Abstract
Nucleosomes are multi-component macromolecular assemblies which present a formidable obstacle to enzymatic activities that require access to the DNA, e.g. DNA and RNA polymerases. The mechanism and pathway(s) by which nucleosomes disassemble to allow DNA access are not well understood. Here we present evidence from single molecule FRET experiments for a previously uncharacterized intermediate structural state before H2A–H2B dimer release, which is characterized by an increased distance between H2B and the nucleosomal dyad. This suggests that the first step in nucleosome disassembly is the opening of the (H3–H4)2 tetramer/(H2A–H2B) dimer interface, followed by H2A–H2B dimer release from the DNA and, lastly, (H3–H4)2 tetramer removal. We estimate that the open intermediate state is populated at 0.2–3% under physiological conditions. This finding could have significant in vivo implications for factor-mediated histone removal and exchange, as well as for regulating DNA accessibility to the transcription and replication machinery.
INTRODUCTION
Chromatin is the large nucleo-protein complex which folds the eukaryotic genome in the nucleus. Nucleosomes, its basic repeating unit, consist of 147 bp of DNA wrapped around an octameric histone core: a modular complex of two H2A–H2B dimers and a (H3–H4)2 tetramer (1,2).
Nucleosome assembly and disassembly profoundly affects all nuclear processes that require DNA as a template, such as transcription, replication, and repair. Various studies have probed how nucleosomes are formed from their components, and how they dissociate. In vivo, nucleosomes are assembled by chromatin assembly factors and histone chaperones, which together orchestrate the ordered deposition of histones as well as prevent improper interactions between histones and DNA (3). Nucleosome disassembly is promoted by ATP-dependent chromatin remodelers and is likely aided by histone chaperones through largely unknown mechanisms.
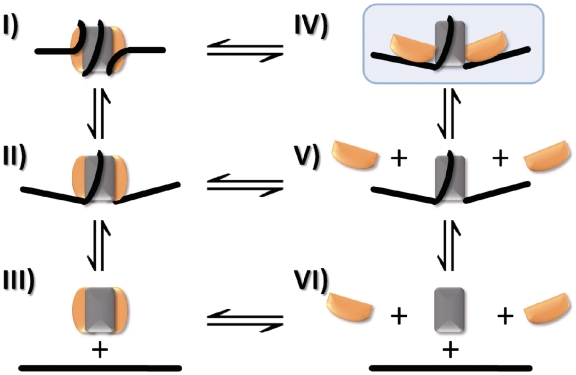
In vitro, buffer salt composition modulates the electrostatic interactions of the nucleosome and has been used to mimic the activity of various factors in vivo (4). Nucleosomes can be assembled reliably by combining DNA and histone octamer at high salt and then decreasing the salt concentration sequentially from 2 M to physiological conditions (5). Conversely, nucleosomes can be disassembled by increasing salt concentration. While (H3–H4)2 deposition has been shown to start nucleosome assembly, followed by H2A–H2B association (5), the mechanism of [NaCl] induced nucleosome disassembly is not known precisely. At least two pathways are discussed: a sequential mechanism, starting with dissociation of H2A–H2B followed by dissociation of (H3–H4)2 (state I→V→VI, Figure 1) (6–9), or dissociation of the histone octamer as a whole (state I→III→VI, Figure 1) (10–13). This latter model forms the basis for many computational studies of nucleosome positioning (14,15). Irrespective of the pathway, information about (dis)assembly intermediates—which may exhibit unique structural properties with profound implications for the in vivo function of chromatin—remains missing.
Figure 1.
Pathways for nucleosome (dis)assembly. A reaction mechanism depicting possible intermediate steps in the transition between nucleosome (I) and free DNA, H2A–H2B dimer and (H3–H4)2 tetramer (VI). DNA breathing (II), as first described by Widom and colleagues (38) increases the accessibility of DNA to protein binding. Disassembly has been proposed to occur either through a sequential mechanism (I→V→VI) or a one-step release of the histone octamer (I→III→VI). State IV (shaded in blue) represents a previously undetected open state in which the interactions between the H2A–H2B dimer and (H3–H4)2 tetramer are partially lost.
Recently, much progress has been made toward analyzing nucleosome conformation and dynamics using fluorescence techniques, specifically Förster resonance energy transfer (FRET) (16,17). This approach is particularly powerful when combined with single molecule techniques, allowing one to observe subpopulations of heterogeneous samples (18–21). Additionally, diffusion properties and conformational fluctuations of nucleosomes have been studied by fluorescence correlation spectroscopy (FCS) (20). Our own previous single pair FRET (spFRET) studies on DNA-labeled nucleosomes have provided evidence for stepwise disassembly, from which we inferred that H2A–H2B might first dissociate from the nucleosome, followed ultimately by complete dissociation into unbound proteins and DNA (22).
Here, we probe the conformational changes that occur during salt-induced assembly and disassembly of nucleosomes by spFRET analysis and FCS. Our results show unambiguously that H2A–H2B dissociates before (H3–H4)2, eliminating a disassembly pathway in which the histone octamer is removed in one step. We provide evidence for a previously uncharacterized first step in nucleosome disassembly where the distance between H2B and the nucleosomal dyad is increased, suggesting that the interface between the (H3–H4)2 tetramer and H2A–H2B dimers opens up before H2A–H2B dissociation from the DNA. Nucleosome assembly follows the reverse pathway with the same intermediates.
MATERIALS AND METHODS
Fluorescently labeled nucleosomes
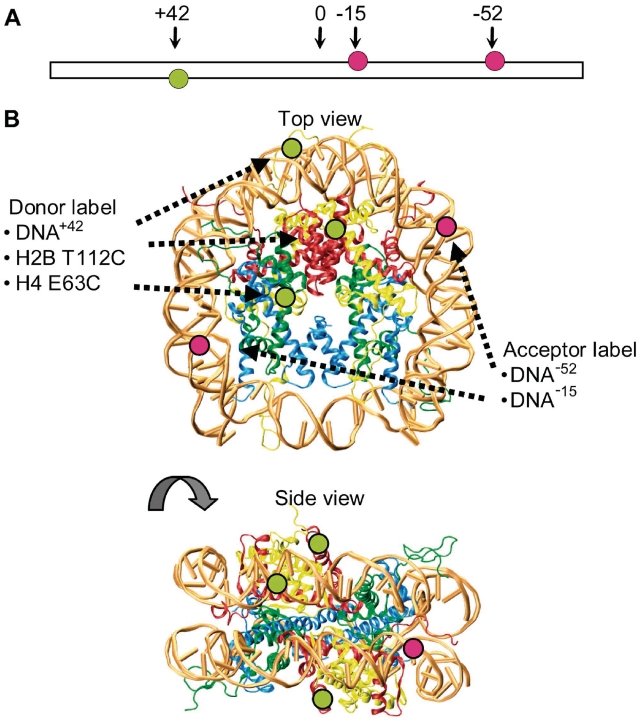
Mononucleosomes were reconstituted by salt dialysis from labeled DNA fragments containing the Systematic Evolution of Ligands by EXponential enrichment (SELEX) generated 601 positioning sequence (170 bp unless stated otherwise) and unlabeled or labeled recombinant Xenopus laevis histone octamers. DNA was labeled via a thymine with C6-linker at −52 or −15 bp (alexa 594) and +42 bp (alexa 488) from the positioning center (Figure 2A and B). Histones were separately expressed and purified as detailed in ref. (23) and labeled prior octamer refolding using histone mutants H2B T112C (9) and H4 E63C (24) and alexa 488-maleimide (Figure 2B). To minimize the occurrence of octamers carrying two donor fluorophores, we purposely labeled only 10% of the histones. The full details of the sample preparation are given in Supplementary Data 1.1.
Figure 2.
Fluorescently labeled nucleosomes. (A) Illustration of the extended 170 bp nucleosomal DNA showing the positions of the donor fluorophore alexa 488 (green circles) and acceptor fluorophores alexa 594 (red circles) relative to the nucleosome dyad axis. (B) Top and side views of the nucleosome crystal structure for visualization of fluorophore positions and FRET distances. H2A is shown in yellow, H2B in red, H3 in blue, H4 in green. The fluorophores are depicted with green and red circles corresponding to donor (alexa 488) and acceptor (alexa 594). Depending on perspective, some fluorophore positions are hidden and therefore not labeled in the crystal structure. In the top view, only fluorophores on one of the two H2B and on one of the two H4 are marked. In the side view, the fluorophore on position −15 on the DNA and the fluorophores on both H4 are not marked.
Single molecule fluorescence spectroscopy
Experiments were carried out using a confocal microscope (18) illuminated with a continuous wave Ar/Kr laser (Melles Griot, Darmstadt, Germany) at 488 nm for excitation. Fluorescence emission was split in two spectral detection windows for donor and acceptor defined by appropriate filters and collected from two avalanche photodiodes (APD, Perkin Elmer Optoelectronics). In FCS experiments, the signal from the detectors was read out by an ALV5000/E autocorrelator (ALV GmbH, Langen, Germany) and autocorrelation functions were fitted as described in ref. (25). In spFRET the signal was read out by a TCSPC board (TimeHarp200, Picoquant GmbH, Berlin, Germany). Software developed in house was used to extract single molecule events and calculate the proximity ratio P(t). P(t) is closely related to the FRET efficiency and provides information on the distance between fluorophores, following Equation (1):
| (1) |
(for detailed description of the setup, the experimental procedure and data evaluation see Supplementary Data 1.2 and 1.3).
RESULTS
FCS reveals that histone complexes are released in a stepwise manner during salt-dependent nucleosome disassembly
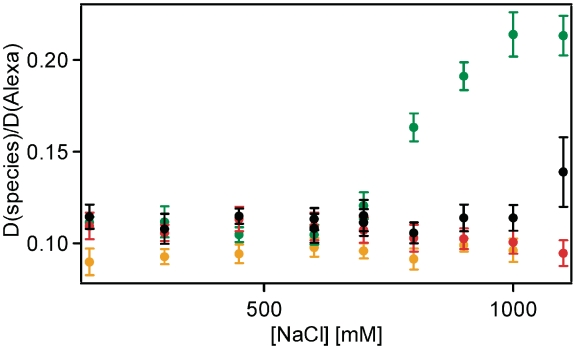
Initially, we studied subunit dissociation during salt induced nucleosome disassembly by measuring the diffusion coefficient D of fluorescently labeled nucleosomes with FCS, as a measure for their overall size and shape. Dissociation of a fluorescent subunit from a large complex can be detected by the increase of its diffusion coefficient. We prepared nucleosomes fluorescently labeled with alexa 488 on either the (H2A–H2B) dimer (H2B T112C), (H3–H4)2 tetramer (H4 E63C), or the DNA (position +42 from the dyad; Figure 2A and B; see Supplementary Data 2.1 for characterization of the constructs). We then measured the diffusion coefficients of these nucleosomes at increasing salt concentrations by FCS.
For DNA-labeled samples, the value of D started to decrease slightly at elevated ionic strength (800 mM) (Figure 3, red symbols), until it approached that of free DNA, which is lower than that of compact nucleosomes because of its extended shape, and in agreement with disassembly. The D-value for H2B-labeled samples increased above 600 mM NaCl, reaching a plateau at 1000 mM NaCl (Figure 3, green symbols). In Supplementary Data 2.3, we identify the diffusing particle at 1100 mM as free fluorescent H2A–H2B. We also show evidence that by increasing and re-decreasing [NaCl] we can induce H2A–H2B exchange between nucleosomes (Supplementary Data 2.4), verifying its dissociation. For H4-labeled samples (Figure 3, black symbols), the D-value increased only at 1100 mM NaCl, indicating (H3–H4)2 release from the DNA at higher [NaCl] than H2A–H2B.
Figure 3.
(H2A–H2B) dimers dissociate before (H3–H4)2 tetramer as shown by FCS. Diffusion coefficients of H2B-, H4- and DNA-labeled nucleosome samples (green, black and red, respectively) and labeled DNA (orange) relative to a standard (alexa 488) as a function of [NaCl] (error bars represent standard deviation from 10 measurements each). The sequential increase of the diffusion coefficients of H2B- and H4-labeled nucleosome samples indicates that H2A–H2B dissociates from the nucleosomal complex at lower [NaCl] than (H3–H4)2.
Nucleosome disassembly intermediates revealed by spFRET: octamer opening precedes H2A–H2B dimer dissociation
The FCS data demonstrate that H2A–H2B dissociates from the nucleosome at lower salt concentrations than (H3–H4)2, corresponding to the mechanism in Figure 1 (state I→V→VI). In order to obtain a more detailed view of nucleosome disassembly, we measured spFRET between different sites within a single nucleosome. Since the amount of energy transferred between a donor and acceptor fluorophore decreases with distance, the judicious placement of donor–acceptor pairs to histones and DNA allows us to monitor the structural changes induced by increasing [NaCl]. We used the following five FRET pairs: H2B T112C and position −52 on DNA close to the nucleosome entry site (H2B–DNA−52); H2B T112C and position −15 on DNA close to the nucleosome dyad axis (H2B–DNA−15); H4 E63C and position −15 on DNA (H4–DNA−15); H4 E63C and position −52 on DNA (H4–DNA−52); and two internal positions on the DNA (+42 and −52) that come into in close proximity if the DNA is wrapped around the octamer (DNA+42–DNA−52) (Figure 2A and B). Due to the presence of two identical copies of every histone within the octamer, specific labeling of one histone was not possible. We purposely labeled only 10% of the histones (using the donor fluorophore), thereby minimizing the presence of two labeled histones on one nucleosome (Supplementary Data 1.1). The acceptor fluorophore was placed on the DNA where we could ensure close to 100% labeling at one specific site. Position −52 was selected because it would best represent any internal structural changes within the nucleosome and prevented confusion of our results with those from DNA end ‘breathing’ as proposed in (20,26,27).
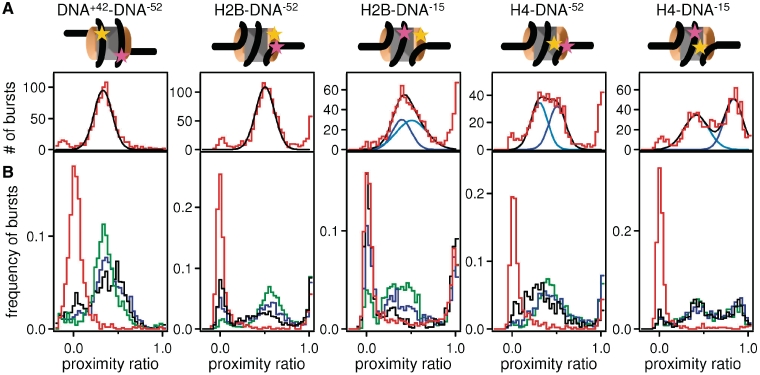
As a measure of energy transfer efficiency E, we determined the proximity ratio P, which is directly related to E and therefore to the interfluorophore distance (Supplementary Data 1.2 for further detail). We measured photon bursts from single nucleosomes, calculated P for each burst and analyzed its distribution in histograms (Figure 4A, example histograms for all constructs). In most cases, three populations could be distinguished (Supplementary Figure S2). Bursts with intermediate P-values, referred to as FRET population, correspond to nucleosomes where the dyes are close to one another, such that energy transfer can occur. The peak centered at 0 (NoFRET population) arises from nucleosomes—or their dissociation products—that show no energy transfer. The peak centered at 1 arises from acceptor-only labeled nucleosomes that are excited directly and were neglected during further analysis (see sample labeling strategy Supplementary Data 1.2).
Figure 4.
spFRET can be used to measure intranucleosomal conformational changes. (A) Example of a proximity ratio distribution for DNA+42–DNA−52, H2B–DNA−52, H2B–DNA−15, H4–DNA−52 and H4–DNA−15 at 150 mM NaCl. Cartoons of nucleosomes indicate the relative locations of labels on the nucleosome. The FRET population represents intact nucleosomes. The number of NoFRET bursts is low. The fit of FRET population is shown in black, fits for subpopulations of the FRET fraction (if present) are shown in different shades of blue. In intact nucleosomes, subpopulations arise if the distances between the fluorophore on the DNA and the fluorophores on the two copies of the labeled histones differ (sSupplementary Data 1.2). (B) Examples of proximity ratio distributions for each construct (as above) at 400 mM (green), 600 mM (blue), 800 mM (black) and 1200 mM (red) NaCl. Elevated [NaCl] causes an increase of the NoFRET population at the cost of the FRET population indicating that the distance between the fluorophores increased above the distance for energy transfer. Other than H4–DNA−15, all constructs displayed changes in the proximity ratio distribution of the FRET population upon increase of [NaCl] which are related to structural changes within the nucleosome.
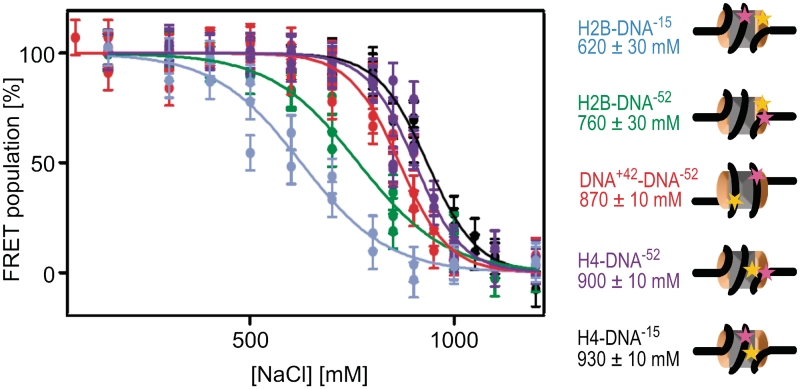
At 150 mM NaCl, the FRET population represents intact nucleosomes. At elevated [NaCl], the NoFRET population increases at the expense of the FRET population for all nucleosomes tested (Figure 4B). Photophysical artifacts at elevated [NaCl] could be excluded (Supplementary Data, section 2.1). Quantifying the NoFRET and FRET-fractions for each experiment (Supplementary Material 1.2) showed a sigmoidal behavior of the decay of the FRET fraction with increasing [NaCl] (Figure 5). The salt concentration at which the FRET fraction has decreased by 50% is denoted as c1/2. Together, our data determine the following order of events during disassembly: at the lowest salt concentrations, FRET disappears between H2A–H2B and the DNA dyad position −15 (c1/2 = 620 mM), followed by loss of FRET between H2A–H2B and the adjacent DNA at position −52 (c1/2 = 760 mM). At significantly higher salt concentrations, FRET between the two internal positions on the DNA (42 and −52) disappears with c1/2 = 870 mM. Lastly, FRET between (H3–H4)2 and DNA disappears (c1/2 = 900 mM for H4–DNA−52 and c1/2 = 930 mM for H4–DNA−15), indicating that nucleosome disassembly is complete at this salt concentration. In contrast to our previous studies (18,22,28), here we used Nonidet P40 to prevent surface adsorption of labeled histones. In the presence of this detergent, which is known to stabilize nucleosomes (29), we observed dissociation at higher [NaCl]. Importantly, in addition to the decreased FRET population, changes in the proximity ratio distributions were also observed, which can be related to structural changes within the nucleosome. Due to technical limitations, only a qualitative analysis of the proximity factor distribution is possible. Nevertheless, this allows us to gain further insights into the structural changes within the nucleosome and further strengthens the proposed mechanism for disassembly. These changes are discussed in detail in Supplementary Data 2.5. In contrast to bulk FRET experiments, in which only the average transfer efficiency of all labeled particles is determined, spFRET analysis allows us to observe and distinguish loss of FRET from shifts in the proximity ratio distribution. Only by using single molecule techniques can the sequential loss of FRET be investigated and a mechanism derived.
Figure 5.
spFRET reveals an intermediate open conformation before H2A–H2B dimer dissociation from the DNA. Plot showing fraction of the FRET population as a function of the [NaCl] for H2B–DNA−15 (blue), H2B–DNA−52 (green), DNA+42–DNA−52 (red), H4–DNA−52 (violet) and H4–DNA−15 (black). Each point represents an independent experiment. Errors were estimated based on the quality of the Gaussian fits of the proximity ratio distributions. Cartoons of nucleosomes indicating the relative locations of labels on the nucleosome, together with the c1/2 values derived from the sigmoidal curves are also shown, using the same color scheme. Donor labels are shown in yellow, acceptor labels are shown in magenta. From the sequence of the loss of FRET between the different nucleosome subunits, a model for disassembly can be derived (see text).
The distance between H2B and DNA position −15, near the dyad axis, increases at the lowest salt concentration before any of the other distance changes. This suggests that the interface between the (H3–H4)2 tetramer and H2A–H2B dimer opens up, while all histones remain bound to DNA. Direct measurement of spFRET to analyze the distance between H2A–H2B and (H3–H4)2 is, unfortunately, impractical: the two copies of each histone present in each octamer would result in four different FRET pairs, leading to a multitude of FRET populations that would be impossible to separate. Measuring FRET between a single labeled histone and two different positions on the DNA is the best alternative to circumvent this complication and to discriminate between the opening of the H2A–H2B/(H3–H4)2 interface and H2A–H2B dissociation. After H2A–H2B dissociation, at least the central ∼100 bp of the DNA are still wrapped around the remaining histones, indicated by the presence of FRET between the two internal DNA positions. We concluded that the disassembly pathway occurs through the following intermediate steps: state I→ IV→ V→ VI (Figure 1).
Nucleosome assembly is a reversal of the disassembly pathway
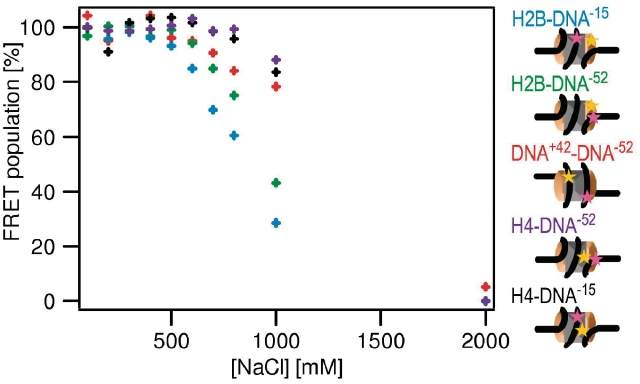
To test the reversibility of the disassembly process described above, we monitored nucleosome assembly using the same FCS and spFRET methods. We reconstituted nucleosomes from octamer and DNA by reducing the salt concentration in a stepwise manner from 2000 to 300 mM NaCl with 300 nM sample concentration. Using FCS, we measured the diffusion coefficients of the labeled subunits [H2A–H2B, (H3–H4)2 or DNA as in the disassembly study] at each step (Supplementary Figure S4). These experiments were carried out directly after dilution to 20 nM, before reaching equilibrium, in order to be as close as possible to the state of the sample during the reconstitution (Supplementary Data 2.6). Our results confirmed that even if the reconstitution is started with histone octamer, (H3–H4)2 binds the DNA at higher ionic strength than H2A–H2B (state V) (30).
For a more detailed analysis, energy transfer between the different positions on the octamer and the nucleosomal DNA was measured at each step of the reconstitution using spFRET (Figure 6). The FRET fractions for each construct increased at the cost of the NoFRET fraction in a reversal of the disassembly process. It can be seen that (H3–H4)2 binds the DNA first, coincident with DNA wrapping around bound (H3–H4)2 indicated by the appearance of FRET between the fluorophores on the DNA (42 and −52) (state V, Figure 1). Next, H2A–H2B is observed to bind to the tetrasome complex, in agreement with our FCS studies. Most importantly, energy transfer between H2A–H2B and the position on the DNA close to the dyad axis (−15) appeared at the lowest [NaCl]. This can only be explained by the presence of an intermediate state in which (H3–H4)2 and H2A–H2B are bound to the DNA, while the H2A–H2B/(H3–H4)2 interface is disrupted. This is the same step observed earliest during nucleosome disassembly (state IV, Figure 1). Only upon further reduction of [NaCl] can the nucleosome adopt a compact form.
Figure 6.
Nucleosome assembly follows the reverse pathway as disassembly. The fraction of FRET population as a function of [NaCl] measured during reconstitution of H2B–DNA−15 (blue), H2B–DNA−52 (green), DNA+42–DNA−52 (red), H4–DNA−52 (violet) and H4–DNA−15 (black). Cartoons of nucleosomes indicate the relative locations of labels on the nucleosome. The experimental error is estimated to be <6%. For clearness of the representation, error bars are not depicted. Upon lowering the [NaCl], FRET appears in the reverse order as it disappears during NaCl induced disassembly, indicating that the reconstitution follows the same mechanism as disassembly.
Analysis of the ionic dependence of nucleosome dissociation reveals an intermediate open nucleosome state
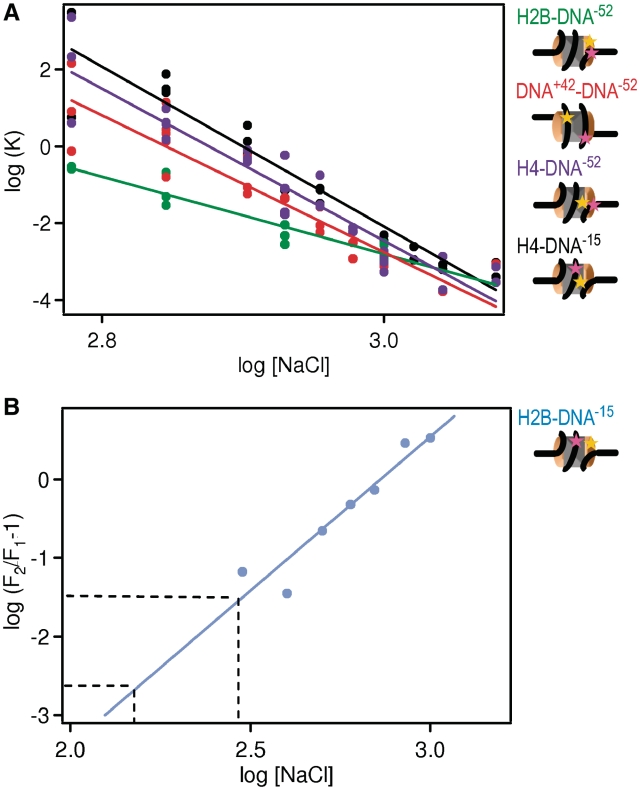
By analyzing the salt dependence of histone dissociation according to ref. (31) we are able to determine the number of ion pairs between histones and DNA (Supplementary Data 2.8). A log–log plot of equilibrium constants for histone dissociation calculated from our spFRET data (Figure 7A) yielded the number of ionic interactions through a linear fit.
Figure 7.
The number of ion pairs between histones and DNA can be derived from the salt dependence of the FRET fraction. (A) Equilibrium constants (K) for histone dissociation as a function of NaCl concentration for H2B–DNA−52 (green), DNA+42–DNA−52 (red), H4–DNA−52 (violet) and H4–DNA−15 (black). Cartoons of nucleosomes indicate the relative locations of labels on the nucleosome. K-values were calculated from the fraction of intact nucleosomes (Figure 5). The number of ions pairs between histones and DNA can be derived from curve fitting (as described in Supplementary Data 2.8, results see Supplementary Table S3). Approximately twice as many ion pairs are broken upon (H3–H4)2 than upon H2A–H2B dissociation. (B) Plot for the determination of the number of ion pairs involved in the opening of the H2A–H2B/(H3–H4)2 interface [see SupplementaryData, Equation (7)]. Data points were calculated from the fraction of intact nucleosomes of H2B–DNA−15 and H2B–DNA−52 (Figure 5). The transition requires the breaking of 4 ± 1 ion pairs as required from the slope of the fit. From extrapolation to physiological salt concentrations (150–300 mM NaCl, indicated by the dashed lines) the fraction of open nucleosomes occupied at physiological salt can be estimated to 0.2–3%.
For the H2B–DNA−52 combination, we derive 11 ± 1 ion pairs. We conclude that H2A–H2B dissociation from the DNA, to which we assigned the loss of FRET between H2B and DNA−52, involves the breakage of 11 ± 1 ion bonds. The number of ion pairs derived for H4–DNA−52, H4–DNA−15 and DNA+42–DNA−52 is nearly identical (23 ± 2, 24 ± 2, 20 ± 2, respectively); almost exactly twice the number of ion bonds as for H2A–H2B dissociation. The data for H4–DNA−52 and H4–DNA−15 constructs are consistent with a release of (H3–H4)2 tetramer from the DNA. The number of ions required for loss of FRET between DNA+42–DNA−52 reflects complete loss of both H2A–H2B dimers from the DNA.
The loss of FRET between H2B–DNA−15 is not assigned to a dissociation event, but to a monomolecular transition corresponding to the opening of the nucleosome structure. The fraction of the population in the open state can be isolated. By mathematically treating the data sets obtained for constructs H2B–DNA−15 and H2B–DNA−52 as described in Supplementary Data 2.8, we can calculate the fraction of closed nucleosome (F1), and the sum of closed and open nucleosomes (F2). Calculating the ratio between F1 and F2 obtains the fraction of the open state. Plotting the log[Na+] versus log(F2/F1−1) generates a plot which can be linearly fit and extrapolated to physiological salt concentrations (150–300 mM NaCl). From these data, we calculate 4 ± 1 ion pairs are broken during this initial transition (Figure 7B). This is significantly fewer than the 11 ions lost for H2A–H2B release, indicating it is electrostatically much weaker. Most importantly, we estimate the fraction of open nucleosomes occupied at physiological salt ranges from 0.2% to 3% resulting in a free energy change of 3.5–2 kcal/mol. The implications of this finding for biological processes acting upon the nucleosome could be highly significant.
DISCUSSION
The structure of the nucleosome shows that the entire 145–147 bp of DNA are in tight contact with the histone octamer (32). Yet, it is a biological necessity that nucleosomes disassemble dynamically to allow for DNA access during transcription, replication and repair (33). To date, these structural transitions have not been characterized at the molecular level.
By monitoring diffusion of fluorescent nucleosomes and FRET within single nucleosomes, we have derived a detailed sequence of events that occur during salt-dependent nucleosome disassembly and assembly. We have identified a previously undetected intermediate state during salt-dependent disassembly precluding H2A–H2B release that is populated at 0.2–3% under physiological conditions. This state corresponds to an open form of the nucleosome in which all components are bound to the DNA, while the distances between H2A–H2B dimer and the DNA near the nucleosomal dyad are increased (state IV, Figure 1), suggesting a separation at the H2A–H2B/(H3–H4)2 interface. Upon further increases in ionic strength, dissociation of H2A–H2B from the DNA is observed, while the DNA remains loosely wrapped around the remaining histones after H2A–H2B dissociation. Last, the dissociation of the (H3–H4)2 tetramer from the DNA completes nucleosome disassembly (I→IV →V →VI, Figure 1). We further demonstrate that salt-dependent nucleosome assembly follows the same, but reverse pathway as observed during disassembly.
While many earlier studies have inferred (10–13) that the histone octamer is released as one entity, our data are consistent with experiments which have demonstrated that H2A–H2B is released from the nucleosome before (H3–H4)2 (6–9), and that H2A/H2B is exchanged at a much higher rate than H3/H4 in vivo (34–36). Here, we present direct evidence for the step-wise dissociation of histone subcomplexes in response to increased ionic strength. In theory, the loss of FRET between H2B and DNA-15 could arise from mobility of the N-terminal helix of H2B to which the donor fluorophore is attached. However, we believe this not to be the case, due to structural constraints and geometric limitations imposed upon the helix [explained in detail in the Supplementary Data 2.5].
Comparing the salt dependence of FRET between different fluorophore pairs has allowed us to determine the number of ion pairs involved in each specific transition. For the dissociation of H2A–H2B from DNA, we find that 11 ion pairs are broken; exactly half the number lost upon (H3–H4)2 release from the DNA. This finding supports our view that H2A/H2B dissociates as a dimer and H3/H4 as a tetramer. It also agrees with all-atom molecular dynamics simulations, which predict the release of 45 counterions upon binding of the octamer (as a whole) to the DNA (37). In contrast, the disruption between H2A–H2B dimer and (H3–H4)2 tetramer requires the breaking of only four ion pairs.
Intermediate states with increased DNA accessibility have been postulated from various in vitro assays (26,38–43). Ultimately, the consensus contends that the ends of the nucleosomal DNA are generally accessible to binding, but the DNA becomes less accessible beyond the terminal 15–25 bp. At the time, these studies provided valuable insight into the dynamic properties of nucleosomes; however, they were technically limited by bulk analysis of rather heterogeneous endogenous nucleosome preparations. Thus, discriminate intermediate states might not have been isolated. Recent technical advances have allowed investigations of the transient accessibility of nucleosomal DNA; reviewed in ref. (44). In contrast to these studies, we chose the locations of the fluorophores on the DNA to specifically monitor structural transitions involving regions beyond those previously observed at the very ends of the DNA. DNA end opening is consistent with our data considering that the penultimate 15 bp of DNA are organized by an N-terminal α helix in histone H3 and that this interaction is likely lost in the open nucleosome state. A loss of this DNA/histone interaction could result in the rapid spontaneous ‘nucleosome breathing’ of the penultimate 10–20 bp of DNA and may likely precede opening of the H2A–H2B/(H3–H4)2 interface (20,26,27). During the opening of the H2A–H2B/(H3–H4)2 interface, all components remain attached to the DNA and consequently nucleosome accessibility is observed, where DNA accessibility remains unaffected. However steric inhibition to protein binding near the ends of the DNA would be reduced, while the remaining H2A–H2B bound to the DNA would prevent accessibility deeper into the nucleosome core.
The possibility that an intermediate open nucleosome state is populated significantly under physiological conditions presents an alternative interpretation for experiments probing nucleosomal DNA accessibility. For example, previous single molecule studies have investigated the conformational changes that occur in nucleosomes when they are pulled apart from the ends (45,46). These studies show the transition from a closed nucleosome to linear DNA as defined by two major steps, the first being reversible and highly sensitive to salt concentration. This was interpreted as the breaking of histone–DNA contacts while the DNA is being pulled off the histone octamer. However, these data can also be explained by the proposed reversible disruption of the H2A–H2B/(H3–H4)2 interface, followed by the irreversible disruption of the (H3–H4)2/DNA complex.
There is indirect evidence that the transitions described in this manuscript and by the Widom and van Noort labs are important in vivo. For example, DNA damage is repaired more efficiently when located in peripheral DNA regions as opposed to near the dyad axis, in support of the presence of these states (47). Transcription elongation has long been known to be inhibited by the barrier imposed by nucleosomes, but the molecular details of the events while this barrier is overcome are only now being elucidated. In vitro experiments detailing the progression of RNA polymerase through nucleosomes show strong pausing at positions 15 and 45 bp, with few intermediates (48–51). In light of our results, these pauses can be explained by the presence of the open nucleosome state, into which the polymerase can easily progress up to the first H2A/H2B–DNA interaction (+15bp). At this point the polymerase must pause until H2A/H2B is lost, and then continues until it reaches the first contact made by histone fold region of H3/H4, 45 bp into the nucleosome, the site of the second strong pause. Our interpretation is further supported by the finding that imposing mechanical torsion on a single chromatin fiber results in an initial opening of the nucleosome without the loss of histones (52), in agreement with the proposed open state, and, that in vivo, this mechanical torsion can be induced by polymerase progression (53).
Our values for a free energy change in the range 3.5–2 kcal/mol fit well with other nucleosome related data. For example, competitive reconstitution assays show ΔΔG values from −3 to 1.5 kcal/mol relative to 5s DNA (54), while the H3K56 acetylation alters the (H3–H4)2–DNA interaction energy by 1.8 kcal/mol. Moreover, typical bimolecular protein–DNA interactions (55), e.g. IHF (from −5.6 to 10.6 kcal/mol), LexA (from −12.6 to −15 kcal/mol) and H2B (from −8.6 to −14.5 kcal/mol), have interaction energies significantly stronger than what we observe for nucleosome opening, suggesting that the energy required for H2A–H2B dimer release is significantly higher than the energy required for the intermediate state to form (8,56–58). Alternatively, the energy required for nucleosome opening more closely reflects small changes in the nucleosome assembly process, which can be altered by differences in DNA sequence or histone modifications.
The equilibrium between the closed and open nucleosome states can be shifted in several intuitively obvious ways. Histone variants are non-allelic versions of histones (mostly H3 and H2A) that modulate nucleosome accessibility and thus transcriptional output through largely unknown mechanisms. Intriguingly, many histone variants differ from their major-type counterparts in regions that form the H2A–H2B/(H3–H4)2 interface [e.g. H2A.Z, and the centromeric H3 variant cenH3 (59)], or in their ability to organize the penultimate 15 bp of DNA [H2A.Bbd (60)]. Thus, histone variants might exhibit subtle changes in the equilibrium between the open and closed conformations of the nucleosome, with profound implications on nucleosome accessibility and nucleosome remodeling (61). Similarly, posttranslational modifications of histones may also modulate this equilibrium. For example, the phosphorylation of H3Y41 (in the context of a nucleosome) stimulates transcription (62). This side chain is buried in the penultimate minor groove of nucleosomal DNA, requiring a partially open state for its modification by the JAK2 kinase. Furthermore, phosphorylation of H3Y41 would almost certainly prevent rebinding of the DNA, thereby stabilizing the open conformation. Bulk acetylation of histone tails also has moderate effects on nucleosome conformation (63,64). Finally, while some protein factors, such as linker histone H1, might stabilize nucleosomes in the closed state, other chromatin architectural proteins might trap or force open the nucleosome.
It is conceivable that the open nucleosome state is specifically recognized by histone chaperones and ATP-dependent chromatin remodelers. For example, the addition of FACT to nucleosomes has been shown to alter their conformation and increase DNA accessibility without losing H2A–H2B (65). FACT, and possibly other chromatin remodeling factors, might stabilize the open state, thereby facilitating the subsequent loss of H2A–H2B dimer. Similarly, an open nucleosome state might be a point of attack for the histone chaperone Asf1, which interacts with a region of H3 and H4 that is occluded in a properly folded ‘closed’ nucleosome (66,67).
The reversible pathway of nucleosome disassembly/assembly proposed here redefines our view of nucleosome accessibility. The stability of the H2A–H2B/(H3–H4)2 interface, so far only marginally considered, seems to play an important role in the regulation of nucleosome accessibility and, consequently, many other nuclear processes. Our approach provides a unique opportunity for further investigating these mechanisms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Science foundation (DFG) in the priority program ‘Optical analysis of the structure and dynamics of supra-molecular biological complexes’ (grant SE 843/9-3, SFB 590, to J.L.); National Institutes of Health (grant GM061909, to K.L., F32GM083532, to A.J.A.); Howard Hughes Medical Institute (to K.L.); DKFZ and HBIGS graduate schools (to V.B.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
V.B. acknowledges support by the DKFZ and HBIGS graduate schools. Histones were obtained from the W.M. Keck protein expression and purification facility at Colorado State University.
REFERENCES
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Van Holde KE. Chromatin. New York: Springer; 1988. [Google Scholar]
- 3.Akey CW, Luger K. Histone chaperones and nucleosome assembly. Curr. Opin. Struct. Biol. 2003;13:6–14. doi: 10.1016/s0959-440x(03)00002-2. [DOI] [PubMed] [Google Scholar]
- 4.Lusser A, Kadonaga JT. Strategies for the reconstitution of chromatin. Nat. Methods. 2004;1:19–26. doi: 10.1038/nmeth709. [DOI] [PubMed] [Google Scholar]
- 5.Wilhelm FX, Wilhelm ML, Erard M, Duane MP. Reconstitution of chromatin: assembly of the nucleosome. Nucleic Acids Res. 1978;5:505–521. doi: 10.1093/nar/5.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton DR, Butler MJ, Hyde JE, Phillips D, Skidmore CJ, Walker IO. The interaction of core histones with DNA: equilibrium binding studies. Nucleic Acids Res. 1978;5:3643–3663. doi: 10.1093/nar/5.10.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoch DA, Stratton JJ, Gloss LM. Protein-protein Förster resonance energy transfer analysis of nucleosome core particles containing H2A and H2A.Z. J. Mol. Biol. 2007;371:971–988. doi: 10.1016/j.jmb.2007.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oohara I, Wada A. Spectroscopic studies on histone-DNA interactions. II. Three transitions in nucleosomes resolved by salt-titration. J. Mol. Biol. 1987;196:399–411. doi: 10.1016/0022-2836(87)90700-5. [DOI] [PubMed] [Google Scholar]
- 9.Park YJ, Dyer PN, Tremethick DJ, Luger K. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. J. Biol. Chem. 2004;279:24274–24282. doi: 10.1074/jbc.M313152200. [DOI] [PubMed] [Google Scholar]
- 10.Ausio J, Borochov N, Seger D, Eisenberg H. Interaction of chromatin with NaCl and MgCl2. Solubility and binding studies, transition to and characterization of the higher-order structure. J. Mol. Biol. 1984;177:373–398. doi: 10.1016/0022-2836(84)90291-2. [DOI] [PubMed] [Google Scholar]
- 11.Brower-Toland BD, Smith CL, Yeh RC, Lis JT, Peterson CL, Wang MD. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc. Natl Acad. Sci. USA. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagerman TA, Fu Q, Molinié B, Denvir J, Lindsay S, Georgel PT. Chromatin stability at low concentration depends on histone octamer saturation levels. Biophys. J. 2009;96:1944–1951. doi: 10.1016/j.bpj.2008.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulaeva OI, Gaykalova DA, Studitsky VM. Transcription through chromatin by RNA polymerase II: histone displacement and exchange. Mutat. Res. 2007;618:116–129. doi: 10.1016/j.mrfmmm.2006.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santis PD, Morosetti S, Scipioni A. Prediction of nucleosome positioning in genomes: limits and perspectives of physical and bioinformatic approaches. J. Biomol. Struct. Dyn. 2010;27:747–764. doi: 10.1080/07391102.2010.10508583. [DOI] [PubMed] [Google Scholar]
- 15.Teif VB, Rippe K. Predicting nucleosome positions on the DNA: combining intrinsic sequence preferences and remodeler activities. Nucleic Acids Res. 2009;37:5641–5655. doi: 10.1093/nar/gkp610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Förster T. Zwischenmolekulare Energiewanderung und Fluoreszenz. Annalen der Physik. 1947;437:55–75. [Google Scholar]
- 17.Stryer L. Fluorescence energy transfer as a spectroscopic ruler. Annu. Rev. Biochem. 1978;47:819–846. doi: 10.1146/annurev.bi.47.070178.004131. [DOI] [PubMed] [Google Scholar]
- 18.Gansen A, Toth K, Schwarz N, Langowski J. Structural variability of nucleosomes detected by single-pair Forster resonance energy transfer: histone acetylation, sequence variation, and salt effects. J. Phys. Chem. B. 2008:2604–2613. doi: 10.1021/jp7114737. [DOI] [PubMed] [Google Scholar]
- 19.Kelbauskas L, Chan N, Bash R, DeBartolo P, Sun J, Woodbury N, Lohr D. Sequence-dependent variations associated with H2A/H2B depletion of nucleosomes. Biophys. J. 2008;94:147–158. doi: 10.1529/biophysj.107.111906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koopmans WJ, Buning R, Schmidt T, van Noort J. spFRET using alternating excitation and FCS reveals progressive DNA unwrapping in nucleosomes. Biophys. J. 2009;97:195–204. doi: 10.1016/j.bpj.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomschik M, van Holde K, Zlatanova J. Nucleosome dynamics as studied by single-pair fluorescence resonance energy transfer: a reevaluation. J. Fluoresc. 2009;19:53–62. doi: 10.1007/s10895-008-0379-1. [DOI] [PubMed] [Google Scholar]
- 22.Gansen A, Valeri A, Hauger F, Felekyan S, Kalinin S, Tóth K, Langowski J, Seidel CAM. Nucleosome disassembly intermediates characterized by single-molecule FRET. Proc. Natl Acad. Sci. USA. 2009;106:15308–15313. doi: 10.1073/pnas.0903005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 24.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Meth. Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 25.Weidemann T, Wachsmuth M, Tewes M, Rippe K, Langowski J. Analysis of ligand binding by two-colour fluorescence cross-correlation spectroscopy. Single Mol. 2002;3:49–61. [Google Scholar]
- 26.Li G, Widom J. Nucleosomes facilitate their own invasion. Nat. Struct. Mol. Biol. 2004;11:763–769. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- 27.Zlatanova J, Seebart C, Tomschik M. The linker-protein network: control of nucleosomal DNA accessibility. Trends Biochem. Sci. 2008;33:247–253. doi: 10.1016/j.tibs.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 28.Gansen A, Hauger F, Toth K, Langowski J. Single-pair fluorescence resonance energy transfer of nucleosomes in free diffusion: optimizing stability and resolution of subpopulations. Anal. Biochem. 2007;368:193–204. doi: 10.1016/j.ab.2007.04.047. [DOI] [PubMed] [Google Scholar]
- 29.Claudet C, Angelov D, Bouvet P, Dimitrov S, Bednar J. Histone octamer instability under single molecule experiment conditions. J. Biol. Chem. 2005;280:19958–19965. doi: 10.1074/jbc.M500121200. [DOI] [PubMed] [Google Scholar]
- 30.Chakravarthy S, Luger K. The histone variant macro-H2A preferentially forms ‘hybrid nucleosomes’. J. Biol. Chem. 2006;281:25522–25531. doi: 10.1074/jbc.M602258200. [DOI] [PubMed] [Google Scholar]
- 31.Record MT, Lohman ML, Haseth PD. Ion effects on ligand-nucleic acid interactions. J. Mol. Biol. 1976;107:145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- 32.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 33.Bucceri A, Kapitza K, Thoma F. Rapid accessibility of nucleosomal DNA in yeast on a second time scale. EMBO J. 2006;25:3123–3132. doi: 10.1038/sj.emboj.7601196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jamai A, Imoberdorf RM, Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol. Cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 35.Kimura H. Histone dynamics in living cells revealed by photobleaching. DNA Repair. 2005;4:939–950. doi: 10.1016/j.dnarep.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 36.Kimura H, Cook PR. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Materese CK, Savelyev A, Papoian GA. Counterion atmosphere and hydration patterns near a nucleosome core particle. J. Am. Chem. Soc. 2009;131:15005–15013. doi: 10.1021/ja905376q. [DOI] [PubMed] [Google Scholar]
- 38.Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 39.Godde JS, Nakatani Y, Wolffe AP. The amino-terminal tails of the core histones and the translational position of the TATA box determine TBP/TFIIA association with nucleosomal DNA. Nucleic Acids Res. 1995;23:4557–4564. doi: 10.1093/nar/23.22.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imbalzano AN, Kwon H, Green MR, Kingston RE. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 41.Protacio RU, Polach KJ, Widom J. Coupled-enzymatic assays for the rate and mechanism of DNA site exposure in a nucleosome. J. Mol. Biol. 1997;274:708–721. doi: 10.1006/jmbi.1997.1440. [DOI] [PubMed] [Google Scholar]
- 42.Ausio J, van Holde KE. Histone hyperacetylation: its effects on nucleosome conformation and stability. Biochemistry. 1986;25:1421–1428. doi: 10.1021/bi00354a035. [DOI] [PubMed] [Google Scholar]
- 43.McGhee JD, Felsenfeld G. The number of charge-charge interactions stabilizing the ends of nucleosome DNA. Nucleic Acids Res. 1980;8:2751–2769. doi: 10.1093/nar/8.12.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buning R, van Noort J. Single-pair FRET experiments on nucleosome conformational dynamics. Biochimie. 2010 doi: 10.1016/j.biochi.2010.08.010. doi:10.1016/j.biochi.2010.08.010 [Epub ahead of print, 25 August 2010] [DOI] [PubMed] [Google Scholar]
- 45.Kruithof M, van Noort J. Hidden Markov analysis of nucleosome unwrapping under force. Biophys. J. 2009;96:3708–3715. doi: 10.1016/j.bpj.2009.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mihardja S, Spakowitz AJ, Zhang Y, Bustamante C. Effect of force on mononucleosomal dynamics. Proc. Natl Acad. Sci. USA. 2006;103:15871–15876. doi: 10.1073/pnas.0607526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suter B, Thoma F. DNA-repair by photolyase reveals dynamic properties of nucleosome positioning in vivo. J. Mol. Biol. 2002;319:395–406. doi: 10.1016/S0022-2836(02)00291-7. [DOI] [PubMed] [Google Scholar]
- 48.Bondarenko VA, Steele LM, Ujvári A, Gaykalova DA, Kulaeva OI, Polikanov YS, Luse DS, Studitsky VM. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol. Cell. 2006;24:469–479. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 49.Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science. 2009;325:626–628. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin J, Bai L, Johnson DS, Fulbright RM, Kireeva ML, Kashlev M, Wang MD. Synergistic action of RNA polymerases in overcoming the nucleosomal barrier. Nat. Struct. Mol. Biol. 2010;17:745–752. doi: 10.1038/nsmb.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kulaeva OI, Gaykalova DA, Pestov NA, Golovastov VV, Vassylyev DG, Artsimovitch I, Studitsky VM. Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat. Struct. Mol. Biol. 2009;16:1272–1278. doi: 10.1038/nsmb.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bancaud A, Wagner G, Conde E, Silva N, Lavelle C, Wong H, Mozziconacci J, Barbi M, Sivolob A, Le Cam E, et al. Nucleosome chiral transition under positive torsional stress in single chromatin fibers. Mol. Cell. 2007;27:135–147. doi: 10.1016/j.molcel.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 53.Bécavin C, Barbi M, Victor J-M, Lesne A. Transcription within condensed chromatin: Steric hindrance facilitates elongation. Biophys. J. 2010;98:824–833. doi: 10.1016/j.bpj.2009.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thastrom A, Lowary PT, Widom J. Measurement of histone-DNA interaction free energy in nucleosomes. Methods. 2004;33:33–44. doi: 10.1016/j.ymeth.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 55.Andrews AJ, Chen X, Zevin A, Stargell LA, Luger K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol. Cell. 2010;37:834–842. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aeling KA, Opel ML, Steffen NR, Tretyachenko-Ladokhina V, Hatfield GW, Lathrop RH, Senear DF. Indirect recognition in sequence-specific DNA binding by Escherichia coli integration host factor: the role of DNA deformation energy. J. Biol. Chem. 2006;281:39236–39248. doi: 10.1074/jbc.M606363200. [DOI] [PubMed] [Google Scholar]
- 57.Holbrook JA, Tsodikov OV, Saecker RM, Record MT., Jr Specific and non-specific interactions of integration host factor with DNA: thermodynamic evidence for disruption of multiple IHF surface salt-bridges coupled to DNA binding. J. Mol. Biol. 2001;310:379–401. doi: 10.1006/jmbi.2001.4768. [DOI] [PubMed] [Google Scholar]
- 58.Mohana-Borges R, Pacheco AB, Sousa FJ, Foguel D, Almeida DF, Silva JL. LexA repressor forms stable dimers in solution. The role of specific dna in tightening protein-protein interactions. J. Biol. Chem. 2000;275:4708–4712. doi: 10.1074/jbc.275.7.4708. [DOI] [PubMed] [Google Scholar]
- 59.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 60.Bao Y, Konesky K, Park Y-J, Rosu S, Dyer PN, Rangasamy D, Tremethick DJ, Laybourn PJ, Luger K. Nucleosomes containing the histone variant H2A.Bbd organize only 118 base pairs of DNA. EMBO J. 2004;23:3314–3324. doi: 10.1038/sj.emboj.7600316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang EY, Ferreira H, Somers J, Nusinow DA, Owen-Hughes T, Narlikar GJ. MacroH2A allows ATP-dependent chromatin remodeling by SWI/SNF and ACF complexes but specifically reduces recruitment of SWI/SNF. Biochemistry. 2008;47:13726–13732. doi: 10.1021/bi8016944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dawson MA, Bannister AJ, Göttgens B, Foster SD, Bartke T, Green AR, Kouzarides T. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Simpson RT. Structure of chromatin containing extensively acetylated H3 and H4. Cell. 1978;13:691–699. doi: 10.1016/0092-8674(78)90219-2. [DOI] [PubMed] [Google Scholar]
- 64.Lucia FD, Alilat M, Sivolob A, Prunell A. Nucleosome dynamics. III. Histone tail-dependent fluctuation of nucleosomes between open and closed DNA conformations. Implications for chromatin dynamics and the linking number paradox. A relaxation study of mononucleosomes on DNA minicircles. J. Mol. Biol. 1999;285:1101–1119. doi: 10.1006/jmbi.1998.2382. [DOI] [PubMed] [Google Scholar]
- 65.Xin H, Takahata S, Blanksma M, McCullough L, Stillman DJ, Formosa T. yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol. Cell. 2009;35:365–376. doi: 10.1016/j.molcel.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.English CM, Adkins MW, Carson JJ, Churchill MEA, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.