Abstract
Genome-scale mapping of pre-replication complex proteins has not been reported in mammalian cells. Poor enrichment of these proteins at specific sites may be due to dispersed binding, poor epitope availability or cell cycle stage-specific binding. Here, we have mapped sites of biotin-tagged ORC and MCM protein binding in G1-synchronized populations of Chinese hamster cells harboring amplified copies of the dihydrofolate reductase (DHFR) locus, using avidin-affinity purification of biotinylated chromatin followed by high-density microarray analysis across the DHFR locus. We have identified several sites of significant enrichment for both complexes distributed throughout the previously identified initiation zone. Analysis of the frequency of initiations across stretched DNA fibers from the DHFR locus confirmed a broad zone of de-localized initiation activity surrounding the sites of ORC and MCM enrichment. Mapping positions of mononucleosomal DNA empirically and computing nucleosome-positioning information in silico revealed that ORC and MCM map to regions of low measured and predicted nucleosome occupancy. Our results demonstrate that specific sites of ORC and MCM enrichment can be detected within a mammalian intitiation zone, and suggest that initiation zones may be regions of generally low nucleosome occupancy where flexible nucleosome positioning permits flexible pre-RC assembly sites.
INTRODUCTION
The replication of DNA once, and only once, per cell cycle is essential for the maintenance of genome stability and cell survival and is ensured by tight regulation over the assembly of pre-replication complexes (pre-RCs) at replication origins. Although budding yeast replication origins have been well defined and contain a consensus sequence (ACS) that is necessary but not sufficient for origin activity (1,2), origin consensus sequences have not been identified in any other eukaryotic organisms including fission yeast, which can initiate within any sufficiently extensive stretch of AT-rich DNA (3,4). Metazoan origins are determined by a complex and poorly understood set of structural and topological features of DNA and chromatin in which DNA sequence motifs do not have a major role (5–7). In some contexts, any DNA sequence can function as an origin (8,9). This complexity may allow for a greater flexibility of origin selection to coordinate DNA replication with other cellular processes such as transcription (10), metabolism (11,12) and differentiation (13–15). Nonetheless, replication does initiate at specific chromatin sites in most eukaryotes and some origin sequences can direct initiation of replication when inserted into certain ectopic locations (8,16–20). In mammals, these sites can be either highly localized (21,22) or part of a broad de-localized ‘initiation zone’ (23–26).
Clearly, our understanding of replication origins, particularly in mammalian cells, would be greatly improved if genome-scale methods could be applied to identify large numbers of origins (27). Recently, several studies have isolated small nascent DNA strands and mapped their positions across segments of the human (28–30) and mouse (31) genomes. However, due to their extremely low abundance, small nascent strands must be carefully prepared to avoid contamination from random breaks in DNA generated during sample preparation (27,32) which may explain the poor overlap between published data sets (28). A complementary method is to map the sites of pre-RC protein binding using chromatin immunoprecipitation (ChIP) followed by microarray analysis. Pre-RC assembly begins with the binding of the hetero-hexameric origin recognition complex (ORC) to origin DNA followed by the Cdc6 and Cdt1 dependent recruitment of the hetero-hexameric MCM2-7 helicase complex. ORC is highly conserved but its origin recognition mechanisms are highly divergent. Budding yeast ORC preferentially binds to the ACS, while fission yeast ORC binds AT stretches through a specialized AT-hook in the ORC4 subunit (4,33,34) and Drosophila ORC may recognize DNA partly through its ORC6 subunit (35). In general, metazoan ORC does not show measurable sequence specificity in vitro although it has an increased affinity for negatively supercoiled DNA (6,36) suggesting that processes that generate superhelical tension such as nucleosome removal may contribute to origin specification. In fact, ORC binds to Nucleosome free regions (NFRs) in both budding yeast and Drosophila (1,37,38), suggesting that nucleosome organization may be a defining feature of origins in all eukaryotes. In the case of the Saccharomyces cerevisiae ARS1 sequence, the presence of a well-positioned nucleosome adjacent to the origin NFR was shown to be essential for ORC to nucleate pre-RC assembly and for origin activity (39,40).
To date, there have been no reports in the literature of genome-scale ChIP for mammalian pre-RC proteins (27). The reasons for these difficulties are not clear, but could include the lack of high quality antibodies or availability of epitopes within chromatin, a large number of pre-RC binding sites making enrichment in the complex mammalian genome difficult, transient non-specific chromatin interactions, or a small cell cycle window for site-specific pre-RC binding (27,41,42). Here, we have taken novel combination of approaches to identify sites of mammalian ORC and MCM binding that address several of these possible explanations. First, to address the problem of genome complexity, we have examined sites within the well-characterized Chinese hamster ovary (CHO) dihydrofolate reductase (DHFR) locus using cells containing 1000 copies of this locus (43). Replication within the CHO DHFR locus initiates throughout a broad ∼50 Kb initiation zone located in the intergenic region between the DHFR and 2BE2121 genes (25,44,45) with several preferential initiation sites (oriβ, oriβ′ and oriγ) (46–48). We employed the high-affinity biotin tag to avoid concerns about antibody quality and took advantage of the high degree of cell synchronization we can achieve in CHO cells in order to map pre-RC proteins during late G1 phase when they are most likely to be stably bound at their functional positions. Co-precipitated DNA was hybridized to a high-density oligonucleotide microarray such that each 500 bp chromatin fragment is recognized by approximately 100 unique probes (50 per strand). We also evaluated the efficiency of initiation at various sites across the DHFR locus in these same cells using DNA fiber methods. Finally, we compared the distribution of pre-RC proteins to nucleosome occupancy. Together, our results demonstrate significant enrichment of pre-RC proteins at regions of generally low nucleosome occupancy that are found within a de-localized region of initiation (‘initiation zone’).
MATERIALS AND METHODS
Cell culture and synchronization
All cells were grown in Dulbecco's Modified Eagle Medium (DMEM, Gibco) supplemented with 10% fetal bovine serum, Penicillin Streptomycin (Cellgro) and non-essential amino acids (Cellgro). For synchronization, cells were plated at 80–90% confluence and incubated for 4 h in the presence of 50 ng/ml Nocodazole. Mitotic cells were collected by shake off. A minimum of 95% mitotic cells were obtained, as determined by metaphase spread analysis. To evaluate the release of mitotic cells, aliquots were plated on glass coverslips and pulse labeled with 10 µM of BrdU for 30 min. Cells were fixed in 70% EtOH and stained for BrdU as previously described (49).
Cell lines
All cell lines described in this paper were based on our previously generated CHOC400 cells expressing the tetracycline transactivator (50). Cells were stably transfected with a plasmid expressing the Escherichia coli Biotin ligase (BirA) (51) gene linked to the Blasticidin resistance gene (Bsr) by an IRES sequence, under the control of the eEF1α promoter. The resulting cell line was transfected with different plasmids carrying subunits of the pre-RC tagged with the biotin ligase target sequence (BLT) (52) at the N-terminus (for ORC and mCherry) or with a fluorescent protein at the C-terminus for MCM, followed by the BLT at the C-terminus of the fluorescent protein. All tagged pre-RC subunits were under the control of a tetracycline-regulated promoter (tet-off) and were cultured in the presence of 6 µg/ml Doxycycline, 500 µg/ml HygromycinB, 400 µg/ml Zeocin and 2.5 µg/ml Blasticidin.
Immunoprecipitation and Avidin pulldown
MCM co-IP was done using a nuclear complex co-IP kit (Active motif cat#54001) according to manufacturer protocol. ORC avidin-pulldown was preformed as described in Vashee et al. (53). Complexes were precipitated using Dynabeads M-280 streptavidin magnetic beads (Invitrogen) and detected using the following antibodies: HRP-Streptavidin (ZYMED, 417449), anti-ORC1 (54), anti-ORC2 (Santa Cruz, sc-13238), anti-ORC4 (BD biosciences, 611170), anti-MCM2 (BM28, BD Transduction Laboratories, 610700), anti-MCM3 (Cell Signaling, 4012), anti-MCM4 (Santa Cruz, sc-28317), anti-MCM5 (Santa Cruz, sc-22780), anti-MCM7 (Santa Cruz, sc-9966) and anti-β-tubulin (Sigma T4026).
Chromatin avidin pulldown and microarray hybridization
Chromatin was prepared from 2 × 107 cells as previously described (55). DNA–protein complexes were precipitated using Streptavidin beads. The purified precipitated and input chromatin were amplified using a whole genome amplification kit (Sigma), labeled with Cy3 and Cy5, respectively, using the NimbleGen hybridization and sample tracking control kits according to the manufacturer recommended protocol, and hybridized to a NimbleGen tiling array covering the 121 Kb of the Chinese hamster DHFR locus (Accession no. BR000241) with a 45–60 nt Tm-matched oligo every 10 bp, (three replicates per strand). The arrays were scanned using the GenePix 4000B scanner (Molecular Devices). Array data was normalized using MA2C (56,57) and peaks were called by false discovery rate with threshold set at 10%.
Nucleosome mapping
Nuclei were prepared by extraction in buffer A (Tris–Cl pH 8, 15 mM, NaCl 15 mM, KCl 60 mM, EDTA 1 mM, EGTA 0.5 mM, Spermidine 0.5 mM) supplemented with 0.2% NP-40 (58). Nuclei were resuspended to a concentration of 108 nuclei/ml in reaction buffer (CaCl2 60 mM, NaCl 750 mM, Diluted 10× in buffer A) supplemented with 30 units/µl MNase (Warthington) and incubated at 25°C for 5 min. The reaction was terminated by addition of equal volume of 2× stop buffer (Tris–Cl pH 8, 50 mM, NaCl 100 mM, SDS 0.1%, EDTA 100 mM, RNaseA 10 µg/ml, Spermidine 1 mM, Spermine 0.3 mM). The sheared chromatin was incubated with proteinase K and DNA was extracted with Phenol/Chloroform. The purified DNA was separated on 2% agarose gel and the band representing mono-nucleosomal DNA was cut from the gel and extracted using gel extraction kit (Qiagen) (59). The mono-nucleosomal DNA was labeled and co-hybridized to the same array described above with total randomly sheared genomic DNA as control. Array data was smoothed by LOESS regression and scaled in R.
For southern hybridization MNase digested DNA was prepared as described above (digestion was with 15, 30 or 60 U/µl) and separated on agarose gel. DNA was transferred to Hybond-XL membrane (Amersham). Probes were labeled with 32P-dCTP using the random primer DNA labeling kit (Invitrogen). Sequence based nucleosome prediction was preformed using a support vector machine (SVM) model (60). High scores represent sequences with high nucleosome positioning.
DNA combing and morse code detection
Nascent DNA was labeled as previously described (12). Genomic DNA was extracted and combing was performed as described (61). Production of silanized coverslips for combing was performed as described (62). Morse code probes were designed as described (63). Primer pairs we used for PCR (using CHOC400 DNA as template) were as follows: probe 1, 5′-CAGCCTGATCCTTACACAAC-3′ and 5′-CAACTAGGGACCAAGCATTC-3′; probe 2, 5′-AGTGGGAGCTGGTATAGATG-3′ and 5′-AGCAGCGTTCAGACTGTT-3′; probe 3, 5′-AGAGGTGGCCGTAAAGTATC-3′ and 5′-TCCCTGCACCAGGCTATATC-3′; probe 4, 5′-GAGCGAATACCAGCATCAAC-3′ and 5′-CTGTCACAATCTGTGCCTACTC-3′; probe 5, 5′-CCCACCACACAGACACATTATC-3′ and 5′-GTGACACCACCTCTCATGAA-3′. In situ hybridization was performed as described (12). Immunodetection was performed as described (63), with the following succession of layers for the detection of nascent DNA and DHFR probes: (i) Alexa-488-conjugated streptavidin (Invitrogen); (ii) Biotin-conjugated rabbit anti-streptavidin (Rockland); (iii) Alexa 488-conjugated streptavidin (Invitrogen), mouse anti bromodeoxyuridine (BrdU) (BD Biosciences) and rat anti-bromodeoxyuridine (AbD Serotec); (iv) Biotin-conjugated rabbit anti-streptavidin (Rockland), Alexa 350-conjugated goat anti-mouse (Invitrogen) and Texas Red-conjugated donkey anti-rat (Jackson ImmunoResearch); (v) Alexa 488-conjugated streptavidin (Invitrogen), Alexa 350-conjugated donkey anti-goat (Invitrogen) and mouse anti-single-stranded DNA (Millipore); (vi) Cy5.5-conjugated goat anti-mouse (Abcam); (vii) Cy5.5-conjugated donkey anti-goat (Abcam). Images were acquired with an Eclipse 90i (Nikon) epifluorescence microscope connected to a CoolSNAP HQ CCD camera and run by Metamorph software (Molecular Devices) with a 60× objective. Image analyses were performed with Photoshop and Illustrator (Adobe).
RESULTS
Generation of BirA expressing cells for specific protein biotinylation
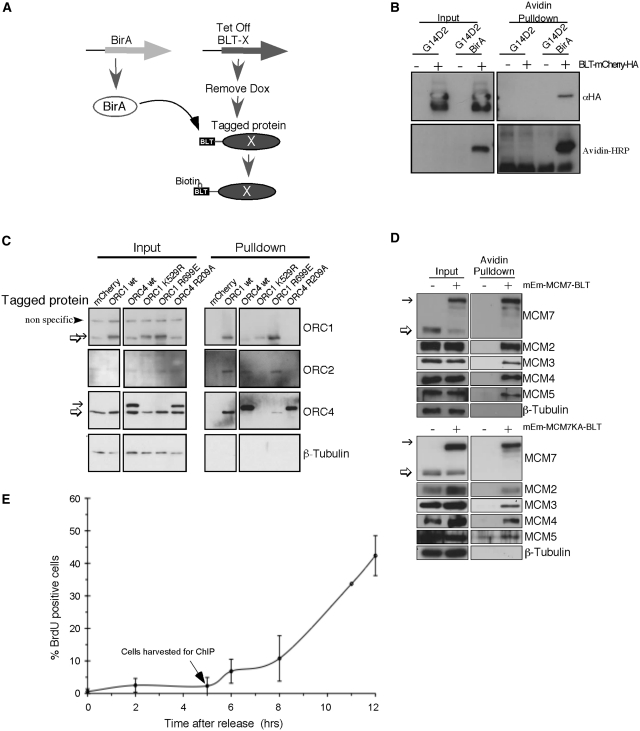
We employed a previously developed system (64–67) that takes advantage of the extremely high affinity avidin-biotin interaction (dissociation coefficient of 10−15 M (68)) to avoid concerns about the quality of antibodies for immunoprecipitation of pre-RC proteins. Hyg16 (a CHO cell line expressing the Tetracycline transactivator) (50) was stably transfected with the sequence specific E. coli BirA biotin ligase and the resulting line was then transfected with different pre-RC components tagged with an optimized peptide substrate for BirA, herein referred to as the biotin ligase target (BLT) (52). We have previously demonstrated that toxicity of pre-RC proteins due to over-expression can be overcome by silencing the expression of the proteins during selection of stable cell lines using the tet-repressible promoter in the presence of Doxycycline (Dox). Once cell lines are established, proteins can be induced by Dox removal and clones expressing low non-toxic and homogeneous levels of the proteins can be selected for further study (54,69). Cell lines expressing tagged ORC subunits were also stably transfected with GFP under the control of the tet-promoter to assist in monitoring induction, while the tagged MCM subunits were expressed as fluorescent fusion proteins that allowed for direct monitoring of induction. Upon expression, BLT-tagged proteins (Protein X, Figure 1A) are biotinylated by the constitutively expressed BirA enzyme. To verify the specificity of biotinylation, BirA-expressing (G14D2 BirA) and non-expressing parental (G14D2) cell lines were transiently transfected with fluorescent mCherry protein tagged with both BLT and HA. Expression of mCherry was detected in both cell lines using an anti- HA antibody (Figure 1B, top left), while BLT-tagged mCherry was detected by Avidin-HRP only in the BirA expressing cells (Figure 1B, bottom left) and could be pulled down by Streptavidin conjugated beads only from the BirA expressing cells (Figure 1B right).
Figure 1.
Establishment of in vitro protein biotinylation system. (A) Schematic of the experimental system used to generate specifically biotinylated proteins. Block arrows represent genes and ovals represent the expressed transgenic protein (X). The Biotin Ligase Target (BLT) tag is shown as a black rectangle. (B) Parental cell line (G14D2) and E. coli biotin ligase (BirA) expressing cells (G14D2 BirA) were transfected with a BLT and HA-tagged mCherry. The tagged protein was expressed in both cell lines (top panel left) but was biotinylated (left bottom panel) and could be pulled down by Streptavidin beads only in the BirA expressing cell line (right panels). The lower molecular weight band is most likely a degraded protein lacking the N-terminus but retaining the C-terminal HA tag. (C) Cells expressing various tagged ORC subunits (indicated above each lane) were used for avidin pulldown and probed with antibodies against ORC1 (top), ORC2 (second panel) and ORC4 (third panel). β-Tubulin (bottom) was used as loading control. Note that due to the large size of ORC1, the tagged protein migrates only slightly slower than the endogenous protein. (D) Cells expressing BLT-tagged MCM7 (+) or no tagged protein (−) were used for avidin pulldown and probed with antibodies against MCM7 (top), MCM2, MCM3, MCM4 and MCM5. β-Tubulin (bottom) was used as a loading control. In (C) and (D), the thin arrow indicates the tagged subunits while the thick arrow indicates the endogenous protein. (E) Mitotic cells were released and pulsed labeled with BrdU for 30 min at different time points. The fraction of BrdU positive cells was determined by immunofluorescence (error bars represent the standard deviation of three independent experiments). The arrow indicates the point in late G1 when cells were collected for pulldown.
BLT tagged ORC and MCM assemble into native complexes
To verify that the BLT and/or fluorescent tags did not interfere with the ability of the tagged proteins to form complexes with native proteins, we precipitated the tagged subunits using Streptavidin conjugated beads and monitored complex formation by immunoblotting with antibodies against the endogenous subunits. Tagged proteins were precipitated from nuclear extracts following induction and the resulting precipitated proteins were separated by SDS–PAGE. As expected, none of the endogenous ORC subunits was precipitated in cells expressing BLT-tagged mCherry, which was used as a negative control. Tagged wt ORC1 and ORC4 each co-precipitated with two other endogenous subunits, but not with their endogenous counterparts, indicating they were assembled into the complex by replacing the endogenous protein (Figure 1C). The tagged MCM7 similarly pulled down the endogenous MCM2, MCM3, MCM4 and MCM5 but not the endogenous MCM7 (Figure 1D, top).
A prior report found that specific binding sites for the chromatin remodeling complex Isw2 were difficult to identify using ChIP-chip due to a non-specific scanning mode for this protein. However an Isw2 mutant that could bind but not hydrolyze ATP was enriched at Isw2-regulated promoters, probably due to trapping of the ATP-bound form at sites where it is functionally engaged (70). Since both ORC and MCM contain AAA-type ATPase domain that bind and hydrolyze ATP and this activity is required for their function (71–82), we reasoned that similar ATPase mutants in these subunits might enrich pre-RC proteins at functional binding sites. We engineered a point mutation in the walker A motif of ORC1 (K529R) that is equivalent to the Isw2 mutation, as well as mutations in the ORC1 (R699E) and ORC4 (R209A) arginine finger shown to effect ORC ATP hydrolysis but not ATP or DNA binding in yeast (71,83). However, the ORC1 walker A and ORC4 arginine finger mutants abolished interaction with other subunits under our experimental conditions (Figure 1C), and therefore could not be used for pre-RC mapping, whereas the ORC1 arginine finger mutation retained the ability to form a complex with ORC. In addition, we generated an ATPase deficient MCM mutation (MCM7KA) similar to one that was shown to support pre-RC assembly but not origin activation in Xenopus egg extracts (78). This mutant was incorporated into the complex similar to the wild-type protein (Figure 1D, bottom).
Pre-RCs are enriched in the initiation zone
Pre-RCs are assembled during telophase (69,84,85), followed by the selection of a subset of these pre-RCs to function as origins at the origin decision point (ODP) (86). Cells that receive the appropriate mitogenic signals subsequently activate Cdk activity, phosphorylate the petinoblastoma (pRb) tumor suppressor protein, pass through the restriction point and rapidly initiate S phase (87,88). During S phase, pre-RCs are either converted to active replication complexes upon initiation or are cleared from chromatin during passage of the replication forks (3). Hence, we reasoned that the post-ODP stages of G1 phase, after origin selection but prior to origin activation during S phase, would be the best cell cycle window to find chromatin enriched with positioned pre-RCs. For this reason, chromatin avidin pulldown (ChAP) was performed with cells synchronized at 5 h after mitosis, previously demonstrated to represent the post-ODP, pre-R-point stage of G1 phase (88,89). Immunostaining for BrdU positive cells was used to verify cell synchrony (Figure 1E). Input and precipitated DNA were labeled and co-hybridized to a high-density NimbleGen tiling array (one probe every 10 bp). Normalization and peak determination was performed using ‘model-based analysis of two color arrays’ (MA2C), a method that normalizes log2 fold enrichment by probe sequence and signal distribution between ChIP (or ChAP in our case) and control, and assigns each probe a normalized, window-smoothed MA2C score (57).
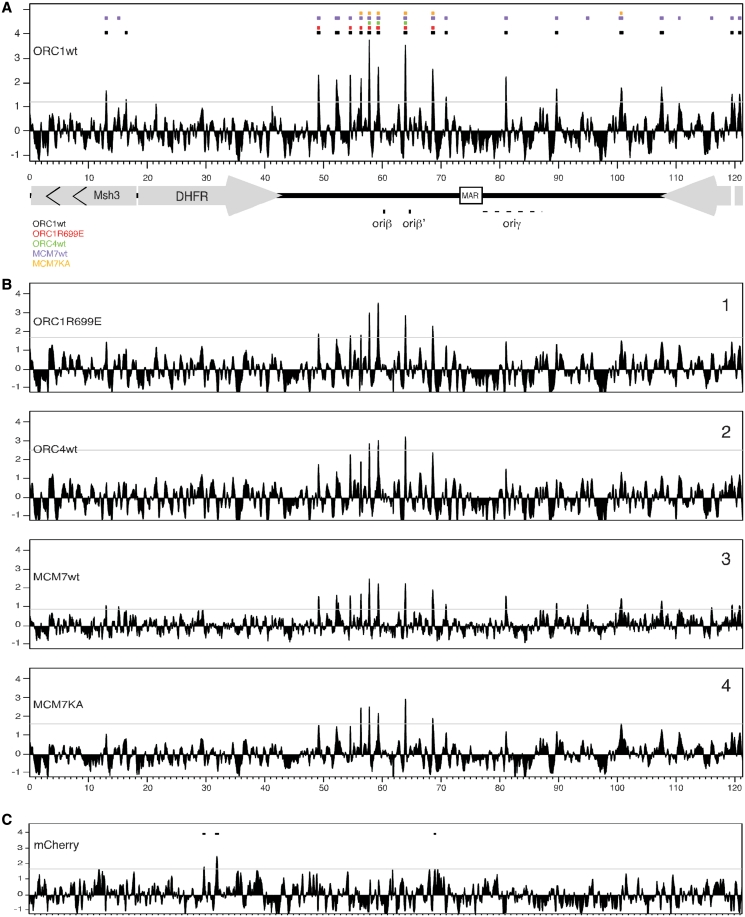
Results for wtORC1 are shown in Figure 2A with peak calls for all other subunits shown at the top by the colored rectangles. The majority of peaks for all pre-RC proteins were identified in the intergenic region between the DHFR and the 2BE2121 genes, in the region of the previously mapped initiation zone (25,44,45), consistent with a prior mapping of Mcm binding sites using a cosmid array (90). The highest density of pre-RC peaks was found around position 60 000 in the region that was shown to contain the major initiation sites (oriβ and oriβ′). Additional peaks for wtORC1 and wtMCM7 were detected throughout the initiation zone. However, ATPase mutants of these proteins and ORC4 were detected at a subset of these peaks. Many of the additional peaks found with wtORC1 and wtMCM7 were visible with the ATPase mutants and ORC4 but were below the peak determination threshold (Figure 2B), suggesting that these sites are also likely to bind ORC and MCM but with lower efficiency. Three sites were identified where all five proteins co-localized near this region. Cell lines expressing BLT-tagged mCherry, which is not expected to bind DNA were used as negative controls for ChAP. Only three peaks were called in the mCherry ChAP, none of which overlap with the pre-RC peaks (Figure 2C). Enrichment at one of the peaks for each protein was verified by qPCR (Supplementary Figure S1)
Figure 2.
ChAP-chip of pre-RC subunits. Cross-linked chromatin from cells expressing tagged pre-RC subunits was precipitated using Streptavidin conjugated beads and the extracted DNA was hybridized to a custom high-density NimbleGen tiling array. Array results were analyzed using MA2C. Peaks were called using a false discovery rate with the threshold set at FDR <10% (marked by a horizontal line). The previously described preferred initiation sites oriβ and oriβ′ are marked by black rectangles on the map of the DHFR region at the bottom while oriγ is marked by a dashed line to indicate the fact that it was not mapped at high resolution. (A) The MA2C scores for wtORC1 are presented as a histogram. Colored rectangles on top indicate peak calls for each subunit detected in (B). (B) MA2C scores for ORC1R699E (1), ORC4wt (2), MCM7wt (3) and MCM7KA (4) mapped by ChAP. (C) ChAP results for the BLT-tagged mCherry used as negative control. Peak calls are indicated by rectangles.
Initiation sites surround pre-RC binding sites
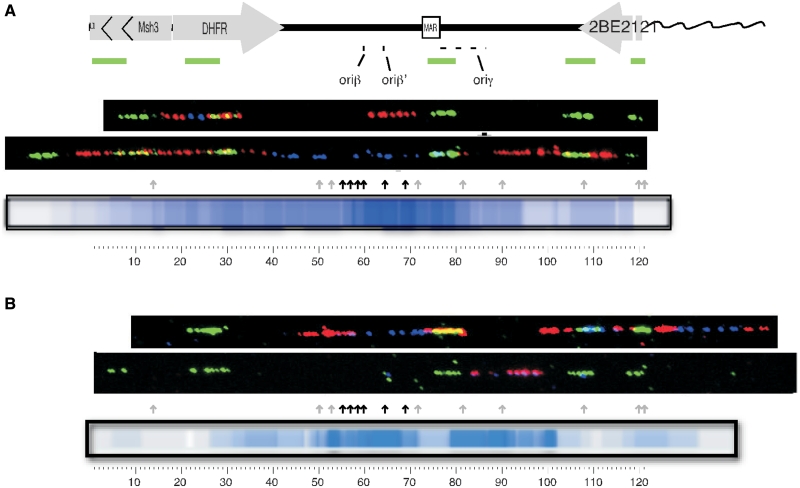
Although the CHO DHFR locus is one of the most well-studied initiation zones, different results have provided different interpretations of the frequency with which replication initiates at different sites within the zone, often referred to as ‘origin efficiency’ (30,42). The most direct means to evaluate origin efficiency is by examining the location and frequency of initiation sites on individual DNA fibers, and this method has not been applied to study initiation at the CHO DHFR locus. Hence, to evaluate how frequently replication initiates near pre-RC assembly sites, DNA fibers were used to examine the pattern of replication initiation at the DHFR locus (Figure 3 shows representative fibers; all fibers can be seen in Supplementary Figure S2). Both asynchronously growing cells, and cells that were synchronized at the G1/S boundary with aphidicolin were pulse labeled with the nucleotide analog IdU for 20 min and then chased with CldU for an additional 20 min. DNA molecules were stretched and the labels were detected with IdU- and CldU-specific antibodies (12). Fibers were aligned to the DHFR locus using the ‘Morse Code’ method, in which a series of unevenly spaced FISH probes creates a characteristic linear pattern (63). In fibers prepared from asynchronous cells the majority of initiation events were found in the same location as the strongest pre-RC ChAP peaks (Figure 3A) close to the previously described main initiation region (oriβ/β′). However, there was clearly a broad distribution of sites within the entire initiation zone and some initiation sites were even detected within the genes. Similar results were obtained with cells synchronized with aphidicolin (11,47,48), although initiation in aphidicolin-arrested cells appears more delocalized, potentially due to the induction of dormant origins upon fork arrest (11).
Figure 3.
Replication initiates near sites of ORC ChAP peaks: either asynchronously growing cells (A) or cells synchronized at the G1/S border with aphidicolin (B) were pulse labeled with IdU (blue) for 20 min and then chased with CldU (red) for 20 min each. DNA fibers were prepared and aligned to each other and to the DHFR map using a combination of unevenly spaced FISH probes (green). Initiation sites for replication within each fiber were identified as patterns of either red–blue–red (initiation during the IdU label and elongation during CldU), red–blue–empty–blue–red (initiation prior to the IdU label and elongation during both labels) or red only (initiation during the CldU label). A heat map consolidating the amount of label associated with each initiation pattern is shown. Arrows mark the position of ORC peaks (grey arrows denotes peaks containing only the wt ORC and MCM while black arrows denote peaks containing both the wt and the ATPase mutants) the locations of oriβ, oriβ′ and oriγ are shown. The appearance of a more de-localized set of initiations in the aphidicolin-arrested cells may result from the induction of dormant origins upon fork arrest (11). Low initiation activity near the MAR in aphidicolin-arrested cells has been reported before (48). Data represent the results from 32 and 27 individual fibers for asynchronous and aphidicolin arrested cells respectively, shown in Supplementary Figure S1.
Pre-RCs localize to regions of low nucleosome occupancy throughout G1 phase
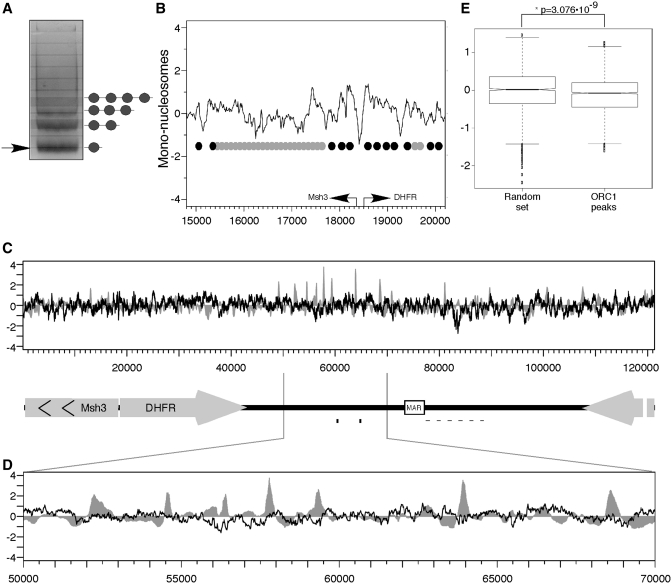
In mammalian cells, chromatin organization is a strong candidate for the determinant of ORC binding since it allows the generation of very complex and hereditary patterns that are not completely dependent on the actual DNA sequence. In fact, both budding yeast and Drosophila ORC bind to NFRs and in budding yeast the NFR has been shown to be necessary for ORC binding (1,37,38). To examine the relationship between pre-RC and NFRs in mammalian cells we mapped nucleosome positions along the locus. Chromatin was digested with microccocal nuclease (MNase), which preferentially cleaves linker DNA between nucleosomes. MNase digestion results in DNA digested into distinct sizes corresponding to oligonucleosomes (Figure 4A). The band corresponding to the mono-nucleosomal fraction (Figure 4A, arrow) was extracted and hybridized to the same microarray used for ChAP experiments. The DHFR locus contains a well-defined active bi-directional promoter between the DHFR and Msh3 genes that is DNaseI hypersensitive (91–93) and served as an internal control for a known NFR. Hybridization of mono-nucleosomes to the array revealed a prominent NFR at the promoter flanked by well-positioned nucleosomes (Figure 4B), confirming the quality of our mono-nucleosome preparations. No differences in nucleosome digestion patterns were observed before or after the ODP (Supplementary Figure S3), demonstrating that origin specification at the ODP is not the result of changes in nucleosome positioning.
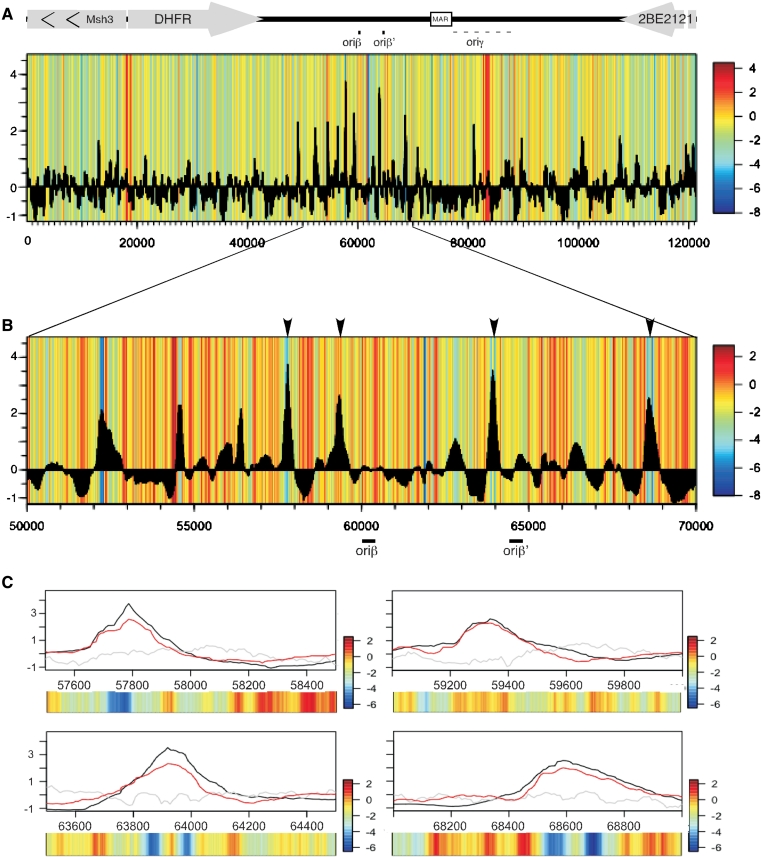
Figure 4.
Mapping nucleosome positions. (A) Chromatin was digested with Microccocal nuclease. Digested DNA was separated on 2% agarose gel and the mono-nucleosome fraction was extracted and hybridized to NimbleGen tiling array. (B) Nucleosome positions mapped by hybridization of mono-nucleosome DNA in the vicinity of the DHFR-Msh3 bi-directional promoter are shown. Arrows represent the transcription start sites of the DHFR and Msh3 genes. Circles indicate a suggested nucleosome arrangement. Black circles indicate positioned nucleosomes (distinct peak with a size of ≈150 bp) while grey circles indicate non-positioned nucleosomes. (C) Alignment of ORC1 ChAP (grey) with mono-nucleosomal DNA (black). (D) Blowup of the region between 50 000 and 70 000 containing the majority of the pre-RC's. (E) The nucleosome scores in the regions of wtORC1 peaks were compared to those of 10 random sets of the same size; the score in the peaks was significantly lower than the random set (*P = 3.076 × 10−9).
We next compared the relative positions of pre-RCs and nucleosomes (Figure 4C). Inspection of the oriβ/β′ region containing the highest amount of pre-RC binding and initiation activity (Figure 4D) revealed that pre-RCs do not align with NFRs but they do assemble near regions of generally low nucleosome occupancy. In order to verify the significance of pre-RC alignment to regions of low nucleosome occupancy, the nucleosome scores of the regions starting 500 bp upstream and ending 500 bp downstream of each wtORC1 peak were averaged and compared to a random set containing the same number of probes. The random set was generated 10 times and the average of the random sets was compared to the ORC peaks set using the Welch t-test (Figure 4E). The mean score for the ORC set was −0.11740 while the random mean was −0.009934. The two are significantly different (confidence interval = 0.95, t = −5.9445, df = 3158.999, P = 3.076 × 10−9) indicating that pre-RCs are indeed associated with regions of low nucleosome occupancy.
There is a growing appreciation for the role of DNA sequences in the positioning of nucleosomes and the ability of sequence information to predict in silico the positions of nucleosomes (60,94,95). We aligned the scores of a nucleosome positioning algorithm (60) along the DHFR locus to the positions of pre-RCs as determined by ChAP (Figure 5). Consistent with our nucleosome-mapping results, pre-RCs align with regions with low nucleosome positioning information, where nucleosomes are not restricted from binding to a specific position. The ability of nucleosomes to be moved might be important for origin assembly and/or activity. Combined with the actual nucleosome mapping data (Figure 4), these data suggest the possibility that initiation zones—regions of many scattered inefficient replication origins—are regions of low nucleosome positioning information where ORC can more easily compete with nucleosome binding to assemble pre-RCs at many positions, with only a few positions favorable enough to be enriched in ChIP-chip experiments.
Figure 5.
Pre-RCs align with region of low nucleosome positioning information. (A) The calculated nucleosome positioning value of sequences along the DHFR locus is presented as a color map with red indicating high values of positioning information and blue low values. The heat map was aligned with ORC1 ChAP data (black histogram). (B) Nucleosome predictions for the oriβ/β′ region (50 000–70 000) aligned to the ORC1 ChAP. The positions of oriβ and oriβ′ are marked by black lines. (C) A 1 kb region surrounding the peaks marked by arrows in (B). The ChAP results for ORC1wt (black) are overlaid with the ChAP results for MCM7wt (red) and mono-nucleosomes (gray). The nucleosome positioning scores for each region are shown as a heatmap.
An unusual chromatin configuration near ori-gamma
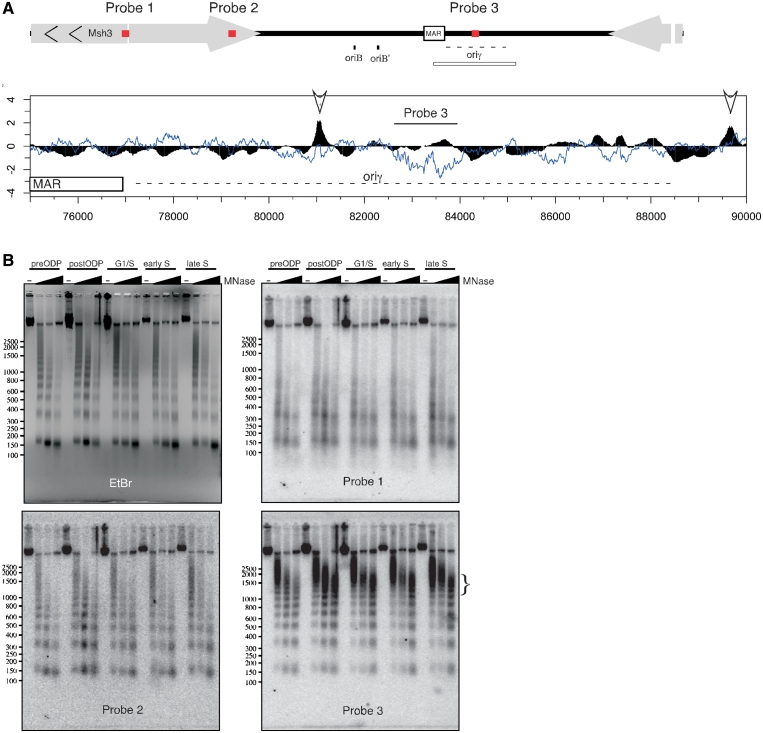
Mono-nucleosome mapping revealed a very weakly-hybridizing region located between position 82 000 and 84 000, previously designated as oriγ (Figure 6A). Oriγ was originally defined as one of the earliest replicating regions in the DHFR locus but was never precisely characterized (47,96). Regions hybridizing poorly to mono-nucleosome preparations can result from either very open and accessible chromatin or very protected chromatin that is highly inaccessible to MNase, either of which will be under-represented in mono-nucleosome preparations. To distinguish between these possibilities, DNA prepared from MNase digested chromatin at different time points during the cell cycle was separated by gel electrophoresis and hybridized to probes representing three different locations along the DHFR locus: (i) the bidirectional DHFR/Msh3 promoter region that is known to be nucleosome-free (93); (ii) a segment of the DHFR coding sequence that should be typically arranged into non-positioned but phased nucleosomes; and (iii) the oriγ segment (Figure 6B). The MNase digestion patterns were compared to that of bulk chromatin visualized by ethidium bromide staining of the gel prior to Southern transfer (Figure 6B, EtBr). Relative to bulk chromatin, the DHFR/Msh3 promoter displayed the expected accessible chromatin pattern visible as a smear with the low molecular weight bands appearing at low concentrations of MNase (Figure 6B, probe 1). The DHFR coding region (Figure 6B, probe 2) gave a pattern indistinguishable from bulk chromatin, also as expected. However, the large NFR region was highly protected, remaining largely undigested even at the highest MNase concentrations (Figure 6B, probe 3). The bulk of the DNA detected by probe 3 is ∼1500 bp in size, in agreement with the size of the mono-nucleosome-free region detected on the array (Figure 6A, 82 500–84 000). The relatively small amount of low molecular weight DNA that was digested by MNase within this region retained a nucleosome ladder pattern, indicating the region can be packaged into nucleosomes in some cells or copies of the locus. Since the ChAP data do not show enrichment of pre-RC proteins, we conclude that a large unidentified complex is strongly bound to the DNA close to the region previously defined as oriγ.
Figure 6.
The oriγ region is highly protected by a non-nucleosome complex throughout the cell cycle. (A) Schematic of probe positions (red rectangles) relative to the entire DHFR region in the top diagram. All probes were 1300 bp long. Below is an alignment of ORC1 ChAP (black) to mono-nucleosomal DNA (blue) along the region of the DHFR locus containing the matrix attachment region (MAR) and the secondary initiation zone, oriγ (indicated by the open rectangle below the diagram at the top). Arrows indicate the significant ORC peaks in this region. (B) CHOC400 cells were collected at different stages of the cell cycle and nuclei were treated with increasing amount of MNase. DNA was extracted and separated on 2% agarose gel. Southern blots performed using probes against the DHFR promoter (probe 1, position 17 816–19 115), the oriγ region (probe 3, position 82 621–83 949) and a control region in the DHFR CDS (probe 2, position 37 623–38 973). The enriched, high molecular weight fraction, seen with probe 3 is marked by the vertical bracket on the side of the panel.
DISCUSSION
The mechanism(s) responsible for the selection of replication origins in metazoa is one of the greatest fundamental mysteries in molecular biology. Unlike bacteria and single cell eukaryotes such as yeast that contain a specific replicator sequence element that binds the initiator complex, metazoan ORC shows no sequence specificity in vitro and no conserved origin sequence has yet been identified. Moreover, initiation in metazoa can occur at many sites distributed throughout large initiation zones, with each cell utilizing a different cohort of sites. Here, we show that pre-RC proteins are preferentially enriched at several specific sites of low nucleosome occupancy within a broad zone of de-localized initiation activity.
Despite the lack of a specific replicator sequence, the initiation of replication in most eukaryotic systems does not occur at random sites. This indicates that there is some mechanism by which pre-RC assembly and/or activation is focused to certain chromosomal regions but not others. Chromatin structure is a likely candidate as it can generate a unique, yet sequence independent, DNA signature. During metazoan development the genome undergoes large-scale reorganization as embryonic cells differentiate to give rise to all the cell types of the adult organism. Differentiation is also accompanied by changes in the pattern of replication (13,97,98). The lack of a specific replicator sequence may allow increased flexibility of origin positioning to coordinate replication with other chromosomal functions that differ between cell types (99). We find that ORC preferentially binds to regions of low nucleosome occupancy, as previously shown in yeast and Drosophila (1,31,37,38). At the same time, many NFRs, including a prominent NFR at the Msh3/DHFR promoter, were not enriched for pre-RCs, suggesting that, as for yeast and Drosophila (1,37), only NFRs associated with specific additional features are permissive for pre-RC assembly. Moreover, our results demonstrate that, even though many replication origins map to transcription start sites in yeast, Drosophila, mouse and human (1,31,32,37), a strong promoter-associated NFR is not sufficient to constitute a strong ORC binding site or an efficient origin of replication.
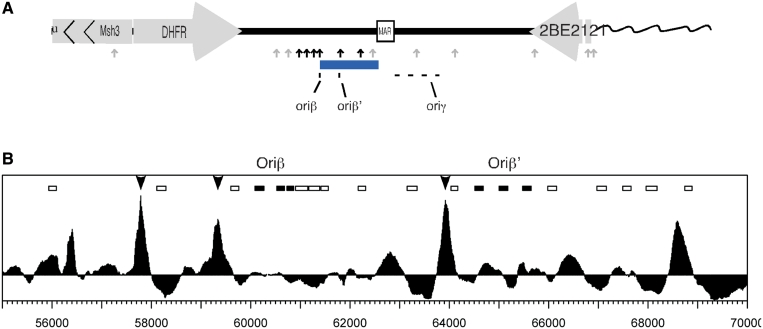
Our study focuses on a highly de-localized initiation zone. Previous origin mapping results at the CHO DHFR locus have concluded that replication can initiate at many sites distributed throughout ∼50 kb, with the oriβ/β′ and oriγ regions being somewhat preferred (48,96,100). Our data agrees with this view of de-localized initiation with both the highest density of ChIP peaks and the highest abundance of initiation sites found in the same region as oriβ/β′ (Figure 7A). Multiple binding sites provide enough flexibility that deletion of 40 Kb out of the 50 Kb of the DHFR initiation zone had no effect on the overall kinetics of replication in the region (101). Presumably, pre-RCs in the remaining 10 Kb, some of which are below the level of our detection, can compensate for the loss of other pre-RC binding sites, including the two major sites. A lack of strong positioning information throughout the initiation zone permits nucleosomes to occupy different positions in different cells with equal thermodynamic stability (102). We suggest that this flexibility in nucleosome positioning may facilitate ORCs competition with nucleosomes, allowing ORC to bind many sites throughout the region. A few of these sites are occupied at a sufficient frequency as to be detected by ChAP, for reasons that we still do not understand. Hence, ORC may not need a strongly positioned NFR in regions where nucleosome positions are highly flexible. Highly flexible ORC binding may lead to highly flexible initiation. Since it is clear that there also exist more localized sites of initiation in mammalian cells (21,22,31,32), it will be interesting to determine whether broad regions of low nucleosome positioning information are a frequent feature of de-localized initiation zones, while NFRs may characterize highly localized origins.
Figure 7.
pre-RC ChIP peaks are in close proximity to the sites of initiation. (A) Summary of results. Arrows indicate the ChAP peaks. Black arrows represent peaks that contain both wt and mutant ORC and MCM while grey arrows represent peaks with only wt present. The blue rectangle indicates the area with highest density of labeled fibers. The previously described preferred initiation sites are shown at the bottom. (B) ChAP results for the region containing the previously described major initiation site. The arrows indicate the peaks in which all tested pre-RC subunits were detected. The rectangles represent the probes shown in Figure 3 of Kobayashi et al. (46). Among them, the filled rectangles represent the probes that were mapped to the two initiation sites oriβ and oriβ′.
Why don't the strongest sites of pre-RC assembly predict the precise sites of preferred initiation? The only high-resolution mapping of initiation sites at the DHFR locus, restricted to ∼10 Kb around the major initiation site (46) identified two initiation sites lying 5 Kb apart (oriβ and oriβ′). While these regions do not overlap our ChAP peaks, both lay within a 1 Kb distance from one of the peaks where all subunits were detected (Figure 7B). Based on studies in yeasts, it is generally assumed that replication initiates near the sites of ORC binding (103,104). However, it is possible that replication may initiate at sites that are not precisely aligned with pre-RCs, as suggested by studies at the Drosophila chorion locus (20) and at some fission yeast origins (105). Initiation sites may also be partially determined by structural proteins that cause folding and partial melting of the DNA (106). The interpretation of our data described above suggests yet another possibility. Given that replication initiates at only a subset of pre-RC assembly sites by an as yet undefined mechanism, the frequency with which ORC occupies different sites need not reflect the frequency with which replication initiates at those sites. It is now of considerable interest to identify more comprehensively the sites of pre-RC assembly in regions of more localized initiation of replication versus de-localized initiation zones.
The mapping of pre-RC proteins to specific sites in mammalian cells has not previously been reported. In this work we describe a system for mapping pre-RC assembly sites in a mammalian system. At present we do not know whether the biotinylated tag, the synchronization, or the high copy number of the DHFR locus in our cells was the primary factor allowing us to detect pre-RC enrichment at specific sites. However, this method should be easily adaptable to other mammalian cells or in conjunction with next-generation sequencing to map pre-RCs at single copy sites or across the entire genome.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute for General Medical Sciences grant RO1GM083337 (to D.M.G.) and R01 CA41644 (to M.M.L.B.). Funding for open access charge: the GM083337 grant.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank T. Ryba for assistance with array analysis and to the members of the Gilbert lab for helpful discussion. We thank D. MacAlpine for helpful comments on the article.
REFERENCES
- 1.Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM. Conserved nucleosome positioning defines replication origins. Genes Dev. 2010;24:748–753. doi: 10.1101/gad.1913210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kearsey S. Analysis of sequences conferring autonomous replication in baker's yeast. EMBO J. 1983;2:1571–1575. doi: 10.1002/j.1460-2075.1983.tb01626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M. Eukaryotic chromosome DNA replication: where, when, and how? Annu. Rev. Biochem. 2010;79:89–130. doi: 10.1146/annurev.biochem.052308.103205. [DOI] [PubMed] [Google Scholar]
- 4.Chuang RY, Kelly TJ. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc. Natl Acad. Sci. USA. 1999;96:2656–2661. doi: 10.1073/pnas.96.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilbert DM. In search of the holy replicator. Nat. Rev. Mol. Cell. Biol. 2004;5:848–855. doi: 10.1038/nrm1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, Walter JC. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 2003;17:1894–1908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aladjem MI. Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat. Rev. Genet. 2007;8:588–600. doi: 10.1038/nrg2143. [DOI] [PubMed] [Google Scholar]
- 8.Lin HB, Dijkwel PA, Hamlin JL. Promiscuous initiation on mammalian chromosomal DNA templates and its possible suppression by transcription. Exp. Cell Res. 2005;308:53–64. doi: 10.1016/j.yexcr.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 9.Krysan PJ, Calos MP. Replication initiates at multiple locations on an autonomously replicating plasmid in human cells. Mol. Cell. Biol. 1991;11:1464–1472. doi: 10.1128/mcb.11.3.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mirkin EV, Castro Roa D, Nudler E, Mirkin SM. Transcription regulatory elements are punctuation marks for DNA replication. Proc. Natl Acad. Sci. USA. 2006;103:7276–7281. doi: 10.1073/pnas.0601127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Courbet S, Gay S, Arnoult N, Wronka G, Anglana M, Brison O, Debatisse M. Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature. 2008;455:557–560. doi: 10.1038/nature07233. [DOI] [PubMed] [Google Scholar]
- 12.Anglana M, Apiou F, Bensimon A, Debatisse M. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003;114:385–394. doi: 10.1016/s0092-8674(03)00569-5. [DOI] [PubMed] [Google Scholar]
- 13.Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schubeler D, Gilbert DM. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Norio P, Kosiyatrakul S, Yang Q, Guan Z, Brown NM, Thomas S, Riblet R, Schildkraut CL. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol. Cell. 2005;20:575–587. doi: 10.1016/j.molcel.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 15.Zhou J, Ashouian N, Delepine M, Matsuda F, Chevillard C, Riblet R, Schildkraut CL, Birshtein BK. The origin of a developmentally regulated Igh replicon is located near the border of regulatory domains for Igh replication and expression. Proc. Natl Acad. Sci. USA. 2002;99:13693–13698. doi: 10.1073/pnas.212392399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paixao S, Colaluca IN, Cubells M, Peverali FA, Destro A, Giadrossi S, Giacca M, Falaschi A, Riva S, Biamonti G. Modular structure of the human lamin B2 replicator. Mol. Cell. Biol. 2004;24:2958–2967. doi: 10.1128/MCB.24.7.2958-2967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altman AL, Fanning E. Defined sequence modules and an architectural element cooperate to promote initiation at an ectopic mammalian chromosomal replication origin. Mol. Cell. Biol. 2004;24:4138–4150. doi: 10.1128/MCB.24.10.4138-4150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu G, Malott M, Leffak M. Multiple functional elements comprise a Mammalian chromosomal replicator. Mol. Cell. Biol. 2003;23:1832–1842. doi: 10.1128/MCB.23.5.1832-1842.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan Z, Hughes CM, Kosiyatrakul S, Norio P, Sen R, Fiering S, Allis CD, Bouhassira EE, Schildkraut CL. Decreased replication origin activity in temporal transition regions. J. Cell Biol. 2009;187:623–635. doi: 10.1083/jcb.200905144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Austin RJ, Orr-Weaver TL, Bell SP. Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes Dev. 1999;13:2639–2649. doi: 10.1101/gad.13.20.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdurashidova G, Deganuto M, Klima R, Riva S, Biamonti G, Giacca M, Falaschi A. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science. 2000;287:2023–2026. doi: 10.1126/science.287.5460.2023. [DOI] [PubMed] [Google Scholar]
- 22.Romero J, Lee H. Asymmetric bidirectional replication at the human DBF4 origin. Nat. Struct. Mol. Biol. 2008;15:722–729. doi: 10.1038/nsmb.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ina S, Sasaki T, Yokota Y, Shinomiya T. A broad replication origin of Drosophila melanogaster, oriDalpha, consists of AT-rich multiple discrete initiation sites. Chromosoma. 2001;109:551–564. doi: 10.1007/s004120000112. [DOI] [PubMed] [Google Scholar]
- 24.Dijkwel PA, Mesner LD, Levenson VV, d'Anna J, Hamlin JL. Dispersive initiation of replication in the Chinese hamster rhodopsin locus. Exp. Cell Res. 2000;256:150–157. doi: 10.1006/excr.2000.4809. [DOI] [PubMed] [Google Scholar]
- 25.Dijkwel PA, Hamlin JL. The Chinese hamster dihydrofolate reductase origin consists of multiple potential nascent-strand start sites. Mol. Cell. Biol. 1995;15:3023–3031. doi: 10.1128/mcb.15.6.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Little RD, Platt TH, Schildkraut CL. Initiation and termination of DNA replication in human rRNA genes. Mol. Cell. Biol. 1993;13:6600–6613. doi: 10.1128/mcb.13.10.6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert DM. Evaluating genome-scale approaches to eukaryotic DNA replication. Nat. Rev. Genet. 2010;11:673–684. doi: 10.1038/nrg2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karnani N, Taylor CM, Malhotra A, Dutta A. Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Mol. Biol. Cell. 2010;21:393–404. doi: 10.1091/mbc.E09-08-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lucas I, Palakodeti A, Jiang Y, Young DJ, Jiang N, Fernald AA, Le Beau MM. High-throughput mapping of origins of replication in human cells. EMBO Rep. 2007;8:770–777. doi: 10.1038/sj.embor.7401026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cadoret JC, Prioleau MN. Genome-wide approaches to determining origin distribution. Chromosome Res. 2010;18:79–89. doi: 10.1007/s10577-009-9094-2. [DOI] [PubMed] [Google Scholar]
- 31.Sequeira-Mendes J, Diaz-Uriarte R, Apedaile A, Huntley D, Brockdorff N, Gomez M. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet. 2009;5:e1000446. doi: 10.1371/journal.pgen.1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cadoret JC, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, Prioleau MN. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc. Natl Acad. Sci. USA. 2008;105:15837–15842. doi: 10.1073/pnas.0805208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JK, Moon KY, Jiang Y, Hurwitz J. The Schizosaccharomyces pombe origin recognition complex interacts with multiple AT-rich regions of the replication origin DNA by means of the AT-hook domains of the spOrc4 protein. Proc. Natl Acad. Sci. USA. 2001;98:13589–13594. doi: 10.1073/pnas.251530398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong D, DePamphilis ML. Site-specific DNA binding of the Schizosaccharomyces pombe origin recognition complex is determined by the Orc4 subunit. Mol. Cell. Biol. 2001;21:8095–8103. doi: 10.1128/MCB.21.23.8095-8103.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balasov M, Huijbregts RP, Chesnokov I. Role of the Orc6 protein in origin recognition complex-dependent DNA binding and replication in Drosophila melanogaster. Mol. Cell. Biol. 2007;27:3143–3153. doi: 10.1128/MCB.02382-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Remus D, Beall EL, Botchan MR. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J. 2004;23:897–907. doi: 10.1038/sj.emboj.7600077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacAlpine HK, Gordan R, Powell SK, Hartemink AJ, MacAlpine DM. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 2010;20:201–211. doi: 10.1101/gr.097873.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berbenetz NM, Nislow C, Brown GW. Diversity of eukaryotic DNA replication origins revealed by genome-wide analysis of chromatin structure. PLoS Genet. 2010;6:e1001092. doi: 10.1371/journal.pgen.1001092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin S, Deng W, Hu L, Kong X. The impact of nucleosome positioning on the organization of replication origins in eukaryotes. Biochem. Biophys. Res. Commun. 2009;385:363–368. doi: 10.1016/j.bbrc.2009.05.072. [DOI] [PubMed] [Google Scholar]
- 40.Lipford JR, Bell SP. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol. Cell. 2001;7:21–30. doi: 10.1016/s1097-2765(01)00151-4. [DOI] [PubMed] [Google Scholar]
- 41.Schepers A, Papior P. Why are we where we are? Understanding replication origins and initiation sites in eukaryotes using ChIP-approaches. Chromosome Res. 2010;18:63–77. doi: 10.1007/s10577-009-9087-1. [DOI] [PubMed] [Google Scholar]
- 42.Hamlin JL, Mesner LD, Dijkwel PA. A winding road to origin discovery. Chromosome Res. 2010;18:45–61. doi: 10.1007/s10577-009-9089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milbrandt JD, Heintz NH, White WC, Rothman SM, Hamlin JL. Methotrexate-resistant Chinese hamster ovary cells have amplified a 135-kilobase-pair region that includes the dihydrofolate reductase gene. Proc. Natl Acad. Sci. USA. 1981;78:6043–6047. doi: 10.1073/pnas.78.10.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dijkwel PA, Hamlin JL. Initiation of DNA replication in the dihydrofolate reductase locus is confined to the early S period in CHO cells synchronized with the plant amino acid mimosine. Mol. Cell. Biol. 1992;12:3715–3722. doi: 10.1128/mcb.12.9.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vaughn JP, Dijkwel PA, Hamlin JL. Replication initiates in a broad zone in the amplified cho dihydrofolate-reductase domain. Cell. 1990;61:1075–1087. doi: 10.1016/0092-8674(90)90071-l. [DOI] [PubMed] [Google Scholar]
- 46.Kobayashi T, Rein T, DePamphilis ML. Identification of primary initiation sites for DNA replication in the hamster dihydrofolate reductase gene initiation zone. Mol. Cell. Biol. 1998;18:3266–3277. doi: 10.1128/mcb.18.6.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leu TH, Hamlin JL. High-resolution mapping of replication fork movement through the amplified dihydrofolate-reductase domain in cho cells by in-gel renaturation analysis. Mol. Cell. Biol. 1989;9:523–531. doi: 10.1128/mcb.9.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S, Dijkwel PA, Hamlin JL. Lagging-strand, early-labelling, and two-dimensional gel assays suggest multiple potential initiation sites in the Chinese hamster dihydrofolate reductase origin. Mol. Cell. Biol. 1998;18:39–50. doi: 10.1128/mcb.18.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilbert DM, Miyazawa H, DePamphilis ML. Site-specific initiation of DNA replication in Xenopus egg extract requires nuclear structure. Mol. Cell. Biol. 1995;15:2942–2954. doi: 10.1128/mcb.15.6.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Izumi M, Gilbert DM. Homogeneous tetracycline-regulatable gene expression in mammalian fibroblasts. J. Cell Biochem. 1999;76:280–289. doi: 10.1002/(sici)1097-4644(20000201)76:2<280::aid-jcb11>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 51.de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc. Natl Acad. Sci. USA. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vashee S, Simancek P, Challberg MD, Kelly TJ. Assembly of the human origin recognition complex. J. Biol. Chem. 2001;276:26666–26673. doi: 10.1074/jbc.M102493200. [DOI] [PubMed] [Google Scholar]
- 54.McNairn AJ, Okuno Y, Misteli T, Gilbert DM. Chinese hamster ORC subunits dynamically associate with chromatin throughout the cell-cycle. Exp. Cell Res. 2005;308:345–356. doi: 10.1016/j.yexcr.2005.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lubelsky Y, Reuven N, Shaul Y. Autorepression of rfx1 gene expression: functional conservation from yeast to humans in response to DNA replication arrest. Mol. Cell. Biol. 2005;25:10665–10673. doi: 10.1128/MCB.25.23.10665-10673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu XS, Meyer CA. ChIP-Chip: algorithms for calling binding sites. Methods Mol. Biol. 2009;556:165–175. doi: 10.1007/978-1-60327-192-9_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song JS, Johnson WE, Zhu X, Zhang X, Li W, Manrai AK, Liu JS, Chen R, Liu XS. Model-based analysis of two-color arrays (MA2C) Genome Biol. 2007;8:R178. doi: 10.1186/gb-2007-8-8-r178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, Cao H, Yu M, Rosenzweig E, Goldy J, Haydock A, et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat. Methods. 2006;3:511–518. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- 59.Ozsolak F, Song JS, Liu XS, Fisher DE. High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- 60.Gupta S, Dennis J, Thurman RE, Kingston R, Stamatoyannopoulos JA, Noble WS. Predicting human nucleosome occupancy from primary sequence. PLoS Comput. Biol. 2008;4:e1000134. doi: 10.1371/journal.pcbi.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Michalet X, Ekong R, Fougerousse F, Rousseaux S, Schurra C, Hornigold N, van Slegtenhorst M, Wolfe J, Povey S, Beckmann JS, et al. Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science. 1997;277:1518–1523. doi: 10.1126/science.277.5331.1518. [DOI] [PubMed] [Google Scholar]
- 62.Labit H, Goldar A, Guilbaud G, Douarche C, Hyrien O, Marheineke K. A simple and optimized method of producing silanized surfaces for FISH and replication mapping on combed DNA fibers. Biotechniques. 2008;45:649–652, 654, 656–648. doi: 10.2144/000113002. [DOI] [PubMed] [Google Scholar]
- 63.Lebofsky R, Heilig R, Sonnleitner M, Weissenbach J, Bensimon A. DNA replication origin interference increases the spacing between initiation events in human cells. Mol. Biol. Cell. 2006;17:5337–5345. doi: 10.1091/mbc.E06-04-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Folter S, Urbanus SL, van Zuijlen LG, Kaufmann K, Angenent GC. Tagging of MADS domain proteins for chromatin immunoprecipitation. BMC Plant Biol. 2007;7:47. doi: 10.1186/1471-2229-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Werven FJ, Timmers HT. The use of biotin tagging in Saccharomyces cerevisiae improves the sensitivity of chromatin immunoprecipitation. Nucleic Acids Res. 2006;34:e33. doi: 10.1093/nar/gkl003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 67.Kim J, Cantor AB, Orkin SH, Wang J. Use of in vivo biotinylation to study protein-protein and protein-DNA interactions in mouse embryonic stem cells. Nat. Protoc. 2009;4:506–517. doi: 10.1038/nprot.2009.23. [DOI] [PubMed] [Google Scholar]
- 68.Hsu SM, Raine L, Fanger H. Use of avidin-biotin-peroxidase complex (ABC) in immunoperoxidase techniques: a comparison between ABC and unlabeled antibody (PAP) procedures. J. Histochem. Cytochem. 1981;29:577–580. doi: 10.1177/29.4.6166661. [DOI] [PubMed] [Google Scholar]
- 69.Okuno Y, McNairn AJ, den Elzen N, Pines J, Gilbert DM. Stability, chromatin association and functional activity of mammalian pre-replication complex proteins during the cell cycle. EMBO J. 2001;20:4263–4277. doi: 10.1093/emboj/20.15.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gelbart ME, Bachman N, Delrow J, Boeke JD, Tsukiyama T. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 2005;19:942–954. doi: 10.1101/gad.1298905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takehara M, Makise M, Takenaka H, Asano T, Mizushima T. Analysis of mutant origin recognition complex with reduced ATPase activity in vivo and in vitro. Biochem. J. 2008;413:535–543. doi: 10.1042/BJ20070484. [DOI] [PubMed] [Google Scholar]
- 72.Brewster AS, Wang G, Yu X, Greenleaf WB, Carazo JM, Tjajadia M, Klein MG, Chen XS. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc. Natl Acad. Sci. USA. 2008;105:20191–20196. doi: 10.1073/pnas.0808037105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bochman ML, Bell SP, Schwacha A. Subunit organization of Mcm2-7 and the unequal role of active sites in ATP hydrolysis and viability. Mol. Cell. Biol. 2008;28:5865–5873. doi: 10.1128/MCB.00161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Speck C, Stillman B. Cdc6 ATPase activity regulates ORC x Cdc6 stability and the selection of specific DNA sequences as origins of DNA replication. J. Biol. Chem. 2007;282:11705–11714. doi: 10.1074/jbc.M700399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moreau MJ, McGeoch AT, Lowe AR, Itzhaki LS, Bell SD. ATPase site architecture and helicase mechanism of an archaeal MCM. Mol. Cell. 2007;28:304–314. doi: 10.1016/j.molcel.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 76.Jenkinson ER, Chong JP. Minichromosome maintenance helicase activity is controlled by N- and C-terminal motifs and requires the ATPase domain helix-2 insert. Proc. Natl Acad. Sci. USA. 2006;103:7613–7618. doi: 10.1073/pnas.0509297103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Costa A, Pape T, van Heel M, Brick P, Patwardhan A, Onesti S. Structural studies of the archaeal MCM complex in different functional states. J. Struct. Biol. 2006;156:210–219. doi: 10.1016/j.jsb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 78.Ying CY, Gautier J. The ATPase activity of MCM2-7 is dispensable for pre-RC assembly but is required for DNA unwinding. EMBO J. 2005;24:4334–4344. doi: 10.1038/sj.emboj.7600892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Speck C, Chen Z, Li H, Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat. Struct. Mol. Biol. 2005;12:965–971. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pape T, Meka H, Chen S, Vicentini G, van Heel M, Onesti S. Hexameric ring structure of the full-length archaeal MCM protein complex. EMBO Rep. 2003;4:1079–1083. doi: 10.1038/sj.embor.7400010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gomez EB, Catlett MG, Forsburg SL. Different phenotypes in vivo are associated with ATPase motif mutations in Schizosaccharomyces pombe minichromosome maintenance proteins. Genetics. 2002;160:1305–1318. doi: 10.1093/genetics/160.4.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bochman ML, Schwacha A. The Saccharomyces cerevisiae Mcm6/2 and Mcm5/3 ATPase active sites contribute to the function of the putative Mcm2-7 ‘gate’. Nucleic Acids Res. 2010;38:6078–6088. doi: 10.1093/nar/gkq422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bowers JL, Randell JC, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol. Cell. 2004;16:967–978. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 84.Dimitrova DS, Prokhorova TA, Blow JJ, Todorov IT, Gilbert DM. Mammalian nuclei become licensed for DNA replication during late telophase. J. Cell Sci. 2002;115:51–59. doi: 10.1242/jcs.115.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dimitrova DS, Gilbert DM. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol. Cell. 1999;4:983–993. doi: 10.1016/s1097-2765(00)80227-0. [DOI] [PubMed] [Google Scholar]
- 86.Wu JR, Gilbert DM. A distinct G1 step required to specify the Chinese hamster DHFR replication origin. Science. 1996;271:1270–1272. doi: 10.1126/science.271.5253.1270. [DOI] [PubMed] [Google Scholar]
- 87.Wu JR, Keezer SM, Gilbert DM. Transformation abrogates an early G1-phase arrest point required for specification of the Chinese hamster DHFR replication origin. Embo J. 1998;17:1810–1818. doi: 10.1093/emboj/17.6.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu JR, Gilbert DM. The replication origin decision point is a mitogen-independent, 2-aminopurine-sensitive, G1-phase event that precedes restriction point control. Mol. Cell. Biol. 1997;17:4312–4321. doi: 10.1128/mcb.17.8.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sasaki T, Ramanathan S, Okuno Y, Kumagai C, Shaikh SS, Gilbert DM. The Chinese hamster dihydrofolate reductase replication origin decision point follows activation of transcription and suppresses initiation of replication within transcription units. Mol. Cell. Biol. 2006;26:1051–1062. doi: 10.1128/MCB.26.3.1051-1062.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Alexandrow MG, Ritzi M, Pemov A, Hamlin JL. A potential role for mini-chromosome maintenance (MCM) proteins in initiation at the dihydrofolate reductase replication origin. J. Biol. Chem. 2002;277:2702–2708. doi: 10.1074/jbc.M108118200. [DOI] [PubMed] [Google Scholar]
- 91.Pemov A, Bavykin S, Hamlin JL. Proximal and long-range alterations in chromatin structure surrounding the Chinese hamster dihydrofolate reductase promoter. Biochemistry. 1995;34:2381–2392. doi: 10.1021/bi00007a034. [DOI] [PubMed] [Google Scholar]
- 92.Shimada T, Inokuchi K, Nienhuis AW. Chromatin structure of the human dihydrofolate reductase gene promoter. Multiple protein-binding sites. J. Biol. Chem. 1986;261:1445–1452. [PubMed] [Google Scholar]
- 93.Azizkhan JC, Vaughn JP, Christy RJ, Hamlin JL. Nucleotide sequence and nuclease hypersensitivity of the Chinese hamster dihydrofolate reductase gene promoter region. Biochemistry. 1986;25:6228–6236. doi: 10.1021/bi00368a059. [DOI] [PubMed] [Google Scholar]
- 94.Segal E, Widom J. What controls nucleosome positions? Trends Genet. 2009;25:335–343. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rando OJ, Ahmad K. Rules and regulation in the primary structure of chromatin. Curr. Opin. Cell Biol. 2007;19:250–256. doi: 10.1016/j.ceb.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 96.Anachkova B, Hamlin JL. Replication in the amplified dihydrofolate-reductase domain in cho cells may initiate at 2 distinct sites, one of which is a repetitive sequence element. Mol. Cell. Biol. 1989;9:532–540. doi: 10.1128/mcb.9.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hyrien O, Maric C, Mechali M. Transition in specification of embryonic metazoan DNA replication origins. Science. 1995;270:994–997. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- 98.Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20:761–770. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gilbert DM. Origins go plastic. Mol. Cell. 2005;20:657–658. doi: 10.1016/j.molcel.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 100.Handeli S, Klar A, Meuth M, Cedar H. Mapping replication units in animal cells. Cell. 1989;57:909–920. doi: 10.1016/0092-8674(89)90329-2. [DOI] [PubMed] [Google Scholar]
- 101.Mesner LD, Li X, Dijkwel PA, Hamlin JL. The dihydrofolate reductase origin of replication does not contain any nonredundant genetic elements required for origin activity. Mol. Cell. Biol. 2003;23:804–814. doi: 10.1128/MCB.23.3.804-814.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pugh BF. A preoccupied position on nucleosomes. Nat. Struct. Mol. Biol. 2010;17:923. doi: 10.1038/nsmb0810-923. [DOI] [PubMed] [Google Scholar]
- 103.Bielinsky AK, Gerbi SA. Where it all starts: eukaryotic origins of DNA replication. J. Cell. Sci. 2001;114:643–651. doi: 10.1242/jcs.114.4.643. [DOI] [PubMed] [Google Scholar]
- 104.Bielinsky AK, Gerbi SA. Chromosomal ARS1 has a single leading strand start site. Mol. Cell. 1999;3:477–486. doi: 10.1016/s1097-2765(00)80475-x. [DOI] [PubMed] [Google Scholar]
- 105.Ogawa Y, Takahashi T, Masukata H. Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Mol. Cell. Biol. 1999;19:7228–7236. doi: 10.1128/mcb.19.10.7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schroll AL, Heintz NH. Chemical footprinting of structural and functional elements of dhfr oribeta during the CHOC 400 cell cycle. Gene. 2004;332:139–147. doi: 10.1016/j.gene.2004.02.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.