Abstract
Nucleoside Monophosphate Kinases (NMPKs) family are key enzymes in nucleotide metabolism. Bacterial UMPKs depart from the main superfamily of NMPKs. Having no eukaryotic counterparts they represent attractive therapeutic targets. They are regulated by GTP and UTP, while showing different mechanisms in Gram(+), Gram(–) and archaeal bacteria. In this work, we have characterized the mycobacterial UMPK (UMPKmt) combining enzymatic and structural investigations with site-directed mutagenesis. UMPKmt exhibits cooperativity toward ATP and an allosteric regulation by GTP and UTP. The crystal structure of the complex of UMPKmt with GTP solved at 2.5 Å, was merely identical to the modelled apo-form, in agreement with SAXS experiments. Only a small stretch of residues was affected upon nucleotide binding, pointing out the role of macromolecular dynamics rather than major structural changes in the allosteric regulation of bacterial UMPKs. We further probe allosteric regulation by site-directed mutagenesis. In particular, a key residue involved in the allosteric regulation of this enzyme was identified.
INTRODUCTION
The growing number of infectious bacteria with multiple drug resistance mechanisms urges the discovery of new drugs with original mechanism of action (1). This is particularly the case for Mycobacterium tuberculosis, which remains a major public health issue. Current treatment requires a therapy involving four different drugs and for a period of 6–9 months. This often leads to poor patient compliance and subsequently to drug resistance (2). Indeed, multi-drug resistant tuberculosis (MDR-TB) emerged in the early 1990s and extensively drug-resistant tuberculosis (XDR-TB) cases were first reported in 2005 (3,4). Consequently, a renewed effort to develop new anti-tuberculosis drugs is necessary (5–7). Accordingly, new therapeutic targets are to be precisely characterized to uncover their specific structural and functional features (8–10).
Large-scale structural genomic projects have been launched to provide X-ray or nuclear magnetic resonance (NMR)-derived structures to help functional annotation and virtual screening (11). Nevertheless, the extent to which each target from a given pathogen should be characterized has to be defined. This is particularly true for 3D-structure determination. NMR and X-ray crystallography are efficient and can provide structures at atomic resolution. However, various bottlenecks exist, such as over-expression, stabilization or crystallisation that slow down the experimental characterization of many targets. In the case of M. tuberculosis, despite some significant improvement in the last decade, many proteins remain refractory to structural analysis (11). Sequence comparisons and molecular modelling have been suggested as an alternative for quicker structural annotation at the expense of the overall quality and precision. However, functional properties may diverge or rapidly evolve despite good sequence conservation. These variations are still difficult to predict.
The multi-faceted limits of functional and structural annotations by means of sequence comparisons can be illustrated with the studies of nucleoside monophosphate kinases (NMPKs). The NMPKs are essential enzymes and represent attractive therapeutic targets. They catalyze the phosphorylation of nucleoside monophosphate into nucleoside diphosphate using ATP as the main phosphoryl group donor. Structure determination and sequence comparisons led to the definition of two main classes of NMPKs. Members from four out of the five functional families (AMP kinase (AMPK), GMP kinase (GMPK), CMP kinase (CMPK) and TMP kinase (TMPK)) belong to class-I. All the enzymes of this class share a common overall fold related to the Rossmann fold (12) and contain a strictly conserved P-loop (13). These similarities suggested that the first structures of AMPKs solved would be useful for modelling most other NMPKs. These class-I enzymes are usually small (∼200 residue long) and monomeric, sharing a structurally common catalytic centre (14,15). This common binding site could be recognized from sequence comparisons despite large sequence divergence (16,17) and they appear to all accommodate their phospho-donor in a similar orientation. However, some structural variations exist and have usually evolved according to substrate specificity. The particular sub-domain organization observed in CMPK could be also predicted (18,19) but the specific dimerization of TMPK was only revealed by resolution of an X-ray structure (20). Surprisingly, eukaryotic UMP/CMPK are bifunctional type-I NMPKs as they efficiently phosphorylate CMP as well as UMP. Conversely, many of the structural features of type-I NMPKs could not be found in the bacterial UMPKs, which constitute the second class of NMPKs.
Bacterial UMPKs do not belong to the P-loop containing kinases but rather to the superfamily of amino acid kinases (AA-kinases, PF00696) (21). They feature a Kx38-42GggN motif instead of a classical P-loop motif (Gx4GKt). They also differ from class-I NMPKs in their overall topology and multimeric organization. Slightly larger (∼240 residues) and usually hexameric, they still adopt a Rossman fold decorated with additional secondary structures or subdomains. A model of the monomer structure was first deduced from the distantly related and mainly dimeric AA-kinases (22,23). A common binding site accommodating the phospho-donor (ATP) could be detected and the rough orientation of the UMP was also extrapolated. A more precise view of the substrate binding and quaternary structure came from the first crystal structures of an UMPK (24–26). Sequence comparisons allowed extending these structural features to all bacterial UMPKs.
Within this context, the studies of the mycobacterial NMPKs revealed peculiar and surprising variations. Sequence analyses suggest that a recent divergence occurred from a bacterial genetic background (27). The mycobacterial genomes (28–31) encode five NMPKs with distinct and original sequence evolution trends. AMPK from Mycobacterium tuberculosis (AMPKmt, Rv0733) represents a new subfamily of short bacterial enzymes (32). Nevertheless, AMPKmt is ∼47% identical to its bacterial orthologues while eukaryotic enzymes are slightly more distant in sequence (∼36%). This sequence conservation was confirmed at the structural level by NMR (33,34). On the contrary, GMPK from M. tuberculosis (Rv1389) is (slightly) more closely related to the eukaryotic enzymes than their bacterial orthologues. Its crystal structure revealed that it is similar in several but not all respects to its eukaryotic counterparts. Indeed its GMP binding site is unique (35). A more complex evolution is illustrated with the TMPK from M. tuberculosis (TMPKmt, Rv3247c), which was shown to be a chimera of prokaryotic and eukaryotic enzymes (36). In this case, modelling at low level of sequence identity (23%) was sufficient to identify a new substrate, although the enzyme inhibition by AZT could not be explained by this method. Based on the X-ray structures, different families of nucleosidic and non-nucleosidic analogues were derived to target TMPKmt (37–43). Some of them proved to have inhibitory activity on mycobacterium growth without cytotoxic effects on human cells (40,43). These data confirm the importance of NMPKs as potential therapeutic targets for antimycobacterial compounds.
The last two mycobacterial NMPKs (CMPK and UMPK) are clearly prokaryotic enzymes. Indeed, a functional CMPK (Rv1712) closely related to its bacterial orthologues (∼40%) was recently described in M. tuberculosis (44). It efficiently phosphorylates CMP (and dCMP) but UMP very poorly. In parallel, the mycobacterial genome encodes for Rv2883c (named herein, UMPKmt) exhibiting ∼50% identity with bacterial UMPKs.
Here, we describe the catalytic and regulatory properties of recombinant UMPKmt overexpressed in Escherichia coli. We have combined biochemical characterization, comparative modelling and, subsequently, X-ray crystallography with directed mutagenesis to unravel the functional features of this enzyme.
MATERIALS AND METHODS
Chemicals
Restriction enzymes, T4 DNA polymerase and coupling enzymes for determination of UMPK activity were purchased from Roche Applied Science. T4 DNA ligase and Vent DNA polymerase were purchased from New England Biolabs, Inc. BD TALON metal affinity resin was purchased from BD Biosciences Clontech. ATP, GTP, UTP, dUTP, dGTP and dTTP were purchased from Sigma. 5F-UMP, 5Br-UMP, 5F-UTP, 5I-UTP, aminoallyl UTP, 8Br-GTP, Mant-GTP, 6-methylthio GTP, GMPPCP and GMPPNP were purchased from Jena Bioscience. NDP kinase was kindly provided by I. Lascu.
Bacterial strains and growth conditions
The E. coli NM554 (45) and BL21(DE3)/pDIA17 (46) strains were used for DNA purification and for production of recombinant wild-type and variant UMPKmt, respectively. Cultures were grown in 2YT medium (47) supplemented with 100 µg/ml ampicillin or 70 µg/ml kanamycin and 30 µg/ml chloramphenicol. Production of recombinant UMPKmt was induced with isopropyl-1-thio-β-d-thiogalactoside (1 mM final concentration) when cultures reached an absorbance of 1.5 at 600 nm. Bacteria were harvested by centrifugation 3 h after induction.
Plasmids
The 786-bp fragment corresponding to the pyrH gene of M. tuberculosis (Rv2883c gene in Tuberculist) was amplified by polymerase chain reaction (PCR (47)) using M. tuberculosis genomic DNA as the matrix. During amplification, NdeI and HindIII restriction sites were created at the ends of the amplified fragment. After digestion by NdeI and HindIII, the amplified pyrH gene was inserted into the pET22b and pET28a plasmids (Novagen, Inc.) digested with the same enzymes. Three clones containing the M. tuberculosis pyrH gene and overexpressing UMPKmt with or without a N-terminal end his-tag were characterized and one of them (harbouring plasmid named pHL60-3 and pHL60, respectively) was kept for further studies.
Site-directed mutagenesis
Site-directed mutagenesis was performed by PCR on plasmid pHL60-3 in two steps using for each variant the T7 promotor and terminator primers and two mutant oligonucleotides. Sequences of mutated oligonucleotides were: 5′- CGGCAACTTTTGGCGGGGCGCA-3′ and 5′-TGCGCCCCGCCAAAAGTTGCCG-3′ for the F81W variant (corresponding plasmid: pHL60-303), 5′-GGCAACTTTTTCCATGGCGCACAGC-3′ and 5′-GCTGTGCGCCATGGAAAAAGTTGCC-3′ for the R82H variant (corresponding plasmid: pHL60-301), 5′-CCGTACCTGTGGTTGCGGGCC-3′ and 5′-GGCCCGCAACCACAGGTACGG-3′ for the P139W variant (corresponding plasmid: pHL60-302), 5′-CCGTACCTCCATTTGCGGGCC-3′ and 5′-GGCCCGCAAATGGAGGTACGG-3′ for the P139H variant (corresponding plasmid: pHL60-304) and, 5′-CCGTACCTGGCCTTGCGGGCC-3′ and 5′-GGCCCGCAAGGCCAGGTACGG-3′ for the P139A variant (corresponding plasmid: pHL60-305). They were designed either to create or to destroy a restriction site. The double mutant F81W S96A (corresponding plasmid pHL60-306) was obtained using the same procedure but with the pHL60-303 plasmid as the matrix and the following mutated oligonucleotides 5′-GAGCGCACCAGGGCGGACTATATGGGA-3′ and 5′-TCCCATATAGTCCGCCCTGGTGCGCTC-3′. The last two mutants were constructed by the one-tube PCR-based mutagenesis method (48) with the following mutagenic oligonucleotides: 5′-CTCCAGGAAGGCTTGCAGTGCAAG-3′ for the D113A variant (corresponding plasmid: pHL60-307) and 5′-CACCTGGAGAAGGGAGCGGTGGTGATCTTCG-3′ for the R150A variant (corresponding plasmid: pHL60-308). For each mutagenesis, the pyrH gene was cloned into the pET28a vector and sequenced to check for the absence of any other mutation.
Purification and activity assay of UMPK from M. tuberculosis
Bacterial pellets expressing recombinant UMPKmt were suspended in 50 mM sodium phosphate pH 8, 0.1 M NaCl and 5 mM imidazole (buffer A) and disrupted by sonication. After centrifugation at 10 000g for 45 min, the soluble extract was added to BD TALON resin (1 ml of resin equilibrated in buffer A per 10 mg of recombinant protein) and gently stirred at room temperature on a platform shaker. The further steps are described by BD Biosciences in the Batch/gravity-Flow column purification section (49). Fractions containing UMPKmt were immediately dialyzed against 20 mM sodium phosphate pH 8 and 0.1 M NaCl (buffer B) using Spectra/Por membrane with a molecular weight cut-off of 12–14 kDa. The same protocol was used for the variants of UMPKmt, which were also expressed as histidine-tagged proteins on their N-termini. When used for crystallization assays, UMPKmt was further purified by gel filtration chromatography on a HiLoad™ 26/60 Superdex™ 200 prep grade (GE Healthcare). The purified protein was concentrated up to 12 mg/ml in buffer B using a 50K stirred cell system (Pall Life Sciences) and stored at −20°C.
UMPKmt activity was determined at 30°C using a coupled spectrophotometric assay (0.5 ml final volume) on an Eppendorf ECOM 6122 photometer (50). The reaction medium contains 50 mM Tris–HCl pH 7.4, 50 mM KCl, 1 mM phosphoenolpyruvate, 0.2 mM NADH, 2 U of each lactate dehydrogenase, pyruvate kinase and NDP kinase, and various concentrations of ATP, GTP, UTP and UMP. A 2 mM excess of MgCl2 over the concentrations of NTPs was selected to ensure over 95% of NTPs appeared in the Mg-complexed form and that the free metal was held at a sufficiently high (between 1.8 and 2.8 mM) but not inhibitory concentration (51). UMPKmt diluted in buffer B was then added and the decrease in absorbance recorded at 340 nm. One unit of enzyme activity corresponds to 1 µmol of the product formed in 1 min at 30°C and pH 7.4. Experimental data were fitted using the Kaleidagraph software according to the Michaelis–Menten equation v = Vm [S]/(Km + [S]) or to the Hill equation v = Vm [S]nH /(K0.5nH + [S]nH), where v is the reaction rate, Vm the maximal rate, [S] the ATP concentration, Km the Michaelis–Menten constant, K0.5 the ATP concentration at half-saturation, and nH the Hill number, indicating the cooperativity index.
The thermal stability was studied by incubating aliquots of 1 mg/ml protein in buffer B at a temperature between 35 and 80°C for 10 min, after which residual activity was determined. Tm corresponds to the temperature at which the protein exhibits 50% of its activity.
Equilibrium sedimentation
The experiments were performed at 20°C on a Beckmann Optima XL-A analytical centrifuge using an An-60 Ti rotor and a cell with a 12 mm optical path length. Samples (150 µl) in buffer B at ∼0.5 mg/ml were centrifuged at 20 000 and 25 000 r.p.m. Radial scans of absorbance at 280 nm were taken at 2 h intervals. Equilibrium was achieved after 14 h. Data were analysed by the XL-A program supplied by Beckman. The partial specific volume of UMPKmt was calculated from the sequence (52).
Molecular modelling
UMPKs were manually aligned using ViTO (53). Crude models were obtained using SCWRL3.0 (54) for the monomers in various configurations depending on the template complexation state. Hexameric models were built using the same templates and the program MODELLER 7v7 (55). The same procedure was applied for the wild-type and the mutant enzymes. Various templates were combined to build models of UMPKmt in complex with its natural ligands. The whole procedure (comparative modelling and ligand docking by similarity) can now be performed on the fly using the Web-server @TOME-2 (56) as exemplified with UMPKmt (http://atome.cbs.cnrs.fr/AT2/EG/3675/atome.html).
Crystallization and X-ray data collection
Crystallization was obtained in the presence of a high concentration of sodium/potassium tartrate (1.2 M) at pH 7.4, and of 10 mM GTP. X-ray diffraction data sets were collected from frozen single crystals at the European Synchrotron Radiation Facility (Grenoble, France, beamline ID14-4) and processed with the programs MOSFLM, SCALA and TRUNCATE from the CCP4 program suite (57).
The structure was solved by molecular replacement using the program MolRep (58) and the crystal structure of UMPK from Streptococcus pyogenes (PDB1Z9D; to be published) as a search model. Model refinement was performed using the program COOT (59) and the program REFMAC5 (60), using a translation/liberation/screw model (61) against the GTP-bound data set at 2.8 Å and then at 2.5 Å resolution (Table 1). In the final model, short segments of the protein were not clearly visible in the electron density map. Most of the histidine-tag, the first 28 residues, as well as a short stretch lying in the ATP-binding site (residues 206–210) could not be modelled in the electron density.
Table 1.
Data collection and refinement statistics for UMPKmt–GTP complexes
| UMPKmt |
||
|---|---|---|
| GTP | GTP-UDP | |
| Data collection | ||
| Space group | P212121 | P21212 |
| Cell dimension a, b, c (Å) | 123.4, 139.3, 183.7 | 136.7, 175.5, 65.4 |
| No. molecules in a.u | 1 | 1 |
| Wavelength (Å) | 0.9334 | 0.91850 |
| Resolution (Å)a | 48.45–2.89 (3.01–2.89) | 87.69–2.47 (2.60–2.47) |
| Rmerge (%)a,b | 7.9 (57.0) | 4.4 (58.3) |
| I/σIa | 8.5 (1.5) | 12.2 (0.8) |
| Completeness (%)a | 100. (100.) | 90.8 (61.4) |
| Redundancy a | 3.9 (4.0) | 4.2 (2.5) |
| B-wilson | 35.2 | 53.4 |
| Refinement | ||
| Resolution (Å) | 2.9 | 2.49 |
| No. Reflections | 18 685 | 43 188 |
| Rwork/Rfree (%)c,d | 23.0/26.2 | 22.0/27.3 |
| No. protein atoms | 4175 | 2065 |
| No. water molecules | – | 98 |
| Ligand type | 12 GTP | 6 GTP 1 UDP |
| B-factors (Å2) | ||
| Protein | 38.7 | 39.5 |
| Ligand | 31.1 | 38.2 |
| Water | 30.6 | 20.4 |
| R.m.s deviationse | ||
| Bond lengths (Å) | 0.009 | 0.009 |
| Bond angles (°) | 1.40 | 1.6 |
aValues in parentheses refer to the outermost resolution shell.
bRmerge = ΣhklΣi∣Ihkl,i – Iaverage,hkl∣/∣ΣhklΣi∣Ihkl,i∣ x100.
cRwork = Σhkl∣Fobs – Fcalc∣/Σhkl ∣Fobs∣ x100.
dRfree is calculated in the same way on a subset of reflections that are not used in the refinement (5%).
eDeviation from ideal values.
Small-angle X-ray scattering experiments and data analysis
Synchrotron X-ray scattering data from solutions of UMPKmt were collected on the SWING beamline of the Soleil Synchrotron (Saint-Aubin, France) using a PCCD-170170 detector at a wavelength of 1.03 Å. The scattering patterns were measured by merging 10–20 data recordings with 1-s exposure time each, for two solute concentrations (1.825 and 5.25 mg/ml). To check for radiation damage, all successive exposures were compared, and no changes were detected. Using the sample-detector distance of 1.8 m, a range of momentum transfer of 0.0065 < s < 0.6 Å−1 was covered (s = 4psin(q)/λ, where 2q is the scattering angle, λ is the X-ray wavelength). The data were processed using standard procedures and extrapolated to infinite dilution using the program PRIMUS (62). The forward scattering, I(0), and the radius of gyration, Rg, were evaluated using the Guinier approximation, assuming that at very small angles (s < 1.3/Rg) the intensity is represented as I(s) = I(0) exp(−s2Rg2)/3). The values of I(0) and Rg, as well as the maximum dimension, Dmax, and the interatomic distance distribution functions, [p(r)], were also computed using the program GNOM (63).
Other analytical procedures
Protein concentration was measured according to Bradford (64), using a Bio-Rad kit or by amino acid analysis on a Beckman system 6300 high-performance analyser after 6 N HCl hydrolysis for 22 h at 110°C. SDS-PAGE was performed as described by Laemmli (65).
RESULTS
Overproduction, purification and molecular characterization of UMPKmt
The wild type UMPKmt was overproduced in large quantities in E. coli strain BL21(DE3)/pDIA17 (46) and it amounted between 25% and 30% of soluble E. coli proteins. Since control experiments showed no significant differences in the specific activity between His-tagged and non-tagged forms of UMPKmt, the His-tagged enzyme purified by affinity chromatography on BD TALON resin (BD Biosciences) was used exclusively in further experiments. Similar conclusions were reached with recombinant UMPKs from E. coli, Haemophilus influenzae or Bacillus subtilis (51). The purity of UMPKmt was over 95% as indicated by SDS-PAGE.
The molecular mass of the native protein, determined by sedimentation equilibrium ultracentrifugation (173 170 ± 3161 Da), corresponds to a hexamer. This is consistent with the quaternary structure observed for other bacterial UMPKs (51,66,67). The pure enzyme was stored at 4°C in 20 mM phosphate (pH 7) and 100 mM NaCl with no apparent loss of activity for ∼2 months. Thermal denaturation experiments indicated Tm values of 60°C in the absence of nucleotides and 65°C and 75°C with 1 mM GTP and UTP, respectively. In this respect UMPKmt exhibits a lower thermal stability than the other NMPKs from the same organism (32,36).
Dependance of UMPKmt activity on various nucleotides acting as substrates or as allosteric effectors
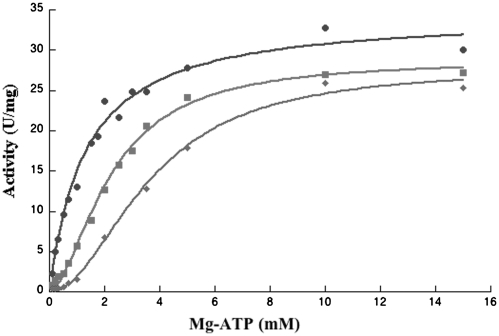
The plot of UMPKmt activity at variable concentrations of ATP and saturating concentrations of UMP (1 mM or over) was sigmoidal (Figure 1). Ten separate experiments using different preparations of UMPKmt yielded an average value of Vm of 30 ± 1 U/mg of protein, K0.5 of 2.2 ± 0.1 mM and nH of 1.8 ± 0.2 (Table 2). These numbers compare well with those observed with UMPK from Staphylococcus aureus, a Gram-positive bacterium (51). Among the other nucleoside triphosphates, only dATP acted as a phosphate donor with reaction rates similar to that exhibited by ATP. On the contrary, CTP, UTP or dTTP were not substrates, while the reaction rates with GTP represented only 0.1% of that with ATP or dATP. At saturating concentrations of Mg-ATP (5 mM or over), the reaction rate of UMPKmt as a function of UMP concentration obeys the Michaelis–Menten equation with an average value from 10 separate experiments of KmUMP = 19.6 ± 1.8 µM and Vm = 34.8 ± 0.8 U/mg (Table 3). Addition of a fluoro group at the 5-position of the base moiety does not affect the Km and the Vm of UMPKmt. On the other hand, 5Br-UMP and dUMP were not substrates of UMPKmt, but acted as weak competitive inhibitors (Ki > 1 mM).
Figure 1.
UMPKmt activity versus Mg-ATP concentration. Enzyme activity was determined at a fixed concentration of UMP (2 mM), and, in the absence (filled square) or in the presence of effectors (0.5 mM GTP: filled circle and 0.1 mM UTP: diamond-shaped). The curves correspond to the fit of the experimental data to the Hill equation and the calculated parameters are displayed in Table 2.
Table 2.
Kinetic parameters of wild-type and variants UMPKmt with Mg-ATP as variable substrate
| Enzyme variant | No effector |
With GTP |
With UTP |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Vm (U/mg) | K0.5 (mM) | nH | Vm (U/mg) | K0.5 (mM) | nH | Vm (U/mg) | K0.5 (mM) | nH | |
| Wild type (1.0; 0.2) | 30.0 ± 1.0 | 2.2 ± 0.1 | 1.80 ± 0.15 | 30.7 ± 1.3 | 1.2 ± 0.1 | 1.02 ± 0.08 | 28.4 ± 0.6 | 4.7 ± 0.1 | 2.42 ± 0.11 |
| F81W (1.0; 1.0) | 12.1 ± 0.6 | 2.9 ± 0.3 | 1.17 ± 0.07 | 12.4 ± 1.4 | 2.2 ± 0.7 | 1.04 ± 0.19 | 8.7 ± 0.4 | 3.2 ± 0.3 | 1.99 ± 0.22 |
| F81W S96A (2.0; 0.2) | 4.1 ± 0.4 | 4.1 ± 0.6 | 1.91 ± 0.27 | 5.1 ± 0.4 | 2.2 ± 0.4 | 1.16 ± 0.13 | 3.5 ± 0.2 | 4.9 ± 0.3 | 2.05 ± 0.13 |
| R82H (2.0; 0.2) | 0.30 ± 0.01 | 3.3 ± 0.3 | 1.07 ± 0.05 | 0.30 ± 0.01 | 3.6 ± 0.4 | 1.36 ± 0.11 | 0.20 ± 0.01 | 3.9 ± 0.2 | 2.14 ± 0.16 |
| P139W (0.5; 0.5) | 14.1 ± 0.4 | 0.8 ± 0.1 | 0.99 ± 0.07 | 14.2 ± 0.9 | 0.5 ± 0.1 | 1.05 ± 0.20 | 14.4 ± 1.8 | 1.5 ± 0.5 | 1.05 ± 0.23 |
| P139H (2.0; 0.5) | 21.9 ± 3.1 | 10.6 ± 3.1 | 0.96 ± 0.06 | 23.8 ± 0.8 | 0.8 ± 0.1 | 0.96 ± 0.08 | nf | nf | nf |
| P139A (1.0; 1.0) | 35.1 ± 3.8 | 3.3 ± 0.9 | 1.04 ± 0.11 | 39.0 ± 4.0 | 3.3 ± 0.8 | 1.01 ± 0.11 | 26.2 ± 2.1 | 3.1 ± 0.4 | 2.01 ± 0.36 |
| D113A (0.5; 0.5) | 38.4 ± 2.3 | 2.2 ± 0.3 | 1.01 ± 0.07 | 33.2 ± 1.7 | 2.3 ± 0.2 | 1.16 ± 0.07 | 32.5 ± 2.5 | 3.3 ± 0.4 | 1.61 ± 0.17 |
| R150A (0.5; 0.2) | 7.1 ± 0.5 | 1.5 ± 0.2 | 1.42 ± 0.15 | 7.4 ± 0.6 | 0.5 ± 0.1 | 0.92 ± 0.14 | 5.5 ± 0.7 | 4.4 ± 0.9 | 1.38 ± 0.15 |
nf: not fitted
Reaction rates were determined at a constant concentration of UMP (2 mM) and in the absence or in the presence of the effectors (GTP and UTP concentrations are given into brackets near the name of the enzyme variant, respectively) and fitted according to the Hill equation (see under ‘Materials and Methods’ section).
Table 3.
Kinetic parameters of wild-type and variants UMPKmt with UMP as variable substrate
| Enzyme variant | Vm (U/mg) | KmUMP (µM) |
|---|---|---|
| WT | 33.6 ± 0.9 | 17.7 ± 1.8 |
| F81W | 7.7 ± 0.1 | 63.9 ± 2.4 |
| F81W S96A | 3.1 ± 0.2 | 3.4 ± 0.1 |
| R82H | 0.25 ± 0.01 | 89.1 ± 5.7 |
| P139W | 7.7 ± 0.2 | 6.1 ± 0.8 |
| P139H | 6.7 ± 0.6 | 2.3 ± 0.1 |
| P139A | 17.6 ± 0.6 | 6.6 ± 0.1 |
| D113A | 25.0 ± 0.2 | 200.0 ± 10.0 |
| R150A | 5.8 ± 0.2 | 14.0 ± 0.8 |
Reaction rates were determined at a constant concentration of Mg-ATP (5 mM) and fitted according to the Michaelis–Menten equation (see under ‘Materials and Methods’ section).
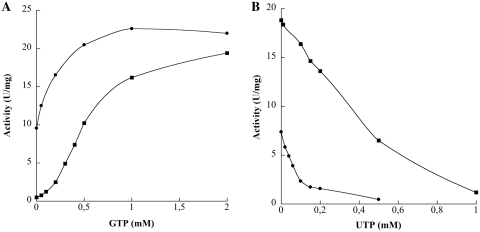
GTP and UTP were explored as potential allosteric effectors. At constant concentrations of ATP and UMP (1 mM each), GTP increased the reaction rate by a factor of 3.5, with half-saturation at 0.1 mM (data not shown). Conversely, UTP behaved as an inhibitor with IC50 at 0.1 mM (data not shown). Neither the activation by GTP nor the inhibition by UTP were affected by the concentrations of UMP (between 5 µM and 2 mM) or MgCl2 (between 3 and 20 mM). As it has been stated that the two effector binding sites were identical or largely overlapping in Gram-positive bacteria (51), the UMPKmt activity was measured at a constant concentration of UTP and at increasing concentrations of GTP (Figure 2A) and vice versa (Figure 2B). The inhibition by 0.5 mM UTP was cancelled in the presence of 2 mM GTP, whereas the activation by 0.5 mM GTP was reversed by addition of 1 mM UTP. In the following experiments, we explored the effect of GTP and UTP on kinetics of UMPKmt with ATP as variable substrate (Figure 1). GTP decreased both the cooperativity and the apparent K0.5 for ATP, whereas UTP exhibited the opposite effect by increasing both the nH and the K0.5 for ATP. Several GTP and UTP analogues were tested for their ability to mimic the action of natural nucleotides. dGTP, Mant-GTP and 6-methylthio GTP exhibited similar properties to GTP in terms of both affinity and extent of activation. GMP-PNP and GMP were much weaker activators (only 50% and 30% increase in activity at 1 and 2.5 mM nucleotide, respectively), whereas GMP-PCP and 8Br-GTP were ineffective. Among UTP analogues, 5I-UTP was the strongest inhibitor (IC50 = 0.02 mM) followed by 5Br-UTP, dUTP and dTTP (IC50 = 0.1, 0.24 and 0.4 mM, respectively). 5F-UTP exhibited a biphasic effect. Below 0.15 mM it increased the activity of UMPKmt by 80%. Over this concentration it became inhibitory. For example, at 0.6 mM 5F-UTP, the enzyme activity is decreased by 40%.
Figure 2.
Effect of the effectors on UMPKmt activity. (A). Enzyme activity was determined at fixed concentrations of substrates (5 mM Mg-ATP and 2 mM UMP) in the absence (filled circle) or in the presence (filled square) of 0.5 mM UTP. Each data point represents a single determination. (B). Same as (A) but with GTP as the effector.
Molecular modelling of UMPKmt
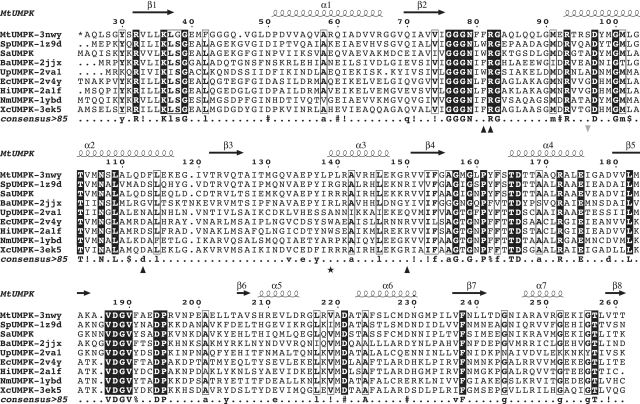
With the exception of a 28-residue long extension at its N-terminus, the UMPKmt sequence appeared highly similar to those of other bacterial UMPKs, showing up to 50% of identity over 240 residues (Figure 3). The specific N-terminal extension is predicted to be flexible and unfolded due to its strong composition bias in small residues (mainly G, A, P). By using templates from other bacterial UMPKs, we have built models of UMPKmt in various configurations (see ‘Materials and Methods’ section). In addition, the conserved quaternary hexameric structure was modelled using multimeric templates and taking into account crystal symmetry when necessary. Using the structures of several bacterial UMPKs in complex with their common ligands (UMP and UDP) (24,68), the ligand-binding mode and the accompanying structural rearrangements could be analysed. Similarly, the mode of recognition of ATP could be deduced using its structure bound to archeal UMPKs (26) or to the more distantly related AA-kinases as previously described (23).
Figure 3.
Sequence-structure alignment of bacterial UMPKs. UMPK sequences from four Gram-positive bacteria (M. tuberculosis (MtUMPK) reported in this work, Streptococcus pyogenes (SpUMPK, PDB code 1Z9D), S. aureus (SaUMPK), B. anthracis (BaUMPK, PDB code 2JJX)), from the small genome size organism Ureaplasma parvum (UpUMPK, PDB code 2VA1) and from four Gram-negative bacteria [E. coli (EcUMPK, PDB codes 2BND, 2BNE, 2BNF and 2V4Y), H. influenzae (HiUMPK, code 2A1F), Neisseiria meningitidis (NmUMPK, PDB code 1YBD), X. campestris (XcUMPK, PDB codes 3EK5 and 3EK6)]. The PDB codes of the 3D-structures used for the UMPKmt modelling are also indicated next to the UMPK acronyms. The secondary elements for UMPKmt are shown over the alignment. The first 23 residues of UMPKmt were omitted as they are not conserved in sequence and not seen in the 3D-structure. Strictly conserved residues in all sequences are boxed in black and conserved residues in eight sequences out of nine are bordered. The consensus sequence (>90%) is provided under the sequence alignment: upper case stands for strictly conserved residue, lower case for conservatively substituted residue, ‘slash’ and ‘hash’ for any hydrophobic or polar residues, respectively. Residues modified by site-directed mutagenesis are indicated by black triangles (single substitutions at positions F81, R82, D113 and R150), asterisk (substitutions at the position P139) and grey triangle (second position of the double mutant F81W, S96A).
The ATP binding site is rather well conserved in sequence and structure among the bacterial UMPKs and also the distantly related aspartokinases and glutamate kinases (25,26,68–70). In this AA-kinase superfamily, it is formed solely by residues from the C-terminal region (residues 170–261 in UMPKmt) from one monomer and contains no residues protruding from other monomers. Several conserved motifs were previously described (23,69) suggesting that the orientation of the phosphate donor is conserved for all these kinases. This conclusion is in agreement with the archaeal UMPK crystal structures determined in complex with ATP or an analogue (25,26). Among these sequence motifs, the DgvyxxDP (upper case stands for strictly conserved residue, lower case for conserved residue and x for any amino acid; 188-DGVFAEDP-195 in UMPKmt) contributes to the specific recognition of the adenosine nucleobase.
The binding site of the phosphate acceptor (UMP) was modelled in two different conformations depending on the presence of a bound ligand in the template. It is mainly composed of residues from the N-terminal part including three different regions. First, a common glycine-rich loop (76-GGGN-79 in UMPKmt) is involved in the recognition of the α-phosphate group of UMP. The conformation of this loop is stabilized by two conserved asparagines (N79 and N107 in UMPKmt). Their side-chains, facing each other, form intertwined hydrogen bonds. The second part of the UMP binding site is composed of the conserved motif 157-GmGxPyFsTD-166 in UMPKmt (the last two residues of this motif participate in the catalytic centre as discussed below) and is involved in the recognition of the base moiety. Two hydrogen bonds are formed between the nucleobase and the protein backbone according to the structure of the E. coli UMPK in complex with UMP (24). In addition, van der Waals interactions are observed between the pyrimidine ring and the backbone of the conserved P161 and F163 of the motif. As in Gram-positive UMPKs, a hydrophobic residue is also present in this region in UMPKmt (M158) while Gram-negative and archaeal enzymes possess a threonine. The latter is hydrogen-bonded to a carbonyl of the uridine moiety. While the conformations of the first two regions of the UMP binding site seem rigid and little affected by the presence UMP or UDP, the last region (81-fRGxxlxxxg-90) seems rather flexible. It is observed in distinct conformations or orientations in the various crystal structures solved so far (see below).
The catalytic site is centred around a conserved and buried lysine (K36 in UMPKmt). This lysine and the three conserved glycines (G76, G77 and G78), which constitute the first region of the UMP binding site, form a sequence motif specific for bacterial UMPKs. The lysine side-chain lies in the vicinity of two strictly conserved polar residues (T165 and D166 in UMPKmt and belonging to the second region of the UMP binding site). A conserved glutamate (E40 in UMPKmt) is located in the vicinity of the transferred phosphate group and interacts in some crystal structures with the conserved R82 (UMPKmt) lying on the top of the UMP binding site. According to this comparative analysis of the known crystal structures, the catalytic centre appears partially flexible and shows a small induced-fit while lying in-between the two substrate binding sites (ATP and UMP, respectively).
Molecular modelling was sufficient to recognize the ATP and UMP binding sites and to explain the specificity of recognition. Conversely, the effector binding sites could not be ascertain although a cluster of positively charged residues was observed at the trimer interface. A similar situation was recently observed in two UMPKs from Gram-negative bacteria (E. coli and Xanthomonas campestris) (71,72). Despite a good overall sequence conservation, local variations suggested a distinct mode of GTP recognition between the E. coli UMPK and UMPKmt. The recent structure of the GTP-bound UMPK from X. campestris (72) showed an alternative conformation of the GTP molecules. The latter resembles that of the ATP molecules in the allosteric binding pocket in the B. anthracis UMPK (68). This new mode of binding seemed more compatible with the expected allosteric pocket in UMPKmt. The observed variations prompted us to solve the experimental structure in complex with one of the allosteric regulators.
Crystal structure of UMPKmt in complex with GTP
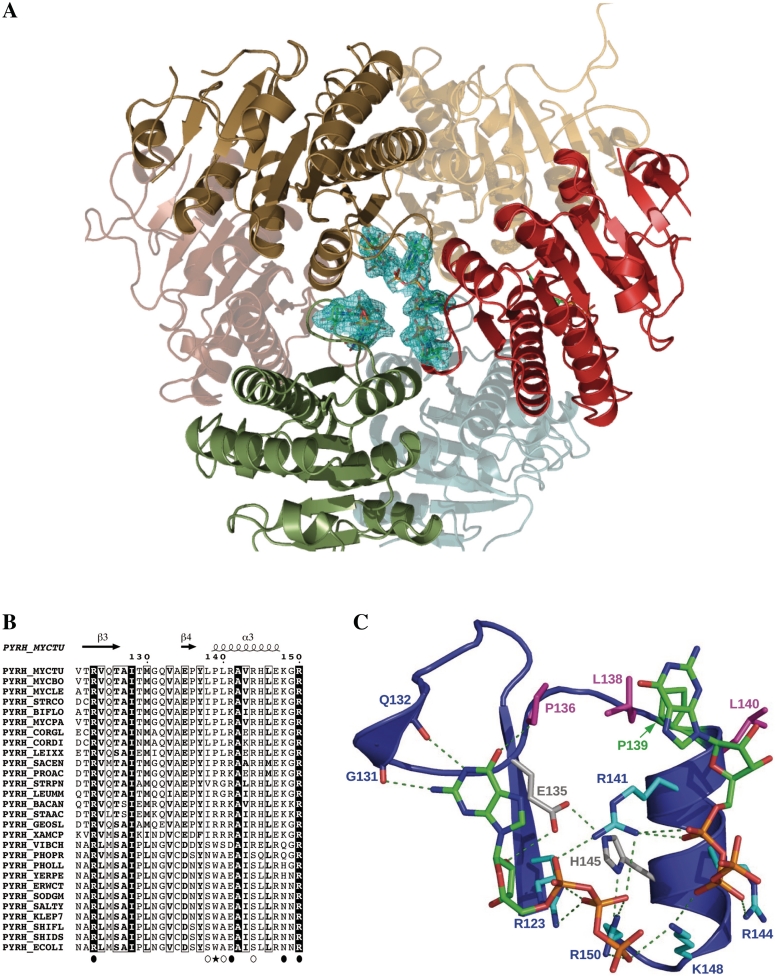
Crystallization was achieved in the presence of GTP, at high concentration of tartrate at a mild basic pH and in the presence of GTP. In the same crystallization conditions UTP prevents crystal growth. The structure was solved by molecular replacement. Two hexamers were observed in the asymetric unit (data not shown) and the electron density map at 2.9 Å resolution was sufficient to identify the position of six GTP molecules bound along the 3-fold axis of the protein. Attempts to soak the crystals with 5I-UTP led to a better diffracting crystal (to 2.5 Å resolution). The latter contains only one hexamer in a half-sized asymmetric unit (Table 1). This new crystal structure was refined to a 2.5-Å resolution using REFMAC5 with TLS simulating rigid-body movement of each monomer. The overall structure is merely identical to that of other known UMPKs (Figures 4A and S1). The monomer showed a conserved topology (RMSD ranging from 0.7 to 1.3 Å) and the hexamer adopted the same overall organization in agreement with the high global sequence identity (40–50% for the closest crystal structures).
Figure 4.
Structure of UMPKmt complexed with GTP. (A) Ribbon representation of the UMPKmt hexamer viewed down the trimeric axis. The three front monomers are shown in red, brown and green, while their rear dimeric counterparts are partially transparent. The GTP molecules are shown as sticks (CPK colors) with the electron density in cyan at a 0.9σ level. (B). Bacterial UMPKs sequences (acronyms provided on the left) are aligned to the UMPKmt region interacting with GTP. Strictly conserved residues in all sequences are boxed in black and conserved residues are bordered. Since each UMPKmt monomer interacts with two GTP molecules, the residues contacting the first GTP molecule are labelled by filled circles, and the residues contacting the second one by opened circles. P139 in UMPKmt is highlighted by a star. (C). Zoom in of the GTP-binding site of one UMPKmt monomer (ribbon representation). The two contacting GTP molecules are shown as sticks. The basic residues contacting the phosphate moiety of the effector are depicted in blue. Residues in grey participate in a hydrogen bond network organizing the GTP binding site. The hydrophobic residues (P136, L138 and L140) contacting the nucleobase are depicted in pink. Residue P139 is shown in green.
The long N-terminal extension (residues 1–28) is not visible in the electron density. The helical segment 81–89 appeared flexible in two monomers and residues 81 to 84 could not be traced convincingly in the electron density. The large loop (residue 192–202) of the complete ATP binding site could only be built in one monomer and its stabilization is likely due to crystal packing. Overall, the six monomers adopt a highly similar conformation with the exception of the helical segment 81–89 and the long ATP-binding loop in the C-terminal domain. The hexameric UMPKmt appeared to bind six GTP and one UDP molecules. No trace of iodide-containing compound could be detected in any nucleotide-binding sites and at any step of the refinement (even at 1σ in the difference map of the electron density). Thus, the crystal structure corresponds to an ‘activated’ form due to the binding of the positive allosteric effector GTP with five monomers in an ‘open’ conformation and one monomer in a ‘closed’ one due to UDP binding. While the ‘opened’ monomers showed an RMSD of ∼0.6 Å with each other, the difference rises to ∼0.8 Å with the closed one.
The UDP molecule is observed in the UMP binding site (Figures S2 and S3) and its orientation perfectly matches that observed in the UDP-bound UMPKec (PDB2BND). The nucleobase is sandwiched by the side-chain of M101 and backbone of S164. In addition, the carbonyl of F163 points toward the uridine ring. The nucleobase recognition is due to a hydrogen-bond network involving carbonyl of L160 (2.9 Å from N3), the amide of M158 (3.0 Å from O4) and the hydroxyl of T168. The latter points toward C5 (3.7 Å), which is also closed to the Cα from G157 (3.5 Å) leaving no space for a halide atom except fluoride, in agreement with our biochemical data. The O2 atom is not hydrogen bonded but is in van der Waals contact with the Cδ of P161 and the Cγ of the aspartate D97. The latter is hydrogen-bonded to the O2′ hydroxyl group of the ribose. In addition, the ribose is in van der Waals contacts with G77, G83 and G100. Finally, the two phosphate groups are hydrogen-bonded to the side chains of K36 (2.6 Å), R82 (Nε1-PαO = 3.3 Å and Nη1-PβO = 2.5 Å) and T165 (Oγ1-PαO = 2.7 Å) and the backbone amines of G39 and G77.
In the five other monomers the UMP sites are totally empty with no trace of any bound nucleotide and the segment 81–89 adopts an ‘open’ conformation. This conformation corresponds mainly to a re-orientation of the helical segment 81–89 as observed in UMPKec (PDB2BND versus PDB2V4Y). The ‘open’ conformation is also observed in most other bacterial UMPKs (usually co-crystallized without UMP, UDP or UTP). The improved diffraction and, simultaneously, the occupation of only one site out of six were rather surprising. The UDP bound to UMPKmt is likely due to 5I-UTP degradation, as no trace of halide could be detected in the electron density in the vicinity of the bound UDP. The absence of iodo substituent is in agreement with the absence of enzyme activity with 5Br-UMP (see above). Meanwhile, several attempts to soak UMPKmt crystals with UMP at millimolar concentration were detrimental for the diffraction power. This suggested that crystal packing constraints were incompatible with simultaneous occupancy of all the six UMP sites in the hexamer while the (very) low concentration of UDP in our successful soaking led to only one bound molecule of this nucleotide per hexamer.
Six GTP molecules (one per monomer) were clearly visible in the electron density (Figure 4A). They adopt a similar orientation and one main conformation although some flexibility in the position of the phosphate groups is evident in two monomers. The observed conformation corresponds to that described for ATP in the B. anthracis UMPK and GTP in X. campestris UMPK (PDB3EK5), while the GTP orientation in E. coli UMPK (PDB2V4Y) appears unique. Most of the interactions between UMPKmt and GTP involved residues from a short stretch in the protein sequence (residues 131–150; Figure 4B and C), although for each GTP molecule there are interacting residues from two monomers related by the three-fold axis. From one monomer, four basic residues (R123, R141, K148 and R150) protrude to interact with the negatively charged phosphate groups and with the ribose (R123). The ribose is also hydrogen bonded to the side chain of E135, whereas the guanine moiety interacts through hydrogen bonds with backbone atoms of G131 and Q132. The Cδ of P136 appears in close contact (∼3.3 Å) with the carbonyl group of the base moiety suggesting that a partial and weak hydrogen bond is also formed. In parallel, a second monomer provides additional interactions. The guanine ring is sandwiched by the two hydrophobic side-chains of L138′ and L140′. Finally, R144′ also points its positively charged side chain toward the phosphate groups of the same GTP molecule.
The conformation of the trinucleotide seems further stabilized by an internal hydrogen bond involving an oxygen of the α-phosphate group and the proton of the carbon C8. This close contact (3.2 Å) explained the selectivity against 8-bromo-GTP. Conversely, few interactions are seen with the hydroxyl groups of the ribose moiety in agreement with the observed binding of dGTP and mantGTP. The presence of numerous positively charged arginines and lysines lining the effector binding site explained the weaker affinity of GMP, GMPPCP and GMPPNP. While other trinucleotides can partially mimic the bound GTP molecules, some of the observed interactions with the nucleobase can discriminate the possible effectors. ATP binding would be prevented in UMPKmt because of a close contact between P136 hydrophobic side-chain and the charged N6 amino group of the adenine. Conversely, there is enough room to accommodate 5-iodo and 5-bromo-UTP although it is not obvious what favourable contact would be formed between the protein and these nucleotides. In the case of UTP, very few contacts are predicted to occur with the nucleobase and the loop G131–P136 without local rearrangement. This structural feature roughly matches the observed affinity for the halide derivatives of the natural negative effector UTP (see above).
This structure provided us with a better insight into the binding mode of the positive effector. In parallel, the conformational changes upon GTP binding appeared to be rather small according to the structural comparisons with the other bacterial UMPKs structures solved so far with and without effectors (RMSD around 1.1 Å ± 0.3). Hexamers of UMPKmt and X. campestris UMPK (PDB3EK5) bound to GTP were superposed with the apo-form of S. pyogenes (PDB1Z9D) and H. influenzae (PDB2A1F) UMPKs. While very few changes are detected near the GTP binding site and the 3-fold axis, the relative orientation of the ATP binding site seemed to be slightly affected (Supplementary Figure S4). To better evaluate this change, we measure the distance between the Cα of a well-conserved threonine (T257 in UMPKmt) buried in the C-terminal domain from two facing monomers. In UMPKmt and X. campestris UMPK, the distance is ∼38.4 and 40.4 Å, respectively compared to 42.4 and 43.6 Å in the apo-forms (in H. influenzae and S. pyogenes UMPKs, respectively). A similar trend can be deduced from the comparison of GTP-bound and UDP-bound forms of E. coli UMPK (PDB2V4Y and PDB2BND, respectively) with distances T237–T237′ of 40.5 and 43.7 Å, respectively. Clearly, a small but common re-orientation of the ATP-binding domain is observed upon GTP binding. This is further confirmed by small-angle X-ray scattering (SAXS) measurements acquired on UMPKmt alone or in presence of GTP, ATP, UTP or 5I-UTP (Supplementary Figure S5). Similar curves were obtained in the range of 0.004 < q < 0.35 and nearly identical curves for small angle (0.01 < q < 0.10) for all the tested samples. These observations extend to higher angles (up to 0.3) for the apo-enzyme and the protein in the presence of the negative effector. Conversely, the ATP and GTP bound protein showed slight differences with the other samples in the range 0.1 < q < 0.3. Little change in the deduced radius of gyration was detected (Rg ∼ 42 ± 2 Å). These data suggested a conservation of the quaternary structure and the global shape of the protein whatever the allosteric effector bound. A small change in domain orientation and/or stability is likely triggered by activator binding. A rather subtle modification in dynamic behaviour is also likely but further experiments would be necessary to shed more light on the allosteric mechanism.
Characterization of UMPKmt variants obtained by site-directed mutagenesis
Site-directed mutagenesis was used to validate the biochemical and structural data. A structural rearrangement affecting the UMP binding site was predicted by our comparative models. Two residues were mutated: R82 and F81 to probe the active site configuration. They were respectively modified to a histidine or a tryptophan, in the light of former studies on the E. coli UMPK (71,73) and of our sequence analysis (see alignment in Figure 3). A third position, S96, was modified to an alanine in the context of the F81W variant, as the model anticipated interactions in the simple mutant that could affect the local conformational change observed upon substrate binding. Finally, to get a better insight into the effector binding sites, three positions (D113, P139 and R150) were selected based on recent reports (71,72) and from our structure. Besides the substitution by a tryptophan found in some Gram-negative bacteria UMPKs, P139 was also mutated to histidine and alanine to discriminate between a removed backbone constraint and a possible occurrence of a hydrogen bond by the side-chain nitrogen of the tryptophan (and the histidine). The two other residues were mutated to alanine.
The different variants expressed in E. coli were purified following the same procedure as described for the wild-type UMPKmt, and their oligomerization state checked by equilibrium sedimentation. Their kinetic parameters were determined as a function of the concentration of either UMP (Table 3), or Mg-ATP with or without the effectors (Figures 5, 6, S6 and S7, Table 2).
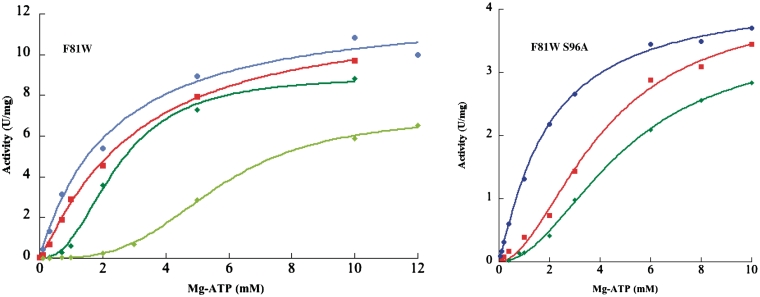
Figure 5.
F81W and F81W S96A variant activity versus Mg-ATP concentration. The concentrations of effectors were 0.5 mM GTP (dark blue), 1 mM GTP (light blue), no effector (red), 0.2 mM UTP (dark green) and 1 mM UTP (light green). Same conditions as Figure 1.
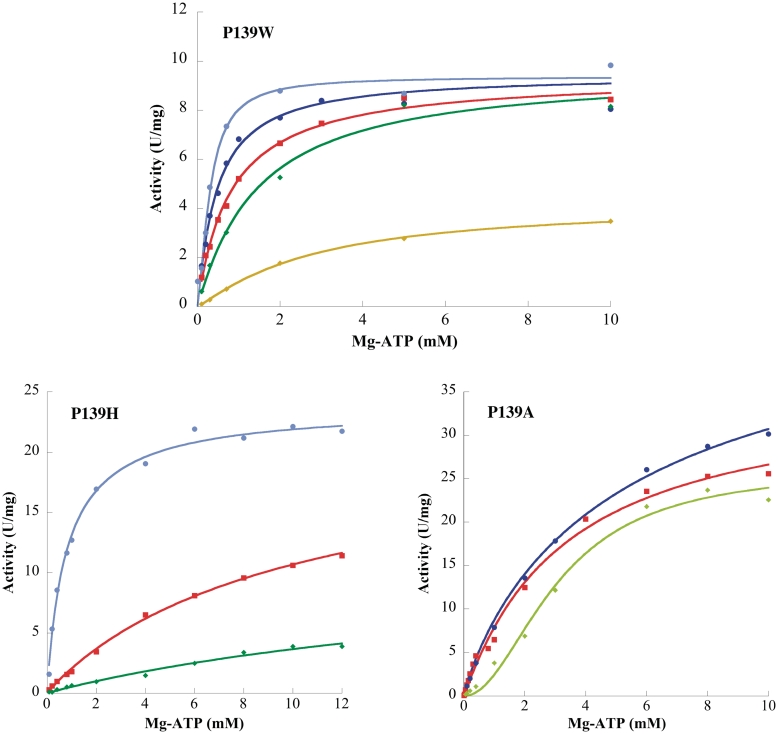
Figure 6.
P139W, P139H and P139A variant activity versus Mg-ATP concentration. The concentrations of effectors were 2 mM GTP (light blue), 0.5 mM GTP (dark blue), no effector (red), 0.5 mM UTP (dark green), 1 mM UTP (light green) and 2 mM UTP (gold green). Same conditions as Figure 1.
As for the wild-type protein, the activity of all the variants versus UMP concentration followed the Michaelis–Menten equation. The R82H specific activity was drastically reduced (0.7% of the wild-type specific activity), whereas for the other variants between 10% and 75% of the wild-type activity was observed. D113A displayed the most altered KmUMP (Table 3). Similarly, the mutant F81W and R82H showed a decreased affinity for UMP (3- and 4.5-fold, respectively). On the other hand, the other mutants exhibit a similar (R150A) or a better (3- to 8.5-fold) affinity for UMP.
Except for the R150A variant (Supplementary Figure S6) which displayed general features similar to the wild-type UMPKmt in any condition, the dependence of the activity as a function of the second substrate concentration revealed striking differences between the mutants (Table 2, Figures 5, 6 and S7). In particular, two P139 variants departed from the other mutants. The P139A variant appeared nearly insensitive to both effectors, whereas the P139W variant exhibit Michaelien kinetics even in the presence of UTP (Figure 6). In the absence of any effector, the curves obtained for the different variants with Mg-ATP as variable substrate were hyperbolic, with nH close to 1, except for the F81W/S96A and R150A variants. However, the affinity for ATP in most of the variants was in the same range as that of the wild-type enzyme (Table 2). The two exceptions were the P139W and the P139H variants with opposite behaviour: a 3-fold decrease and a 5-fold increase of the K0.5 for Mg-ATP. Interestingly, the activation by GTP was abolished for the F81W, R82H, D113A and P139A variants, leading to unchanged K0.5 for ATP in the presence of the effector (Table 2, Figures 5, 6 and S7). The P139W variant was slightly activated by the addition of 1 mM GTP (factor of 1.5 versus 3.5 for the WT at 1 mM ATP and UMP), whereas the D113A was inhibited up to 50 % in the same conditions (data not shown). On the other hand, the inhibition by UTP is retained for most of the variants with IC50 between 0.15 and 0.3 mM (data not shown). Again, the P139W plot (Figure 6) showed a loss of cooperativity and the P139A variant exhibited a weak effect of the effector on the affinity for ATP (Table 2).
DISCUSSION
A multi-disciplinary approach was used to study the last uncharacterized NMPK from M. tuberculosis, namely UMPKmt. As shown by kinetic experiments on the wild-type enzyme, UMPKmt behaves as a Gram-positive bacterial UMPK with cooperativity toward the phosphate donor ATP and an inhibition by UTP, independent of the UMP or divalent ion concentrations (51). The Gram-positive UMPK pattern of the M. tuberculosis enzyme was also confirmed by the effects of effector analogues, in particular the halogenated derivatives. However, no inhibition by excess of UMP was observed as seen for B. subtilis or S. pneumoniae UMPKs. Another point worth mentioning is the minor effect of the activator on the affinity for ATP, as, in the absence of GTP, the K0.5 for ATP of UMPKmt is in the same range than the supposed physiological concentration of this nucleotide in mycobacteria. Therefore, the relative concentration of the pools of purine and pyrimidine nucleotides might be the major factor responsible of the regulation of the UMPKmt activity in mycobacteria. The retro-inhibition of bacterial UMPKs by UTP is not surprising as end products are usually involved in the regulation of metabolic pathways. The identity of the positive effector is more intriguing but might be rationalized by taking into account the general role of GTP and UTP in translation and transcription in bacteria (74).
To better understand the biochemical and kinetic properties of UMPKmt, 3D-structure determination was carried out, along with a validation by mutational analysis. Sequence similarities with well-characterized bacterial orthologues allowed molecular modelling of its structure and active site using X-ray structures solved in various ligand states (apo or substrate bound, inhibitor or activator bound enzymes) (24,25,68,71,72). Among our predictive results, some correspond to fine local dynamical behaviour. A structural rearrangement was predicted to occur around the F81 residue upon binding of UMP. These alternating configurations explain the observed behaviour of the F81W and F81W S96A variants. The double mutant F81W S96A was expected to stabilize one of the two orientations of the tryptophan by removing a potential hydrogen bond between serine 96 and the introduced tryptophan. Indeed, the affinity for UMP of the F81W S96A variant was enhanced by a factor of 7 and 18 compared to the wild-type and the single mutant, respectively. Accordingly, the tryptophan side-chain seems to be stabilized in an orientation favourable to the binding of UMP by the introduction of the polar serine. These results revealed the robustness and the usefulness of our models, despite the fact that they failed to provide additional clues for the identification of the GTP or UTP binding sites. On the other hand, as analysis of the regulation of activity by GTP (activator) and UTP (inhibitor) revealed variations among the bacterial enzymes (22,26,51,66,69,75), the allosteric behaviour of one orthologue appeared barely predictable from another despite an overall high sequence identity.
The activator binding site has recently been identified by X-ray crystallography for three bacterial UMPKs (68,71,72), and now UMPKmt (this study). All these structures revealed that the activator binding pocket is located at the trimer interface, a unique feature among the AA-kinases. In these UMPKs (but not in E. coli UMPK), the allosteric effector interacts with a short region of around 30 residues. The phosphate moieties are in interaction with four positive residues, mainly arginines (R123, R141, K148 and R150 in UMPKmt). Therefore, it is not surprising that the mutation of a single residue (R150A variant) has almost no effect on the enzyme properties. The other residues involved in the binding of the ribose and the nucleobase moieties are also conserved, except in E. coli UMPK (71), whose GTP binding pocket seems to be unique. In this UMPK, the base moiety is sandwiched by two residues (H96 and W119), and the reorientation of the side-chain of the tryptophan is observed upon GTP binding. This tryptophan is replaced by an arginine or a proline in many Gram-positive UMPKs (P139 for UMPKmt). Even if this residue does not directly interact with the bound GTP molecule in UMPKmt, its mutation to an alanine leads to a loss of GTP activation while retaining normal enzymatic activity level. Two other modifications have detrimental effects on the cooperativity properties of the UMPKmt variants (P139W and P139H). Therefore, P139 is a key element in the allosteric regulation of UMPKmt and might serve as a sensor of the effector binding at the trimeric interface. Indeed, it interacts with residues lying on the loop connecting the N- and C-terminal domains (Supplementary Figure S8). This loop is predicted by normal mode analysis to correspond to a hinge region (data not shown). Furthermore, it also corresponds to the end of the α-helix bearing polar residues conserved in most UMPKs and pointing into the catalytic centre, especially T165 and D166. This helix also bears two conserved (R149 and E152 in UMPKmt) residues directly in contact with the conserved Y137 to form an intricate network of interactions midway from the GTP binding site, the UMP binding site and the catalytic centre. Slight rearrangement or stabilization of the GTP-binding segment and the nearby secondary structures may affect the entire active site without dramatic change of the global structure.
Nature has developed various strategies to control and regulate the activity of proteins. Cooperativity between different subunits of an oligomeric protein is one of them (76,77). Ligand binding at one site induced structural changes that are transmitted to distal active site(s) and enable fine tuning of activity in response to slight changes in substrate concentration. In addition to this long-range cross-talk communication between subunits, recent work highlighted the role of dynamics in allosteric regulation (78). Indeed, the allosteric mechanism of the related AA-kinases has been recently connected to slow motions by normal mode analysis (79). This study pointed out a hinge between the N- and C-terminal domains in agreement with our present results (Supplementary Figure S8). Further analysis might be necessary to gain further insight into the mechanism of allosteric regulation of the bacterial UMPKs as the overall structure seems little affected by the binding of its positive effectors and only subtle changes can be described. Meanwhile, this suggested that the particular evolution of UMPKs is connected to the appearance of distinct allosteric regulation.
ACCESSION NUMBER
PDB ID code 3NWY.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Centre National de la Recherche Scientifique (CNRS); the Institut National de la Santé Et de la Recherche Médicale (INSERM); the Institut Pasteur (grant GPH tuberculose); and the Région Ile de France (Chemical Library Project, grants n° I 06-222/R and I 09-1739/ R, which included a fellowship for I.S.-A.). Funding for open access charge: Institut Pasteur; CNRS.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Dr Edward Seclaman is acknowledged for performing kinetics experiments on some variants, Evelyne Turlin for participation in the site-directed mutagenesis, Roland Nageotte and Dr Thierry Rose for performing the equilibrium sedimentation experiments, Dr Martin Cohen-Gonsaud for helpful discussion, Dr Octavian Barzu for his mentorship, and Dr Aude Echalier and Dr Yves Janin for their critical readings of the manuscript. We would also like to thank the referees for their thorough and useful comments on the manuscript. We thank the ESRF staff for their help in recording diffraction data on beamline ID14-4. SAXS experiments were recorded on the beamline SWING in SOLEIL (Saint-Aubin, France) with the kind help of Javier Perez.
REFERENCES
- 1.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tuberculosis. Scientific blueprint for tuberculosis drug development. Tuberculosis (Edinb) 2001;81(Suppl. 1):1–52. doi: 10.1054/tube.2001.0288. [DOI] [PubMed] [Google Scholar]
- 3.WHO. Global Tuberculosis Control: Epidemiology, Strategy, Financing: WHO report 2009. WHO Press; 2009. [Google Scholar]
- 4.Jassal M, Bishai WR. Extensively drug-resistant tuberculosis. Lancet Infect. Dis. 2009;9:19–30. doi: 10.1016/S1473-3099(08)70260-3. [DOI] [PubMed] [Google Scholar]
- 5.Janin YL. Antituberculosis drugs: ten years of research. Bioorg. Med. Chem. 2007;15:2479–2513. doi: 10.1016/j.bmc.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 6.Rivers EC, Mancera RL. New anti-tuberculosis drugs in clinical trials with novel mechanisms of action. Drug Discov. Today. 2008;13:1090–1098. doi: 10.1016/j.drudis.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Sacchettini JC, Rubin EJ, Freundlich JS. Drugs versus bugs: in pursuit of the persistent predator Mycobacterium tuberculosis. Nat. Rev. Microbiol. 2008;6:41–52. doi: 10.1038/nrmicro1816. [DOI] [PubMed] [Google Scholar]
- 8.Mdluli K, Spigelman M. Novel targets for tuberculosis drug discovery. Curr. Opin. Pharmacol. 2006;6:459–467. doi: 10.1016/j.coph.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y. The magic bullets and tuberculosis drug targets. Annu. Rev. Pharmacol. Toxicol. 2005;45:529–564. doi: 10.1146/annurev.pharmtox.45.120403.100120. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Post-Martens K, Denkin S. New drug candidates and therapeutic targets for tuberculosis therapy. Drug Discov. Today. 2006;11:21–27. doi: 10.1016/S1359-6446(05)03626-3. [DOI] [PubMed] [Google Scholar]
- 11.Ioerger TR, Sacchettini JC. Structural genomics approach to drug discovery for Mycobacterium tuberculosis. Curr. Opin. Microbiol. 2009;12:318–325. doi: 10.1016/j.mib.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 12.Rossmann MG, Moras D, Olsen KW. Chemical and biological evolution of nucleotide-binding protein. Nature. 1974;250:194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- 13.Saraste M, Sibbald PR, Wittinghofer A. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 14.Schulz GE. Structural and functional relationships in the adenylate kinase family. Cold Spring Harbor Symp. Quant. Biol. 1987;52:429–439. doi: 10.1101/sqb.1987.052.01.050. [DOI] [PubMed] [Google Scholar]
- 15.Vonrhein C, Schlauderer GJ, Schulz GE. Movie of the structural changes during a catalytic cycle of nucleoside monophosphate kinases. Structure. 1995;3:483–490. doi: 10.1016/s0969-2126(01)00181-2. [DOI] [PubMed] [Google Scholar]
- 16.Bârzu O, Gilles A-M. Ann. Inst. Pasteur/Actualités. Vol. 3. Elsevier, Paris; 1993. pp. 121–132. [Google Scholar]
- 17.Fukami-Kobayashi K, Nosaka M, Nakazawa A, Gõ M. Ancient divergence of long and short isoforms of adenylate kinase: molecular evolution of the nucleoside monophosphate kinase family. FEBS Lett. 1996;385:214–220. doi: 10.1016/0014-5793(96)00367-5. [DOI] [PubMed] [Google Scholar]
- 18.Briozzo P, Golinelli-Pimpaneau B, Gilles A-M, Gaucher J-F, Burlacu-Miron S, Sakamoto H, Janin J, Bârzu O. Structures of Escherichia coli CMP kinase alone and in complex with CDP: a new fold of the nucleoside monophosphate binding domain and insights into cytosine nucleotide specificity. Structure. 1998;6:1517–1527. doi: 10.1016/s0969-2126(98)00150-6. [DOI] [PubMed] [Google Scholar]
- 19.Bucurenci N, Sakamoto H, Briozzo P, Palibroda N, Serina L, Sarfati RS, Labesse G, Briand G, Danchin A, Bârzu O, et al. CMP kinase from Escherichia coli is structurally related to other nucleoside monophosphate kinases. J. Biol. Chem. 1996;271:2856–2862. doi: 10.1074/jbc.271.5.2856. [DOI] [PubMed] [Google Scholar]
- 20.Lavie A, Konrad M, Brundiers R, Goody RS, Schlichting I, Reinstein J. Crystal structure of yeast thymidylate kinase complexed with the bisubstrate inhibitor P1-(5′-adenosyl) P5-(5'-thymidyl) pentaphosphate (TP5A) at 2.0 Å resolution: implications for catalysis and AZT activation. Biochemistry. 1998;37:3677–3686. doi: 10.1021/bi9720787. [DOI] [PubMed] [Google Scholar]
- 21.Coggill P, Finn RD, Bateman A. Identifying protein domains with the Pfam database.Curr. Protoc. Bioinformatics. 2008 doi: 10.1002/0471250953.bi0205s23. Chapter 2, Unit 2 5. [DOI] [PubMed] [Google Scholar]
- 22.Serina L, Blondin C, Krin E, Sismeiro O, Danchin A, Sakamoto H, Gilles AM, Barzu O. Escherichia coli UMP-kinase, a member of the aspartokinase family, is a hexamer regulated by guanine nucleotides and UTP. Biochemistry. 1995;34:5066–5074. doi: 10.1021/bi00015a018. [DOI] [PubMed] [Google Scholar]
- 23.Labesse G, Bucurenci N, Douguet D, Sakamoto H, Landais S, Gagyi C, Gilles AM, Barzu O. Comparative modelling and immunochemical reactivity of Escherichia coli UMP kinase. Biochem. Biophys. Res. Commun. 2002;294:173–179. doi: 10.1016/S0006-291X(02)00450-3. [DOI] [PubMed] [Google Scholar]
- 24.Briozzo P, Evrin C, Meyer P, Assairi L, Joly N, Barzu O, Gilles AM. Structure of Escherichia coli UMP kinase differs from that of other NMP kinases and sheds new light on enzyme regulation. J. Biol. Chem. 2005;280:25533–25540. doi: 10.1074/jbc.M501849200. [DOI] [PubMed] [Google Scholar]
- 25.Marco-Marin C, Gil-Ortiz F, Rubio V. The crystal structure of Pyrococcus furiosus UMP kinase provides insight into catalysis and regulation in microbial pyrimidine nucleotide biosynthesis. J. Mol. Biol. 2005;352:438–454. doi: 10.1016/j.jmb.2005.07.045. [DOI] [PubMed] [Google Scholar]
- 26.Jensen KS, Johansson E, Jensen KF. Structural and enzymatic investigation of the Sulfolobus solfataricus uridylate kinase shows competitive UTP inhibition and the lack of GTP stimulation. Biochemistry. 2007;46:2745–2757. doi: 10.1021/bi0618159. [DOI] [PubMed] [Google Scholar]
- 27.Jang J, Becq J, Gicquel B, Deschavanne P, Neyrolles O. Horizontally acquired genomic islands in the tubercle bacilli. Trends Microbiol. 2008;16:303–308. doi: 10.1016/j.tim.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 28.Brosch R, Gordon SV, Garnier T, Eiglmeier K, Frigui W, Valenti P, Dos Santos S, Duthoy S, Lacroix C, Garcia-Pelayo C, et al. Genome plasticity of BCG and impact on vaccine efficacy. Proc. Natl Acad. Sci. USA. 2007;104:5596–5601. doi: 10.1073/pnas.0700869104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 30.Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, Honore N, Garnier T, Churcher C, Harris D, et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 31.Garnier T, Eiglmeier K, Camus JC, Medina N, Mansoor H, Pryor M, Duthoy S, Grondin S, Lacroix C, Monsempe C, et al. The complete genome sequence of Mycobacterium bovis. Proc. Natl Acad. Sci. USA. 2003;100:7877–7882. doi: 10.1073/pnas.1130426100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munier-Lehmann H, Burlacu-Miron S, Craescu CT, Mantsch HH, Schultz CP. A new subfamily of short bacterial adenylate kinases with the Mycobacterium tuberculosis enzyme as model: a predictive and experimental study. Proteins. 1999;36:238–248. [PubMed] [Google Scholar]
- 33.Miron S, Munier-Lehmann H, Craescu CT. 1H, 13C and 15N resonance assignment and secondary structure of Mycobacterium tuberculosis adenylate kinase. J. Biomol. NMR. 2001;19:89–90. doi: 10.1023/a:1008392515781. [DOI] [PubMed] [Google Scholar]
- 34.Miron S, Munier-Lehmann H, Craescu CT. Structural and dynamic studies on ligand-free adenylate kinase from Mycobacterium tuberculosis revealed a closed conformation that can be related to the reduced catalytic activity. Biochemistry. 2004;43:67–77. doi: 10.1021/bi0355995. [DOI] [PubMed] [Google Scholar]
- 35.Hible G, Christova P, Renault L, Seclaman E, Thompson A, Girard E, Munier-Lehmann H, Cherfils J. Unique GMP-binding site in Mycobacterium tuberculosis guanosine monophosphate kinase. Proteins. 2006;62:489–500. doi: 10.1002/prot.20662. [DOI] [PubMed] [Google Scholar]
- 36.Munier-Lehmann H, Chaffotte A, Pochet S, Labesse G. Thymidylate kinase of Mycobacterium tuberculosis: a chimera sharing properties common to eukaryotic and bacterial enzymes. Protein Sci. 2001;10:1195–1205. doi: 10.1110/ps.45701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pochet S, Dugue L, Douguet D, Labesse G, Munier-Lehmann H. Nucleoside analogues as inhibitors of thymidylate kinases: possible therapeutic applications. ChemBioChem. 2002;3:108–110. doi: 10.1002/1439-7633(20020104)3:1<108::AID-CBIC108>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 38.Pochet S, Dugué L, Labesse G, Delepierre M, Munier-Lehmann H. Comparative study of purine and pyrimidine nucleoside analogues acting on the thymidylate kinases of Mycobacterium tuberculosis and of humans. ChemBioChem. 2003;4:742–747. doi: 10.1002/cbic.200300608. [DOI] [PubMed] [Google Scholar]
- 39.Vanheusden V, Munier-Lehmann H, Froeyen M, Busson R, Rozenski J, Herdewijn P, Van Calenbergh S. Discovery of bicyclic thymidine analogues as selective and high-affinity inhibitors of Mycobacterium tuberculosis thymidine monophosphate kinase. J. Med. Chem. 2004;47:6187–6194. doi: 10.1021/jm040847w. [DOI] [PubMed] [Google Scholar]
- 40.Van Daele I, Munier-Lehmann H, Froeyen M, Balzarini J, Calenbergh SV. Rational design of 5'-thiourea-substituted alpha-thymidine analogues as thymidine monophosphate kinase inhibitors capable of inhibiting mycobacterial growth. J. Med. Chem. 2007;50:5281–5292. doi: 10.1021/jm0706158. [DOI] [PubMed] [Google Scholar]
- 41.Familiar O, Munier-Lehmann H, Negri A, Gago F, Douguet D, Rigouts L, Hernandez AI, Camarasa MJ, Perez-Perez MJ. Exploring acyclic nucleoside analogues as inhibitors of Mycobacterium tuberculosis thymidylate kinase. ChemMedChem. 2008;3:1083–1093. doi: 10.1002/cmdc.200800060. [DOI] [PubMed] [Google Scholar]
- 42.Douguet D, Munier-Lehmann H, Labesse G, Pochet S. LEA3D : a computer-aided ligand design for structure-based drug design. J. Med. Chem. 2005;48:2457–2468. doi: 10.1021/jm0492296. [DOI] [PubMed] [Google Scholar]
- 43.Gasse C, Douguet D, Huteau V, Marchal G, Munier-Lehmann H, Pochet S. Substituted benzyl-pyrimidines targeting thymidine monophosphate kinase of Mycobacterium tuberculosis: synthesis and in vitro anti-mycobacterial activity. Bioorg. Med. Chem. 2008;16:6075–6085. doi: 10.1016/j.bmc.2008.04.045. [DOI] [PubMed] [Google Scholar]
- 44.Thum C, Schneider CZ, Palma MS, Santos DS, Basso LA. The Rv1712 Locus from Mycobacterium tuberculosis H37Rv codes for a functional CMP kinase that preferentially phosphorylates dCMP. J. Bacteriol. 2009;191:2884–2887. doi: 10.1128/JB.01337-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raleigh EA, Murray NE, Revel H, Blumenthal RM, Westaway D, Reith AD, Rigby PW, Elhai J, Hanahan D. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. [erratum appears in. Nucleic Acids Res. 1995 23, 3612] Nucleic Acids Res. 1988;16:1563–1575. doi: 10.1093/nar/16.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Munier H, Gilles A-M, Glaser P, Krin E, Danchin A, Sarfati RS, Bârzu O. Isolation and characterization of catalytic and calmodulin-binding domains of Bordetella pertussis adenylate cyclase. Eur. J. Biochem. 1991;196:469–474. doi: 10.1111/j.1432-1033.1991.tb15838.x. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 48.Picard V, Bock SC. Rapid and efficient one-tube PCR-based mutagenesis method. Methods Mol. Biol. 1997;67:183–188. doi: 10.1385/0-89603-483-6:183. [DOI] [PubMed] [Google Scholar]
- 49. (2003) BD TALONTM Metal Affinity Resins User Manual, BD Biosciences, Vol. PT1320-1 (PR34731) [Google Scholar]
- 50.Blondin C, Serina L, Wiesmüller L, Gilles A-M, Bârzu O. Improved spectrophotometric assay of nucleoside monophosphate kinase activity using the pyruvate kinase/lactate dehydrogenase coupling system. Anal. Biochem. 1994;220:219–221. doi: 10.1006/abio.1994.1326. [DOI] [PubMed] [Google Scholar]
- 51.Evrin C, Straut M, Slavova-Azmanova N, Bucurenci N, Onu A, Assairi L, Ionescu M, Palibroda N, Barzu O, Gilles AM. Regulatory mechanisms differ in UMP kinases from gram-negative and gram-positive bacteria. J. Biol. Chem. 2007;282:7242–7253. doi: 10.1074/jbc.M606963200. [DOI] [PubMed] [Google Scholar]
- 52.Zamyatnin AA. Amino acid, peptide, and protein volume in solution. Annu. Rev. Biophys. Bioeng. 1984;13:145–165. doi: 10.1146/annurev.bb.13.060184.001045. [DOI] [PubMed] [Google Scholar]
- 53.Catherinot V, Labesse G. ViTO: tool for refinement of protein sequence–structure alignments. Bioinformatics. 2004;20:3694–3696. doi: 10.1093/bioinformatics/bth429. [DOI] [PubMed] [Google Scholar]
- 54.Canutescu AA, Shelenkov AA, Dunbrack RL., Jr A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003;12:2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 56.Pons JL, Labesse G. @TOME-2: a new pipeline for comparative modeling of protein-ligand complexes. Nucleic Acids Res. 2009;37:W485–W491. doi: 10.1093/nar/gkp368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 58.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 59.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 60.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 61.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 62.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 63.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 64.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye-binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 65.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 66.Fassy F, Krebs O, Lowinski M, Ferrari P, Winter J, Collard-Dutilleul V, Salahbey Hocini K. UMP kinase from Streptococcus pneumoniae: evidence for co-operative ATP binding and allosteric regulation. Biochem. J. 2004;384:619–627. doi: 10.1042/BJ20040440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Serina L, Bucurenci N, Gilles A-M, Surewicz WK, Fabian H, Mantsch HH, Takahashi M, Petrescu I, Batelier G, Bârzu O. Structural properties of UMP-kinase from Escherichia coli : modulation of protein solubility by pH and UTP. Biochemistry. 1996;35:7003–7011. doi: 10.1021/bi960062v. [DOI] [PubMed] [Google Scholar]
- 68.Meier C, Carter LG, Sainsbury S, Mancini EJ, Owens RJ, Stuart DI, Esnouf RM. The crystal structure of UMP kinase from Bacillus anthracis (BA1797) reveals an allosteric nucleotide-binding site. J. Mol. Biol. 2008;381:1098–1105. doi: 10.1016/j.jmb.2008.06.078. [DOI] [PubMed] [Google Scholar]
- 69.Gagyi C, Bucurenci N, Sirbu O, Labesse G, Ionescu M, Ofiteru A, Assairi L, Landais S, Danchin A, Barzu O, et al. UMP kinase from the Gram-positive bacterium Bacillus subtilis is strongly dependent on GTP for optimal activity. Eur. J. Biochem. 2003;270:3196–3204. doi: 10.1046/j.1432-1033.2003.03702.x. [DOI] [PubMed] [Google Scholar]
- 70.Ramon-Maiques S, Marina A, Gil-Ortiz F, Fita I, Rubio V. Structure of acetylglutamate kinase, a key enzyme for arginine biosynthesis and a prototype for the amino acid kinase enzyme family, during catalysis. Structure. 2002;10:329–342. doi: 10.1016/s0969-2126(02)00721-9. [DOI] [PubMed] [Google Scholar]
- 71.Meyer P, Evrin C, Briozzo P, Joly N, Barzu O, Gilles AM. Structural and functional characterization of Escherichia coli UMP kinase in complex with its allosteric regulator GTP. J. Biol. Chem. 2008;283:36011–36018. doi: 10.1074/jbc.M802614200. [DOI] [PubMed] [Google Scholar]
- 72.Tu JL, Chin KH, Wang AH, Chou SH. Unique GTP-binding pocket and allostery of uridylate kinase from a gram-negative phytopathogenic bacterium. J Mol Biol. 2009;385:1113–1126. doi: 10.1016/j.jmb.2008.11.030. [DOI] [PubMed] [Google Scholar]
- 73.Bucurenci N, Serina L, Zaharia C, Landais S, Danchin A, Bârzu O. Mutational analysis of UMP kinase from Escherichia coli. J. Bacteriol. 1998;180:473–477. doi: 10.1128/jb.180.3.473-477.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328:504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Egeblad-Welin L, Welin M, Wang L, Eriksson S. Structural and functional investigations of Ureaplasma parvum UMP kinase-a potential antibacterial drug target. FEBS J. 2007;274:6403–6414. doi: 10.1111/j.1742-4658.2007.06157.x. [DOI] [PubMed] [Google Scholar]
- 76.Koshland DE, Jr, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 77.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 78.Kern D, Zuiderweg ER. The role of dynamics in allosteric regulation. Curr. Opin. Struct. Biol. 2003;13:748–757. doi: 10.1016/j.sbi.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 79.Marcos E, Crehuet R, Bahar I. On the conservation of the slow conformational dynamics within the amino acid kinase family: NAGK the paradigm. PLoS Comput Biol. 2010;6:e1000738. doi: 10.1371/journal.pcbi.1000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.