Abstract
Various chemical modifications are currently being evaluated for improving the efficacy of short interfering RNA (siRNA) duplexes as antisense agents for gene silencing in vivo. Among the 2′-ribose modifications assessed to date, 2′deoxy-2′-fluoro-RNA (2′-F-RNA) has unique properties for RNA interference (RNAi) applications. Thus, 2′-F-modified nucleotides are well tolerated in the guide (antisense) and passenger (sense) siRNA strands and the corresponding duplexes lack immunostimulatory effects, enhance nuclease resistance and display improved efficacy in vitro and in vivo compared with unmodified siRNAs. To identify potential origins of the distinct behaviors of RNA and 2′-F-RNA we carried out thermodynamic and X-ray crystallographic analyses of fully and partially 2′-F-modified RNAs. Surprisingly, we found that the increased pairing affinity of 2′-F-RNA relative to RNA is not, as commonly assumed, the result of a favorable entropic contribution (‘conformational preorganization’), but instead primarily based on enthalpy. Crystal structures at high resolution and osmotic stress demonstrate that the 2′-F-RNA duplex is less hydrated than the RNA duplex. The enthalpy-driven, higher stability of the former hints at the possibility that the 2′-substituent, in addition to its important function in sculpting RNA conformation, plays an underappreciated role in modulating Watson–Crick base pairing strength and potentially π–π stacking interactions.
INTRODUCTION
Fluorine, thanks to unusual properties such as a small size paired with high electronegativity, has found numerous applications in medicinal chemistry (1,2). In the oligonucleotide realm, fluorine was used as a substitute of the RNA’s 2′-hydroxyl group that locks the sugar predominantly in the C3′-endo conformation and leads to significant increases in target affinity (3–5). Initially tested in antisense applications and shown to stabilize the modified strand against nucleases (6), the 2′-deoxy-2′-fluoro-RNA (2′-F-RNA) modification along with a host of others is now being evaluated in the context of RNA interference (RNAi) and in terms of its compatibility with the RNA-induced silencing complex (RISC) that mediates cleavage of the RNA targets (7,8). Key steps in this process include ATP-dependent unwinding of the short interfering RNA (siRNA) duplex composed of sense (passenger) and antisense (guide) strands, pairing of the guide strand to mRNA and subsequently ATP-independent cleavage of the latter between nucleotides paired to the central guide residues (7,9,10).
In addition to conferring protection against degradation and prolonging the half-life of oligonucleotides compared with their unmodified counterparts (11–18), the 2′-F modification also leads to a reduction in immunostimulatory effects (19,20). However, these favorable properties did not necessarily translate into higher in vivo efficacy of 2′-F-modified siRNAs. On the one hand, siRNAs with incorporated 2′-F-RNA residues exhibited potent anti-HBV activity in mice when they were co-injected in a liposome particle (19). By contrast, in two other in vivo studies, 2′-F-modified RNAs did not produce an enhanced or prolonged reduction in target gene expression despite higher bioavailability (16,21).
We recently evaluated several RNA 2′-modifications, including 2′-F-RNA, 2′-O-methyl-RNA (2′-O-Me-RNA), 2′-O-(2-methoxyethyl)-RNA (2′-O-MOE-RNA) and locked nucleic acid (LNA) in terms of improvements of siRNA in vitro and in vivo efficacy in an established endogenous system (coagulation factor VII, FVII) (22). FVII is a single-chain glycoprotein of 48 kDa that circulates in the plasma, has a short 3–6 h half-life and interacts with tissue factor (TF). The FVII:TF complex activates factors X and IX and initiates the blood clotting pathway (23). In cancer, the FVII:TF complex has been found in abundance in lung, breast, pancreatic, gastric and colon tumors and facilitates tumor growth and invasion (24). Animal models suggest that inhibitors of FVII may block tumor growth and metastases (25–27). All pyrimidines in the tested siRNA duplex were replaced by 2′-F-RNA nucleotides. The activities of mixed sequences with other modifications were also assessed relative to the corresponding native siRNA because in some cases siRNAs containing multiple modifications (19,28) were found to be more potent than those with an individual analog (18,29). For in vivo trials with mice, delivery of the FVII siRNA at various doses to liver via IV was achieved using the recently developed LNP01 liposomal formulation (30).
In vitro silencing was assessed in HeLa SS6 cells stably expressing mouse FVII with transfections performed using lipofectamine 2000. The siRNA with 2′-F-RNA-U and -C residues in both the guide and passenger strands was roughly 2-fold more potent compared with the native siRNA (IC50 values of 0.95 and 0.50 nM, respectively). Remarkably, this siRNA also triggered similar gains in silencing in vivo and comparisons between the FVII protein levels following injection of the 2′-F-modified siRNA to those achieved with siRNAs bearing other 2′-modifications illustrated a clear advantage for 2′-F-RNA (22). The best activity was observed with the compound containing 2′-F-modified pyrimidines on both strands. Conversely, the 2′-O-Me and LNA modifications appeared to be tolerated only when placed on the sense strand but not on both the sense and antisense strands. Lastly, the 2′-O-MOE modification was not tolerated on either sense or antisense strand, most likely a consequence of its steric bulk in the minor groove (22).
Although the 2′-F-modification increases the melting temperature (Tm) of duplexes considerably, LNA is significantly more stabilizing (22). This indicates that raising the duplex stability beyond a certain point does not provide any benefits with regard to siRNA efficacy. Therefore, it is likely that other properties than simply increased pairing affinity contribute to the unique in vitro and in vivo activity of 2′-F-modified siRNAs. In order to gain insight into unusual structural and thermodynamic features potentially underlying the above favorable properties of 2′-F-RNA, we conducted detailed UV melting, calorimetric, osmotic stress and X-ray crystallographic investigations of all-2′-F-RNA and mixed 2′-F-/2′-OH-RNA duplexes. One surprising finding brought to light by these studies is that the stability increases afforded by the 2′-F modification are almost exclusively due to a favorable enthalpy. Osmotic stress and crystallography both provide evidence that replacement of 2′-hydroxyl groups by fluorine leads to diminished hydration of the duplex. Thus, proteins such as the Piwi domain of Ago2 (‘Slicer’) that interact with the minor groove of RNA will encounter a very different environment as a consequence of 2′-F modification. Differences in the water structures of RNA and 2′-F-RNA duplexes will likely affect all steps along the RNAi pathway, from uptake, unwinding and incorporation of the guide strand into the RISC to cleavage of the targeted mRNA opposite the guide RNA.
MATERIALS AND METHODS
Oligoribonucleotide synthesis and purification
The 2′-F-modified ribonucleoside phosphoramidite building blocks for incorporation into oligonucleotides were prepared as previously described (5). We synthesized the RNA, mixed 2′-F/2′-OH and fully 2′-F modified versions of octamers CCCCGGGG and CGAAUUCG as well as the dodecamer UAUAUAUAUAUA (Table 1). All oligonucleotides were synthesized by the solid phase approach on a 394 ABI synthesizer and applying extended coupling times in some cases. Following cleavage from the CPG and simultaneous deprotection of base and phosphate groups using a mixture of ammonia and ethanol (3:1) for 16 h at 55°C, the fully deprotected all-2′-F oligonucleotides were precipitated from anhydrous methanol. With all-RNA or mixed 2′-F/2′-OH oligonucleotides, the solution was decanted, lyophilized and re-suspended in triethylamine trihydrofluoride (TEA.3HF, Aldrich) followed by incubation at 65°C for 90 min to remove the t-butyldimethylsilyl (TBDMS) 2′-hydroxyl protection groups. As in the case of the fully 2′-F-modified sequences this was followed by precipitation from anhydrous methanol. All oligonucleotides were purified by PAGE and the appropriate bands were cut out and shaken overnight in a 100 mM sodium acetate solution. Desalting was accomplished with C18 Sep-Pak cartridges (Waters). All oligonucleotides were characterized by electrospray mass spectrometry as well as analytical anion-exchange HPLC and/or capillary gel electrophoresis.
Table 1.
Sequences and nomenclature of analyzed RNAs, 2′-F-RNAs and mixed 2′-OH-/2′-F-RNAs
| Sequence | Chemistry | Abbreviated name |
|---|---|---|
| r(CCCCGGGG) | RNA | rC4G4 (reference) |
| fCrCfCrCfGrGfGrG | Alternating 2′-F/OH-RNA | f/rC4G4 |
| f(CCCCGGGG) | 2′-F-RNA | fC4G4 |
| r(CGAAUUCG) | RNA | rA2U2 |
| fCrGfArAfUrUfCrG | Alternating 2′-F/OH-RNA | f/rA2U2 |
| f(CGAAUUCG) | 2′-F-RNA | fA2U2 |
| r(UAUAUAUAUAUA) | RNA | rUA6 |
| fUrAfUrAfUrAfUrAfUrAfUrA | Alternating 2′-F/OH-RNA | f/rUA6 |
| f(UAUAUAUAUAUA) | 2′-F-RNA | fUA6 |
UV thermal melting
Melting of each oligonucleotide (2 µM) was done in 10 mM sodium cacodylate (pH = 7.4), 0.1 mM EDTA and 300 mM NaCl in the presence of 0, 5, 10, 15 and 20% wt/vol of each of the three organic co-solutes (Tables 2 and 3). Oligonucleotide concentrations were calculated from the nearest-neighbor approximation (31). Absorbance versus temperature profiles were measured at 260 nm on a Varian Bio 100 UV-Visible spectrometer equipped with a six-position Peltier temperature controller or a Shimadzu 800 UV-Visible spectrometer equipped with an eight-position Peltier temperature controller. The temperature was increased at 0.5°C per minute. The melting temperatures were obtained using the Varian Cary and Shimadzu LabSolution TmAnalysis software. The experimental absorbance versus temperature curves were converted into a fraction of strands remaining hybridized (α) versus temperature curves by fitting the melting profile to a two-state transition model, with linearly sloping lower and upper base lines. The melting temperatures (Tm) were obtained directly from the temperature at α = 0.5. The final Tm was an approximation of usually five to eight measurements.
Table 2.
Thermodynamic stability analysis for RNA and 2′-F-RNA
| UV melting | van't Hoff analysis of UV melting curves |
UV melting from δα/δ (tM−1) | UV melting, concentration dependence |
DSC | ||||
|---|---|---|---|---|---|---|---|---|
| Column #1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
| Oligonucleotide (abbreviation) | Tm (°C) | −ΔH (kcalmol−1) | −ΔS (eu) | −ΔG (kcalmol−1) | −ΔH (kcalmol−1) | −ΔH (kcalmol−1) | −ΔS (eu) | −ΔH (kcalmol−1) |
| 5′-r(CGAAUUCG)-3′ (rA2U2) | 34.1 ± 0.6 | 63.4 ± 6.6 | 180.0 ± 21.6 | 7.5 ± 0.2 | 55.6 ± 4.6 | 58.0 ± 9.4 | 189.7 ± 30.6 | 39.2 ± 1.4 |
| 5′-fCrGfArAfUrUfCrG-3′ (f/rA2U2) | 43.7 ± 0.4 | 63.8 ± 3.9 | 175.2 ± 12.0 | 9.4 ± 0.2 | 58.4 ± 3.7 | 58.3 ± 12.9 | 185.8 ± 41.0 | 48.8 ± 0.3 |
| 5′-f(CGAAUUCG)-3′ (fA2U2) | 53.3 ± 0.3 | 69.5 ± 2.0 | 186.9 ± 6.2 | 11.6 ± 0.1 | 62.1 ± 2.9 | 62.3 ± 9.8 | 192.1 ± 30.2 | 53.5 ± 0.7 |
| 5′-r(UAUAUAUAUAUA)-3′ (rUA6) | 31.7 ± 0.4 | 70.9 ± 5.9 | 206.5 ± 19.1 | 6.9 ± 0.1 | 66.0 ± 2.1 | 82.5 ± 7.8 | 271.6 ± 25.8 | 49.5 ± 0.5 |
| 5′-fUrAfUrAfUrAfUrAfUrAfUrA-3′ (f/rUA6) | 44.1 ± 0.4 | 83.5 ± 4.8 | 237.2 ± 15.1 | 10.0 ± 0.1 | 78.4 ± 5.1 | 75.9 ± 9.5 | 241.5 ± 30.4 | 54.3 ± 0.4 |
| 5′-f(UAUAUAUAUAUA)-3′ (fUA6) | 53.1 ± 0.5 | 83.7 ± 4.1 | 230.8 ± 12.6 | 12.2 ± 0.2 | 80.2 ± 5.4 | 79.1 ± 14.0 | 244.7 ± 43.4 | 56.6 ± 0.7 |
Table 3.
Osmotic stress analysis for RNA and 2′-F-RNA
| Oligonucleotide (abbreviation) | Δnw (ethylene glycol) | Δnw (glycerol) | Δnw (acetamide) |
|---|---|---|---|
| 5′-r(CGAAUUCG)-3′ (rA2U2) | 18.8 ± 4.9 | 22.5 ± 6.1 | 37.9 ± 5.8 |
| 5′-fCrGfArAfUrUfCrG-3′ (f/rA2U2) | 14.8 ± 4.2 | 11.2 ± 5.1 | 21.4 ± 3.7 |
| 5′-f(CGAAUUCG)-3′ (fA2U2) | 1.2 ± 3.8 | 3.0 ± 6.9 | 14.8 ± 3.3 |
| 5′-r(UAUAUAUAUAUA)-3′ (rUA6) | 22.4 ± 4.6 | 35.9 ± 8.4 | 42.0 ± 5.7 |
| 5′-fUrAfUrAfUrAfUrAfUrAfUrA-3′ (f/rUA6) | 17.0 ± 3.9 | 28.8 ± 4.2 | 30.3 ± 3.2 |
| 5′-f(UAUAUAUAUAUA)-3′ (fUA6) | 6.3 ± 4.6 | 18.9 ± 6.1 | 21.2 ± 4.9 |
The thermodynamic parameters (ΔH, ΔS and ΔG) were obtained by van’t Hoff analysis of melting curves using Varian Cary software (Table 2, columns 3–5). In addition, the enthalpy of the melting was determined by three independent methods as follows:
From the width at the half-height of the differentiated melting curve (Table 2, column 6). The fraction of strands remaining hybridized (α) versus temperature curves were converted into differentiated melting curves [δα/δ(Tm−1) versus Tm] using Varian Cary software. The width of the differentiated melting curve at the half-height is inversely proportional to the van’t Hoff transition enthalpy; for a bimolecular transition ΔH = 10.14/(T1−1−T2−1), where T1 is the lower temperature and T2 is the upper temperature (both in K) at one-half of [δα/δ(Tm−1)] (32).
From the concentration dependence of the melting temperature (Table 2, columns 7 and 8). Melting experiments were done in pure buffer over a concentration range of 1 to 16 µM (or 32 µM) of oligonucleotides. For a bimolecular association of self-complementary strands 1/Tm = (R/ΔH)lnC + ΔS/ΔH where R is the universal gas constant (1.986 cal/molK) and C is the total strand concentration (1 × 10−6 to 16 × 10−6 M) (32). The plot of 1/Tm versus lnC is linear with R/ΔH as the slope. The final −ΔH was obtained by linear fitting using KaleidaGraph software (Version 3.51). Uncertainties in the slopes of the 1/Tm versus lnC plots were estimated as described previously (33).
From the Differential scanning calorimetry (DSC) experiments (Table 2, column 9) as described below.
Differential scanning calorimetry
DSC experiments were run in 10 mM phosphate buffer (pH = 7.4) containing 300 mM NaCl. RNA samples (144 nmols) were dissolved in phosphate buffer (1 ml) and dialyzed against the same buffer (3 × 300 ml) using a Float-A-Lyzer G2 1 ml dialysis bag (Spectrum Laboratories). The buffer was changed after 2 h, then again after another 2 h and the last dialysis was done overnight. The sample and the last dialysis buffer were degassed by stirring under medium vacuum (80 torr) for 30 min and then loaded into the sample and reference cells (capillary) of a TA Instruments NanoDSC, respectively. The DSC sample chamber was pressurized to 3 atm. and the DSC run was performed at a heating rate of 1°C/min. Typically, five to six cycles of heating (see Supplementary Figures S7, S10, S13, S16, S18 and S22) and cooling were done to obtain reproducible data. The base line runs were done with phosphate buffer only in both sample and reference cells and the obtained base lines were subtracted from the RNA data before analysis. Integration of the calorimetric peak using TA Instruments NanoAnalyze software (Version 1.0) gave model-independent ΔH (Table 2, column 9), which was used in calculations of ΔnW from osmotic stress data.
Osmotic stress
The changes in the number of water molecules associated with the melting process ΔnW were determined as originally described by Spink and Chaires (34) and later used by us in studies of other modified RNA and DNA fragments (33,35–38) (Table 3).
where −ΔH is the enthalpy determined in the DSC experiment and R is the universal gas constant (1.986 cal/mol/K). The experimentally determined values of water activity (lnaw) at given co-solute concentrations were provided by Professors Spink and Chaires. The slope of the plot of reciprocal temperature (in K) of melting versus the logarithm of water activity (lnaw) at different concentrations (0, 5, 10, 15 and 20%) of small co-solutes gave the value of d(Tm−1)/d(lnaw). The final Δnw were obtained by linear fitting the data using KaleidaGraph software (Version 3.51). The experimental uncertainties were obtained as previously reported (33,38).
Crystallization
Crystals of 5′-f(CGAAUUCG)-3′ were grown by the hanging-drop vapor diffusion technique using the Nucleic Acid Miniscreen (Hampton Research, Aliso Viejo, CA) (39). Droplets (2 µl) containing oligonucleotide (0.6 mM), sodium cacodylate (20 mM, pH 6.0), strontium chloride (40 mM), magnesium chloride (10 mM), spermine tetrahydrochloride (6 mM) and 2-methyl-2,4-pentanediol [MPD; 5% (v/v)] were equilibrated against a reservoir of MPD (1 ml, 35%). The mixed octamer 5′-fCrGfArAfUrUfCrG-3′ crystallized from two hanging-drop vapor diffusion conditions: crystals were obtained from (1) droplets (2 µl) containing oligonucleotide (0.6 mM), sodium cacodylate (20 mM, pH 6.0), sodium chloride (40 mM), barium chloride (10 mM), spermine tetrahydrochloride (6 mM) and 2-methyl-2,4-pentanediol [MPD; 5% (v/v)] that were equilibrated against a reservoir of MPD (1 ml, 35%) (f/rA2U2-P3 form) and (2) from droplets (2 µl) containing oligonucleotide (0.6 mM), sodium cacodylate (20 mM, pH 6.0), lithium chloride (20 mM), strontium chloride (40 mM), spermine tetrahydrochloride (6 mM) and 2-methyl-2,4-pentanediol [MPD; 5% (v/v)] that were equilibrated against a reservoir of MPD (1 ml, 35%) (f/rA2U2-R32 form). The mixed octamer 5′-fCrCfCrCfGrGfGrG-3′ was also crystallized by the hanging-drop vapor diffusion technique. Droplets (2 µl) containing oligonucleotide (0.6 mM), sodium cacodylate (20 mM, pH 6.0), sodium chloride (40 mM), spermine tetrahydrochloride (6 mM) and 2-methyl-2,4-pentanediol [MPD; 5% (v/v)] were equilibrated against a reservoir of MPD (1 ml, 35%). All crystals were mounted in nylon loops without further cryo-protection and frozen in liquid nitrogen.
X-ray data collection, phasing and refinement
Diffraction data were collected on the 21-ID-D beam line of the Life Sciences Collaborative Access Team (LS-CAT) at the Advanced Photon Source (APS) located at Argonne National Laboratory (Argonne, IL). The wavelength was tuned to 0.769 Å (absorption edge for Sr2+ ion) and the crystal was kept at 110 K during data collection using a MARCCD 300 detector. Diffraction data were integrated, scaled and merged with HKL2000 (40). A summary of selected crystal data and data collection parameters is provided in Table 4. The structure fA2U2 with three duplexes per crystallographic asymmetric unit was phased by the single-wavelength anomalous dispersion (SAD) technique using the program HKL2MAP (41). The resulting experimental electron density map allowed visualization of all 48 ribonucleotides per asymmetric unit. Building of the models was performed using the program TURBO-FRODO (42). The initial orientations of the three duplex models were optimized by several rounds of rigid body refinement while gradually increasing the resolution of the diffraction data. The refinements were carried out using the program CNS (43) by performing simulated annealing, gradient minimization and refinement of individual isotropic temperature factors. The later refinements were done using the program SHELX (44), keeping aside 5% of the reflections to compute the R-free (45). After a few cycles of refinement in SHELX, the 2′-hydroxyl groups were substituted with fluorine atoms and the dictionary was adapted to account for the substitution of all 2′-OH groups with 2′-F. Ions and water molecules were added on the basis of Fourier 2Fo−Fc sum and Fo−Fc difference electron density maps, and accepted on the basis of standard distance and B-factor criteria. Final refinement parameters and deviations from ideal geometries are listed in Table 4 and examples of the quality of the final electron density are depicted in Figure 1. Helical parameters were calculated with the program CURVES (46).
Table 4.
Crystal data, data collection parameters and structure refinement statistics
| Oligonucleotide | fA2U2 | f/rA2U2-P3 | f/rA2U2-R32 | f/rC4G4 |
|---|---|---|---|---|
| Space group | Trigonal, P3 | Trigonal, P3 | Rhombohedral, R32 | Rhombohedral, R32 |
| Unit cell constants (Å) | a = b = 43.25 c = 60.79 | a = b = 43.38 c = 60.99 | a = b = 40.55 c = 117.22 | a = b = 41.63 c = 126.49 |
| Resolution (Å) | 1.20 | 1.45 | 1.15 | 1.85 |
| Outer shell (Å) | 1.24–1.20 | 1.48–1.45 | 1.17–1.15 | 1.92–1.85 |
| No. of unique reflections | 39 940 | 22 442 | 13 343 | 3824 |
| Completeness (outer shell) (%) | 99.7 (99.3) | 98.5 (81.8) | 97.9 (72.9) | 99.5 (99.7) |
| R-merge (outer shell) | 0.05 (0.35) | 0.07 (0.47) | 0.095 (0.41) | 0.077 (0.43) |
| R-work (%) | 17.0 | 19.7 | 18.2 | 20.9 |
| R-free (%) | 23.4 | 25.1 | 24.9 | 27.1 |
| No. of RNA atoms | 1002 | 1002 | 355a (334) | 338 |
| No. of waters / Sr2+ / Mg2+ | 297/9/2 | 142/2b/0 | 93/3/0 | 28/0/0 |
| R.m.s.d. bonds (Å) | 0.010 | 0.022 | 0.010 | 0.011 |
| R.m.s.d. angles (1–3 dist. Å; deg) | 0.026 | 2.07 | 0.027 | 2.12 |
| Avg. B-factor, RNA atoms (Å2) | 15.0 | 17.3 | 13.4 | 24.1 |
| Avg. B-factor, solvent (Å2) | 30.7 | 26.2 | 41.2 | 28.7 |
| Average B-factor, ions (Å2) | 22.5 | 37.4 | 28.1 | – |
| PDB entry code | 3P4A | 3P4B | 3P4C | 3P4D |
aDual occupancy of G8 (Figure 1B), bBa2+.
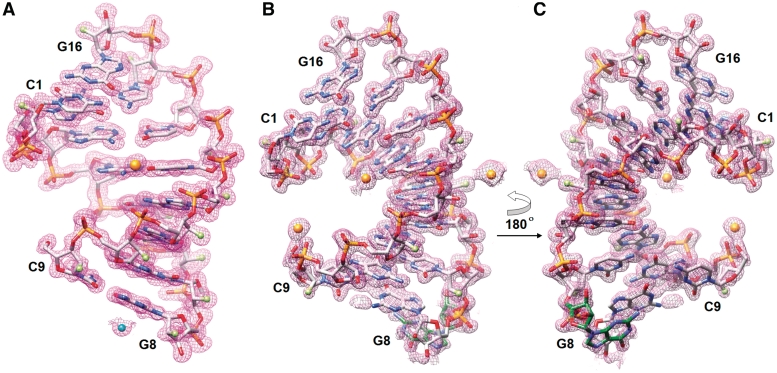
Figure 1.
Examples of the quality of the final Fourier (2Fo−Fc) sum electron density (∼1.3σ threshold). (A) The fA2U2 octamer duplex. A Sr2+ and a Mg2+ ion are shown as spheres, colored in orange and cyan, respectively. (B) The f/rA2U2-R32 octamer duplex in which the terminal G8 exhibits alternative conformations. (C) The f/rA2U2 duplex after a rotation of 180° around the vertical; the looped-out G8 (carbon atoms of this residue are highlighted in green) which has an occupancy of 0.76 is clearly visible in this view. Atoms are colored silver, red, blue, orange and green for carbon, oxygen, nitrogen, phosphorus and fluorine, respectively, and Sr2+ ions are shown as orange spheres.
The structure of f/rA2U2-P3 was solved by molecular replacement using the program MOLREP (47) (score in MOLREP was 0.68) and the refined structure of fA2U2, consisting of all the three duplexes (without the ions/water molecules), as the search model. Initial refinements in CNS were carried out in a similar fashion as described for fA2U2 after replacing the 2′-F atoms with 2′-OH atoms. TLS (48) and restrained refinement were carried out using Refmac5 (49). Final refinements were carried out using anisotropic temperature factors for all nucleic acid and solvent atoms.
The structure of f/rA2U2-R32 was solved by SAD using Sr2+ as the anomalous scatterer. The asymmetric unit in the R32 space group contains a single RNA octamer duplex, and fourteen of the sixteen residues (G8 and C9 were missing) could be traced in the anomalous map. Initial gradient minimization refinements were carried out in CNS, keeping aside 5% of the randomly chosen reflections to compute the R-free (45). After the addition of water molecules and metal ions and replacement of alternating 2′-F with 2′-OH atoms as well as adaptation of the SHELX dictionary files, refinements were carried out using anisotropic temperature factors for all nucleic acid atoms.
The structure of f/rC4G4 was solved by molecular replacement using the program MOLREP (47) and the native RNA octamer, PDB ID:1RXB (50), as the search model. Initial refinements were carried out using the program CNS (43) by performing simulated annealing and gradient minimization. The structure was then subjected to refinements with Refmac5 (49) and the strategies pursued were similar to those used in the refinement of f/rA2U2-P3 above. However, only isotropic temperature factor refinement was applied because of the lack of high-resolution data.
Coordinates
Final coordinates and structure factors for all four octamers have been deposited in the Protein Data Bank (http://www.rcsb.org.) The PDB ID codes are 3P4A (fA2U2), 3P4B (f/rA2U2-P3), 3P4C (f/rA2U2-R32) and 3P4D (f/rC4G4).
RESULTS
Stabilizing effects of the 2′-F-RNA modification
To study the consequences of 2′-F modification on the thermodynamic stability and hydration of RNA duplexes we chose the octamers CCCCGGGG and CGAAUUCG as well as the dodecamer UAUAUAUAUAUA. For all three the native RNAs as well as the corresponding fully 2′-F-modified sequences and the alternating 2′-F/2′-OH versions (fCrCfCrCfGrGfGrG, fCrGfArAfUrUfCrG and fUrAfUrAfUrAfUrAfUrAfUrA) were synthesized. An overview of the sequences and chemical composition of all analyzed oligonucleotides and the simplified names we will use for them throughout this paper is given in Table 1.
As previously observed (3,5), 2′-F modification leads to a significant increase in the stability of duplexes as assessed by UV melting experiments (Table 2). Thus, under the conditions used (2 µM single strand, 300 mM NaCl, pH 7.4; see ‘Materials and Methods’ section), both the octamer f(CCCCGGGG) (fC4G4) and the mixed octamer fCrCfCrCfGrGfGrG (f/rC4G4) melted beyond 90°C and the C4G4 sequence was therefore not included in a detailed comparison of thermodynamic parameters of duplex formation. An earlier analysis of the thermodynamics of the native RNA CCCCGGGG (rC4G4) had shown a Tm of 70.1°C at an 8 µM strand concentration in 100 mM NaCl (50,51). Complete modification of the rA2U2 octamer and rUA6 dodecamer sequences raises the melting temperature by ca. 20 and 22°C, respectively. The additive nature of the stability boost is indicated by the ∼10 and 12°C increases in Tm observed for the f/rA2U2 octamer and f/rUA6 dodecamer in which half the ribonucleotides were 2′-F-modified (Table 2, column 2).
Whereas the stabilizing effect of the 2′-F modification has long been known, it is noteworthy that the precise origins of the stability gains seem to have never been explored. We used DSC and several methods based on UV thermal melting to measure the contributions of entropy and enthalpy to the superior Gibbs free energy exhibited by 2′-F-RNA relative to RNA (Table 2). As with the melting temperatures, the calorimetrically determined changes in enthalpy (ΔH) are strongly favorable for 2′-F-RNA (Table 2, column 9). A similar picture emerges from the van’t Hoff analysis of the melting curves (Table 2, column 3) and analysis of the differentiated melting curve (Table 2, column 6), although the increases for the octamers with incorporated 2′-F-modified residues relative to their native counterparts differ in magnitude from those based on DSC. The changes in enthalpy values obtained from concentration dependency of UV melting temperatures (Table 2, column 7) are too small compared to the relatively large errors to show any trend. In general the values based on DSC can be considered more reliable, because the data derived from melting curves hinge on the assumption of an ideal two-state transition that often doesn’t represent the actual situation. Nevertheless, the trends in the enthalpy changes obtained from DSC or UV melting in the rA2U2–f/rA2U2–fA2U2 octamer series are overall the same. Unlike the enthalpic changes accompanying duplex formation or melting that differ considerably between RNA and 2′-F-RNA, the entropic contributions are almost identical within the established error margins (Table 2, columns 4 and 8). Although we only studied the thermodynamic parameters of duplex formation with two RNA sequences, it seems unlikely that the particular sequence will influence the higher stability of 2′-F-RNA and the relative contributions of ΔH and ΔS to a significant extent. Therefore, we conclude that the stabilization afforded by the 2′-F modification is chiefly the result of a favorable contribution to enthalpy.
Osmotic stress measurements
In order to gain insight into changes in RNA hydration as a result of replacement of half or all of the 2′-hydroxyl groups by fluorine we applied the osmotic stress method (33,34,38). We carried out UV melting experiments with the various oligonucleotides in the presence of different concentrations of ethylene glycol, glycerol or acetamide, whereby the addition of these small co-solutes typically lowers the melting temperature. The changes in the number of water molecules associated with the melting process were subsequently determined following the approach by Spink and Chaires (34), as described in detail in the ‘Materials and Methods’ section. The osmotic stress approach allows a qualitative estimate of the number of water molecules bound to a nucleic acid duplex and is based on the assumption that the co-solutes change the water activity but do not directly interact with the RNA.
As indicated by the smaller Δnw values (Table 3), the presence of 2′-fluorine in the f/rA2U2 and fA2U2 octamer duplexes dramatically reduces the hydration relative to the rA2U2 duplex. The difference in hydration between rA2U2 and fA2U2 is particularly striking in the case of the ethylene glycol and glycerol co-solutes (−94 and −87%, respectively), and still amounts to a 61% reduction in the case of acetamide (rA2U2, 37.9 versus fA2U2, 14.8). As in earlier studies reporting the outcomes of osmotic stress experiments (33,35,36), the ΔnW values obtained in the presence of acetamide are somewhat higher than those in the ethylene glycol or glycerol series (Table 3). This difference can be attributed to residual interactions with nucleic acids or proteins exhibited by small organic molecules (36,52). In fact, it was shown that the majority of organic co-solutes (including glycerol) were not completely excluded from the surface of the protein bovine serum albumin and that the use of such compounds in osmotic stress experiments can be expected to underestimate the magnitude in change of hydration (53).
Compared with the octamer sequence, the reductions in hydration seen with the fUA6 and f/rUA6 dodecamers relative to rUA6 are still striking. They amount to changes of −72% (ethylene glycol), −53% (glycerol) and −50% (acetamide). The somewhat divergent magnitudes in water loss seen with the two sequences are most likely caused by the different CG contents of the two RNAs that we tested (50% in the octamer and 0% in the dodecamer). The more extensive water structure around C:G pairs (54) may be disturbed to a more significant degree than that around A:U pairs by the 2′-fluoro modification. For now, we note that substituting the 2′-hydroxyl group with fluorine goes along with a loss in hydration. Interestingly, this change appears to be of no consequence for the entropic portion of the free energy of duplex melting as shown in the previous section.
X-ray crystallography of 2′-F-modified RNA octamers
To analyze the potential changes in RNA conformation and water structure as a result of the replacement of the 2′-hydroxyl group by fluorine, we conducted crystallographic studies of modified octamers. In all, five duplexes, f/rC4G4, fC4G4, rA2U2, f/rA2U2 and fA2U2, were included in the crystallization experiments (Table 1). The crystal structure of the sixth octamer, rC4G4, had previously been determined at 1.4 Å resolution and serves as a reference here (50). All of the above native and 2′-modified strands yielded crystals. However, crystals of the native RNA octamer rA2U2 and the fully 2′-F-modified fC4G4 diffracted only to low resolution and the diffraction limits of both remained below 3 Å despite extensive efforts to improve the initial crystals. All attempts directed at identifying alternative crystal forms for the two duplexes unfortunately remained unsuccessful. Crystal structures were subsequently determined at high resolution for fA2U2 and the mixed f/rA2U2 (two crystal forms) and f/rC4G4 (Table 4 and ‘Materials and Methods’ section). Examples of the quality of the final electron density are depicted in Figure 1.
The structure of fA2U2 with three duplexes per crystallographic asymmetric unit was phased by the SAD technique, using Sr2+ as the anomalously scattering center (Figure 1A). The structure of f/rA2U2-P3 was determined by molecular replacement using the refined structure of fA2U2 consisting of all three duplexes as the search model. The structure of f/rA2U2-R32 was also phased by Sr2+-SAD. All residues except for the terminal G8:C9 base pair were well resolved in the anomalous map. Residue G8 adopts two different orientations one of which is looped out (Figure 1B). Finally, the structure of f/rC4G4 was solved by molecular replacement using the native RNA octamer with PDB ID code 1RXB (50) as the search model.
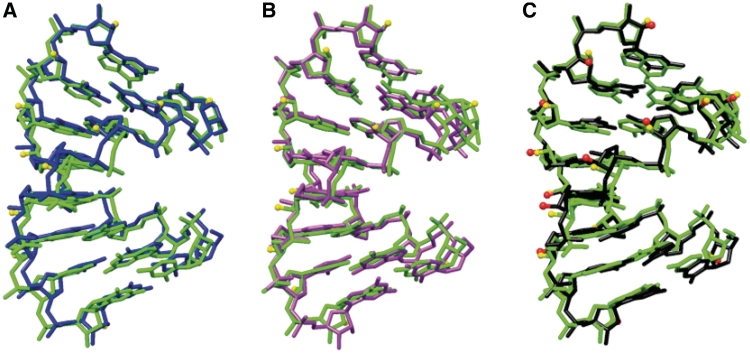
The comparison between the conformations of RNA and partially or fully 2′-F-modified RNA duplexes based on X-ray crystallography shows that substitution of the 2′-hydroxyl group with fluorine is of little consequence in terms of the local and overall helix geometry. Superimpositions of the symmetry-independent helices in the structure of fA2U2 as well as of an individual fA2U2 helix and the reference helix rC4G4 are depicted in Figure 2. From these alignments it is obvious that the geometrical variations among fA2U2 duplexes (panels A and B) are of the same order as those between 2′-F-RNA and RNA (panel C). The calculated helical parameters (46), among them rise, and twist, as well as sugar puckers and backbone torsion angles confirm the similar conformations of the two duplex types (data not shown). In particular, all 2′-deoxy-2′-fluororiboses in the fA2U2, f/rA2U2 and f/rC4G4 duplexes exhibit the C3′-endo pucker with a pseudoaxial orientation of the fluorine substituent (Figure 1). One slight geometrical difference between the chemically modified sugar and the standard ribose is the shorter distance between C2′ and F2′ in the former relative to the C2′-O2′ distance (1.33 and 1.43 Å, respectively). With regard to an interpretation of the aforementioned thermodynamic differences between RNA and 2′-F-RNA, we note for now that conformational differences that are easily detectable in crystal structures, such as divergent degrees of sliding between adjacent base pairs that would affect stacking, cannot be responsible for the favorable enthalpy displayed by the 2′-F-modified analog.
Figure 2.
Comparison between the conformations of 2′-F-RNA and RNA duplexes. Superimpositions of (A) fA2U2 duplexes 1 (green) and 2 (blue), (B) fA2U2 duplexes 1 (green) and 3 (magenta) and (C) fA2U2 duplex 1 (green) and the RNA duplex rC4G4 (50) (black). The views are across the major and minor grooves, fluorine atoms of the fA2U2 duplex 1 are shown as yellow spheres and RNA 2′-hydroxyl oxygen atoms are shown as red spheres (C).
Hydration
In light of the negligible conformational differences between RNA and 2′-F-RNA seen in their crystal structures, we turned our attention to the water structure. The hydration of rC4G4 had been analyzed in exceptional detail and at high resolution (1.4 Å) some 15 years ago (50). Striking hydration patterns were detected both in the major and the minor groove. Thus, water molecules linking adjacent O6 keto groups in the center of the major groove created a spine that was only interrupted at the central CpG step (Figure 3A). 2′-Hydroxyl groups served as anchor points for bridges involving two water molecules across the minor groove (Supplementary Figure S1). The latter feature in particular led us to postulate that the significantly more favorable enthalpy and, in turn, the clearly unfavorable entropy of pairing between rC4G4 RNA strands relative to pairing between dC4G4 DNA strands was to a considerable degree the result of the more extensive hydration of the RNA duplex (50). This hypothesis largely ignored the possibility that although rC4G4 obviously adopted an A-form geometry, the DNA duplex dC4G4 of the same sequence was not necessarily locked in the A-form, but could prefer the B-form geometry in solution {in the crystal dC4G4 displays a canonical A-form geometry [(55) and references therein]}.
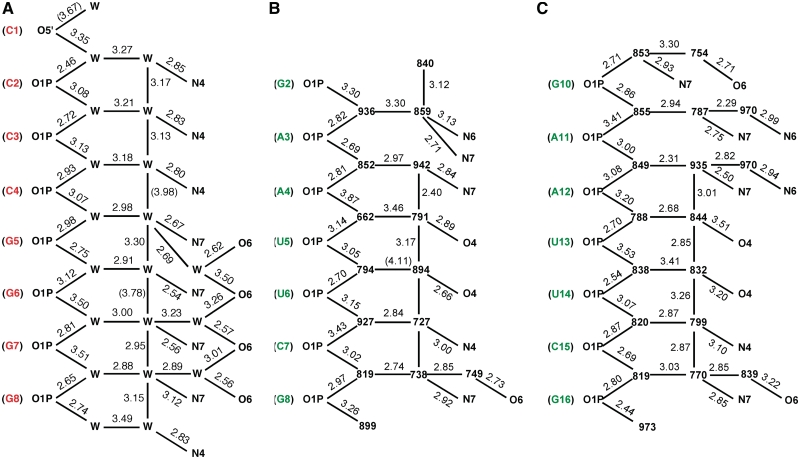
Figure 3.
Major groove hydration pattern in the crystal structures of (A) rC4G4 (50) (one duplex per asymmetric unit; only one strand shown) and (B and C) fA2U2 (three duplexes per asymmetric unit; only the two strands of the first duplex are shown). Residues are highlighted in red (RNA) and green (2′-F-RNA), water molecules are either shown as ‘W’ (A) or with their numbers in the coordinate file (B and C), and hydrogen bonds are indicated with thin solid lines, with distances given in Å. Replacement of the RNA 2′-hydroxyl group by fluorine in the minor groove has no effect on the formation of regular pentagons consisting of a phosphate oxygen and four water molecules along both sides of the major groove. As well this hydration pattern appears to be independent of the nucleotide sequence.
Unlike in the case of DNA and RNA, for which a meaningful correlation between water structure and thermodynamic stability hinges on the assumption of similar duplex geometries, we now know for a fact that the 2′-F-RNA and RNA duplexes are virtually indistinguishable as far as conformational properties are concerned (Figure 2; see also the comparison between solution CD spectra for octamer and dodecamer sequences depicted in Supplementary Figure S2). Because the crystal structures of the reference duplex and the three 2′-F-modified duplexes were all determined at relatively high resolution, most of the first-shell water molecules can be visualized and distances between water and RNA atoms as well as between water molecules can be established with some confidence. Because 2′-fluorine atoms jut out into the minor groove one would not expect the water structure in the major groove of 2′-F-RNA to be significantly affected by the modification. Indeed, an analysis of the hydration patterns in the major groove of the fully 2′-F-modified fA2U2 duplex (Figure 3B and C) reveals close similarities to the major groove hydration pattern in the rC4G4 RNA duplex (Figure 3A). Water molecules coordinated to the edges of nucleobases and those bridging adjacent phosphate groups form regular pentagons that are fused by one side and run down along the edges of the major groove. The only changes in this hydration motif between the rC4G4 and fA2U2 duplexes are directly related to the divergent sequences and not to the replacement of 2′-hydroxyl groups with fluorine in the minor groove. Thus, the zigzag-like hydrogen bonding pattern connecting waters and O6 functions of Gs in both halves of the rC4G4 major groove (Figure 3A) is absent in fA2U2 except for a few hydrogen bonds that link water molecules closer to the edges of the major groove to either O6 of G or N6 of A in some cases (Figure 3B and C). Therefore, it is unlikely that the poorer hydration of 2′-F-RNA relative to RNA evident from the osmotic stress data is a consequence of changes in the major groove water structure.
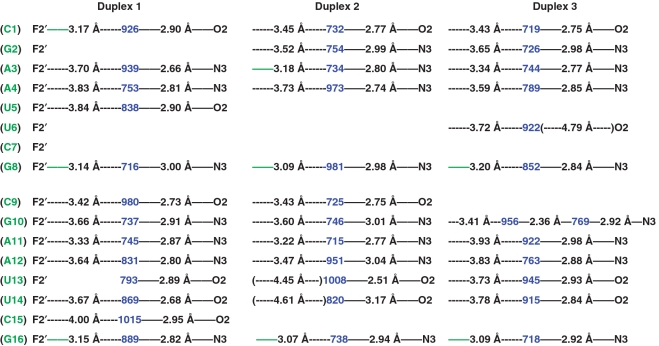
Next we inspected the water structures in the minor groove of the fA2U2, f/rA2U2 and f/rC4G4 duplexes and compared them with the arrangement of water molecules in the rC4G4 minor groove. In the structure of the latter, the 2′-hydroxyl substituents were typically surrounded by three water molecules and the donor and acceptor abilities of the OH moiety were thus satisfied (50). Water molecules not only bridged the 2′-OH groups to the phosphate-sugar backbone, but also to the base functions and a view into the minor groove revealed water tandems that linked 2′-hydroxyls from opposite strands (Supplementary Figure S1). It became clear from this analysis that the hydroxyl groups act as bridgeheads and stitch together backbone and base as well as the pairing strands across the periphery of the minor groove. Thus, water molecules are basically shared between nucleobase acceptors, such as O2 (C and U) or N3 (G or A) and 2′-OH. By comparison, an examination of the distribution of water molecules in the minor groove of the fA2U2 duplex demonstrates that the water molecules associated with base edges with a few exceptions are completely separated from 2′-fluorines (Figure 4). Although the acceptors of most bases (in some cases the pattern is interrupted by lattice interactions) are still hydrated, the seamless backbone-base-base-backbone connection across the minor groove and mediated by 2′-hydroxyl groups is severed as a consequence of the 2′-F modification (Supplementary Figures S1 and Figure 4).
Figure 4.
Minor groove hydration pattern in the crystal structure of fA2U2 (three duplexes per asymmetric unit). 2′-F-RNA residues and water molecules (as numbered in the coordinate file) are highlighted in green and blue, respectively. Hydrogen bonds (≤3.3 Å distance between potential acceptor and donor atoms) are indicated with thin solid lines, with distances given in Å. Distances below 3.3 Å between fluorine atoms and water are highlighted in green. Please note the typically much longer distances (dashed lines in black) between 2′-fluorine atoms and water molecules in the minor groove of 2′-F-RNA compared with distances between nucleobase atoms and water or those between 2′-OH groups and water in RNA (Supplementary Figure S1).
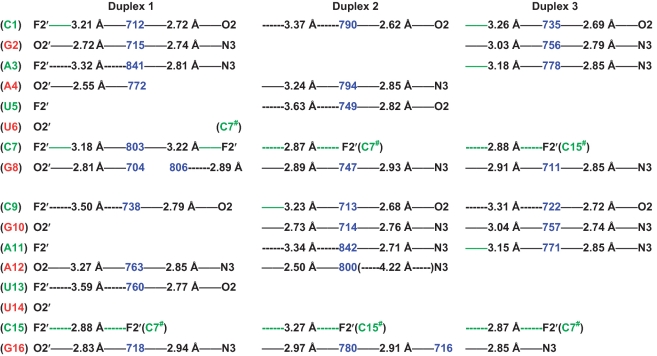
Inspection of the minor groove hydration patterns in the alternating 2′-F/2′-OH duplex f/rA2U2 in two crystal lattices (four individual duplexes; Figure 5 and Supplementary Figure S3) confirms the distance trends for fluorine and hydroxyls seen in the fully modified fA2U2 duplex. The structure of f/rC4G4 is of lower resolution and does not allow a detailed analysis of the minor groove hydration as only relatively few water molecules could be visualized (Supplementary Figure S4). Virtually every 2′-hydroxyl group is engaged in a hydrogen bond to a water molecule that itself H-bonds to either O2 of a pyrimidine or N3 of a purine. The O2′(H)…OH2 [or (H)O2′…H-O-H] distances are mostly well below 3 Å. Conversely, the majority of 2′-fluorines are relatively far removed from water molecules in the minor groove and distances are in the 3.5 Å range in most cases (a third of the interactions display distances ≥3.5 Å; Figures 4 and 5, Supplementary Figure S3). However, there are a few examples of more tightly spaced F2′/water pairs. The shortest of these is found in duplex 2 of the fA2U2 structure (3.07 Å) and there are others that lie in the 3.1–3.3 Å distance range in both the fA2U2 and f/rA2U2-P3 structures (Figures 4 and 5; solid green lines). Nevertheless, the distributions of water molecules seen in the minor grooves of the fA2U2 and f/rA2U2 structures demonstrate that 2′-hydroxyl groups are significantly better hydrated than 2′-fluorines. The superior hydration of RNA relative to 2′-F-RNA revealed by the osmotic stress data may to a large degree be the result of water networks in the minor groove and backbone that require the participation of the 2′-substituent.
Figure 5.
Minor groove hydration pattern in f/rA2U2-P3 (three duplexes per asymmetric unit). 2′-F-RNA and RNA residues are highlighted in green and red, respectively, and water molecules are highlighted in blue. Hydrogen bonds (≤3.3 Å distance between potential acceptor and donor atoms) are indicated with thin solid lines, with distances given in Å. Distances below 3.3 Å between fluorine atoms and water are highlighted in green and van der Waals contacts between fluorine atoms are dashed and green. Please note the typically much longer distances (dashed lines in black) between 2′-fluorine atoms and water molecules compared with distances between 2′-OH groups and water.
In the f/rA2U2-P3 crystal, lattice interactions between adjacent duplexes also include van der Waals contacts between fluorines, the shortest of which measures 2.87 Å (Figure 5). This distance is consistent with a van der Waals radius of fluorine of ca. 1.4 Å (56–59). Therefore, F2′ and OH2 pairs separated by a distance significantly below the sum of their van der Waals radii [ΣvdW = F 1.4)(1.2 H–1.0–O ≈ 3.6 Å], i.e. 3.1 Å, can be considered hydrogen bonded.
DISCUSSION
A recent comparison of the in vitro and in vivo activities of siRNA duplexes with different 2′-sugar modifications in the sense and antisense strands demonstrated the unique properties of 2′-F-RNA (22) and provided the motivation for the detailed thermodynamic, osmotic stress and structural investigations described in the present manuscript. Unlike modifications such as LNA, 2′-O-methyl-RNA (2′-OMe-RNA) and 2′-O-(2-methoxyethyl)-RNA (2′-O-MOE-RNA) that exhibit similar or higher gains in RNA affinity than 2′-F-RNA, the latter modification was tolerated in both strands of siRNA duplexes and lacked immunostimulatory effects. Compared with native siRNAs, their counterparts with all pyrimidine ribonucleotides replaced by 2′-F-modified residues displayed improved efficacy both in vitro and in vivo (22). Interestingly, in the FDA-approved RNA aptamer Macugen® (pegaptanib sodium injection) that selectively antagonizes vascular endothelial growth factor (VEGF) and is used clinically for treating wet age-related macular degeneration (wet AMD), every U and C nucleotide is substituted with the corresponding 2′-F-modified analog (60).
The 2′-F-modification is by no means new—fluorine-modified nucleotides incorporated into antisense oligonucleotides and ribozymes were studied more than 15 years ago (3–6). However, our interest in 2′-F-RNA stems from its recently established favorable properties in RNAi applications (18–20,22) and the fact that the precise origins of the increased RNA affinity afforded by the modification as well as RNA-processing enzymes’ tolerance of it have not been established to date. Thus, our thermodynamic studies based on calorimetry and UV melting and the accompanying osmotic stress experiments have yielded some surprising findings. First, the common assumption that the enhanced RNA affinity displayed by 2′-F-modified RNA is mainly the result of conformational preorganization may not be correct because the differences between the ΔS values measured for RNA, mixed 2′-F-/2′-OH RNA and 2′-F-RNA are very small. Second, despite the similar magnitude of the entropic change, melting of an RNA duplex releases significantly more water molecules then the melting of the corresponding 2′-F-RNA duplex. Third, the gains in duplex stability (Tm, Gibbs free energy) seen with 2′-F-RNA compared to the native species are almost entirely based on a favorable enthalpic contribution, which suggests that the origins of the stabilizing effect of the 2′-F substituent are more complex than just preorganization.
It is puzzling that the entropy components of the Gibbs free energy for RNA and 2′-F-RNA should be so similar. This is because the increased electronegativity of the 2′-fluorine relative to the 2′-oxygen (OH) substituent is expected to render the sugar more rigid in the case of 2′-F-RNA. Provided this frozen-out conformation of the single strand is compatible with the A-form RNA duplex type, it should provide 2′-F-RNA with an entropic advantage. Moreover, the 2′-F-RNA duplex is dryer than the RNA duplex and the immobilization of fewer water molecules inside the grooves and around the backbones in the former should constitute a further advantage in terms of entropy. There is no indication from the crystallographic data that the C3′-endo pucker uniformly adopted by the sugar moieties in the 2′-F-RNA-modified duplexes will differ significantly from the sugar conformation in the single-stranded state. The osmotic stress data provide a measure for the amount of water that is expelled when the two strands separate and show that there is a clear difference between the number of water molecules hydrating RNA and 2′-F modified duplexes. Looking at the structures we note that 2′-fluorines are not well hydrated but 2′-hydroxyls are (Figures 4–6, Supplementary Figures S1 and S3), with the latter supporting water bridges across the minor groove (Supplementary Figure S1) and likely enhancing the ordered water structure around the backbones of RNA duplexes (50). To potentially rationalize the similar entropies, one may conclude that waters bridging the minor groove in RNA are inherently more flexible, thus minimizing the entropic loss as the duplex melts and the difference to 2′-F-RNA where fluorine atoms don't seem to stabilize the minor groove water structure to the same extent (Figures 4 and 5, Supplementary Figure S3). But then the considerably more favorable enthalpy for 2′-F-RNA pairing relative to RNA still begs an explanation. We had previously concluded that the favorable enthalpy and unfavorable entropy of [r(C4G4)]2 duplex formation compared with that of the [d(C4G4)]2 duplex was in part due to the increased hydration of the former: more water → more hydrogen bonds but less mobility → ΔH and ΔS both negative (50). But the case of 2′-F-RNA suggests that this explanation while logical in principal is too simplistic.
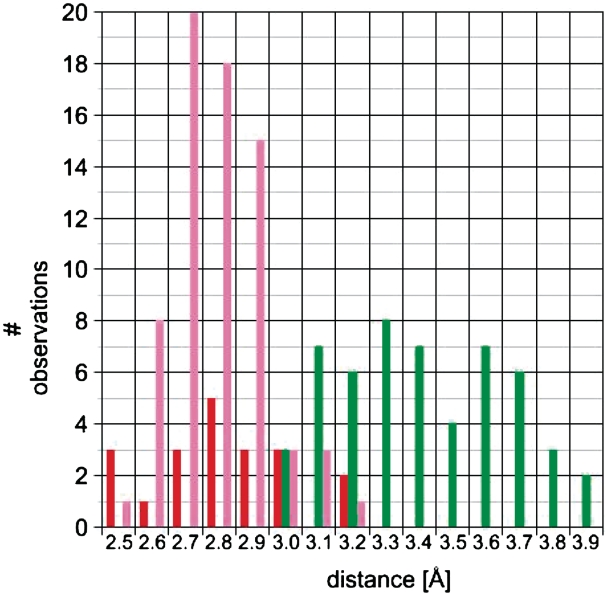
Figure 6.
Distance distribution for water interactions in the minor grooves of fully 2′-F-modified RNA (fA2U2) and mixed 2′-F/-OH RNA (f/rA2U2-P3 and f/rA2U2-R32) duplexes. Color code: green, 2′-F···H2O; red, 2′-OH···H2O and pink, O2(C,U)/ N3(G,A)···H2O.
The above assumptions that reduced hydration at the duplex level and higher rigidity of the sugar and concomitant conformational preorganization are both favorable in terms of entropy are not unreasonable. Unlike in the case of the collagen triple helix where the higher stability as a result of the presence of 4-hydroxyproline is no longer attributed to enhanced hydrogen bonding and hydration but to stereoelectronic effects of the substituent as a result of studies with collagen containing 4-fluoroproline (61), the effects of hydration on nucleic acid duplex stability and conformation are well established. However, other consequences of the substitution of the 2′-hydroxyl group with fluorine could offset the relative contributions of ΔH and ΔS to the Gibbs free energy. It is possible that the F2′ substituent is able to influence pairing strength not simply by conformationally preorganizing the single strand, but by actually modulating the electronic state of the nucleobase more significantly than anticipated hitherto. Thus, the strongly electronegative and poorly polarizable fluorine could increase the Watson–Crick (W–C) hydrogen bonding strengths in the modified duplex relative to RNA and possibly also improve stacking. There is experimental evidence based on NMR for the increased strength of W–C hydrogen bonds in RNA relative to DNA (62–64). Fluorine will certainly polarize the base more significantly than the natural 2′-hydroxyl substituent and hence further strengthen W–C hydrogen bonds. At the same time, polarization of the nuclebase could favorably affect stacking (65–67) and together these effects may explain the enthalpic benefits of the 2′-fluoro modification. Obviously, there is a trade-off in terms of ΔH and ΔS, and gains in enthalpy would likely trigger a loss in entropy. If this change in ΔS roughly neutralizes the aforementioned changes in ΔS due to altered hydration and conformational preorganization, the overall entropic changes associated with duplex melting may turn out to be similar for RNA and 2′-F-RNA as indicated by our experimental data.
The analysis of the crystallographic environments of fluorine in the structures of the fully modified fA2U2 duplex and the f/rA2U2 duplex with alternating 2′-F-modified and 2′-OH nucleotides in two space groups at high resolution demonstrates that fluorine only acts as a hydrogen bond acceptor in a few cases, in line with an assessment by others (68). Unlike in previous investigations of the ability of fluorine to participate in hydrogen bonds that typically focused on one or a few atoms per structure, our crystal structures contain a much larger number of fluorines per asymmetric unit (48 in the case of fA2U2). Several of the interactions with water molecules by fluorine atoms are consistent with the formation of a hydrogen bond as judged by F···O distances significantly below 3.5 Å (Figures 4–6). But such interactions are hardly more common than van der Waals contacts between fluorine atoms from neighboring duplexes. The wide and shallow minor groove that is exposed to solvent in many places probably does not promote the formation of hydrogen bonds in which fluorine acts as an acceptor. Conversely, the interior of a duplex (69,70) or an enzyme active site (71) may be more conducive to fluorine engaging in a hydrogen bond.
Fluorine has a number of unique attributes that render it a promising modification in siRNAs [(22) and references therein]. At the 2′-ribose position, fluorine increases the RNA affinity of modified siRNA guide strands for their targets and enhances the stability of siRNA duplexes. However, the gains in affinity and pairing strength are not detrimental to the function of siRNAs and their interactions with proteins participating in the RNAi pathway. Thus, the modification is tolerated in both strands and does not apparently interfere with auxiliary factors that unwind the duplex, load the guide strand into the RISC or the Ago2 slicer that cleaves the mRNA target. The small size and chimeric nature of fluorine—neither hydrophobic nor hydrophilic—are likely at the roots of the remarkable lack of adverse consequences of this modification. Unlike more bulky 2′-modifications such as LNA, 2′-OMe-RNA or 2′-O-MOE-RNA that interfere with protein binding in the minor groove (22), fluorine atoms lining the oligonucleotide backbone seem to be remarkably good mimics of the ribose 2′-hydroxyl group. The 2′-RNA analog that we analyzed here is not the only fluorine modification that was found to be tolerated by the RNAi machinery, even when positioned adjacent to the Ago2 mRNA cleavage site. Thus, substitutions of one or two thymidines by the isostere ribo-2,4-difluorotoluene neither significantly affected the siRNA affinity of modified strands nor did they alter their structural properties in a major way (72,73).
ACCESSION NUMBERS
PDB ID: 3P4A, 3P4B, 3P4C, 3P4D.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US National Institutes of Health (R01 GM055237 to M.E. and R01 GM071461 to E.R.); US Department of Energy, Basic Energy Sciences, Office of Science, APS (under Contract No. W-31-109-Eng-38). Funding for open access charge: US NIH (grant R01 GM055237).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Emma Schweizer for initial osmotic stress experiments. Vanderbilt University is a member institution of LS-CAT at the Advanced Photon Source (Argonne, IL).
REFERENCES
- 1.Strunecká A, Patocčka J, Connett P. Fluorine in medicine. J. Appl. Biomed. 2004;2:141–150. [Google Scholar]
- 2.Hagmann WK. The many roles for fluorine in medicinal chemistry. J. Med. Chem. 2008;51:4359–4369. doi: 10.1021/jm800219f. [DOI] [PubMed] [Google Scholar]
- 3.Pieken WA, Olsen DB, Benseler F, Aurup H, Eckstein F. Kinetic characterization of ribonuclease-resistant 2′-modified hammerhead ribozymes. Science. 1991;253:314–317. doi: 10.1126/science.1857967. [DOI] [PubMed] [Google Scholar]
- 4.Williams DM, Benseler F, Eckstein F. Properties of 2′-fluorothymidine-containing oligonucleotides: interaction with restriction endonuclease EcoRV. Biochemistry. 1991;30:4001–4009. doi: 10.1021/bi00230a027. [DOI] [PubMed] [Google Scholar]
- 5.Kawasaki AM, Casper M, Freier SM, Lesnik EA, Zounes MC, Cummins LL, Gonzalez C, Cook PD. Uniformly modified 2′-deoxy-2′-fluoro-phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J. Med. Chem. 1993;36:831–841. doi: 10.1021/jm00059a007. [DOI] [PubMed] [Google Scholar]
- 6.Monia BP, Lesnik EA, Gonzalez C, Lima WF, McGee D, Guinosso CJ, Kawasaki AM, Cook PD, Freier SM. Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem. 1993;268:14514–14522. [PubMed] [Google Scholar]
- 7.Pratt AJ, MacRae IJ. The RNA-induced silencing complex: a versatile gene-silencing machine. J. Biol. Chem. 2009;284:17897–17901. doi: 10.1074/jbc.R900012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deleavey GF, Watts JK, Damha MJ. Chemical modification of siRNA. Curr. Protoc. Nucleic Acid Chem. 2009;39:16.3.1–16.3.22. doi: 10.1002/0471142700.nc1603s39. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 10.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 11.Braasch DA, Jensen S, Liu Y, Kaur K, Arar K, White MA, Corey DR. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- 12.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harborth J, Elbashir SM, Vandenburgh K, Manninga H, Scaringe SA, Weber K, Tuschl T. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 2003;132:83–105. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- 14.Prakash TP, Allerson CP, Dande P, Vickers TA, Sioufi N, Jarres R, Baker BF, Swayze EE, Griffey RH, Bhat B. Competition for RISC binding predicts in vitro potency of siRNA. J. Med. Chem. 2005;48:4247–4253. doi: 10.1021/jm050044o. [DOI] [PubMed] [Google Scholar]
- 15.Muhonen P, Tennilä T, Azhayeva E, Parthasarathy RN, Janckila AJ, Väänänen HK, Azhayev A, Laitala-Leinonen T. RNA interference tolerates 2′-fluoro-modifications at the Argonaute2 cleavage site. Chem. Biodivers. 2007;4:858–873. doi: 10.1002/cbdv.200790073. [DOI] [PubMed] [Google Scholar]
- 16.Layzer JM, McCaffrey AP, Tanner AK, Huang Z, Kay MA, Sullenger BA. In vivo activity of nuclease-resistant siRNAs. RNA. 2004;10:766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrissey DV, Blanchard K, Shaw L, Jensen K, Lockridge JA, Dickinson B, McSwiggen JA, Vargeese C, Bowman K, Shaffer CS, et al. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology. 2005;41:1349–1356. doi: 10.1002/hep.20702. [DOI] [PubMed] [Google Scholar]
- 18.Allerson CR, Sioufi N, Jarres R, Prakash TP, Naik N, Berdeja A, Wanders L, Griffey RH, Swayze EE, Bhat B. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- 19.Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 20.Deleavey GF, Watts JK, Alain T, Robert F, Kalota A, Aishwarya V, Pelletier J, Gewirtz AM, Sonenberg N, Damha MJ. Synergistic effects between analogs of DNA and RNA improve the potency of siRNA-mediated gene silencing. Nucleic Acids Res. 2010;38:4547–4557. doi: 10.1093/nar/gkq181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viel T, Boisgard R, Kuhnast B, Jego B, Siquier-Pernet K, Hinnen F, Dolle F, Tavitian B. Molecular imaging study on in vivo distribution and pharmacokinetics of modified small interfering RNAs (siRNAs) Oligonucleotides. 2008;18:201–212. doi: 10.1089/oli.2008.0133. [DOI] [PubMed] [Google Scholar]
- 22.Manoharan M, Akinc A, Pandey RK, Qin J, Hadwiger P, John M, Mills K, Charisse K, Maier MA, Nechev L, et al. Unique gene-silencing and structural properties of 2′-F-modified siRNAs. Angew. Chem. 2010 doi: 10.1002/anie.201006519. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konigsberg W, Kirchhofer D, Riederer MA, Nemerson Y. The TF:VIIa complex: clinical significance, structure-function relationships and its role in signaling and metastasis. Thromb. Haemost. 2002;86:757–771. [PubMed] [Google Scholar]
- 24.Mousa SA. Tissue factor VIIa in thrombosis and cancer. Methods Mol. Med. 2004;93:119–132. doi: 10.1385/1-59259-658-4:119. [DOI] [PubMed] [Google Scholar]
- 25.Hembrough TA, Swartz GM, Papathanassiu A, Vlasuk GP, Rote WE, Green SJ, Pribluda VS. Tissue factor VIIa inhibitors block angiogenesis and tumor growth through a nonhemostatic mechanism. Cancer Res. 2003;63:2997–3000. [PubMed] [Google Scholar]
- 26.Amirkhosravi A, Meyer T, Amaya M, Davila M, Mousa SA, Robson T, Francis JL. The role of tissue factor pathway inhibitor in tumor growth and metastasis. Semin. Thromb. Hemost. 2007;33:643–652. doi: 10.1055/s-2007-991531. [DOI] [PubMed] [Google Scholar]
- 27.Zhao J, Aguilar G, Palencia S, Newton E, Abo A. rNAPc2 inhibits colorectal cancer in mice through tissue factor. Clin. Cancer Res. 2009;15:208–216. doi: 10.1158/1078-0432.CCR-08-0407. [DOI] [PubMed] [Google Scholar]
- 28.Koller E, Propp S, Murra H, Lima W, Bhat B, Prakash TP, Allerson CR, Swayze EE, Marcusson EG, Dean NM. Competition for RISC binding predicts in vitro potency of siRNA. Nucleic Acids Res. 2006;34:4467–4476. doi: 10.1093/nar/gkl589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dande P, Prakash TP, Siouf N, Gaus H, Jarre R, Berdeja A, Swayze EE, Griffey RH, Bhat B. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 2006;49:1624–1634. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- 30.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, Bacallado SA, Nguyen DN, Fuller J, Alvarez R, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nature Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puglisi JD, Tinoco I., Jr Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- 32.Breslauer KJ. Extracting thermodynamic data from equilibrium melting curves for oligonucleotide order-disorder transitions. Methods Enzymol. 1995;259:221–242. doi: 10.1016/0076-6879(95)59046-3. [DOI] [PubMed] [Google Scholar]
- 33.Rozners E, Moulder J. Hydration of short DNA, RNA, and 2′-OMe oligonucleotides determined by osmotic stressing. Nucleic Acids Res. 2004;32:248–254. doi: 10.1093/nar/gkh175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spink CH, Chaires JB. Effects of hydration, ion release, and excluded volume on the melting of triplex and duplex DNA. Biochemistry. 1999;38:496–508. doi: 10.1021/bi9820154. [DOI] [PubMed] [Google Scholar]
- 35.Kolarovic A, Schweizer E, Greene E, Gironda M, Pallan PS, Egli M, Rozners E. Interplay of structure, hydration and thermal stability in formacetal modified oligonucleotides: RNA may tolerate nonionic modifications better than DNA. J. Am. Chem. Soc. 2009;131:14932–14937. doi: 10.1021/ja904926e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li F, Pallan PS, Maier MA, Rajeev KG, Mathieu SL, Kreutz C, Fan Y, Sanghvi J, Micura R, Rozners E, et al. Crystal structure, stability and in vitro RNAi activity of oligoribonucleotides containing the ribo-difluorotoluyl nucleotide: insights into substrate requirements by the human RISC Ago2 enzyme. Nucleic Acids Res. 2007;35:6424–6438. doi: 10.1093/nar/gkm664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rozners E, Smicius R, Uchiyama C. Expanding functionality of RNA: synthesis and properties of RNA containing imidazole modified tandem G-U wobble base pairs. Chem. Commun. 2005:5778–5780. doi: 10.1039/b510846b. [DOI] [PubMed] [Google Scholar]
- 38.Rozners E. Determination of nucleic acid hydration using osmotic stress. Curr. Protoc. Nucleic Acid Chem. 2010:7.14.1–7.4.13. doi: 10.1002/0471142700.nc0714s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berger I, Kang CH, Sinha N, Wolters M, Rich A. A highly efficient 24-condition matrix for the crystallization of nucleic acid fragments. Acta Cryst. D. 1996;52:465–468. doi: 10.1107/s0907444995013564. [DOI] [PubMed] [Google Scholar]
- 40.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 41.Pape T, Schneider TR. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Cryst. 2004;37:843–844. [Google Scholar]
- 42.Cambillau C, Roussel A. Turbo Frodo, Version OpenGL.1. Marseille, France: Université Aix-Marseille II; 1997. [Google Scholar]
- 43.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Cryst. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 44.Sheldrick GM, Schneider TR. SHELXL: high resolution refinement. Meth. Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 45.Brünger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 46.Lavery R, Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- 47.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 48.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Cryst. D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 49.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 50.Egli M, Portmann S, Usman N. RNA hydration: a detailed look. Biochemistry. 1996;35:8489–8494. doi: 10.1021/bi9607214. [DOI] [PubMed] [Google Scholar]
- 51.Egli M. Structural aspects of nucleic acid analogs and antisense oligonucleotides. Angew. Chem. Int. Ed. 1996;35:1894–1909. [Google Scholar]
- 52.Timasheff SN. In disperse solution, ‘‘osmotic stress’’ is a restricted case of preferential interactions. Proc. Natl Acad. Sci. USA. 1998;95:7363–7367. doi: 10.1073/pnas.95.13.7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Courtenay ES, Capp MW, Anderson CF, Record MT., Jr Vapor pressure osmometry studies of osmolyte-protein interactions: implications for the action of osmoprotectants in vivo and for the interpretation of ‘‘osmotic stress’’ experiments in vitro. Biochemistry. 2000;39:4455–4471. doi: 10.1021/bi992887l. [DOI] [PubMed] [Google Scholar]
- 54.Auffinger P, Westhof E. Hydration of RNA base pairs. J. Biomol. Struct. Dyn. 1998;16:693–707. doi: 10.1080/07391102.1998.10508281. [DOI] [PubMed] [Google Scholar]
- 55.Portmann S, Usman N, Egli M. The crystal structure of r(CCCCGGGG) in two distinct lattices. Biochemistry. 1995;34:7569–7575. doi: 10.1021/bi00023a002. [DOI] [PubMed] [Google Scholar]
- 56.Pauling L. The Nature of the Chemical Bond. 2nd edn. 1939. Chapter 5. Cornell University Press, Ithaca, N.Y. [Google Scholar]
- 57.Bondi A. van der Waals volumes and radii. J. Phys. Chem. 1964;68:441–451. [Google Scholar]
- 58.Kitaigorodsky AI. Molecular Crystals and Molecules. 1973. Chapter 1. Academic Press, New York. [Google Scholar]
- 59.Dunitz JD, Schweizer WB. Molecular pair analysis: C-H.F interactions in the crystal structure of fluorobenzene? And related matters. Chem. Europ. J. 2006;12:6804–6815. doi: 10.1002/chem.200600092. [DOI] [PubMed] [Google Scholar]
- 60.Gragoudas ES, Adamis AP, Cunningham ET, Jr, Feinsod M, Guyer DR. Pegaptanib for neovascular age-related macular degeneration. N. Engl. J. Med. 2004;351:2805–2816. doi: 10.1056/NEJMoa042760. [DOI] [PubMed] [Google Scholar]
- 61.Raines RT. 2005 Award address, Emil Thomas Kaiser award. Protein Sci. 2006;15:1219–1225. doi: 10.1110/ps.062139406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vakonakis I, LiWang AC. N1···N3 hydrogen bonds of A:U base pairs of RNA are stronger than those of A:T base pairs of DNA. J. Am. Chem. Soc. 2004;126:5688–5689. doi: 10.1021/ja048981t. [DOI] [PubMed] [Google Scholar]
- 63.Manalo MN, Kong X, LiWang AC. 1JNH values show that N1…N3 hydrogen bonds are stronger in dsRNA A:U than dsDNA A:T base pairs. J. Am. Chem. Soc. 2005;127:17974–17975. doi: 10.1021/ja055826l. [DOI] [PubMed] [Google Scholar]
- 64.Swart M, Fonseca Guerra C, Bickelhaupt FM. Hydrogen bonds of RNA are stronger than those of DNA, but NMR monitors only presence of methyl substituent in uracil/thymine. J. Am. Chem. Soc. 2004;126:16718–16719. doi: 10.1021/ja045276b. [DOI] [PubMed] [Google Scholar]
- 65.Guckian KM, Schweitzer BA, Ren RX-F, Sheils CJ, Paris PL, Tahmassebi DC, Kool ET. Experimental measurement of aromatic stacking affinities in the context of duplex DNA. J. Am. Chem. Soc. 1996;118:8182–8183. doi: 10.1021/ja961733f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guckian KM, Schweitzer BA, Ren RX-F, Sheils CJ, Tahmassebi DC, Kool ET. Factors contributing to aromatic stacking in water: evaluation in the context of DNA. J. Am. Chem. Soc. 2000;122:2213–2222. doi: 10.1021/ja9934854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manalo MN, Pérez LM, LiWang A. Hydrogen bonding and π-π base-stacking interactions are coupled in DNA, as suggested by calculated and experimental trans-Hbond deuterium isotope shifts. J. Am. Chem. Soc. 2007;129:11298–11299. doi: 10.1021/ja0692940. [DOI] [PubMed] [Google Scholar]
- 68.Dunitz JD. Organic fluorine: odd man out. Chembiochem. 2004;5:614–621. doi: 10.1002/cbic.200300801. [DOI] [PubMed] [Google Scholar]
- 69.Pallan PS, Egli M. The pairing geometry of the hydrophobic thymine analog 2,4-difluorotoluene in duplex DNA as analyzed by X-ray crystallography. J. Am. Chem. Soc. 2009;131:12548–12549. doi: 10.1021/ja905739j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li F, Sarkhel S, Wilds CJ, Wawrzak Z, Prakash TP, Manoharan M, Egli M. 2′-Fluoroarabino- and arabinonucleic acid show different conformations, resulting in deviating RNA affinities and processing of their heteroduplexes with RNA by RNase H. Biochemistry. 2006;45:4141–4152. doi: 10.1021/bi052322r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Irimia A, Eoff RL, Pallan PS, Guengerich FP, Egli M. Structure and activity of Y-class DNA polymerase Dpo4 from Sulfolobus solfataricus with templates containing the hydrophobic thymine analog 2,4-difluorotoluene. J. Biol. Chem. 2007;282:36421–36433. doi: 10.1074/jbc.M707267200. [DOI] [PubMed] [Google Scholar]
- 72.Xia J, Noronha A, Toudjarska I, Li F, Akinc A, Braich R, Rajeev KG, Egli M, Manoharan M. siRNAs with a ribo-difluorotoluyl nucleotide: structure, RISC-mediated recognition, and silencing. ACS Chem. Biol. 2006;1:176–183. doi: 10.1021/cb600063p. [DOI] [PubMed] [Google Scholar]
- 73.Li F, Pallan PS, Maier MA, Rajeev KG, Mathieu SL, Kreutz C, Fan Y, Sanghvi J, Micura R, Rozners E, et al. Crystal structure, stability and in vitro RNAi activity of oligoribonucleotides containing the ribo-difluorotoluyl nucleotide: insights into substrate requirements by the human RISC Ago2 enzyme. Nucleic Acids Res. 2007;35:6424–6438. doi: 10.1093/nar/gkm664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.