Abstract
The XPA (Xeroderma pigmentosum A) protein is one of the six core factors of the human nucleotide excision repair system. In this study we show that XPA is a rate-limiting factor in all human cell lines tested, including a normal human fibroblast cell line. The level of XPA is controlled at the transcriptional level by the molecular circadian clock and at the post-translational level by a HECT domain family E3 ubiquitin ligase called HERC2. Stabilization of XPA by downregulation of HERC2 moderately enhances excision repair activity. Conversely, downregulation of XPA by siRNA reduces excision repair activity in proportion to the level of XPA. Ubiquitination and proteolysis of XPA are inhibited by DNA damage that promotes tight association of the protein with chromatin and its dissociation from the HERC2 E3 ligase. Finally, in agreement with a recent report, we find that XPA is post-translationally modified by acetylation. However, contrary to the previous claim, we find that in mouse liver only a small fraction of XPA is acetylated and that downregulation of SIRT1 deacetylase in two human cell lines does not affect the overall repair rate. Collectively, the data reveal that XPA is a limiting factor in excision repair and that its level is coordinately regulated by the circadian clock, the ubiquitin–proteasome system and DNA damage.
INTRODUCTION
Nucleotide excision repair (excision repair) is the primary repair system for removing bulky base adducts from DNA in organisms ranging from Escherichia coli to humans (1–4). The repair system has a wide substrate spectrum, ranging from lesions such as thymine glycols that cause a modest helical distortion to the benzo[a]pyrene-guanine adducts that induce major changes in DNA structures. Intrinsic to such a repair system of low specificity is the ability to process undamaged DNA at a low but finite rate. Indeed, it has been found that both human and bacterial excision repair systems attack undamaged DNA at a measurable rate (5) and excise undamaged DNA fragments leading to gratuitous excision and repair synthesis. This gratuitous DNA repair may result in spontaneous mutagenesis (6,7) with undesirable consequences. Thus, from an evolutionary perspective, it would be beneficial if the levels of repair factors were under tight regulation such that repair activity increased at times of genotoxic stress or times of potential genotoxic stress and were downregulated when the damage is eliminated or the probability of damage occurring is relatively low. Indeed, in prokaryotes the levels of excision repair proteins UvrA and UvrB, which are involved in damage recognition, are tightly controlled by the SOS response system and the ClpXP protease: the levels of UvrA and UvrB increase in response to DNA damage by SOS-mediated transcriptional upregulation and, upon elimination of DNA damage, the transcription of the SOS genes including UvrA and UvrB is turned off and the accumulated UvrA and possibly UvrB are degraded by ClpXP protease (8,9).
In mammalian cells, DNA damage excision is carried out by six repair factors comprising 16 polypeptides, RPA, XPA, XPC, TFIIH, XPG and XPF•ERCC1 (1,2). Additionally, DDB2 (the XPE gene product) in complex with DDB1, UV-DDB, may stimulate excision repair (10). There is no known SOS response in mammalian cells in terms of coordinated transcriptional response to UV and UV-mimetic agents that promotes cell survival by upregulating excision-, recombination- and post-replication repair/recovery pathways. Instead, there is a frequently discussed, but mechanistically ill-defined, system involving transcriptional induction and post-translational ubiquitination of XPC and DDB2. In short, while there is convincing evidence that DDB2 and XPC transcription is induced by UV and that both proteins are subject to regulation by proteolysis, there is no convincing evidence that DDB2 (in the form of UV-DDB) plays any direct role in excision repair or that transcriptional induction and ubiquitination of XPC affect the level or activity of the protein (10–12). In contrast to DDB2, which may or may not play a role in repair and XPC that is required for transcription independent but not for transcription-coupled repair, XPA is an essential component of both pathways for excision repair and its transcriptional and post-transcriptional regulation may have significant effects on cellular repair and survival following exposure to UV and UV-mimetic agents.
Recently, we reported that excision repair in mice is regulated by the circadian clock such that the repair activity changed by ∼10-fold over the course of the day (13,14). We reported that this oscillation was accomplished by transcriptional control of the XPA gene by the core clock transcription–translation feedback loop and presented preliminary data that the putative mammalian HERC2 E3 ligase contributed to the robust high amplitude oscillatory pattern of XPA and hence of excision repair. In this article, we expand on the previous findings to demonstrate that the XPA level is rate limiting in vivo in normal human fibroblasts and that manipulating XPA levels by gene knockdown of XPA or HERC2 using siRNA affects the rate of excision repair in vivo. We further show that the HECT domain of HERC2 is necessary and sufficient for ubiquitination of XPA. Finally, we show that XPA is subject to acetylation in mouse liver but that only a small percent of XPA is acetylated and the acetylation pattern does not correlate with excision repair activity. Furthermore, downregulation of NAD+-dependent SIRT1 deacetylase in two human cell lines has no measurable effect on the overall rate of repair by human excision nuclease. Taken together, our data indicate that XPA and hence excision repair in mammals is regulated by the circadian clock at the transcriptional level and by HERC2 ubiquitin ligase at the post-translational level.
MATERIALS AND METHODS
Cell culture and siRNA transfection
HEK293T, A549, HeLa (obtained from ATCC), NHF-1(15) and XP-A (XP2OS) cell lines [Coriell Institute, (16)] were grown in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) supplemented with 10% fetal bovine serum and 100 U/ml penicillin G and 0.1 mg/ml streptomycin. For the analysis of protein stability, cells were treated with 20 µg/ml cycloheximide (Sigma) for the indicated times. DharmaFECT reagent (Dharmacon) was used according to the manufacturer’s directions for transfection of ON-TARGET plus SMARTpool siRNA duplexes obtained from Dharmacon [XPA (L-005067-00-0050), HERC1 (L-007181-00-0005), HERC2 (L-007180-00-0020), RNF8 (L-006900-00-0005) or Cyclophilin B (D-001820-01-20) as a control]. siRNA (100 nM) was used for transfection unless otherwise indicated.
UV-damage repair assay
Cells grown in 12-well plates to near confluency were washed once with pre-warmed Hank’s balanced salt solution (Gibco) prior to UV irradiation at the indicated J/m2 from a germicidal lamp (GE) emitting primarily UV-C light. After UV irradiation, culture media was added back to the cells and repair was allowed for the indicated times before harvesting. Genomic DNA preparation and transfer to membranes were described in a previous report (14). Cyclobutane thymine dimer (CPD) and 6–4 photoproduct [(6–4)PP] UV damages were detected by immunoslot blot using anti-CPD (Kamiya) and anti-(6–4)PP (Cosmo Bio) monoclonal antibodies, respectively. After the immunoslot blot assay, total DNA amounts loaded onto the membrane were visualized by Sybr-gold staining, and these values were used to normalize the values.
Immunoblotting
Protein levels from whole cell lysates or fractionated extracts were determined by immunoblot assay. Antibodies used in this study include XPA (Kamiya), XPF and RNF8 (Abcam), GAPDH (Cell Signaling Technology), Cyclophilin B and PCNA (Santa Cruz Biotechnology), HERC1 and HERC2 (Bethyl Laboratory), Flag (Sigma), histone H2AX, H3, SIRT1 and Acetyl-lysine (Millipore) and Cry1 (17).
RT–PCR
Total RNA preparation and reverse transcription were done as described previously (18). The following primers were used for PCR (for RNF8 5′: CTACCTCTAGGCATGTTTCA and 3′: ATTGTGACCAATGGCAGATC; for HERC2 5′: CTGCCCTTCACAGTGCCAAG and 3′: GGTCTGGAGGGTTGTATTTA; for XPA 5′ CAGCCCCAAAGATAATTGAC and 3′: CGCTGCTTCTTACTGCTCGC; and for β-ACTIN 5′ GTTCCGATGCCCTGAGGCTC and 3′: CACTTGCGGTGCACGATGGA).
Recombinant protein purification
Flag-tagged XPA and HECT domain of HERC2 (wild-type and Cys→Ala mutant) were immunopurified using anti-Flag agarose (Sigma). Flag-XPA was prepared as reported previously (14). Human HECT domain of HERC2 was amplified by RT–PCR and subcloned into a Topo-TA vector (Invitrogen). This DNA was used as PCR template to introduce appropriate restriction sites and Flag and 6X His epitope tags at the 5′- and 3′-ends, respectively, and subcloned into the pcDNA3 vector. The C4762A mutation at the active site was introduced by site-directed mutagenesis (Stratagene QuikChange). DNA constructs were sequenced prior to use. The HECT domain of HERC2 was transiently expressed in HEK293T cells using Lipofectamine 2000 reagent (Invitrogen). After 24 h cells were harvested and lysed with TBS buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl) containing 0.5% NP-40, and purified with anti-Flag-agarose. After extensive washing with TBS buffer, the bound proteins were eluted with TBS buffer containing 0.3 mg/ml Flag-peptide (Sigma).
Ubiquitination assay
Either immunoprecipitated-HERC2 from HEK293T whole cell lysate (Figure 3C) or purified HECT domain of HERC2 (Figure 3D) was used in the ubiquitination assay with Flag-XPA purified from HEK293T cells as the substrate. Briefly 1.5 ng of E1 (UBE1), 10 ng of E2 (UbcH5a), 500 ng of HA-ubiquitin and 500 ng of purified Flag-XPA were incubated in a buffer containing 50 mM Tris pH 7.6, 5 mM MgCl2, 0.6 mM DTT and 2 mM ATP for 30 min at 30°C. Ubiquitinated-XPA was detected by immunoblot assay using either anti-Flag or anti-XPA antibody. Recombinant UBE1, GST-UbcH5a and HA-ubiquitin were purchased from Boston Biochem.
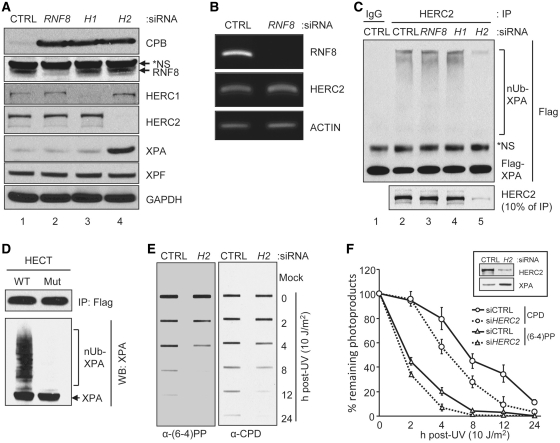
Figure 3.
Regulation of DNA excision repair by HERC2 via XPA stability control. (A) Protein levels were analyzed by immunoblotting with the indicated antibodies from A549 cells transfected with the indicated siRNAs for 96 h. Asterisk in the RNF8 blot indicates non-specific (NS) signal. CPB, cyclophilin B. (B) cDNA was prepared from cells transfected with the indicated siRNAs and was used for the amplification of the indicated genes using PCR. (C) HERC2 was immunoprecipitated from cells transfected with the indicated siRNAs and was used for the ubiquitination assay. Human Flag-XPA purified from 293T cells was used as a substrate. Total and ubiquitinated XPA were detected by immunoblotting with the anti-Flag antibody. The amount of HERC2 immunoprecipitated from each siRNA treated cell was analyzed by immunoblotting with the anti-HERC2 antibody. Asterisk in the Flag blot indicates NS signal. (D) Wild-type (WT) and mutant (Mut) HECT domain of HERC2 were transiently expressed in 293T cells, immuno-purified using Flag-agarose beads and used for the ubiquitination assay. Flag-XPA was used as a substrate. Total and ubiquitinated XPA were detected by immunoblotting with the anti-XPA antibody. The amount of HECT immunoprecipitated was analyzed by immunoblotting with anti-Flag antibody. (E) Residual (6–4)PP or CPD damage in the genomic DNA was detected by immunoslot blotting using damage specific monoclonal antibodies. (F) Average values and standard deviations from three independent experiments are shown. Protein levels were analyzed from cells transfected with siRNAs with the indicated antibodies (inset).
Mouse liver extracts
Cry1−/−Cry2−/− mice in C57BL/6J background were generated in our laboratory (17,19), and wild-type control animals (10-weeks-old C57BL/6J male mice) were purchased from the Jackson Laboratory (Bar Harbor, ME). Mice were maintained on a light/dark 12:12 schedule for at least 2 weeks before sacrifice. ZT0 is the time of light-on and ZT12 is the time of light-off. The mice were handled in accordance with the guidelines of the NIH and the University of North Carolina School of Medicine. At the indicated times, the mice were sacrificed by carbon dioxide exposure, liver tissues were removed, washed extensively with cold phosphate buffered saline (PBS), diced into <1 mm3 pieces and incubated with red blood cell lysing buffer (Sigma) for 5 min at 30°C. The liver tissues were homogenized in hypotonic cytosol extraction buffer [10 mM HEPES (pH 7.6), 40 mM KCl, 3 mM MgCl2, 2 mM DTT, 5% glycerol (vol/vol), 0.5% NP-40 and protease inhibitor cocktail (Roche)]. After centrifugation (1250g) for 5 min at 4°C, supernatant (cytosol extract) was removed and nuclei were washed extensively with cytosol buffer and then extracted with high salt nuclear extraction buffer [10 mM HEPES (pH 7.9), 450 mM NaCl, 1.5 mM MgCl2, 0.1 mM EGTA, 0.5 mM DTT, 25% glycerol (vol/vol) and protease inhibitor cocktail] to obtain soluble fraction (nuclear extract).
XPA acetylation levels in mouse liver tissue
Total acetylated proteins from liver extracts were immunoprecipitated with an anti-acetyl lysine antibody, and acetylated-XPA was detected by immunoblotting with anti-XPA antibody. For second immunoprecipitation (2° IP) analysis (Figure 5D), lysates from the first immunoprecipitation (1° IP) were subjected to a second round of precipitation to ensure that all acetylated XPA was identified.
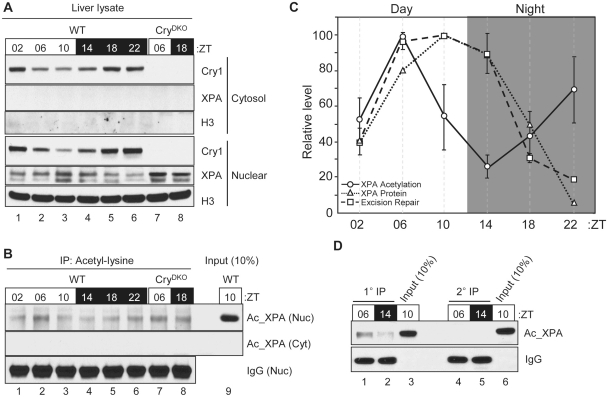
Figure 5.
Circadian oscillation of the acetylation of XPA. (A) Protein levels in liver cytosol and nuclear extracts were analyzed by immunoblotting with the indicated antibodies including histone H3 (H3). (B) Acetylated proteins from mouse liver cytosol and nuclear extracts were immunoprecipitated using acetyl-lysine antibody and the level of XPA was analyzed by immunoblotting for XPA. (C) Quantitative analysis of the acetylated XPA in the liver nuclear extract. Average values and standard deviations are shown from two independent experiments with two technical measurements on each together with XPA protein expression and DNA excision repair activity data from a previous report (14). (D) Acetylated proteins from mouse liver nuclear extracts were immunoprecipitated with anti-acetyl-lysine antibodies (1° IP). The immunodepleted-lysate from 1° IP was used for second round of immunoprecipitation (2° IP). The level of acetylated XPA from each IP was analyzed and compared with the input signal (10% of nuclear extracts used for IP) by immunoblotting for XPA.
RESULTS
XPA is rate limiting in excision repair in human cell lines
Previously, we reported that excision repair in mouse brain and liver extracts harvested at various times of the day exhibited a circadian rhythmicity with the minimum around the biological dawn and the maximum at the biological dusk. To determine the cause of this oscillatory pattern, we analyzed the expression profiles of the six core excision repair factors to determine which were responsible for the cyclic change in repair activity. We found that XPA exhibited a circadian pattern of expression with a maximum at the biological dusk and a minimum at biological dawn both at the mRNA and the protein levels. In contrast, the transcript and the protein levels of the polypeptides of all other five core excision repair factors and the excision repair accessory protein XPE (DDB2) remained essentially constant over the course of the day. Thus, we ascribed the circadian rhythmicity of excision repair activity to the oscillatory pattern of XPA protein level. In support of this interpretation, we found that low excision repair activity in extracts of tissues harvested in the early morning hours could be complemented with purified XPA protein (13).
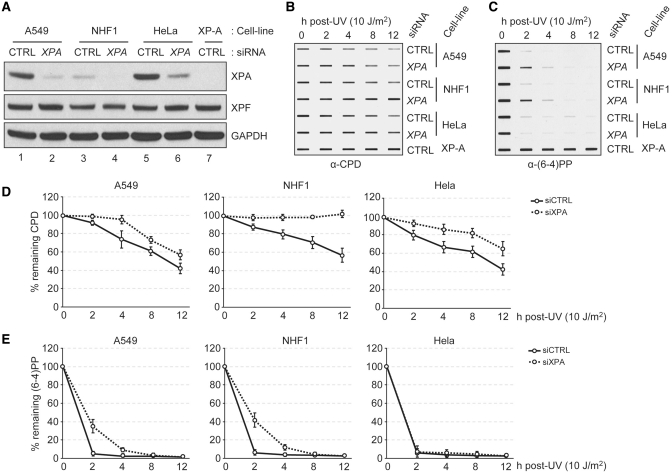
However, it has been reported that in a human cell line expressing XPA under control of the tetracycline on/off system there was no correlation between the XPA level in the cell and the excision repair activity, leading to the conclusion that XPA is not rate limiting for excision repair in human cells (20). Hence, we decided to downregulate XPA by siRNA in various human cell lines and test them for excision repair activity in vivo. The results obtained with a non-transformed cell line NHF-1 (TERT-immortalized normal human fibroblast), a lung cancer cell line with a functional p53 (A549) and a cervical cancer cell line with non-functional p53 (HeLa) are shown in Figure 1. The A549 and HeLa cells, p53 (+) and p53 (–), respectively, were chosen because it has been reported that p53 affects excision repair by an ill-defined mechanism (21,22) and therefore we wished to test the repair rate in both backgrounds, and we chose NHF-1 as representative of normal mammalian tissues. As is apparent from Figure 1A the two cancer cell lines express XPA at considerably higher levels than the normal human fibroblasts. As a consequence, even after downregulation by siRNA, these cell lines contain XPA (normalized to total protein) at a level comparable to that in normal fibroblasts before downregulation. These differences in the XPA levels are reflected in the repair kinetics of the UV photoproducts (Figure 1B–E): Downregulation of XPA to about 1/15th of its original level in NHF-1 abolished CPD repair and reduced the rate of (6–4)PP repair by a factor of ∼2. Repair of the latter lesion is only moderately affected because of its much higher affinity relative to CPD for XPA and the other damage recognition factors, XPC and RPA (23). Proportionally (but not in absolute terms) similar reductions in XPA levels in A549 and HeLa cells have more modest, but statistically significant, effects on CPD repair and even less pronounced (or in the case of HeLa no measurable effect) on the rate of repair of (6–4)PP. It must be noted, however, that these cancer cell lines are polyploid and hence contain more DNA as well as more XPA per cell and it is difficult to deconvolute the opposing effects of these two factors on the photoproduct density and repair rates. Hence, we decided to conduct our studies regarding the relationship between XPA level and excision repair capacity on the NHF-1 cell line which approximates a normal diploid human cell (15), and use other representative cell lines for the generality of our conclusions.
Figure 1.
Protein level of XPA is a determinant of the DNA excision repair activity. (A) Protein levels in human cells were analyzed by immunoblotting with the indicated antibodies from cells transfected with 100 nM siRNA targeting XPA or cyclophylin B (Control, CTRL). (B and C) Residual CPD (B) or (6–4)PP (C) damage in genomic DNA was detected by immunoslot blotting using damage specific monoclonal antibodies. Genomic DNA was prepared from cells transfected with indicated siRNAs for 24 h and repair was allowed for the indicated times. (D and E) Averages and error bars (standard deviations) are shown for data from two independent biological experiments and two technical repeats from each biological experiment.
Proportionality of XPA levels with rate of excision repair in normal human fibroblasts
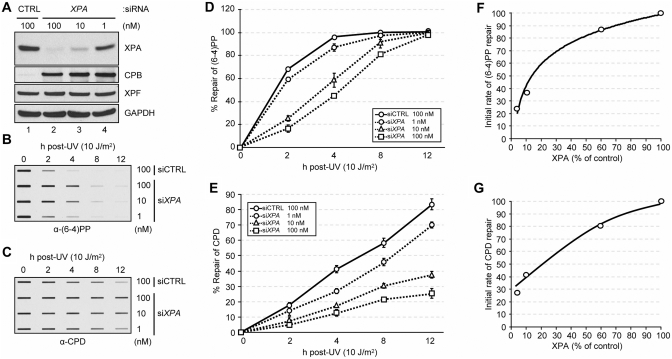
The data in Figure 1 show that drastic reduction in XPA level in NHF-1 leads to a severe reduction in the rate of repair of CPD. However, in mouse tissues, we observed a more gradual change in XPA level over the course of the day and a similarly gradual change in excision repair activity (24). Therefore, we wished to examine the effect of gradual changes in XPA level on the excision repair rate in our model system. To this end, we downregulated the XPA level in NHF-1 to varying degrees by using different amounts of siRNA in transfection, then irradiated the cells with UV and measured the rates of removal of UV photoproducts. The results are shown in Figure 2. As expected, varying the amounts of XPA siRNA resulted in different levels of XPA depletion (Figure 2A) and the reduction in XPA level proportionally affected the repair of both CPD and the (6–4)PP as measured by slot–blot assay (Figure 2B and C). Quantitative analyses of the slot–blot data reveal that there is an approximately linear relationship between the XPA level and the rate of repair of CPDs (Figure 2E and G). The rate of repair of (6–4)PP was only moderately affected when XPA was reduced to 60% of its original level but more severe changes in NHF-1 XPA levels, such as those observed between the zenith and nadir values of XPA in mouse tissues, resulted in reduction in repair rates comparable to those seen with CPDs (Figure 2D and F). These results are consistent with the in vitro (25) and in vivo (23) data that (6–4)PP is a 5- to 10-fold more efficient substrate for human excision nuclease and its rate of repair is better buffered against minor fluctuations of the rate limiting repair factors. Importantly, on the whole these data support the conclusion that rhythmic changes in XPA levels in mouse brain and liver are responsible for rhythmicity of excision repair in these tissues.
Figure 2.
Correlation between the XPA level and DNA excision repair activity in human cells. (A) Protein levels in NHF-1 cells transfected with different amounts of siRNA targeting XPA for 24 h were analyzed by immunoblotting with the indicated antibodies. CPB, cyclophilin B. (B and C) Residual (6–4)PP (B) or CPD (C) damage in genomic DNA was detected by immunoslot blotting using damage specific monoclonal antibodies. (D and E) Average repair values and standard deviations are shown for data from three independent experiments for the (6–4)PP (D) and CPD (E). (F and G) Initial repair rate for (6–4)PP (F) and CPD (G) were measured as a function of XPA protein level which was manipulated by knockdown of XPA using siRNA. The initial rates were calculated from the linear range of the kinetic data in D and E.
Regulation of XPA level by ubiquitination
With few exceptions (26) for a protein to show robust circadian oscillation, the gene encoding it must first, be transcribed with circadian rhythmicity and secondly, the protein must have a relatively short lifetime; even if a gene is transcribed with circadian rhythm it would not show high amplitude oscillation if it is stable (26). We previously reported that XPA transcription is controlled by the circadian clock and presented preliminary evidence that the XPA protein binds to and is ubiquitinated by the putative HERC2 ubiquitin ligase resulting in the degradation of XPA by the ubiquitin–proteasome system and short lifetime that contributes to rhythmicity (14). However, a subsequent study reported that HERC2 (which is a putative HECT domain E3 ligase) functions mainly as a scaffold for the RNF8 enzyme, a RING Finger type E3 ligase, which carries out the actual ubiquitination of some key proteins both in double-strand break repair (27) and in nucleotide excision repair (28). Therefore, we first wished to determine whether the HERC2-dependent ubiquitination of XPA observed in our previous study was actually carried out by HERC2 or by RNF8 that might have co-immunoprecipitated with HERC2 and, second, to assess the functional consequences of this ubiquitination. To this end, we carried out siRNA knockdown of HERC2 and RNF8 and tested the effects of these downregulations on XPA levels, XPA ubiquitination and UV damage repair. The results are shown in Figure 3. As seen in Figure 3A, both RNF8 and HERC2 are efficiently downregulated by the appropriate siRNAs. However, a strong cross-reacting non-specific band in the RNF8 immunoblot raised some concern regarding the efficacy of the RNF8 siRNA. Therefore, we tested the efficiency of this siRNA by RT–PCR. As is clear in Figure 3B the RNF8 siRNA is very effective in downregulating the transcript and presumably the RNF8 protein. Therefore, the effects of siRNAs of HERC2 and RNF8 on XPA ubiquitination are likely to reflect the direct effects of these E3 ligases on XPA. Importantly, Figure 3C shows that neither RNF8 knockdown nor the downregulation of the HERC2-related HECT-domain protein, HERC1, affects XPA ubiquitination. In contrast, downregulation of HERC2 essentially eliminated XPA ubiquitination, indicating that HERC2 is the E3 ligase for XPA.
To confirm this conclusion we expressed the HECT domain or the HECT domain containing the Cys→Ala mutation in the putative active site cysteine of HERC2 in HEK293T cells and immunopurified both the wild-type and the mutant forms. The purified proteins were tested for E3 ligase activity using XPA as a substrate. The results are shown in Figure 3D. As is clear from this figure, the wild-type HERC2 HECT domain, but not the mutant form, ubiquitinates XPA. Taken together the in vivo downregulation and the in vitro ubiquitination data lead us to conclude that HERC2 is the ubiquitin ligase responsible for ubiquitination and proteolytic degradation of XPA and, therefore, HERC2 contributes to the high amplitude circadian rhythmicity of XPA. Next, we wished to test the effect of HERC2 downregulation on XPA level and excision repair. HERC2 was downregulated by siRNA to ∼10% of its level in A549 cells and then the cells were irradiated with 10 J/m2 and the repair kinetics of (6–4)PP and CPD were determined. Representative images of the slot blots are shown in Figure 3E and quantitative analysis of the data is presented in Figure 3F along with the levels of HERC2 and XPA in control and HERC2 siRNA treated cells (Figure 3F, inset). As expected, downregulation of HERC2 leads to elevation of the steady-state level of XPA (Figure 3F, inset). In parallel with this increase in the XPA level the rates of repair of both UV photoproducts are elevated (Figure 3F), albeit not proportionally to the level of elevation of XPA. We conclude that XPA is a rate-limiting factor in human excision nuclease repair and that reducing the XPA level decreases the repair of DNA damage (Figure 2) and likewise, that elevation of XPA level above normal increases the rate of repair.
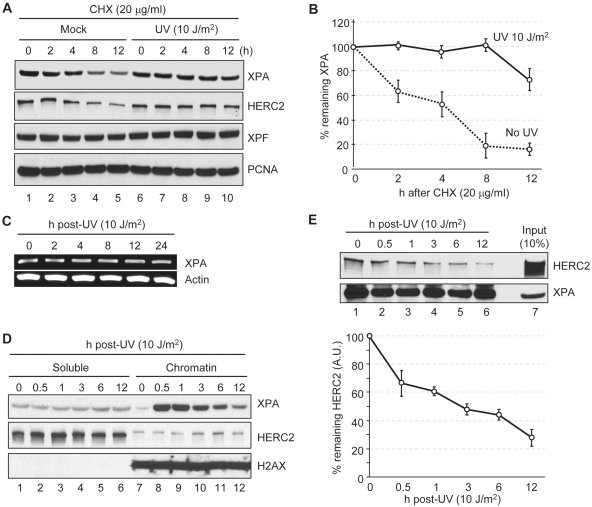
Effect of DNA damage on XPA–HERC2 interaction and XPA stability
To perform its repair function XPA must associate with damaged chromatin, so we asked whether UV damage affects the XPA–HERC2 interaction and therefore the stability of XPA. We used the A549 cell line for this series of experiments because these cells have functional p53, which is known to affect DNA repair by an ill-defined mechanism (21,22), and performing the experiments in a wild-type p53 background eliminates complications arising from the global effects of p53 mutation that affect cell physiology but are not directly involved in repair. To determine the effect of DNA damage on XPA stability, cells were irradiated with UV and incubated with protein synthesis inhibitor cycloheximide for various time periods and the levels of XPA and HERC2 were determined by immunoblotting. As is apparent from Figure 4A and B, and in agreement with previous findings (14), in the absence of DNA damage XPA decays with a half-life of 3–4 h. After UV irradiation, XPA is greatly stabilized, reaching a half-life of >12 h. Interestingly, HERC2 in the absence of DNA damage exhibited decay kinetics similar to that of XPA and was also stabilized by UV damage. It should be noted that, in agreement with previous reports (11), UV does not induce XPA transcription (Figure 4C) and hence the increase in XPA level after UV and in the presence of cycloheximide must be due to the stabilization of XPA protein. To gain some insight into the mechanism of XPA stabilization following UV damage, we analyzed the subcellular localization of XPA before and after DNA damage. All XPA, with and without DNA damage, is known to be localized in the nucleus (29). However, after DNA damage, XPA rapidly associates with the chromatin fraction without a significant change in HERC2 distribution (Figure 4D), suggesting that the DNA damage-promoted physical separation of XPA and HERC2 contributes to the increased stability of XPA following UV irradiation. This suggestion was reinforced by analyzing the XPA–HERC2 interaction by co-immunoprecipitation. The results are presented in Figure 4E. As is apparent from this figure, the fraction of HERC2 remaining associated with XPA linearly decreases with time over the 12 h period of the experiment even though there is no significant change in the overall levels of either XPA or HERC2 over this time span (compare Figure 4D and E, top panel). Thus, it appears that both the association of XPA with chromatin, as well as the dynamic engagement of XPA with other excision repair factors, contributes to the dissociation of XPA from HERC2 and hence the increased stability of XPA following DNA damage.
Figure 4.
Effect of DNA damage on the interaction of XPA and HERC2. (A) Protein levels were analyzed by immunoblotting with the indicated antibodies. A549 cells were treated with UV or mock irradiated and then times were allowed as indicated for repair in the presence of cycloheximide (CHX). (B) Quantitative analysis of the XPA expression from the blot shown in panel (A). Averages and standard deviations were plotted from three independent experiments. (C) cDNA was prepared from cells allowed to repair UV damage for the indicated times and was used for PCR amplification of the genes indicated. (D) UV treated A549 cells were allowed to repair for the indicated times and cells were fractionated into soluble and chromatin fractions. Equal amounts of soluble and chromatin fractions were analyzed by immunoblotting with the indicated antibodies. H2AX was used as a chromatin loading control. (E) HERC2 interaction with XPA was analyzed by immunoprecipitation of XPA from UV treated A549 cells followed by immunoblotting of HERC2. Average values and standard deviations from three independent experiments are shown. HERC2 levels are shown as arbitrary units (AU) with 100% defined as HERC2 in XPA complexes from mock-irradiated cells.
Effect of the circadian clock on XPA acetylation and of XPA acetylation on excision repair activity
Recently, it was reported that XPA activity was inhibited by acetylation at residues K63 and K67, and that deacetylation of XPA by the NAD+-dependent SIRT1 deacetylase was a major determinant of excision repair activity (30). It has also been reported that the NAD+-synthesizing enzyme, nicotinamide phosphoribosyl-transferase (NAMPT) is expressed in a circadian manner (31,32) and the SIRT1 deacetylase itself exhibits circadian rhythmicity in deacetylating the PER2 and BMAL1 proteins (33,34). Hence, we wished to know if XPA, as well, was subject to rhythmic acetylation/deacetylation and if this rhythmicity correlated with the circadian rhythm of excision repair activity. To this end, we first analyzed the effect of the circadian rhythm on XPA acetylation in mouse liver. As seen in Figure 5A, XPA protein is anti-phase with CRY1 with the XPA peak at ZT10 and the CRY1 peak at ZT22, as reported previously (13,14). When the acetyl-XPA was measured as a function of ZT time (ZT = 0 and 12, light on and off, respectively, under a 12 h light : 12 h dark cycle) although a weak rhythmic pattern was observed, with maximum at ZT06 and minimum at ZT14, there was no obvious circadian pattern (Figure 5B and C). Furthermore, the rhythmic pattern was abolished in CRY-null background (Figure 5B, lanes 7 and 8), indicating that the weak non-circadian rhythmicity is controlled by the circadian clock.
When the XPA acetylation pattern was compared with the XPA protein level and the excision repair activity, there was an essentially linear relationship between XPA level and excision repair rate as reported (13,14), but no obvious relation could be discerned between the level of acetylated-XPA and excision repair (Figure 5C). Indeed, when we examined the fraction of acetylated-XPA (presumably inactive) over the course of a circadian cycle it was found that at most 3–4% of XPA is acetylated at any given time point (Figure 5B and D) and hence the presumptive activation of XPA by SIRT1 is not expected to make a measurable contribution to the overall active XPA level and hence excision repair activity.
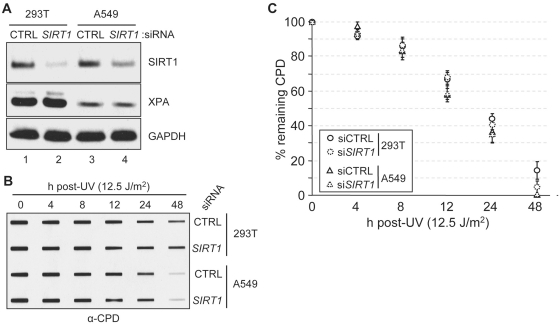
The suggestion that XPA is inactivated by acetylation and that its deacetylation by SIRT1 is a major determinant in the excision repair rate (30) was further tested in tissue culture systems. Human HEK293T and A549 cell lines were transfected with SIRT1 siRNA to reduce the SIRT1 level to 10–30% of its control value (Figure 6A), and then the cells were irradiated with UV and the rate of repair of CPDs was determined by slot blot. The results are shown in Figure 6B and C. As apparent qualitatively from Figure 6B and from the quantitative analysis shown in Figure 6C, downregulation of SIRT1 has no significant effect on the rate of repair of CPDs. Perhaps this is to be expected because >95% of XPA is in the de-acetylated/non-acetylated (active) form in untreated cells and potential inactivation of <5% of the total XPA protein by acetylation in the absence of SIRT1 is not expected to have a significant effect on the overall rate of excision repair.
Figure 6.
Effect of SIRT1 on DNA excision repair. (A) Protein levels in the indicated cells transfected with the indicated siRNAs were analyzed by immunoblotting with the indicated antibodies. (B) Residual CPD damage in genomic DNA from cells transfected with siRNA for 36 h was detected by immunoslot blotting using damage specific monoclonal antibodies. (C) Quantitative analysis of data shown in B. Average values and standard deviations from two independent experiments with two technical repeats for each are plotted.
DISCUSSION
The data in this article, along with our previous reports (13,14), indicate that the XPA protein level in the mouse brain and liver exhibits robust circadian rhythmicity which is achieved by transcriptional control of the XPA gene by the core molecular clock and at the post-translational level by HERC2 E3 ubiquitin ligase. In addition, in this article we present evidence indicating that, at least in normal human fibroblasts, XPA is rate limiting in nucleotide excision repair, providing further support that the circadian rhythmicity observed in mouse brain and liver is caused by the circadian rhythmicity of the XPA protein. Finally, we show that even though XPA exhibits a weak and non-circadian rhythmic acetylation in the mouse liver, because such a small fraction of the protein is acetylated (inactive) at a given time of the day, this acetylation and the deacetylation by SIRT1 do not contribute significantly to the control of excision repair. We wish to briefly discuss these findings in the context of other studies relevant to the topic.
First, whether XPA is rate limiting in excision repair has been the subject of some debate. Some studies have reported that XPA is rate limiting (35,36) while others have claimed that reducing XPA levels to <10% of its original value in WI38-VA fibroblasts or increasing it 10-fold in a testis tumor cell line had no measurable effect on the excision rate of UV photoproducts (20,37). In our study, all cell lines tested, including two commonly used tumor cell lines, HeLa and A549, and a normal human fibroblast line immortalized by telomerase overexpression (NHF-1), exhibited reduced rates of CPD repair when XPA was downregulated by siRNA regardless of the initial number of XPA molecules in a given cell line. Importantly, when XPA was downregulated to 60, 10 and 4% of its original value in the NHF-1 cell line in a controlled manner by titrating the amount of XPA siRNA used in transfection, the rates of repair of both the (6–4)PP and the CPD were proportionally reduced. Although the rate of repair of CPDs was linearly correlated with the level of XPA, the rate of (6–4)PP repair exhibited a parabolic relationship with the XPA level, consistent with the well-established fact that the (6–4)PP is repaired at a 5- to 10-fold faster rate than the CPD both in vivo (23) and in vitro (25). Regardless of the type of relationship between the rate of repair and the level of XPA it is clear, however, that the repair rate of both the efficient substrate, the (6–4)PP, and the inefficient substrate, the CPD, is affected by as little as a 40% reduction in XPA level in normal human fibroblasts, suggesting that XPA is rate limiting in normal human cells. This conclusion is further supported by the finding that downregulation of HERC2 E3 ligase results in ∼2-fold increase in XPA level and causes a proportional increase in the rates of repair of both CPDs and (6–4)PPs. Thus, the combination of the data regarding the correspondence of rhythmicity of excision repair with the rhythmicity of XPA and the data on the effects of altering XPA levels by either XPA siRNA or HERC2 siRNA leads us to conclude that XPA is rate limiting in mouse tissues, including brain and liver, and cultured human fibroblasts. Finally, in further support of this conclusion, we note that testis which lacks a circadian clock (38) expresses XPA at a constant level over the course of the day and in parallel with this expression pattern excision repair activity in testis is constant throughout the day (14). In a similar manner, in Cry1/2 mutant mice XPA expression is derepressed and constant over the circadian period and so is the nucleotide excision repair activity in the liver and brain (14).
A second aspect of regulation of XPA level is the contribution of HERC2 to XPA turnover and hence nucleotide excision repair capacity. HERC2, a putative HECT domain E3 ligase (39), was recently reported to function as a scaffold to recruit a RING Finger E3 ligase to the sites of double-strand breaks to aid in double-strand break repair (27). It was reported that, while knockdown of either HERC2 or RNF8 sensitizes cells to ionizing radiation, an active site HERC2 mutant complemented HERC2 depleted cells but an active site mutant of RNF8 failed to complement RNF8 depleted cells for resistance to ionizing radiation. Therefore, it was concluded that RNF8 was the active E3 ligase within the HERC2–RNF8 complex. In addition, a separate study reported that RNF8 played an equally prominent role in the cellular response to UV-induced DNA damage (28). In light of these reports, our preliminary findings indicating that HERC2 is the E3 ligase responsible for ubiquitinating XPA (14) needed re-evaluation because our in vitro ubiquitination assay was performed with immunoprecipitated HERC2 which conceivably could have been contaminated with RNF8. To address this issue, we expressed the wild-type and putative active site mutant HECT domains of HERC2, purified them and tested them for activity. We found that the HECT domain with the active site Cys→Ala mutation failed to ubiquitinate XPA while the wild-type HECT domain ubiquitinated it very efficiently, supporting our initial report that HERC2 is the E3 ligase responsible for binding and ubiquitinating XPA. Indeed, in a recent report it was shown that HERC2 ubiquitinates BARD1-uncoupled BRCA1 and targets it for degradation and, importantly, the active site Cys→Ala mutation abolishes ubiquitination of BRCA1, leading to its accumulation (40). With these considerations, then, it is safe to conclude that HERC2 plays a direct role both in nucleotide excision repair through its effect on XPA and in homologous recombination/double-strand break repair through its effect on BRCA1.
Third, it was recently reported that XPA is acetylated by an unknown acetyltransferase and that this acetylation significantly reduced XPA activity in excision repair by interfering with the XPA–RPA interaction and possibly with the interaction of XPA with other core excision repair factors (30). It was also reported that SIRT1 bound to XPA and prevented its acetylation or, if XPA were acetylated, SIRT1 deacetylated and thus activated it. In support of this model it was reported that downregulation of SIRT1 significantly reduced both the repair rate of CPDs and the survival of UV-irradiated cells. This report raised the interesting possibility that the circadian rhythm of excision repair (13,14) might be generated by the XPA acetylation/deacetylation rhythm because both SIRT1 (33,34) and the NAD+ synthesizing enzyme, NAMPT (31,32) exhibit circadian rhythms. Thus, it was conceivable that the circadian rhythms of NAD+ and SIRT1 would engender an acetylation–deacetylation cycle of XPA with circadian periodicity, resulting in daily oscillation of excision repair activity. However, our data show that in mouse liver <5% of XPA is acetylated at a given time of the day, which means that >95% of XPA is active at all times and activation of <5% of XPA is not expected to make a substantial contribution to the rate of repair. In support of this prediction, we found that downregulation of SIRT1 in two human cell lines did not affect the rate of excision repair of CPDs. We have no satisfactory explanation for the difference between our data and the previous report (30). We note, however, that there was no quantitative analysis of the apparently minor difference seen in the repair data in the previous study and that downregulation of SIRT1 affects many cellular functions and may indirectly affect cell survival following UV damage by affecting general cellular physiology. In any event, our experiments on XPA acetylation and repair both in mouse liver and in human cell lines yield results consistent with the conclusion that SIRT1 deacetylation of XPA does not contribute to the circadian rhythmicity of excision repair.
Finally, the selective advantage of circadian rhythmicity of excision repair deserves some comment. As has been noted previously the excision repair system is not entirely without side effects (3,9). Biochemical evidence indicates that both in prokaryotes and in eukaryotes nucleotide excision repair systems attack undamaged DNA, removing oligonucleotides free of damage (5). Based on this finding, it was suggested that during filling of the excision gap (gratuitous repair) errors could occur resulting in spontaneous mutagenesis. Indeed, recent studies in prokaryotes have provided some support for this prediction (6,7). Therefore, the levels of excision repair proteins must be controlled within certain limits so as to be able to eliminate DNA lesions that occur during daily wear and tear, and upregulated when there is an increase to the spontaneous mutation load. This fine-tuning is achieved in E. coli by increasing the transcription of the uvrA and uvrB genes by the SOS response in response to genotoxic stress and proteolysis of UvrA by ClpXP protease upon completion of repair (8,9). In eukaryotes, there is no SOS-like damage response. The transcription of two genes, XPC and XPE (DDB2), is induced by DNA damage but the levels of these proteins either do not change or show only a modest increase (10–12) and both proteins are ubiquitinated by the CUL4 complex following DNA damage. Ubiquitination leads to degradation of DDB2; however, the role of ubiquitination of XPC is controversial with some reports suggesting that it increases the specificity of XPC for damaged DNA while others claim that it promotes degradation by the proteasome (12). In any event, XPE is not essential for repair and whether it participates in repair at all is controversial (10,12,41). Here, we show that a key protein in nucleotide excision repair, XPA, is regulated both at transcriptional and post-translational levels in a manner quite similar to the regulation of UvrA, albeit the transcriptional control of UvrA is by the SOS response while the control of XPA transcription is by the molecular clock.
A further question is the physiological and evolutionary relevance of the phasing of the XPA and excision repair rhythmicity with the sleep/wake and anabolic/catabolic rhythms of the mouse. It is difficult to address this issue with any certainty. It is generally accepted that the circadian clock evolved as a mechanism to minimize DNA damage by the high flux UV present in the beginning of evolution of eukaryotic organisms (‘escape from light’ hypothesis) (42,43) and thus the clock and DNA repair pathways are evolutionarily linked. Viewed from this perspective, several models can be advanced. In one model, the phasing is an evolutionary relic with no selective advantage because mice are nocturnal animals, rarely venture into sunlight and therefore the steady increase in excision repair during the daytime and its decline at night does not help mice to cope better with genotoxic stress from chemical agents they are exposed to during their active night phase. Alternatively, it could be argued that even though mice are nocturnal animals they do venture out for short periods during the day and therefore the increased rate of repair of UV photoproducts over the course of the day does confer a selective advantage. Finally, it is also possible that the day phase in mice, the anabolic phase in which reactive metabolites such as the lipid peroxidation product malondialdehyde are generated, attack DNA and cause mutagenic lesions (44) and therefore increased repair rate during this phase helps in rapid removal of potentially mutagenic legions.
FUNDING
Funding for open access charge: National Institutes of Health (Grants No GM31082 and GM32833).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr William Kaufmann (University of North Carolina) for the NHF-1 cell line.
REFERENCES
- 1.Sancar A. DNA excision repair. Annu. Rev. Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 2.Wood RD. Nucleotide excision repair in mammalian cells. J. Biol. Chem. 1997;272:23465–23468. doi: 10.1074/jbc.272.38.23465. [DOI] [PubMed] [Google Scholar]
- 3.Reardon JT, Sancar A. Nucleotide excision repair. Prog. Nucleic Acid Res. Mol. Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 4.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 5.Branum ME, Reardon JT, Sancar A. DNA repair excision nuclease attacks undamaged DNA. A potential source of spontaneous mutations. J. Biol. Chem. 2001;276:25421–25426. doi: 10.1074/jbc.M101032200. [DOI] [PubMed] [Google Scholar]
- 6.Hasegawa K, Yoshiyama K, Maki H. Spontaneous mutagenesis associated with nucleotide excision repair in Escherichia coli. Genes Cells. 2008;13:459–469. doi: 10.1111/j.1365-2443.2008.01185.x. [DOI] [PubMed] [Google Scholar]
- 7.Koskiniemi S, Hughes D, Andersson DI. Effect of translesion DNA polymerases, endonucleases and RpoS on mutation rates in Salmonella typhimurium. Genetics. 2010;185:783–795. doi: 10.1534/genetics.110.116376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pruteanu M, Baker TA. Controlled degradation by ClpXP protease tunes the levels of the excision repair protein UvrA to the extent of DNA damage. Mol. Microbiol. 2009;71:912–924. doi: 10.1111/j.1365-2958.2008.06574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pruteanu M, Baker TA. Proteolysis in the SOS response and metal homeostasis in Escherichia coli. Res. Microbiol. 2009;160:677–683. doi: 10.1016/j.resmic.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 11.Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- 12.Liu L, Lee J, Zhou P. Navigating the nucleotide excision repair threshold. J. Cell Physiol. 2010;224:585–589. doi: 10.1002/jcp.22205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang TH, Reardon JT, Kemp M, Sancar A. Circadian oscillation of nucleotide excision repair in mammalian brain. Proc. Natl Acad. Sci. USA. 2009;106:2864–2867. doi: 10.1073/pnas.0812638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang TH, Lindsey-Boltz LA, Reardon JT, Sancar A. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc. Natl Acad. Sci. USA. 2010;107:4890–4895. doi: 10.1073/pnas.0915085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyer JC, Kaufmann WK, Cordeiro-Stone M. Role of postreplication repair in transformation of human fibroblasts to anchorage independence. Cancer Res. 1991;51:2960–2964. [PubMed] [Google Scholar]
- 16.Levy DD, Saijo M, Tanaka K, Kraemer KH. Expression of a transfected DNA repair gene (XPA) in xeroderma pigmentosum group A cells restores normal DNA repair and mutagenesis of UV-treated plasmids. Carcinogenesis. 1995;16:1557–1563. doi: 10.1093/carcin/16.7.1557. [DOI] [PubMed] [Google Scholar]
- 17.Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl Acad. Sci. USA. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang TH, Park DY, Choi YH, Kim KJ, Yoon HS, Kim KT. Mitotic histone H3 phosphorylation by vaccinia-related kinase 1 in mammalian cells. Mol. Cell. Biol. 2007;27:8533–8546. doi: 10.1128/MCB.00018-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selby CP, Thompson C, Schmitz TM, Van Gelder RN, Sancar A. Functional redundancy of cryptochromes and classical photoreceptors for nonvisual ocular photoreception in mice. Proc. Natl Acad. Sci. USA. 2000;97:14697–14702. doi: 10.1073/pnas.260498597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koberle B, Roginskaya V, Wood RD. XPA protein as a limiting factor for nucleotide excision repair and UV sensitivity in human cells. DNA Repair. 2006;5:641–648. doi: 10.1016/j.dnarep.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Hwang BJ, Ford JM, Hanawalt PC, Chu G. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl Acad. Sci. USA. 1999;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaddameedhi S, Kemp MG, Reardon JT, Shields JM, Smith-Roe SL, Kaufmann WK, Sancar A. Similar nucleotide excision repair capacity in melanocytes and melanoma cells. Cancer Res. 2010;70:4922–4930. doi: 10.1158/0008-5472.CAN-10-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell DL. The relative cytotoxicity of (6-4) photoproducts and cyclobutane dimers in mammalian cells. Photochem. Photobiol. 1988;48:51–57. doi: 10.1111/j.1751-1097.1988.tb02785.x. [DOI] [PubMed] [Google Scholar]
- 24.Kang TH, Sancar A. Circadian regulation of DNA excision repair: implications for chrono-chemotherapy. Cell Cycle. 2009;8:1665–1667. doi: 10.4161/cc.8.11.8707. [DOI] [PubMed] [Google Scholar]
- 25.Reardon JT, Sancar A. Recognition and repair of the cyclobutane thymine dimer, a major cause of skin cancers, by the human excision nuclease. Genes Dev. 2003;17:2539–2551. doi: 10.1101/gad.1131003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levi F, Schibler U. Circadian rhythms: mechanisms and therapeutic implications. Annu. Rev. Pharmacol. Toxicol. 2007;47:593–628. doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- 27.Bekker-Jensen S, Rendtlew Danielsen J, Fugger K, Gromova I, Nerstedt A, Lukas C, Bartek J, Lukas J, Mailand N. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat. Cell Biol. 2010;12:80–86. doi: 10.1038/ncb2008. [DOI] [PubMed] [Google Scholar]
- 28.Marteijn JA, Bekker-Jensen S, Mailand N, Lans H, Schwertman P, Gourdin AM, Dantuma NP, Lukas J, Vermeulen W. Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J. Cell Biol. 2009;186:835–847. doi: 10.1083/jcb.200902150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyamoto I, Miura N, Niwa H, Miyazaki J, Tanaka K. Mutational analysis of the structure and function of the xeroderma pigmentosum group A complementing protein. Identification of essential domains for nuclear localization and DNA excision repair. J. Biol. Chem. 1992;267:12182–12187. [PubMed] [Google Scholar]
- 30.Fan W, Luo J. SIRT1 regulates UV-induced DNA repair through deacetylating XPA. Mol. Cell. 2010;39:247–258. doi: 10.1016/j.molcel.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 31.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 34.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cleaver JE, Charles WC, McDowell ML, Sadinski WJ, Mitchell DL. Overexpression of the XPA repair gene increases resistance to ultraviolet radiation in human cells by selective repair of DNA damage. Cancer Res. 1995;55:6152–6160. [PubMed] [Google Scholar]
- 36.Koberle B, Masters JR, Hartley JA, Wood RD. Defective repair of cisplatin-induced DNA damage caused by reduced XPA protein in testicular germ cell tumours. Curr. Biol. 1999;9:273–276. doi: 10.1016/s0960-9822(99)80118-3. [DOI] [PubMed] [Google Scholar]
- 37.Koberle B, Roginskaya V, Zima KS, Masters JR, Wood RD. Elevation of XPA protein level in testis tumor cells without increasing resistance to cisplatin or UV radiation. Mol. Carcinog. 2008;47:580–586. doi: 10.1002/mc.20418. [DOI] [PubMed] [Google Scholar]
- 38.Miyamoto Y, Sancar A. Circadian regulation of cryptochrome genes in the mouse. Brain Res. Mol. Brain Res. 1999;71:238–243. doi: 10.1016/s0169-328x(99)00192-8. [DOI] [PubMed] [Google Scholar]
- 39.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 40.Wu W, Sato K, Koike A, Nishikawa H, Koizumi H, Venkitaraman AR, Ohta T. HERC2 is an E3 ligase that targets BRCA1 for degradation. Cancer Res. 2010;70:6384–6392. doi: 10.1158/0008-5472.CAN-10-1304. [DOI] [PubMed] [Google Scholar]
- 41.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 42.Gehring W, Rosbash M. The coevolution of blue-light photoreception and circadian rhythms. J. Mol. Evol. 2003;57(Suppl. 1):S286–S289. doi: 10.1007/s00239-003-0038-8. [DOI] [PubMed] [Google Scholar]
- 43.Sancar A. Regulation of the mammalian circadian clock by cryptochrome. J. Biol. Chem. 2004;279:34079–34082. doi: 10.1074/jbc.R400016200. [DOI] [PubMed] [Google Scholar]
- 44.Marnett LJ. Oxy radicals, lipid peroxidation and DNA damage. Toxicology. 2002;181–182:219–222. doi: 10.1016/s0300-483x(02)00448-1. [DOI] [PubMed] [Google Scholar]