be other protein molecules functioning by facilitating the mRNA–sRNA interaction. In view of this point, one important direction for future studies is to clarify whether Hfq-independent trans-acting antisense RNAs act on their own or depend on other factors that substitute for the Hfq protein. It is remarkable that a recent study provided evidence for Hfq-dependent translational repression in the Gram-positive human pathogen L. monocytogenes, indicating that modulation of translation by trans-encoded sRNAs may occur by both Hfq-dependent and independent mechanisms, thus adding another layer of complexity to sRNA-mediated riboregulation in Gram-positive species (29). It is certainly conceivable that this mode of regulation is more widespread in L. monocytogenes and other Gram-positive bacteria.
It is widely acknowledged that although RNA interference (RNAi) has become a powerful gene silencing tool in eukaryotes, the frequency of so-called “off-target effect” is still high to some extent. In our study, the atsRNAs designed following our principle achieved a notable inhibition effect against the targets; however, a similar issue should not be neglected. Because the atsRNA only have a considerably short and incomplete complementary base pairing with the predicted target mRNA, the cross-talk to mRNAs other than the targets seems not to be avoided easily; in other word, just like the siRNA in RNAi, the specificity of atsRNA is hard to be guaranteed. But hitherto there has not been a convincing method to settle this significant problem, so developing new specially analysis software is a promising approach, which would improve the current principle remarkably.
The small RNA has become a standard experimental tool and its therapeutic potential is being aggressively harnessed in eukaryotes. Similarly, the atsRNA-mediated gene silencing will become a popular method for studying gene function and elucidating networks of gene expression in bacteria. Furthermore, recent studies have demonstrated that numerous endogenous trans-encoded sRNAs have crucial roles in bacterial stress responses and virulence regulation (78,79). Therefore, atsRNAs targeting against virulence genes would function effectively in bacterial pathogens and it could potentially serve as antibiotics. Given the emergence and increasing prevalence of bacterial strains that are resistant to available antibiotics, atsRNAs will provide an alternative approach to antimicrobial therapy that offers promising opportunities to inhibit pathogenesis and its consequences without placing immediate life-or-death pressure on the target bacterium.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Basic Research Program of China (973 Program) [2003CB716402]; National High Technology Research and Development Program of China (863 Program) [2007AA09Z418 and 2007AA091506]. Funding for open access charge: National High Technology Research and Development Program of China (863 Program) [2007AA091506].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Gisela Storz (National Institutes of Health), R. Gary Sawers (Martin-Luther University Halle-Wittenberg) and Masaaki Wachi (Tokyo Institute of Technology) for providing plasmid and strains. We thank Guanpin Yang (Ocean University of China) for advice and comments on the article.
REFERENCES
- 1.Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- 3.Andersen J, Forst SA, Zhao K, Inouye M, Delihas N. The function of micF RNA. micF RNA is a major factor in the thermal regulation of OmpF protein in Escherichia coli. J. Biol. Chem. 1989;264:17961–17970. [PubMed] [Google Scholar]
- 4.Møller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 2002;16:1696–1706. doi: 10.1101/gad.231702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Massé E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl Acad. Sci. USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, Morozov I, Baker CS, Georgellis D, Babitzke P, Romeo T. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol. Microbiol. 2003;48:657–670. doi: 10.1046/j.1365-2958.2003.03459.x. [DOI] [PubMed] [Google Scholar]
- 7.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 8.Davis BM, Quinones M, Pratt J, Ding Y, Waldor MK. Characterization of the small untranslated RNA RyhB and its regulon in Vibrio cholerae. J. Bacteriol. 2005;187:4005–4014. doi: 10.1128/JB.187.12.4005-4014.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papenfort K, Vogel J. Regulatory RNA in Bacterial Pathogens. Cell Host Microbe. 2010;8:116–127. doi: 10.1016/j.chom.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Mizuno T, Chou MY, Inouye M. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA) Proc. Natl Acad. Sci. USA. 1984;81:1966–1970. doi: 10.1073/pnas.81.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouché F, Bouché JP. Genetic evidence that DicF, a second division inhibitor encoded by the Escherichia coli dicB operon, is probably RNA. Mol. Microbiol. 1989;3:991–994. doi: 10.1111/j.1365-2958.1989.tb00249.x. [DOI] [PubMed] [Google Scholar]
- 12.Altuvia S, Zhang A, Argaman L, Tiwari A, Storz G. The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J. 1998;17:6069–6075. doi: 10.1093/emboj/17.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urbanowski ML, Stauffer LT, Stauffer GV. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol. Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- 14.Vanderpool CK, Gottesman S. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 2004;54:1076–1089. doi: 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- 15.Møller T, Franch T, Højrup P, Keene DR, Bächinger HP, Brennan RG, Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA–RNA interaction. Mol. Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 16.Christiansen JK, Nielsen JS, Ebersbach T, Valentin-Hansen P, Sogaard-Andersen L, Kallipolitis BH. Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA. 2006;12:1383–1396. doi: 10.1261/rna.49706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 18.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 19.Sukhodolets MV, Garges S. Interaction of Escherichia coli RNA polymerase with the ribosomal protein S1 and the Sm-like ATPase Hfq. Biochemistry. 2003;42:8022–8034. doi: 10.1021/bi020638i. [DOI] [PubMed] [Google Scholar]
- 20.Geissmann TA, Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohanty BK, Maples VF, Kushner SR. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol. Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- 22.Kawamoto H, Koide Y, Morita T, Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- 23.Arluison V, Mura C, Guzmán MR, Liquier J, Pellegrini O, Gingery M, Regnier P, Marco S. Three-dimensional structures of fibrillar Sm proteins: Hfq and other Sm-like proteins. J. Mol. Biol. 2006;356:86–96. doi: 10.1016/j.jmb.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 24.Gönczy P, Echeverri C, Oegema K, Coulson A, Jones SJ, Copley RR, Duperon J, Oegema J, Brehm M, Cassin E, et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- 25.Moll I, Afonyushkin T, Vytvytska O, Kaberdin VR, Bläsi U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA. 2003;9:1308–1314. doi: 10.1261/rna.5850703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jousselin A, Metzinger L, Felden B. On the facultative requirement of the bacterial RNA chaperone, Hfq. Trends Microbiol. 2009;17:399–405. doi: 10.1016/j.tim.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen JS, Lei LK, Ebersbach T, Olsen AS, Klitgaard JK, Kallipolitis PV, Kallipolitis BH. Defining a role for Hfq in Gram-positive bacteria: evidence for Hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic Acids Res. 2009;3:907–919. doi: 10.1093/nar/gkp1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- 31.Lum L, Yao S, Mozer B, Rovescalli A, Von Kessler D, Nirenberg M, Beachy PA. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science. 2003;299:2039–2045. doi: 10.1126/science.1081403. [DOI] [PubMed] [Google Scholar]
- 32.Green PJ, Pines O, Inouye M. The role of antisense RNA in gene regulation. Annu. Rev. Biochem. 1986;55:569–597. doi: 10.1146/annurev.bi.55.070186.003033. [DOI] [PubMed] [Google Scholar]
- 33.Inouye M. Antisense RNA: its functions and applications in gene regulation–a review. Gene. 1988;72:25–34. doi: 10.1016/0378-1119(88)90124-2. [DOI] [PubMed] [Google Scholar]
- 34.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doherty N, Holden MT, Qazi SN, Williams P, Winzer K. Functional analysis of luxS in Staphylococcus aureus reveals a role in metabolism but not quorum sensing. J. Bacteriol. 2006;188:2885–2897. doi: 10.1128/JB.188.8.2885-2897.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wickes BL, Edman JC. The Cryptococcus neoformans GAL7 gene and its use as an inducible promoter. Mol. Microbiol. 1995;16:1099–1109. doi: 10.1111/j.1365-2958.1995.tb02335.x. [DOI] [PubMed] [Google Scholar]
- 38.Ory JJ, Griffith CL, Doering TL. An efficiently regulated promoter system for Cryptococcus neoformans utilizing the CTR4 promoter. Yeast. 2004;21:919–926. doi: 10.1002/yea.1139. [DOI] [PubMed] [Google Scholar]
- 39.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl Acad. Sci. USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aiba H, Matsuyama S, Mizuno T, Mizushima S. Function of micF as an antisense RNA in osmoregulatory expression of the ompF gene in Escherichia coli. J. Bacteriol. 1987;169:3007–3012. doi: 10.1128/jb.169.7.3007-3012.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 42.Repoila F, Majdalani N, Gottesman S. Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: the RpoS paradigm. Mol. Microbiol. 2003;48:855–861. doi: 10.1046/j.1365-2958.2003.03454.x. [DOI] [PubMed] [Google Scholar]
- 43.Brescia CC, Mikulecky PJ, Feig AL, Sledjeski DD. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA. 2003;9:33–43. doi: 10.1261/rna.2570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tu KC, Bassler BL. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev. 2007;21:221–233. doi: 10.1101/gad.1502407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Argaman L, Altuvia S. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol. 2000;300:1101–1112. doi: 10.1006/jmbi.2000.3942. [DOI] [PubMed] [Google Scholar]
- 46.Tsui HC, Leung HC, Winkler ME. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 47.Takada A, Nagai K, Wachi M. A decreased level of FtsZ is responsible for inviability of RNase E-deficient cells. Genes Cells. 2005;10:733–741. doi: 10.1111/j.1365-2443.2005.00872.x. [DOI] [PubMed] [Google Scholar]
- 48.Persson BC, Gustafsson C, Berg DE, Björk GR. The gene for a tRNA modifying enzyme, m5U54-methyltransferase, is essential for viability in Escherichia coli. Proc. Natl Acad. Sci. USA. 1992;89:3995–3998. doi: 10.1073/pnas.89.9.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown ED, Vivas EI, Walsh CT, Kolter R. MurA (MurZ), the enzyme that catalyzes the first committed step in peptidoglycan biosynthesis, is essential in Escherichia coli. J. Bacteriol. 1995;177:4194–4197. doi: 10.1128/jb.177.14.4194-4197.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arigoni F, Talabot F, Peitsch M, Edgerton MD, Meldrum E, Allet E, Fish R, Jamotte T, Curchod ML, Loferer H. A genome-based approach for the identification of essential bacterial genes. Nat. Biotechnol. 1998;16:851–856. doi: 10.1038/nbt0998-851. [DOI] [PubMed] [Google Scholar]
- 51.Ji Y, Zhang B, Van SF, Horn, Warren P, Woodnutt G, Burnham MK, Rosenberg M. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science. 2001;293:2266–2269. doi: 10.1126/science.1063566. [DOI] [PubMed] [Google Scholar]
- 52.Engdahl HM, Hjalt TA, Wagner EG. A two unit antisense RNA cassette test system for silencing of target genes. Nucleic Acids Res. 1997;25:3218–3227. doi: 10.1093/nar/25.16.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Engdahl HM, Lindell M, Wagner EG. Introduction of an RNA stability element at the 5′-end of an antisense RNA cassette increases the inhibition of target RNA translation. Antisense Nucleic Acid Drug Dev. 2001;11:29–40. doi: 10.1089/108729001750072100. [DOI] [PubMed] [Google Scholar]
- 54.Sledjeski DD, Whitman C, Zhang A. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 2001;183:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl Acad. Sci. USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakashima N, Tamura T, Good L. Paired termini stabilize antisense RNAs and enhance conditional gene silencing in Escherichia coli. Nucleic Acids Res. 2006;34:e138. doi: 10.1093/nar/gkl697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 58.Afonyushkin T, Vecerek B, Moll I, Bläsi U, Kaberdin VR. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 2005;33:1678–1689. doi: 10.1093/nar/gki313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nordström K, Wagner EG. Kinetic aspects of control of plasmid replication by antisense RNA. Trends Biochem. Sci. 1994;19:294–300. doi: 10.1016/0968-0004(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 60.Lease RA, Cusick ME, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl Acad. Sci. USA. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Massé E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Levine E, Zhang Z, Kuhlman T, Hwa T. Quantitative characteristics of gene regulation by small RNA. PLoS Biol. 2007;5:e229. doi: 10.1371/journal.pbio.0050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kolb FA, Engdahl HM, Slagter-Jäger JG, Ehresmann B, Ehresmann C, Westhof E, Wagner EG, Romby P. Progression of a loop–loop complex to a four-way junction is crucial for the activity of a regulatory antisense RNA. EMBO J. 2000;19:5905–5915. doi: 10.1093/emboj/19.21.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kolb FA, Malmgren C, Westhof E, Ehresmann C, Ehresmann B, Wagner EG, Romby P. An unusual structure formed by antisense-target RNA binding involves an extended kissing complex with a four-way junction and a side-by-side helical alignment. RNA. 2000;6:311–324. doi: 10.1017/s135583820099215x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kolb FA, Westhof E, Ehresmann C, Ehresmann B, Wagner EG, Romby P. Bulged residues promote the progression of a loop–loop interaction to a stable and inhibitory antisense-target RNA complex. Nucleic Acids Res. 2001;29:3145–3153. doi: 10.1093/nar/29.15.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morita T, Mochizuki Y, Aiba H. Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc. Natl Acad. Sci. USA. 2006;103:4858–4863. doi: 10.1073/pnas.0509638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morfeldt E, Taylor D, von Gabain A, Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14:4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heidrich N, Chinali A, Gerth U, Brantl S. The small untranslated RNA SR1 from the Bacillus subtilis genome is involved in the regulation of arginine catabolism. Mol. Microbiol. 2006;62:520–536. doi: 10.1111/j.1365-2958.2006.05384.x. [DOI] [PubMed] [Google Scholar]
- 71.Heidrich N, Moll I, Brantl S. In vitro analysis of the interaction between the small RNA SR1 and its primary target ahrC mRNA. Nucleic Acids Res. 2007;35:4331–4346. doi: 10.1093/nar/gkm439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jousselin A, Metzinger L, Felden B. On the facultative requirement of the bacterial RNA chaperone, Hfq. Trends Microbiol. 2009;17:399–405. doi: 10.1016/j.tim.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 73.Bohn C, Rigoulay C, Bouloc P. No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus. BMC Microbiol. 2007;7:10. doi: 10.1186/1471-2180-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun X, Zhulin I, Wartell RM. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 2002;30:3662–3671. doi: 10.1093/nar/gkf508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Christiansen JK, Larsen MH, Ingmer H, Sogaard-Andersen L, Kallipolitis BH. The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J. Bacteriol. 2004;186:3355–3362. doi: 10.1128/JB.186.11.3355-3362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 77.Liu Y, Wu N, Dong J, Gao Y, Zhang X, Mu C, Shao N, Yang G. Hfq is a global regulator that controls the pathogenicity of Staphylococcus aureus. PLoS ONE. 2010;5:e13069. doi: 10.1371/journal.pone.0013069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Toledo-Arana A, Repoila F, Cossart P. Small noncoding RNAs controlling pathogenesis. Curr. Opin. Microbiol. 2007;10:182–188. doi: 10.1016/j.mib.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 79.Bejerano-Sagie M, Xavier KB. The role of small RNAs in quorum sensing. Curr. Opin. Microbiol. 2007;10:189–198. doi: 10.1016/j.mib.2007.03.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.