Abstract
We developed two new site-specific recombination systems named VCre/VloxP and SCre/SloxP for genome engineering. Their recognition sites are different from Cre recognition sites because VCre and SCre recombinases share less protein similarity with Cre, even though the basic 13-8-13 structures of their recognition sites are identical. Mutant VloxP and SloxP, which have the same uses as mutant loxP, were also developed. VCre/VloxP and SCre/SloxP in combination with Cre/loxP and Flp/FRT systems can serve as powerful tools for genome engineering, especially when used to genetically modify both alleles of a single gene in mouse and human cells.
INTRODUCTION
Two popular tools used in genomic engineering are the Cre/loxP and Flp/FRT site-specific recombination systems (1), which are often used to develop conditional knockout mice. Cre/loxP is typically used for knocking out exon(s) in genes of interest, whereas Flp/FRT is used for removing neo cassettes of positive selection markers so that normal gene expression remains intact. Cre recombinase originates from P1 phage and recognizes the loxP site, which is composed of an 8-bp sequence flanked on either side by two 13-bp inverted repeats, forming a 13-8-13 structure. Cre recombinase can excise a region of DNA surrounded by two loxP sites. On the other hand, Flp recombinase originates from yeast plasmid 2µ and recognizes distinct FRT sites, which have the same 13-8-13-bp structure as loxP sites. The recombination efficiency of the Flp/FRT system, however, is less than that of the Cre/loxP system (2). To improve recombination efficiency, some have attempted to optimize Flp through mutations, resulting in the conversion of Flp to a thermostable FLP mutant (2) and adaptation for codon usage (3). Many mutant recognition sites for loxP [e.g. lox2272 (4), lox551, lox66 and lox71 (5)] and FRT [e.g. F3 and F5 (6)] have been developed, along with various methods for excising DNA sequences, exchanging cassettes [i.e. recombinase-mediated cassette exchange (RMCE) (7)], and inverting DNA sequences in a genome. Other valuable methods and applications also have been developed, such as conditional gene trap (8) and viral vector packaging (9) using native and mutant recognition sites of these two site-specific recombination systems.
Despite of the usefulness of alternative site-specific recombination systems, which when used in combination with other methods can serve as valuable tools, still only a few unconventional systems (10–13) are available for genome engineering. Although Dre recombinase from phage D6 recognizes a 32-bp DNA site (rox), to our knowledge, mutant rox sites are yet to be developed. The site-specific integrase from bacteriophage phiC31 catalyzes unidirectional recombination between its 39-bp attP and 34-bp attB recognition sites. Because phiC31 integrase recognizes 100–1000 sites (called pseudo attP or attB sites) in human and mouse genomes that possess partial sequence identity to attP or attB, phiC31 integrase is unfit for genome engineering of specific alleles (14). Alternative combinations of recombination systems with various types of mutant recognition sites, in addition to the Cre/loxP and Flp/FRT systems, can be powerful tools for genetically modifying genomes, allowing researchers to perform fine genomic manipulations and to construct more complex systems. Additional recombination tools would especially benefit human-induced pluripotent stem (iPS) cell applications necessitating the genetic modification of both alleles of a gene. In mice, both alleles of a gene are knocked out by mating heterozygote mice with single-allele knockout mice. In humans, however, this strategy is not practical. Just as various restriction enzymes recognizing different sites are valuable biotechnology tools, so too would various site-specific recombination systems serve as valuable genome engineering tools. For this reason, we sought to identify recombinases that recognize different sites.
MATERIALS AND METHODS
In silico prediction of two new site-specific recombinases—VCre and SCre—and their VloxP and SloxP sites
We searched for potential Cre homologs by performing a BLAST search of the public DNA database Genbank/EMBL/DDBJ. The GenBank/EMBL/DDBJ accession numbers of two Cre orthologues, VCre and SCre proteins, are ABX77110 and ABK50591, respectively, and those of the plasmids containing VCre and SCre are CP000756 (15) and CP000470, respectively.
To identify putative site-specific recombination sites for VCre and SCre, we used criteria that would increase the probability that the candidates are indeed true site-specific recombination sites. These criteria are as follows.
Palindromic structure. The candidate site-specific recombination site should have a palindromic structure. The 13-8-13-bp structure of loxP (for Cre) and FRT (for Flp) is palindromic. Thus, the site-specific recombination site of Cre homologs also should be palindromic. At this time, however, we were not concerned about recognition-site sequence similarity, size of inverted repeat (13 bp), and perfect matching of the inverted repeat.
Proximity of the Cre homolog to its recognition site within the plasmid. The site-specific recombination site should be located near the Cre homolog gene and should be contained within the same plasmid, because the loxP site is located near the Cre gene in the P1 phage genome. We speculated that during evolution dynamic insertion and deletion events may have occurred often in the plasmid. Thus, if the recombinase and its recognition site were distantly located, they would be easily dispersed, rendering each to become non-functional.
Not located in the ORF. The site-specific recombination site should not be located in the protein-coding region. We speculated that the recombinase and its site-specific recognition site co-evolved interactively. During evolution, it is unlikely that the prototype site-specific recognition site would have ‘jumped’ into the protein-coding region without destroying protein function.
Candidate recognition sites were confirmed via the experiment described below.
We searched for sequences that were identical or highly similar to those of VloxP and SloxP sites in mouse and human genomes by performing a BLAST search using the specified expectation value (E = 1000) and by performing a FASTA search of the Genbank/EMBL/DDBJ database using VloxP and SloxP as query sequences.
Construction of plasmid expressing VCre tagged with NLS and FLAG
The plasmid pTurboCre was a kind gift from Dr Jon Kratochvil (Washington University, St Louis, MO, USA). pTurboCre constitutively produces Cre recombinase from a CMV enhancer upstream from a chicken β-actin promoter, which drives the untranslated region of the first exon, first intron and part of the second exon of the β-actin gene, as described previously (16). Nuclear localization signal (NLS)-FLAG-VCre and NLS-FLAG-SCre was composed of a SV40 NLS and a FLAG sequence introduced into the N terminus of the VCre ORF or SCre ORF, respectively. The artificially synthesized genes were optimized for mouse codon usage in addition to removing all CpG dinucleotides (Genscript Inc). pTurboVCre and pTurboSCre were constructed by replacing the NLS-Cre gene with NLS-FLAG-VCre or NLS-FLAG-SCre, respectively.
We constructed tester plasmids by inserting two of each recognition site flanked by a stop cassette (blasticidin resistance) in front of the EGFP gene of pEGFP-C3 (Clontech). The following oligomers were used for the PCR amplification of the blasticidin resistance cassette (which contains a bacterial promoter, stop codon and poly-A addition signals) of pcDNA6TR (Invitrogen,): primer-01, primer-02 for VloxP; primer-03, primer-04 for VloxM1; primer-05, primer-06 for VloxM2; primer-07, primer-08 for loxP. All primers are listed online in the Supplementary Data.
To construct tester plasmids pVloxP-EGFP, pVloxM1-EGFP, pVloxM2-EGFP and ploxP-EGFP, we digested the resulting PCR products with NheI and AgeI, and the resulting fragments were ligated into the NheI and AgeI site of pEGFP-C3 (Clontech).
The following oligomers were used to construct tester plasmids pVloxBsd-loxPCm and pVloxBsd-Vlox2272Cm and for the PCR amplification of the chloramphenicol (Cm) resistance gene of pBACe3.6 (Invitrogen): primer-09, primer-10 for loxP; primer-11, primer-12 for Vlox2272. The resulting PCR products were digested with BamHI and XhoI, and the resulting fragments were ligated into the BamHI and XhoI sites of pVloxP-EGFP. All constructs were confirmed by DNA sequencing.
The following oligomers were used to construct tester plasmids pSloxP-EGFP, pSloxBsd-loxPCm and pSloxBsd-Slox2272Cm and for PCR amplification: primer-13, primer-14 for SloxP; primer-15, primer-16 for Slox2272.
Recovery of plasmid DNA from transfected cells
Using FuGENE® 6 (Roche), we co-transfected Flp-in TREx 293 cells (Invitrogen) with a tester plasmid (pVloxP-EGFP, pVloxM1-EGFP, pVloxM2-EGFP or ploxP-EGFP) and pTurboVCre, which encodes VCre recombinase. Twenty-four hours after transiently transfecting the cells, the cells were dissolved with DNA-extraction buffer [100 mM Tris–HCl (pH 8.5), 5 mM EDTA, 200 mM NaCl, 0.2% SDS] containing 100 µg/ml proteinase K, and the DNA from the transfected cells was purified by means of isopropanol precipitation.
Assessing site-specific recombination activity through transient transfection of VCre or SCre expression plasmids
Flp-in TREx 293 cells (Invitrogen), which originate from HEK293 cells, were plated at a density of 2 × 105 cells per dish with appropriate DMEM medium on poly-d-lysine-coated 35-mm glass bottom dishes (MatTek Corporation). The expression plasmids encoding VCre, Cre or SCre and tester plasmids were transfected into HEK293 cells using FuGENE® 6 Transfection Reagent (Roche). Twenty-four hours after transfection, cells were washed with PBS, fixed with 4% paraformaldehyde in 0.1 M phosphate buffer and 3.4% sucrose for 30 min at room temperature. Transfected cells were observed with an Olympus IX71 fluorescence microscope.
Construction of temperature-sensitive plasmids expressing VCre, SCre or Cre under temperature control in Escherichia coli
Plasmid pSIM18 (a kind gift provided by Dr Donald L. Court, National Cancer Institute-Frederick, Fredrick, MD, USA) is a temperature-sensitive, low-copy plasmid (pSC101) that carries an expression system under the control of the λ pL promoter and the cI857 temperature-sensitive repressor (17). Plasmid pSIM18 carries the hygromycin-resistant gene.
Dre gene was synthesized artificially using the original codon-usage pattern of Enterobacteria phage D6 (Genscript Inc.). We constructed expression plasmids by inserting VCre, SCre, Cre and Dre genes under the control of the pL promoter of pSIM18. The following oligomers were used for the PCR amplification of the backbone of pSIM18: primer-17, primer-18.
The following oligomers were used to construct plasmids pSIMVCre, pSIMSCre, pSIMCre and pSIMDre and for the PCR amplification of the VCre, SCre, Cre and Dre genes: primer-19, primer-20 for VCre; primer-21, primer-22 for SCre; primer-23, primer-24 for Cre; primer-25, primer-26 for Dre.
To construct expression plasmids pSIMVCre, pSIMSCre, pSIMCre and pSIMDre, we digested the resulting PCR products with AscI and NotI, and the resulting fragments were ligated into the AscI and NotI site of the PCR product of the backbone of pSIM18. In plasmids pVloxPAmp-loxPCm, pSloxM1Amp-loxPCm and pRoxAmp-loxPCm, we replaced the appropriate recognition site and antibiotics resistance gene using pVloxPBsd-loxPCm. We used the 32-bp rox site, which is recognized by Dre recombinase, and a segment with a two-base innate boundary in D6 phage, (A)TAACTTTAAATAAT TGGC ATTATTTAAAGTTA(G).
Assessing site-specific recombination activity through VCre or SCre expression plasmids in E. coli
Escherichia coli DH10B-bearing expression plasmids pSIMVCre, pSIMSCre, pSIMCre and pSIMDre, which encode VCre, SCre, Cre or Dre, respectively, were grown in LB medium containing 75 µg/ml hygromycin overnight at 32°C under constant shaking. Fifty microliters of the overnight culture was inoculated into 2 ml of fresh LB medium containing 75 µg/ml hygromycin. The culture was shaken for 2 h at 32°C, and then transferred into a 42°C water bath for 15 min. For electroporation, cells were washed three times with cold water and cold 10% glycerol, and then electroporated with tester plasmid (0.2 µg) in 25 µl of 10% glycerol. Transformants were incubated for 2 h at 37°C, and then plated onto LB agar plates containing 50 µg/ml kanamycin (Km). One hundred randomly selected colonies were replicated on LB agar plates containing 100 µg/ml ampicillin (Amp), 50 µg/ml Km or 12.5 µg/ml Cm to test growth activity. The experiments were performed in triplicate. HaloTag was fused to the recombinase in each expression system, labeled with HaloTag-TMR ligand and then scanned with a Fluorescence Image Analyzer. Quantitative analyses revealed no differences in the expression level of any of the recombinases after protein expression in E. coli was induced by shifting temperature (data not shown).
Double Red recombination method for constructing Lin28 targeting vector
The Lin28-targeting vector used in the RMCE method was constructed by using a high-throughput method involving the λ Red recombination system. We previously constructed a BAC library containing ∼40 000 clones derived from the chromosomal DNA of R1 ES cells harvested from the F1 offspring of 129X1/SvJ and 129S1/Sv mice (18). A BAC clone containing Lin28 was screened from this library for targeting one 129 mouse allele. We constructed the targeting vector by carrying out Red recombination two times. First, the DNA cassette for the retrieving vector was produced by PCR amplification using the following primers and pDT-6 plasmid containing the diphtheria toxin A (DT-A) chain as templates: primer-27, primer-28. The first Red recombination was performed in order to construct pDTLin28 plasmid from a BAC clone (No. 68A24) containing Lin28 gene using the DNA cassette for retrieving.
Second, the tag cassette was produced by serial PCR amplification using the following primers and pDTLin28 and pBSloxPEGFPtkNeo plasmids as templates: primer-29, primer-30. To integrate the EGFP-tag cassette into the C terminus of the Lin28-coding sequence, we performed a second Red recombination. The C terminus of Lin28 was fused to the EGFP gene in frame. To increase the efficiency of cassette exchange, we designed the tag cassette to contain loxP and lox2272 sites, which were to be exchanged by using the RMCE method; a neomycin resistance gene for positive selection; and the herpes thymidine kinase gene for negative selection.
The tag-exchanging cassette for DsRed-contained loxP and lox2272 sites, and a DsRed Monomer gene from pDsRedMonomer (Clontech).
First homologous recombination
The linearized knock-in vector was introduced into mouse ES cells (M1 originated from hybrid embryos derived from C57BL/6 and 129Sv mice) by electroporation (GenePulser; Bio-Rad). The ES cells were grown on feeder cells in a medium containing 150 µg/ml G418. Each colony was picked up and expanded, and the genomic DNA of each clone was purified. To identify true homologous recombinant colonies, we analyzed both outer sides of the knock-in vector through PCR amplification and the following oligomers: primer-31, primer-32 for 5′-side testing; primer-33, primer-34 for 3′-side testing. Typically, ∼5% of tested clones were true homologous recombined colonies.
RMCE method
We designed another Cre gene optimized for mouse codon usage; the gene lacked CpG dinucleotides. The artificial sequence was synthesized by Genscript, Inc. pTurboCreopti was constructed by replacing the NLS-Cre gene with an NLS-tagged Creopti gene.
For RMCE, one of the knockin ES cell clones was transfected with 25 µg of pBSloxPDsRedMPuro2272 and 25 µg of pTurboCreopti by electroporation (Genepulser; Bio-Rad). The transfected ES cells were grown on feeder cells in medium for ES cells containing 1 µg/ml puromycin. One day after incubation, the medium was replaced with fresh medium containing 1 µg/ml puromycin and 2 µM of ganciclovir for negative selection. Each ES cell colony was picked up and expanded. The genomic DNA of the ES cells was purified. For typing the genomic DNA for the tag cassette in the ES cell genome, we used the following oligomers for PCR amplification: primer-35, primer-36. The PCR products were confirmed by determining the size of the PCR products by electrophoresis and by DNA sequencing. Typically, ∼70% of the tested clones were engineered colonies showing site-specific recombination via RMCE.
Second homologous recombination
For the modification of the second allele (which originated from a C57BL/6 mouse), we used a BAC clone (RP23-281F6) containing mouse genomic DNA originating from C57BL/6 mice (Invitrogen). We expected to increase the possibility of targeting a different Lin28 allele by using genomic DNA from C57B6 mice, because the ES cells we used originated from hybrid mice with a C57B6 and 129Sv genetic background and because these two alleles can be distinguished by certain single-nucleotide polymorphisms (SNPs). Thus, we constructed a targeting vector containing VloxP sites for the second allele by carrying out Red recombination two times. First, the DNA cassette for the retrieving vector was produced by PCR amplification using the following primers and pDT-6 plasmid as templates: primer-37 and primer-28 for Lin28-site-D. The first Red recombination was performed to construct the pDTLin28-2nd-allele plasmid. Second, the tag cassette was produced by serial PCR amplification using the following primers and pDTLin28-2nd-allele and pENTR-puroBsd plasmid as templates: primer-38, primer-39, primer-40, primer-41, primer-42, primer-43, primer-44.
The linearized targeting vector, pDTLin28V-Puro was introduced into mouse knock-in ES cells (clone No. 80, Lin28 knockin ES cells) by electroporation (GenePulser; Bio-Rad). The cells were grown in medium containing 1 µg/ml puromycin. Each colony was picked up and expanded, and the genomic DNA of each clone was purified. The two outer sides of the knock-in vector were examined by PCR amplification (primer-45, primer-46 for 5′-side testing; primer-47, primer-48 for 3′-side testing), and true homologous recombinant colonies were detected. To confirm modification of both alleles of the Lin28 gene, we performed Southern hybridization using each outside probe.
VCre-dependent knockout of Lin28 gene via the VCre/VloxP system
To remove exon 3 of Lin28 in one allele, we transfected ES cells with 25 µg of pTurboVCre by electroporation. The ES cells were grown on feeder cells in standard medium. Each colony was picked up and expanded, and the genomic DNA of each clone was purified. PCR amplification was performed to identify ES cell colonies lacking exon 3. PCR amplification (primer-49 and primer-50) and direct sequencing were carried out to confirm site-specific recombination.
These clones were also examined by Southern hybridization using outside probes. Typically, ∼80% of the tested clones were engineered colonies showing site-specific recombination.
Fluorescence microscopy analysis
ES cells were plated onto mitomycin C-treated MEF feeder cells on glass bottom dishes (MatTek Corporation). The cells were washed with PBS, fixed with 4% paraformaldehyde in 0.1 M phosphate buffer and 3.4% sucrose for 30 min at room temperature. Cells were examined for expression of EGFP- or DsRed-fusion proteins with an Olympus IX71 fluorescence microscope equipped with UPlanApo 40×/0.85 objectives. Observations were recorded with a Hamamatsu ORCA-ER CCD camera.
Southern-blot analysis
Southern-blot analysis was performed with two DIG-labeled DNA probes located outside the targeting vector. Southern-blot analyses of each ES cell clone were performed using two probes. Each probe was generated by PCR amplification of genomic DNA containing the Lin28 gene with the following oligomers: primer-51 and primer-52 for the 5′ probe of 516 bp; primer-53 and primer-54 for the 3′ probe of 537 bp.
Immunoblotting
ES cells were grown under standard growth conditions. Cell extracts were prepared by washing the cells with PBS and adding 100 µl of lysis buffer [10 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40] containing Complete Protease Inhibitor (Roche). After a 30-min incubation on ice, lysates were centrifuged at 15 000 rpm for 10 min in a microcentrifuge. Immunoblot analysis was performed according to a procedure described previously (19). Briefly, aliquots of extract were analyzed by immunoblotting using anti-Lin28 antibody [Lin28(A177); Cell Signaling Technology, Danvers, MA, USA]. The bands were detected using the ECLplus western-blotting detection system (Amersham Biosciences) according to the manufacturer’s instructions and visualized with a Luminescent Image Analyzer LAS4000 mini (Fujifilm).
RESULTS AND DISCUSSION
VCre specifically recombines the VloxP site
We searched for candidate site-specific recombinases that recognize different sites by performing a BLAST search of the Genbank/EMBL/DDBJ database using the Cre amino acid sequence as a query sequence. The search returned one possible candidate, which we named VCre. VCre showed very weak similarity to Cre, sharing 29% identity to the Cre amino acid sequence (Supplementary Figure S1). We chose to focus our analysis on VCre because we expected that it would recognize a distinct site due to its low-sequence homology to Cre and because it originates from plasmid p0908 of Vibrio sp. (Supplementary Figure S1). The replication of plasmid p0908 of Vibrio sp. is analogous to that of P1 phage, which replicates as a circular plasmid in the lysogenic state within a bacterial cell. From the 81-kbp sequence of p0908 plasmid, we predicted a VCre recognition sequence, VloxP, which has a 13-8-13-bp structure (Figure 1A). However unlike loxP, VloxP has one mismatch present in its 13-bp inverted repeats.
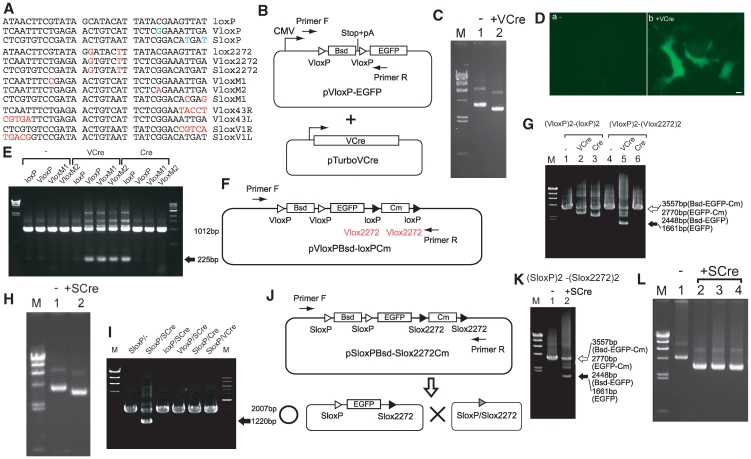
Figure 1.
VCre/VloxP and SCre/SloxP site-specific recombination systems. (A) Comparison of loxP, VloxP and SloxP sites with three mutant sites (lox2272, Vlox2272 and Slox2272) and various other mutant sites. (B) Schematic diagram of expression and tester plasmids. (C) Recovery of the tester plasmid from cells expressing VCre protein. Both expression and tester plasmids shown in (B) were simultaneously transfected into cells. The size of the original tester plasmid and the plasmid eliminated via VloxP were 5545 and 4758 bp, respectively. M, size markers (λ/HindIII; 23, 9.4, 6.6, 4.4, 2.3 and 2.0 kb). (D) EGFP expression was induced by removing a stop codon and poly-A cassette via VCre/VloxP. The VCre-expressing plasmid and tester plasmid shown in (B) were transfected into HEK293 cells. Scale bar, 10 µm. (E) Site-specific recombination using VCre and Cre. Either the VCre- or Cre-expressing plasmid and a tester plasmid harboring loxP, VloxP or mutant VloxP sites were introduced into cells. The arrow indicates the PCR product with the DNA flanked by loxP, VloxP, VloxM1 and VloxM2 eliminated. (F) Schematic diagram of a tester plasmid containing two VloxP sites and two loxP sites. (G) The mutant Vlox2272 site functions as an inconvertible mutant site. The tester plasmids shown in (F) and the VCre- or Cre-expressing plasmids were transfected into cells. The open arrow indicates the PCR product containing DNA flanked by VloxP and Vlox2272. Reaction with only VloxP or loxP(Vlox2272) produced 2770- or 2448-bp PCR products, respectively. The solid arrow indicates the PCR product in which both of the two DNA regions flanked by VloxP and Vlox2272 were eliminated. (H) Recovery of tester plasmid from cells expressing SCre protein. The tester plasmids, pSloxP-EGFP and SCre-expressing plasmid and pTurboSCre were simultaneously transfected into cells. The sizes of the tester plasmid containing SloxP and the tester plasmid without SloxP were 5545 and 4758 bp, respectively. (I) Site-specific recombination using SCre, VCre and Cre. The tester plasmids (pSloxP-EGFP, ploxP-EGFP or pVloxP-EGFP) and SCre-, Cre- or VCre-expressing plasmids were transfected into cells in various combinations. The arrow indicates the PCR product in which DNA flanked by SloxP was eliminated in a SCre-dependent manner. (J) Schematic diagram of a tester plasmid containing two SloxP sites and Slox2272 sites. (K) The mutant Slox2272 site functions as an inconvertible mutant site. The tester plasmid shown in (J) and the SCre-expressing plasmid were transfected into cells. The open arrow indicates the PCR product without the eliminated DNA. Reaction with only SloxP or Slox2272 produced 2770- or 2448-bp PCR products, respectively. The solid arrow indicates the PCR product in which both of the two DNA regions flanked by SloxP and Slox2272 were eliminated. (L) Recovery of the tester plasmid containing two SloxP sites and Slox2272 sites from cells expressing SCre protein. The tester plasmid (pSloxPBsd–Slox2272Cm) and the SCre-expressing plasmid (pTurboSCre) were simultaneously transfected into cells. The sizes of the original tester plasmid and the eliminated plasmid were 6648 and 4752 bp, respectively. Lanes 2–4 contain the plasmids of independent colonies recovered from cells expressing SCre.
To demonstrate that VCre can catalyze a recombination reaction at VloxP sites, we synthesized VCre gene using a DNA sequence optimized for mouse codon usage and removed all CpG sequences, which can convey potential gene-silencing effects. To enhance recombination efficiency, we fused the SV40 NLS to the N terminus of VCre (1). The expression plasmid (pTurboVCre, which expresses VCre) was driven by the strong constitutive CAG promoter. We created a tester plasmid by flanking a blasticidin resistance gene containing a stop codon and poly-A addition signals with two VloxP sites; this cassette was located between a CMV promoter and EGFP gene (Figure 1B). To confirm the site-specific recombination of VCre, we co-transfected HEK293 cells with an expression plasmid expressing VCre recombinase and with the tester plasmid described above. DNA was purified from the co-transfected cells, and then electroporated into E. coli to recover the tester plasmid. The tester plasmid recovered from co-transfected cells was ∼1 kbp shorter than the original tester plasmid from cells not transfected with the expression plasmid (Figure 1C). DNA sequencing of the shortened plasmid confirmed that the 787-bp segment containing the blasticidin resistance cassette flanked by two VloxP sites was eliminated by VCre recombinase. The tester plasmid lacking VCre could not produce EGFP (Figure 1D, panel a), because the CMV promoter and EGFP gene was separated by the blasticidin resistance cassette containing a stop codon and poly-A addition signals flanked by two VloxP sites. When the cassette containing the stop codon and poly-A addition signals was removed by site-specific recombination, EGFP was produced from the shortened tester plasmid. Indeed, brilliant EGFP signals were observed in cells expressing VCre protein (Figure 1D, panel b).
Unlike loxP, VloxP has one mismatch present in its 13-bp inverted repeat. Thus, we wondered whether this mismatch significantly decreases recombination efficiency. To address this issue, we created a perfect 13-bp inverted repeat for VloxP by constructing two mutants, VloxM1 and VloxM2 (Figure 1A). Cells were co-transfected with each tester plasmid series (pVloxP-EGFP, pVloxM1-EGFP or pVloxM2-EGFP), which contained either VloxP or mutant pVlox sites, and an expression plasmid that either expressed or lacked VCre recombinase. DNA from the co-transfected cells was purified, and site-specific recombination was determined by PCR analysis using primers F and R, as illustrated in Figure 1B. All of the original tester plasmids produced 1012-bp PCR products, whereas the tester plasmid recombined via VloxP produced a shorter 225-bp PCR product. Figure 1E shows the site-specific recombination of the VCre and VloxP series in HEK293 cells expressing both VCre recombinase and a tester plasmid (pVloxP-EGFP, pVloxM1-EGFP or pVloxM2-EGFP). The 787-bp segment containing the blasticidin resistance cassette flanked by either VloxP or mutant sites was eliminated by VCre recombinase. The present assay showed no significant difference in the recombination efficiency of VloxP, VloxM1 and VloxM2.
VCre/VloxP and Cre/loxP systems can be used together within the same cell because VCre/VloxP and Cre/loxP do not cross-react
Next, we checked the cross-reactivity between Cre and VCre. VloxP differs from loxP by 16 bases, which translates to 53% homology. Only 7 bases of the 13-bp inverted repeat of VloxP are identical to the 13-bp inverted repeat of loxP. Of course, no similarity was found between VloxP and FRT. As shown in Figure 1E, VCre did not react with the loxP site and Cre did not react with the VloxP, VloxM1 and VloxM2 sites because the shorter 225-bp PCR product was not detected. In contrast, Cre reacted with the loxP site because the shorter 225-bp PCR product was detected.
To confirm that there is no cross-reactivity between VCre/VloxP and Cre/loxP systems, even when VloxP and loxP sites are located very close to each other, we constructed a tester plasmid containing two VloxP and two loxP sites (Figure 1F) and co-transfected it into HEK293 cells with either an expression plasmid-lacking recombinase or with plasmids expressing VCre or Cre recombinase. We then performed PCR analyses using primers F and R (Figure 1F) to detect site-specific recombination. When the tester plasmid was introduced into cells lacking recombinase, we detected a 3557-bp PCR product. Its large size indicates that none of its components were deleted by site-specific recombination. In contrast, when a plasmid expressing VCre recombinase was co-introduced into the cells, we detected a shorter 2770-bp PCR product (Figure 1G, lane 2), which is consistent with the size of a product lacking the blasticidin resistance cassette flanked by two VloxP sites. Other short PCR products were not detected. These results strongly demonstrate that VCre recombinase recombines only VloxP sites but not loxP sites, even when VloxP and loxP sites are located proximally within the same plasmid. When cells were co-transfected with a tester plasmid and expression plasmid expressing Cre recombinase, we detected a short 2448-bp PCR product (Figure 1G, lane 3), which is consistent with the size of a product lacking the Cm resistance cassette flanked by two loxP sites. Again, no other short PCR products were detected. These results strongly show that Cre recombinase recombines only loxP sites but not VloxP sites, even when VloxP and loxP sites are located proximally within the same plasmid. This experiment, which used a tester plasmid containing two VloxP and two loxP sites, confirmed that the recognition sites are not interchangeable and that no cross-reactivity exists between VCre and Cre recombinase.
VloxP and Vlox2272 do not cross-react even though VCre recombines both sites
We examined the mutant sites of the VCre/VloxP system, because mutant loxP sites can serve as very useful tools for genome engineering, significantly increasing the utility of the Cre/loxP system. Previous analysis of loxP mutants demonstrated that the 8-bp core sequence directs the orientation and convertibility of mutant loxP sites (1). Two base substitutions in the 8-bp sequence of VloxP were made to form mutant Vlox2272, which possesses a C-to-G substitution at position no. 2 and an A-to-T substitution at position no. 7. We named this mutant Vlox2272 because its base substitutions were analogous to those of lox2272 and because two other substitutions in the 8-bp sequence of Vlox2272 matched that of lox2272 (Figure 1A).
To confirm that VloxP and Vlox2272 do not cross-react, we constructed a tester plasmid containing two VloxP and two Vlox2272 sites (Figure 1F) and co-transfected it into HEK293 cells along with a plasmid-lacking recombinase or plasmids expressing either VCre or Cre recombinase. We then performed PCR analyses using primers F and R (Figure 1F) to detect site-specific recombination. When the tester plasmid was introduced into cells lacking recombinase, we detected a 3557-bp PCR product. Its large size indicates that none of its components were removed by site-specific recombination. In contrast, when a tester plasmid expressing VCre recombinase was co-introduced, we detected a shorter 1661-bp PCR product (Figure 1G, lane 5), which is consistent with the size of a product lacking both the blasticidin resistance cassette flanked by two VloxP sites and the Cm resistance cassette flanked by two Vlox2272 sites. In addition, two other minor PCR products, 2270 and 2448 bp, were detected. The 2270-bp PCR product corresponded to a segment that lacked the blasticidin resistance cassette flanked by two VloxP sites, whereas the 2448-bp PCR product corresponded to a segment that lacked the Cm resistance cassette flanked by two Vlox2272 sites. The 2270- and 2448-bp PCR products resulted from the reaction of only one VloxP site or only one Vlox2272 site, respectively, and are thought to be intermediate products of the reaction producing the final 1661-bp product, i.e. reaction of the two VloxP sites and two Vlox2272 sites. Since our assay did not detect any other shortened PCR products, we concluded that VCre reacted with the two Vlox2272 sites but did not react to the combination VloxP/Vlox2272 sites (Figure 1G). When the tester plasmid containing two VloxP and two Vlox2272 sites was transfected into cells containing the expression plasmid expressing Cre recombinase, no shortened PCR product was detected (Figure 1G, lane 6), indicating that Cre recombinase does not react with either VloxP or its mutant recognition site, Vlox2272. These results will stimulate a variety of applications for using various mutant VloxP sites, applications that are analogous to those implementing Cre/mutant loxP such as RMCE methods and inversion of DNA cassettes. We also identified lox66/lox71-type mutant sites (20), Vlox43R/Vlox43L, for inversion and integration (Supplementary Figure S2).
SCre specifically recombines the SloxP site
Using the same strategy, we identified a second recombinase, which we named SCre recombinase (Supplementary Figures S3 and S4), and its recognition site, SloxP (Figure 1A), from the 279-kbp sequence of plasmid 1 of Shewanella sp. ANA-3. We constructed a tester plasmid by replacing the two VloxP sites of pVloxP-EGFP with two SloxP sites, and then introduced it into HEK293 cells lacking and HEK293 cells containing an expression plasmid expressing SCre recombinase. DNA purified from the co-transfected cells was then electroporated into E. coli to recover the tester plasmid. The tester plasmid recovered from cells co-transfected with the expression plasmid expressing SCre was ∼1 kbp shorter than the original tester plasmid recovered from cells lacking the expression plasmid (Figure 1H). DNA sequencing of the shortened plasmid confirmed that SCre recombinase had eliminated the 787 bp of the blasticidin resistance cassette flanked by two SloxP sites, demonstrating that SCre can recombine SloxP.
VCre/VloxP, SCre/SloxP and Cre/loxP do not cross-react with each other
To confirm that SCre, VCre and Cre do not cross-react, we transfected tester plasmids containing two SloxP, loxP or VloxP sites into cells lacking or containing the expression plasmid expressing SCre recombinase. Site-specific recombination was assessed by subjecting DNA purified from the transfected cells to PCR analysis. All of the original tester plasmids produced 2007-bp PCR products; however, the tester plasmid recombined via SloxP produced a shorter 1220-bp PCR product (Figure 1I, lanes SloxP/- and SloxP/SCre). The 787-bp PCR product representing the blasticidin resistance cassette flanked by two SloxP sites was eliminated by SCre recombinase. These results indicate that SCre recombinase does not recombine loxP and VloxP (Figure 1I, lanes loxP/SCre and VloxP/SCre). When a tester plasmid containing two SloxP sites was transfected into HEK293 cells containing an expression plasmid expressing either Cre or VCre recombinase, no shortened PCR product was detected (Figure 1I, lanes SloxP/Cre and SloxP/VCre), indicating that both Cre and VCre also failed to recombine SloxP.
SloxP and mutant Slox2272 do not cross-react even though SCre recombines both sites
In addition to mutant VloxP (Vlox2272), we also designed a mutant SloxP, called Slox2272, which contains two base substitutions (a C-to-G substitution at position no. 2 and an A-to-T substitution at position no. 7) in the 8-bp core sequence of SloxP. We named this mutant Slox2272 because its base substitutions were analogous to those of lox2272 and because two other substitutions in the 8-bp core sequence of Slox2272 matched that of lox2272 and Vlox2272 (Figure 1A). To confirm the specificity of mutant Slox2272, we constructed a tester plasmid containing two SloxP and two Slox2272 sites (Figure 1J) and co-transfected it into HEK293 cells along with a plasmid-lacking recombinase or a plasmid-expressing-SCre recombinase. We then performed PCR analyses using primers F and R (Figure 1J) to detect site-specific recombination. When the tester plasmid was introduced into cells lacking recombinase, we detected a 3557-bp PCR product. Its large size indicates that none of its components were removed by site-specific recombination. However in cells expressing SCre recombinase, we detected a shorter 1661-bp PCR product (Figure 1K, lane 2), which is consistent with a product lacking both the blasticidin resistance cassette flanked by two SloxP sites and the Cm resistance cassette flanked by two Vlox2272 sites. In addition, two other minor PCR products, 2270 and 2448 bp were detected. The 2270-bp PCR product corresponded to a segment that lacked the blasticidin resistance cassette flanked by two SloxP sites, whereas the 2448-bp PCR product corresponded to a segment that lacked the Cm resistance cassette flanked by two Slox2272 sites. The 2270- and 2448-bp PCR products resulted from the reaction of only one SloxP site or only one Slox2272 site, respectively, and are thought to be intermediate products of the reaction producing the final 1661-bp product, i.e. reaction of two SloxP sites and two Slox2272 sites. Since our assay did not detect any other shortened PCR products, we concluded that SCre reacted with two Slox2272 sites but did not react to the combination SloxP/Slox2272 sites (Figure 1K).
To confirm the specificity of SloxP and Slox2272, we purified DNA from the co-transfected cells and electroporated the DNA into E. coli to recover the tester plasmid. The tester plasmids recovered from cells co-transfected with the expression plasmid expressing SCre were ∼1.9 kbp shorter than the original tester plasmid from cells lacking SCre (Figure 1L, lanes 1–4). DNA sequencing of the shortened plasmids confirmed that the blasticidin resistance cassette flanked by two SloxP sites and the Cm resistance cassette flanked by two Slox2272 sites were eliminated by SCre recombinase. This conclusively confirmed that SCre reacted specifically with SloxP and mutant Slox2272. We also identified lox66/lox71-type mutant sites, SloxV1R/SloxV1L, for inversion and integration (Supplementary Figure S5).
Comparison of the site-specific recombination efficiencies of VCre and SCre with those of two other recombinases
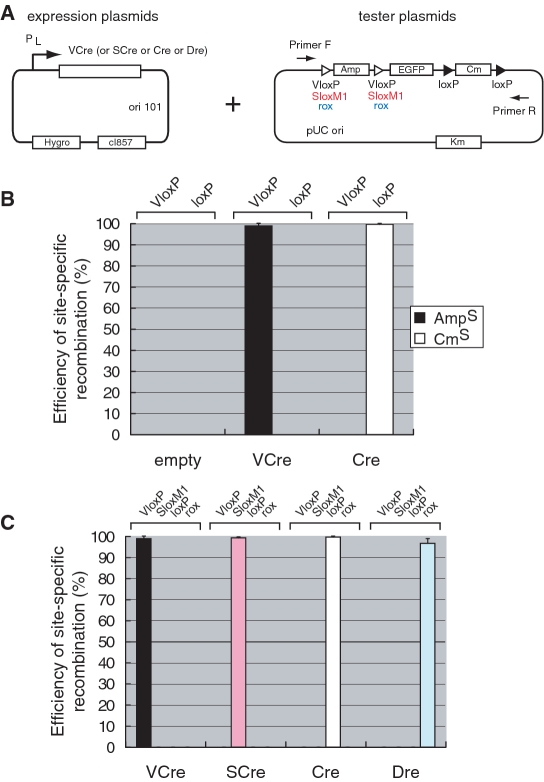
Next, we compared the efficiency of site-specific recombination of VCre with that of Cre by first constructing expression plasmids expressing either VCre or Cre in E. coli. Production of each recombinase was driven by a temperature-shift inducible promoter. The tester plasmid contained an Amp resistance cassette flanked by two VloxP sites and a Cm resistance cassette flanked by two loxP sites (Figure 2A). The negative control was the empty vector containing the plasmid backbone, which does not express recombinase. After electroporation of the tester plasmid containing two VloxP and two loxP sites into E. coli DH10B cells that lacked recombinase or expressed either VCre or Cre, we selected Km-resistant colonies, which were then replicated on Amp, Cm or Km agar plates to test their ability to grow. When VCre recombinase recombines the tester plasmid and the Amp resistance cassette flanked by two VloxP sites is removed, transformants lose their ability to grow on LB plates containing Amp. Similarly, when Cre recombinase recombines the Cm resistance cassette flanked by two loxP sites, transformants lose their ability to grow on LB plates containing Cm. The number of Amp-sensitive transformants and Cm-sensitive transformants reflect the site-specific recombination efficiencies of VCre and Cre, respectively. In our system, the recombination efficiency of VCre was almost the same as that of Cre (Figure 2B). Again, no cross-reactivity of site-specific recombination was detected in this assay in E. coli.
Figure 2.
Site-specific recombination of VCre or SCre recombinase in E. coli. (A) Illustrations of plasmids expressing VCre or SCre recombinase under the control of temperature-shift and tester plasmids for site-specific recombination. (B) Efficiency and specificity of site-specific recombination of VCre and Cre recombinase. After electroporation of the tester plasmid (A) into E. coli DH10B cells that express either VCre or Cre, we selected Km-resistant colonies, which were then replicated on Amp, Cm or Km agar plates to test their ability to grow. The empty vector contains a backbone plasmid that does not express recombinase. (C) Efficiency and specificity of site-specific recombination of VCre, SCre, Cre and Dre recombinases. The efficiency and specificity of four recombinases were examined in E. coli. In experiments testing various pairs of VCre, SCre, Cre and Dre with VloxP, SloxM1, loxP and rox, we detected no cross-reactivity. The recombination efficiencies of VCre and SCre were almost the same as those of Cre and Dre in an E. coli site-specific recombination assay.
We also compared the efficiency and specificity of site-specific recombination of VCre, SCre, Cre and Dre recombinases using the same assay strategy in E. coli. Tester plasmids were constructed by replacing the two VloxP sites of a tester plasmid containing the Amp resistance gene with either two SloxM1 sites (for SCre) or two rox sites (for Dre) (Figure 2A). SloxP is composed of an 8-bp sequence flanked on either side by 13-bp inverted repeats. Unlike loxP, the two 13-bp segments of SloxP do not perfectly match, as there is a two-base mismatch. SloxM1 (CTCGTGTCCGATA ACTGTAAT TATCGGACACGAG) was modified so that the two 13-bp segments would match perfectly but would not contain an ATG, which can be used as an initial Met codon. The recombination efficiencies of VCre and SCre were almost the same as those of Cre and Dre in an E. coli site-specific recombination assay (Figure 2C) [a t-test on the numbers of recombinant cells (three experiments; P > 0.05) produced by these two methods revealed no statistically significant differences]. It should be noted that the codon-usage pattern of VCre and SCre were changed to match that found in mouse and human. The codon-usage pattern of Cre and Dre, however, was the original phage-type pattern, which is suitable for E. coli. In experiments testing various pairs of VCre, SCre, Cre and Dre with VloxP, SloxM1, loxP and rox, we observed no cross-reactivity when testing various heterotypic combinations of recombinases and recognition sites (Figure 2C). Taken together, these findings show that VCre/VloxP and SCre/SloxP are sufficient biotechnology tools, because of their recombination efficiency and because VCre and SCre lack cross-reactivity.
Genome engineering of mouse ES cells: modification of one allele by VCre/VloxP and another allele by Cre/loxP
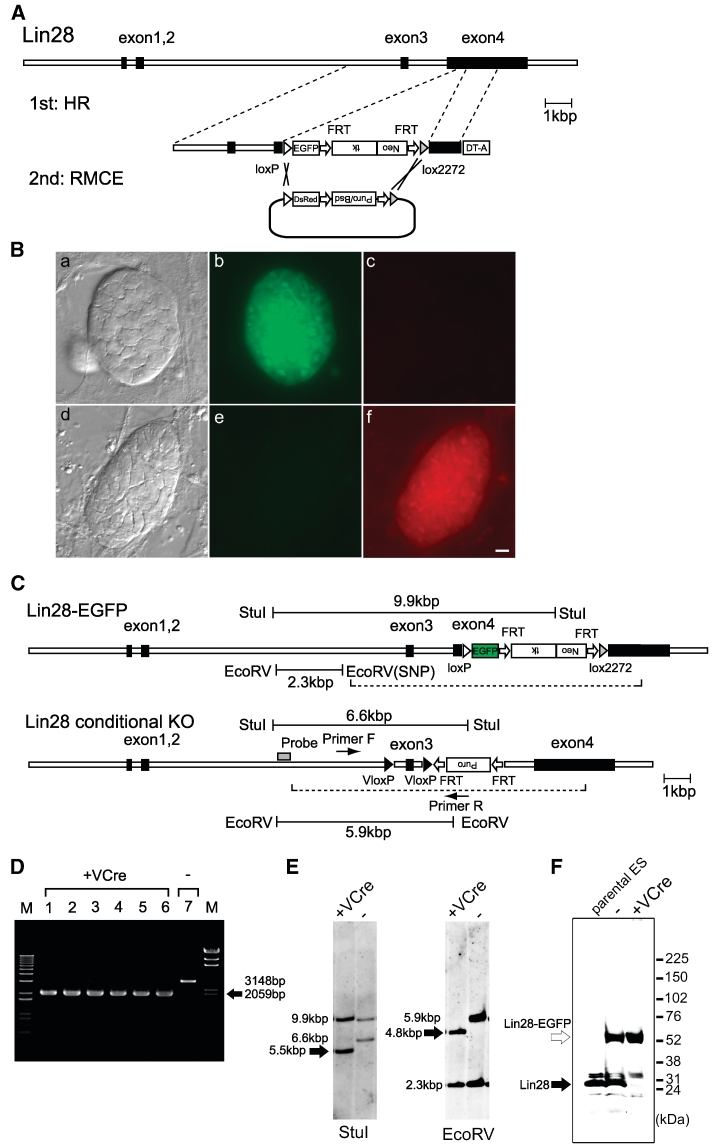
To test the feasibility and utility of VCre/VloxP to modify a gene (in this case Lin28) within a genome, we performed genome engineering on mouse ES cells using VCre/VloxP, which can genetically modify both alleles of a single gene. We demonstrated that one allele was modified by Cre/loxP and another allele was modified by VCre/VloxP. Figure 3A and C depicts the strategy we used to generate two novel Lin28 (also known as Lin28a) alleles. First, we performed a knock-in affecting one Lin28 allele, which could be freely exchanged with a C-terminal protein tag by using the RMCE method and loxP and lox2272. If necessary, one can remove the positive antibiotics selection cassette via the Flp/FRT system in order to enable reliable Lin28 gene expression. The EGFP C-terminus tag of lin28 can be exchanged with a DsRed-dependent tag following RMCE of Cre (see Figure 3B, panels b and f). Second, we performed a conditional knockout of the other Lin28 allele (Figure 3C). In this scenario, exon 3 of Lin28 was flanked by two VloxP sites, allowing the deletion of the gene of interest in a VCre-dependent manner. ES cells were then genetically modified by homologous recombination by introducing the conditional knockout of this particular Lin28 allele.
Figure 3.
Genome engineering using mouse ES cells. (A) C-terminal protein-tag exchange in ES cells using the RMCE method. (B) Mouse ES cells expressing Lin28 with an EGFP tag fused at its C terminus. The EGFP tag was replaced with a DsRed tag. Fluorescence microscopy images showing the intracellular distribution of Lin28-EGFP (b, e) and Lin28-DsRed (c, f). The corresponding differential interference microscopy images are shown in (a, d). Scale bar, 10 µm. (C) Schematic diagram of genetically modified alleles of Lin28. The dashed lines indicate the location of the targeting vectors. (D) PCR analysis of mouse ES cell genome engineered using VCre. The arrow indicates the PCR products in which VloxP-flanked exon 3 was eliminated from ES cells producing VCre. Lanes 1–6 show independently isolated ES colonies after the introduction of VCre. (E) Southern-hybridization analysis of mouse ES cell genome engineered using VCre. The arrows indicate VloxP-eliminated DNA fragments (5.5 or 4.8 kbp, respectively) in StuI- or EcoRV-digested genomic DNA from ES cells producing VCre. The 1.1-kbp DNA fragment containing exon 3-flanked VloxP was eliminated. (F) Immunoblot analysis of conditional knockout ES-cell genome engineered using the VCre/VloxP system. Only EGFP-tagged lin28 was detected in the VCre-mediated conditional knockout ES cells. Blots were probed with anti-lin28 antibody. The solid arrow indicates intact VCre protein; the open arrow indicates Lin28-fused EGFP.
We then performed PCR amplification analysis using primers F and R (Figure 3C) on DNA from ES cells harboring the two modified alleles. In the absence of VCre recombinase, a 3148-bp PCR product was detected. However, in ES cells transfected with an expression plasmid expressing VCre recombinase, a 2059-bp PCR product was detected (Figure 3D). The shorter 2059-bp PCR product resulted from the removal of the 1089-bp segment containing exon 3 of Lin28 flanked by two VloxP sites. Southern hybridization was also performed on StuI- or EcoRV-digested DNA purified from ES cells that lacked or expressed VCre recombinase. The probe shown in Figure 3C distinguished the two LIN alleles: the Lin28-EGFP allele and the Lin28 conditional KO allele. In DNA samples digested with either StuI or EcoRV, Southern hybridization detected bands from samples of cells expressing VCre that were 1.1 kbp shorter than bands from samples of cells lacking VCre: 5.5 versus 6.6 kbp and 4.8 versus 5.9 kbp, respectively (Figure 3E). This result demonstrated that the removal of the 1089-bp segment containing exon 3 of Lin28 flanked by two VloxP sites was dependent on VCre expression. Conclusively, PCR amplification and Southern hybridization verified the site-specific recombination of VCre/VloxP, and direct sequencing analysis of the PCR products also confirmed recombination with VloxP sites in the mouse ES cell genome (data not shown).
Finally, we confirmed the expression of lin28. Immunoblotting analysis revealed two bands representing intact lin28 and EGFP-tagged lin28, which were detected before applying VCre (Figure 3F). However, only EGFP-tagged lin28 was detected after applying VCre. Intact lin28 in cells harboring the conditional knockout allele was completely abolished. If necessary, both puromycin and neomycin resistance cassettes can be removed by using the Flp/FRT system (data not shown). In this study, we did not detect in our experiments cytotoxicity toward mouse ES cells, HEK293 cells or E. coli producing VCre or SCre. According to an in silico search, mouse and human genomes do not contain identical or sequences highly similar to VloxP and SloxP.
In the present study, we developed two novel site-specific recombination systems: VCre/VloxP and SCre/SloxP. VCre/VloxP and SCre/SloxP in combination with both Cre/loxP and Flp/FRT will serve as powerful tools for genome engineering, especially for applications requiring the genetic modification of both alleles of a gene of interest in mouse or human cells containing iPS cells. In this article, we also proposed various potential applications of the VCre/VloxP and SCre/SloxP systems, such as double and triple conditional knockouts, conditional knockout with virus vectors containing a Cre/loxP viral packaging system, and construction of complex BAC clones (see Supplementary Figures S6–S10). The VCre/VloxP and SCre/SloxP systems will strongly motivate novel ideas for genome engineering and BAC engineering.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants-in-Aid for Scientific Research (KAKENHI) from the Japan Society for the Promotion of Science (JSPS); grants from the Kazusa DNA Research Institute. Funding for open access charge: Grants-in-Aid for Scientific Research (KAKENHI) from the Japan Society for the Promotion of Science (JSPS).
Conflict of interest statement. None declared.
Note added in proof
The VloxP in this paper is different from loxP V (shortened name of loxP 2272) appeared in Kondo et al. Nucleic Acids Res. 31:e76, 2003 and Nakano et al. Nucleic acids Res. 33:e76, 2005.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to S. Minorikawa, M. Takayama, K. Toyama and E. Abe for their excellent technical assistance. We also thank Dr Osamu Ohara for his encouragement.
REFERENCES
- 1.Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev. Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 3.Wu Y, Wang C, Sun H, LeRoith D, Yakar S. High-efficient FLPo deleter mice in C57BL/6J background. PLoS ONE. 2009;4:e8054. doi: 10.1371/journal.pone.0008054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee G, Saito I. Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene. 1998;216:55–65. doi: 10.1016/s0378-1119(98)00325-4. [DOI] [PubMed] [Google Scholar]
- 5.Albert H, Dale EC, Lee E, Ow DW. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 1995;7:649–659. doi: 10.1046/j.1365-313x.1995.7040649.x. [DOI] [PubMed] [Google Scholar]
- 6.Schlake T, Bode J. Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry. 1994;33:12746–12751. doi: 10.1021/bi00209a003. [DOI] [PubMed] [Google Scholar]
- 7.Bouhassira EE, Westerman K, Leboulch P. Transcriptional behavior of LCR enhancer elements integrated at the same chromosomal locus by recombinase-mediated cassette exchange. Blood. 1997;90:3332–3344. [PubMed] [Google Scholar]
- 8.Friedel RH, Seisenberger C, Kaloff C, Wurst W. EUCOMM–the European conditional mouse mutagenesis program. Brief. Funct. Genom. Proteom. 2007;6:180–185. doi: 10.1093/bfgp/elm022. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki K, Mitsui K, Aizawa E, Hasegawa K, Kawase E, Yamagishi T, Shimizu Y, Suemori H, Nakatsuji N, Mitani K. Highly efficient transient gene expression and gene targeting in primate embryonic stem cells with helper-dependent adenoviral vectors. Proc. Natl Acad. Sci. USA. 2008;105:13781–13786. doi: 10.1073/pnas.0806976105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartung M, Kisters-Woike B. Cre mutants with altered DNA binding properties. J. Biol. Chem. 1998;273:22884–22891. doi: 10.1074/jbc.273.36.22884. [DOI] [PubMed] [Google Scholar]
- 11.Santoro SW, Schultz PG. Directed evolution of the site specificity of Cre recombinase. Proc. Natl Acad. Sci. USA. 2002;99:4185–4190. doi: 10.1073/pnas.022039799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sauer B, McDermott J. DNA recombination with a heterospecific Cre homolog identified from comparison of the pac-c1 regions of P1-related phages. Nucleic Acids Res. 2004;32:6086–6095. doi: 10.1093/nar/gkh941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anastassiadis K, Fu J, Patsch C, Hu S, Weidlich S, Duerschke K, Buchholz F, Edenhofer F, Stewart AF. Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E. coli, mammalian cells and mice. Dis. Model Mech. 2009;2:508–515. doi: 10.1242/dmm.003087. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Skjorringe T, Gjetting T, Jensen TG. PhiC31 integrase induces a DNA damage response and chromosomal rearrangements in human adult fibroblasts. BMC Biotechnol. 2009;9:31. doi: 10.1186/1472-6750-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hazen TH, Wu D, Eisen JA, Sobecky PA. Sequence characterization and comparative analysis of three plasmids isolated from environmental Vibrio spp. Appl. Environ. Microbiol. 2007;73:7703–7710. doi: 10.1128/AEM.01577-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westervelt P, Lane AA, Pollock JL, Oldfather K, Holt MS, Zimonjic DB, Popescu NC, DiPersio JF, Ley TJ. High-penetrance mouse model of acute promyelocytic leukemia with very low levels of PML-RARalpha expression. Blood. 2003;102:1857–1865. doi: 10.1182/blood-2002-12-3779. [DOI] [PubMed] [Google Scholar]
- 17.Chan W, Costantino N, Li R, Lee SC, Su Q, Melvin D, Court DL, Liu P. A recombineering based approach for high-throughput conditional knockout targeting vector construction. Nucleic Acids Res. 2007;35:e64. doi: 10.1093/nar/gkm163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakayama M, Iida M, Koseki H, Ohara O. A gene-targeting approach for functional characterization of KIAA genes encoding extremely large proteins. FASEB J. 2006;20:1718–1720. doi: 10.1096/fj.06-5952fje. [DOI] [PubMed] [Google Scholar]
- 19.Nakayama M, Nakajima D, Yoshimura R, Endo Y, Ohara O. MEGF1/fat2 proteins containing extraordinarily large extracellular domains are localized to thin parallel fibers of cerebellar granule cells. Mol. Cell Neurosci. 2002;20:563–578. doi: 10.1006/mcne.2002.1146. [DOI] [PubMed] [Google Scholar]
- 20.Oberdoerffer P, Otipoby KL, Maruyama M, Rajewsky K. Unidirectional Cre-mediated genetic inversion in mice using the mutant loxP pair lox66/lox71. Nucleic Acids Res. 2003;31:e140. doi: 10.1093/nar/gng140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.