Abstract
Retrotransposed sequences arise from messenger RNAs (mRNAs) that have been reinserted into genomic DNA by reverse transcription. Usually, these sequences are embedded in dormant regions, collect missense mutations over time and constitute processed, nonfunctional pseudogenes. There are thousands of processed pseudogenes in the mouse and human genome. Here, we report evidence for two paralog genes (termed Arxes1 and Arxes2), which arose by retrotransposition of the signal peptidase Spcs3 followed by a segmental duplication event. They gained a functional promoter that we show to be transactivated by adipogenic transcription factors. We further show that the Arxes mRNAs are highly expressed in adipose tissue and strongly upregulated during adipogenesis in different cell models. Additionally, their expression is elevated by an anti-diabetic agent in vitro and in vivo. Importantly, we provide evidence that the Arxes genes are translated and that the proteins are located in the endoplasmic reticulum. Although the sequence similarity and subcellular location are reminiscent of their parental gene, our data suggest that the Arxes have developed a different function, since their expression is required for adipogenesis, whereas Spcs3 is dispensable. In summary, we report retrotransposed-duplicated genes that evolved from a parental gene to function in a tissue and adipogenesis-specific context.
INTRODUCTION
As obesity and its associated diseases spread worldwide, and can now be termed pandemic, fat cell and fat tissue biology have increasingly gained researchers’ attention. This is based on research over the last 20 years, redefining adipose tissue from a passive fat storage to a highly reactive, interactive and endocrine organ. Growth of adipose tissue is the result of the development of new fat cells from precursor cells (hyperplasia) and/or the increase of existing fat cells in volume (hypertrophy). The process of fat cell development, known as adipogenesis, leads to the accumulation of lipids and an increase in the number of fat cells.
Cumulative evidence describes adipogenesis as a process coordinated by a cascade of transcription factors acting together to turn progenitor cells into lipid-storing, insulin-responsive adipocytes (1–3). Much research focused on the two master regulators of adipogenesis: peroxisome proliferator-activated receptor gamma (PPARγ) and CCAAT/enhancer-binding protein alpha (C/EBPα). The nuclear receptor PPARγ was shown by several loss- and gain-of-function studies to be necessary and sufficient for in vitro adipogenesis (1) as well as for development of adipose tissue in mice (4). Importantly, PPARγ is the target of thiazolidinediones (TZDs), a class of antidiabetic drugs. The C/EBP protein family is comprised of six basic leucine zipper transcription factors (5). While C/EBPβ and C/EBPδ play essential roles in early adipogenesis, C/EBPα is upregulated together with PPARγ at a later point to synergistically act on target genes during maturation of adipocytes (6). More recent evidence shows that C/EBPβ, along with C/EBPα, is also expressed during late adipogenesis and essential for it to occur (6). In addition to PPARγ and the C/EBPs, a number of other transcription factors are reported to be involved in the adipogenic network (1–3). Recently, factors with enzymatic activity have also been described as essential modulators of adipogenesis (7–9). However, the list of adipogenic regulators and the knowledge of the exact ‘wiring’ of its components remain incomplete.
Here, we aimed to identify novel C/EBP target genes during adipogenesis. We used a combined in silico and experimental approach and employed a unique mouse model of dominant-negative C/EBP expression. Within the set of validated candidates, there was an uncharacterized Riken transcript. Bioinformatic analyses identified a paralog sequence 40 kb upstream and suggested that both sequences, which we termed Arxes (Adipocyte-related X-chromosome expressed sequence) arose by retrotransposition from the signal peptidase Spcs3 followed by a segmental duplication. Functional analyses showed completely different expression, regulation and function of the Arxes compared to their parental gene. We could show that the messenger RNA (mRNA) of Arxes1 and Arxes2 is upregulated during adipogenesis in different model systems by C/EBPα and PPARγ/RXRα through proximal promoter sites and possibly a distant enhancer region. Furthermore, we provide evidence for the existence of an endoplasmatic reticulum (ER)-located protein translated from these mRNAs. Knocking down Arxes1 and Arxes2 abolished differentiation of 3T3-L1 preadipocytes, while knockdown of Spcs3 had no effect on adipogenesis. Finally, silencing of Arxes expression in mesenchymal stem cells also attenuated adipogenesis while augmenting differentiation to osteoblasts. This suggests an important role of the Arxes in cell commitment and in a metabolic context that is distinct from that of their parental gene Spcs3.
MATERIAL AND METHODS
Cell culture and gene silencing
Mouse embryonic fibroblast (MEF) cultures were established from wild-type mice, and from transgenic mice expressing a protein that inhibits the DNA binding, and thereby the transactivation potential, of the C/EBP family of transcription factors (10). Transgenic mice (subsequently termed A-C/EBP) were kindly provided by Charles Vinson. These MEFs were harvested and cultured as described elsewhere (11). Briefly, pregnant mice were sacrificed by cervical dislocation at Day 12.5 to 14.5 post coitum and embryos were carefully dissected out and separated from yolk sacs. The heads were used for genotyping to determine transgenic cultures. In phosphate-buffered saline (PBS), extremities and visceral organs were separated from the torso, which was then minced using a scalpel and homogenized by pipetting several times. Three incubation steps with 0.25% trypsin-EDTA (Invitrogen/Gibco) were performed (each 10 min, 37°C) with pipetting in between. After adding culture medium (αMEM, 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin; all from Invitrogen/Gibco) cells were forced through a 20-gauge needle using a syringe. Finally, cells were spun down (3 min, 200 g), supernatant was removed and the cells were resuspended in 20 ml culture media, plated in a 75 cm2 culture flask and cultured at 37°C and 5% CO2. Medium was refreshed every third day and cells were subcultivated (at a 3:1 rate) before they reached confluence. At passage one, cells (2 million per ml per tube) were frozen in liquid nitrogen in medium with a supplementation of 10% DMSO (Sigma). Differentiation experiments were performed at passage three. A standard DMI mix [1 µM dexamethasone, 500 µM 3-isobutyl-1-methylxanthine (IBMX) and 5 µg/ml Insulin; all from Sigma] was supplemented to the medium, along with 1 µM rosiglitazone (Alexis), to initiate differentiation of 2 days post-confluent cells. The addition of 1 µM rosiglitazone during the whole differentiation process enhanced adipogenesis in wild-type MEFs, while not changing the differentiation potential of A-C/EBP MEFs. Medium was changed every other day, leaving dexamethasone and IBMX out and reducing insulin concentration to 1 µg/ml from Day 2 onward.
3T3-L1 cells were propagated and maintained in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen/Gibco) supplemented with 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin (all from Invitrogen/Gibco). Two days post-confluent cells were induced to undergo adipogenesis by addition of a DMI cocktail and 1 µM rosiglitazone when indicated. From Day three on the hormonal cocktail contained only 1 µg/ml insulin. OP9 cells were differentiated with a DMI protocol described in ref .12.
C3H10T1/2 cells were obtained from ATCC and maintained in DMEM supplemented as described above for 3T3-L1 cells. For adipogenic differentiation postconfluent cells were treated with 100 ng/ml BMP-2 (Sigma) for 3 days followed by incubation in DMI cocktail as described above. For osteoblastogenic differentiation 100 ng/ml BMP-2, 10 nM dexamethasone, 10 mM b-glycerophosphat and 50 µg/ml L-ascorbic acid phosphate (all Sigma) were added to medium of postconfluent cells. Medium was changed every other day until harvesting on Day 10.
Lentiviral shRNA particles and a nontargeting control construct were obtained from Sigma (SHVRS-NM_029541 and SHC002V, respectively). The most effective silencing construct that is specifically directed against the Arxes, while showing several mismatches to the Spcs3 coding sequence Figure S5B) was used for further experiments (NCBI probe ID: Pr008727591.1). Infections of 3T3-L1 or C3H10T1/2 cells were followed by puromycin (3 µg/ml) selection. Lentiviral silencing particles targeting Spcs3 were obtained from Santa Cruz Biotechnologies (TCR-5). Infections of 3T3-L1 cells were performed according to manufacturer’s recommendations. To stain for cytoplasmic triglycerides cells were fixed with 4% paraformaldehyde for 1 h and incubated for 30 min with the lipophilic dye oil red O (ICN). ALP activity was measured as previously described (13). Briefly, cells were rinsed and harvested in homogenization buffer (20 mM Tris, 1 mM EDTA, 1 mM β-mercaptoethanol), sonicated and mixed with p-nitrophenyl phosphate (pNPP, Sigma). Reaction kinetics was measured with a spectrophotometer at 405 nm for 3 min. Measurement was normalized to protein contents determined with BCA assay (Pierce).
Microarray analysis
For expression analyses of MEFs an in-house cDNA platform (14) was used. Total RNA from four different preparations from wild-type (WT) and from A-C/EBP transgenic MEFs were harvested at specified time points using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. Total RNA from each two preparations were pooled leaving two biological replicates for each WT and A-C/EBP MEFs. RNA samples were reverse-transcribed and indirectly labeled as described previously (14). Eighty percent confluent cells served as reference sample against which every time point sample was hybridized in a dye-swap configuration. Hybridization was performed in a 42°C water bath for >16 h. Slides were scanned using an Axon scanner. After normalization, the data were stored in the MARS database (15) and exported to ArrayExpress (ID: E-MARS-11). Self-organizing map (SOM) clustering was performed using Genesis 1.6.0 (16). Differentially expressed genes in Figure 1A are defined as more than 2-fold deregulated in at least one time point in transgenic or wild-type MEFs compared to preconfluent MEFs. Genes showing a differential profile (positively regulated in late time points of WT MEFs, while showing a flat profile in late transgenic time points) were selected as follows: Transcripts showing a difference between WT and transgenic log2-ratios of equal to or more than 0.5 in one or both of the late time points (d3 and d8), while being differentially expressed in the wild-type time point (≥1), were included in this differential profile. Potential C/EBP target genes were subjected to Gene Ontology mapping using ClueGO (17).
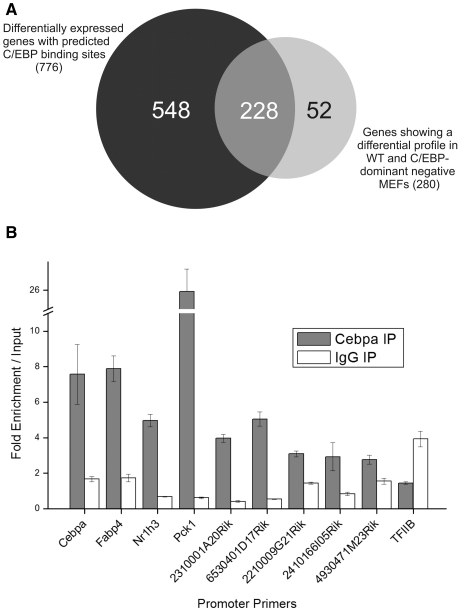
Figure 1.
Combined computational and experimental approaches predict putative targets of C/EBP transcription factors in adipogenesis. (A) Venn diagram of the intersection of the lists of differentially expressed genes containing predicted C/EBP binding sites (776) and C/EBP-dependent genes, identified by expression analysis of wild-type and C/EBP dominant-negative MEFs during adipogenesis (280; see ‘Material and Methods’ section for exact definition of gene groups). The intersection comprises 228 RefSeq-annotated genes, which constitute putative C/EBP targets. (B) A subset of predicted C/EBP targets confirmed by using ChIP-qPCR in MEFs at Day 3 of adipogenesis. Shown are fold-enrichments over input for primer pairs aimed at predicted binding sites in promoter regions of potential C/EBP target genes. Fold-enrichments are normalized to signals from 18S rRNA and related to genomic input DNA. An IgG reaction was performed to account for unspecific antibody binding (white bars). The TFIIB promoter served as negative control. Values are mean values (±standard deviation) from three technical replicates. Shown are values from one representative experiment out of three (Supplementary Figure S1C and S1D).
Bioinformatic analyses
Experimentally derived binding sites for the C/EBP family of transcription factors were assembled in a position weight matrix (PWM) from literature (Table S1). Further, matrices were collected from the public database TRANSFAC (18) to be included in the downstream search (M00109, M00117, M00116, M00159, M00190 and M00201).
Promoter sequences from the mouse genome (version: mm7, Aug. 2005, NCBI Build 35) were obtained directly from the UCSC genome browser (http://genome.ucsc.edu) (19). Chromosomal coordinates of RefSeq genes (20) were used to extract −5500 bp to +2500 bp relative to transcription start sites (TSSs). These extracted regions were scored against the different PWMs by using a MatInspector-based algorithm (21) implemented in Perl. An optimized matrix threshold was calculated for each PWM, which permits a maximum of one hit (in average) in 10 000-bp segments of conjoined (repeat-masked) coding sequences from all RefSeq transcripts. These optimized matrix thresholds balance the differences in match frequencies between short (unspecific) and long (more specific) PWMs, based on the assumption, that hits in nonregulatory (protein coding) sequences are occurring merely by chance (22).
For detection of retrotransposition and duplication events Arxes coding sequences from mouse (mm9) were aligned against the genome versions of 29 organisms [including mouse (mm9), rat (rn4), rabbit (oryCun2), human (hg19) and chimp (panTro2)] using BLAT. Only alignments with score >30 were considered. Spcs3 and Arxes genomic sequences including 1000 bases up- and downstream for mouse, rat, and rabbit (mm9, rn4 and oryCun2) were retrieved from UCSC genome browser. Blast2sequences with discontinuous MegaBLAST were applied to get most extreme 3′ and 5′ genomic positions of the alignment of Spcs3 against Arxes sequences. Upstream sequences of the most extreme 5′ genomic position (–100/+10) [corresponding to 5′ target site duplication (TSD) search space] and downstream of the most extreme 3′ genomic position (–10/+100) (=3′TSD search space) were extracted. To find potential TSDs best local alignments and relative scores (100*score/maximum possible score) for subsequences of the 5′TSD search space (sliding window from 7 to 25 nt) against the 3′TSD search sequence were determined using EMBOSS Matcher based on the lalign algorithm. PolyA sequences between most extreme 3′ genomic position of the alignments and the potential 3′TSD were detected [starting with at least 2 A in a row, stop on the occurrence of two consecutive none-A (C,G,T,N), except if in a look-ahead-window of 10 nt >70% of As are present, and with at least 7 As in total]. Distances and scores were calculated as described elsewhere (23). Analyses were performed using Perl 5.10.1.
In vivo experiments
Male C57/Bl6 mice were kept on a 12-h light/dark cycle and housed according to institutional guidelines and approved by the Austrian Bundesministerium für Wissenschaft und Forschung. Four- to five-month-old mice were fasted overnight and refed for ∼2 h prior to sacrificing to synchronize their nutritional states. Mice were fed with regular chow diet. For the rosiglitazone study, mice were put on either chow diet or chow diet with rosiglitazone (0.01% w/w) immediately after weaning and kept on this diet for 7 weeks. Before harvesting the epididymal fat pads, mice were fasted for 4 h. Statistics were calculated using a Student’s t-test. P < 0.05 was considered as significant.
Western blot analysis
Transfected COS-7 and 3T3-L1 cells (at several differentiation time points) were harvested for protein analysis by scraping in lysis buffer (50 mM Tris–HCl pH 6.8, 10% glycerol, 2.5% SDS, 1× protease inhibitor cocktail, 1 mM PMSF) after two washes with ice-cold TBS. The suspension was incubated on 90°C for 10 min. Benzonase (Merck) was added followed by incubation at room temperature for 1 h. Protein concentration was determined with the BCA protein assay kit from Pierce according to the manufacturer’s instructions. Each lane of a 10% Bis-Tris Gel (NuPAGE, Invitrogen) was loaded with 40 µg of sample. After electophoresis, gels were blotted to Nitrocellulose-membranes (Invitrogen). Blots were blocked and incubated with the following antibodies: anti-Arxes (1:300 dilution), anti-β-actin (1:25 000; Sigma). For chemiluminescent detection horseradish peroxidase-conjugated secondary antibodies were used (anti-rabbit for Arxes and anti-mouse for His and β-actin, both 1:3000, Pierce) and ECL component (Pierce) served as a substrate. Developed films were scanned with HP Scanjet at 600 dpi.
Quantitative real-time reverse transcriptase polymerase chain reaction
For quantitative real-time reverse transcriptase polymerase chain reaction (qPCR) measurement samples totalRNA was isolated using Trizol reagent (Invitrogen) according to the provider’s protocol, employing an extra homogenization step for mouse tissues. Reverse transcription was performed as described above. qPCR assays were performed on 4.5 ng cDNA in a 18 µl SYBR green master mix (Invitrogen) reaction on an ABI Prism 7000 sequence detection system. Data were analyzed using a real-time PCR management and analysis system [http://genome.tugraz.at/qpcr; (24)]. Values were normalized to Uxt and the algorithms from Zhao and Fernald (25) were used to derive Ct values and PCR efficiencies. Primer sequences are to be found in Table S2.
Chromatin immunoprecipitation
MEFs or 3T3-L1 cells were grown in 15-cm dishes and cross-linked at Day 3 of adipogenesis by incubating in 1% formaldehyde for 20 min while shaking at room temperature. Cross-linking was stopped by the addition of 0.125 M glycine in PBS for 5 min. Cells were harvested in ice-cold PBS and subsequently incubated in 800 µl cell lysis buffer [10 mM Tris pH8, 10 mM NaCl, 0.2% NP-40, 1× protease inhibitor cocktail (PIC), 1 mM PMSF] to extract nuclei which were then lysed in 200 µl ice-cold nuclei lysis buffer (50 mM Tris pH 8, 10 mM EDTA, 0.8% SDS, 1× PIC, 1 mM PMSF). Isolated chromatin/DNA mixture was diluted with 200 µl IP dilution buffer (50 mM Tris pH7.5, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 1% Triton X-100, 1× PIC, 1 mM PMSF, 2 mg/ml BSA) and subjected to three 20-s bursts using an HD2070 sonicator (Bandlin) at 40% output. This yielded DNA fragments of 200–600 bp. This material was diluted 1:10 in IP dilution buffer before immunoprecipitation was performed using anti-C/EBPα (Santa Cruz, sc-61), anti-PPARγ (Abcam, ab41928) and anti-IgG antibody (Santa Cruz, sc-2027) and protein A/G plus-agarose beads (Santa Cruz, sc-2003). Washing steps were performed subsequently with IP dilution buffer, low salt buffer (20 mM Tris pH8, 50 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100, 1× PIC, 1 mM PMSF), high salt buffer (20 mM Tris pH8, 250 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100, 1× PIC, 1 mM PMSF), LiCl buffer (10 mM Tris pH8, 0.25 M LiCl, 1 mM EDTA, 1% Na-deoxycholate, 1% NP-40, 1× PIC, 1 mM PMSF) and, finally, TE buffer. Before the first wash, the supernatant was set aside to serve as input control. Immunoprecipitated chromatin and input chromatin were reverse cross-linked, column purified and amplified using a whole-genome amplification kit (WGA2, Sigma). Amplified DNA was subjected to qPCR analysis. Measurements were normalized to an 18S rRNA upstream region and related to input measurements to obtain relative enrichment values. Primer sequences used are given in Table S2.
Fluorescence microscopy
The Arxes CDS was cloned into a pCDNA4/HisMaxC expression vector (Invitrogen) yielding an N-terminally tagged Arxes protein using standard cloning procedures. COS-7 cells and 3T3-L1 preadipocytes were seeded and grown on coverslips (Corning #1.5). At 80% confluence, they were transfected with 1 µg of an Arxes-HisMax construct using 2 µl MetafectenePro (Biontex). For negative controls, an empty vector was transfected. The transfection was performed according to the manufacturer’s manual. Forty-eight hours after transfection, the cells were fixed with 3.5% paraformaldehyde for 15 min. After fixation, the cells were washed three times in PBS, permeabilized for 10 min in ice-cold 0.5% Triton-X 100 in PBS and again washed three times with PBS. Immunodetection of His-tagged Arxes was performed by incubating the cells over night at 4°C, with either a monoclonal antibody against the His-tag raised in mouse (GE Healthcare Life Sciences, 27471001) or the customized polyclonal antibody against the synthetic Arxes peptides raised in rabbit (both 1:200 dilution in 4% BSA/0.1% Tween-20/PBS). The cells were then washed three times with PBS and incubated for 1 h at room temperature with an anti-mouse and an anti-rabbit antibody coupled to AlexaFluor-488 and AlexaFluor-594 (both Invitrogen), respectively. After washing three times with PBS, the nuclei were counterstained with DAPI and the coverslips were mounted on microscopy slides.
For endogenous Arxes detection 3T3-L1 preadipocytes were grown on coverslips and fixed for 15 min with 2% paraformaldehyde/0.4% glutaraldehyde/PBS. Confluent 3T3-L1 adipocytes were detached at Day 7 of differentiation with a trypsin (0.25%)/collagenase (0.5 mg/ml) mix. Cells were then seeded on coverslips in a lower density to be accessible for antibodies before fixation. After quenching for 5 min with 20 mM glycine pH 8.5/PBS the cells were washed three times with PBS, permeabilized with 0.2% Triton-X 100/PBS and again washed three times with PBS. In addition to the Arxes, the ER was detected by a mouse anti-Calnexin antibody (Abcam, ab31290). Both antibodies were diluted 1:200 in 4% BSA/0.1% Tween-20/PBS and applied to the cells. Incubation and subsequent steps were performed as described above. All images were collected on a Zeiss AxioImager Z1 epifluorescence microscope using a 63× 1.4 NA objective lens.
Luciferase reporter assays
Indicated proximal promoter elements were cloned in pGl4.21 luciferase reporter vectors (Promega) using standard procedures. The enhancer regions were cloned in a pGl4.26 luciferase expression vector (Promega) containing a minimal promoter. The Renilla reporter vector pGl4.75 (Promega) was cotransfected in all experiments in a ratio of 1:50 to luciferase reporter vectors, to control for varying transfection efficiencies.
For the cotransfection in COS-7 cells C/EBPα CDS was cloned in a pMSCV vector. Cloning of PPARγ2 and RXRα in pCMX expression vectors has been described elsewhere (26). Cotransfection was done in 96-well plates using MetafectenePro (Biontex) according to the provider’s protocol in ratio of 3:1 (µl MFP:µg DNA). One-hundred nanograms of luciferase reporter vectors were used. For PPARγ, RXRα and C/EBPα, 25 ng, 25 ng and 50 ng of DNA were employed, respectively. After 48 h cells were lysed and assayed according to protocol provided with the Dual-Glo luciferase assay system (Promega). Luminescence read-outs were generated with a Berthold Orion II luminometer. Relative Luciferase activity was calculated by referring Renilla-normalized values to empty Luciferace vector measurements (pGl4.21 or pGl4.26).
To efficiently transfect adipocytes with reporter vectors, electroporation was carried out as follows. Differentiated cells (d7–d9) were detached with a trypsin (0.25%)/collagenase (0.5 mg/ml) mix and 8 × 10−6 cells/ml (in 10 ml electroporation buffer provided by with the kit) were mixed with 1 µg reporter plasmid and pulsed with 1400 V/30 ms pulses using a Neon transfection system (Invitrogen). For comparison, 3T3-L1 preadipocytes were treated in the same way. Three electroporations were reseeded in one well of a 24-well plate. After 48 h, cells were assayed and calculations performed as described above.
RESULTS
Large-scale gene expression analysis during adipogenesis in a wild type and a perturbation system reveals novel C/EBP target genes
To identify novel target genes of C/EBPs during adipogenesis, we employed mouse embryonic fibroblasts (MEFs) from wild-type and C/EBP dominant-negative mice. As evidenced by oil red O staining and western blotting, wild-type MEFs treated with adipogenic cocktail differentiated into adipocytes, whereas dominant-negative MEFs did not (Supplementary Figure S1A, further validation can be found in Supplementary Data). RNA was harvested at several time points during the differentiation process up to Day 8 and subjected to time-series microarray analysis (Supplementary Figure S2A). Genes with a differential expression profile (upregulated in wild-type MEFs and not so in dominant-negative MEFs) were then identified and filtered for RefSeq-annotated genes that contain a predicted C/EBP binding site close to their TSS (Figure 1A; see ‘Material and Methods’ section for detailed procedure). A list of the identified genes can be found in Supplementary Table S1. These 228 RefSeq genes were subjected to Gene Ontology (GO) analysis via ClueGo (17) showing a highly significant enrichment in metabolically important GO terms such as mitochondrion, lipid metabolic processes, fatty acid metabolism, acetyl CoA-related terms and generation of precursor metabolites and energy (Supplementary Figure S3).
To further validate these predictions, we chose a subset of four known C/EBP-bound genes and five yet uncharacterized transcripts (annotated by the Riken consortium as full length transcripts). We could show binding of C/EBPα to the predicted binding sites in the upstream sequence region of the selected transcripts using chromatin immunoprecipitation followed by quantitative real-time PCR (ChIP-qPCR) in MEFs differentiated for 3 days. As shown in Figure 1B, the enrichments of the formerly uncharacterized genes are in general lower compared to that of the bona fide C/EBP target genes, but show at least a 2-fold enrichment over input. Within this set of uncharacterized transcripts, the strongest values for enrichment were shown at regions upstream of two Riken transcripts. The transcript 2310001RikA20 codes for a transmembrane protein and was recently characterized in our lab as an important player in adipogenesis (27). In this study, the transcript 6530401D17Rik was further investigated.
Genomic setup and sequence analyses of Adipocyte-related X-chromosome expressed sequences (Arxes)
Close inspection of 6530401D17Rik identified it as an intron-less gene, which is coded on the forward strand of the X-chromosome of the mouse genome. Its coding sequence (CDS) is identical to another Riken gene (2900062L11Rik), located ∼40 kb upstream (see Figure 5F for genomic overview). As these two genes have more in common than just their CDSs (e.g. tissue expression pattern, regulation; see later) we will refer to them as Arxes, for Adipocyte-related X-chromosome expressed sequences (Arxes1 for 6530401D17Rik; Arxes2 for 2900062L11Rik).
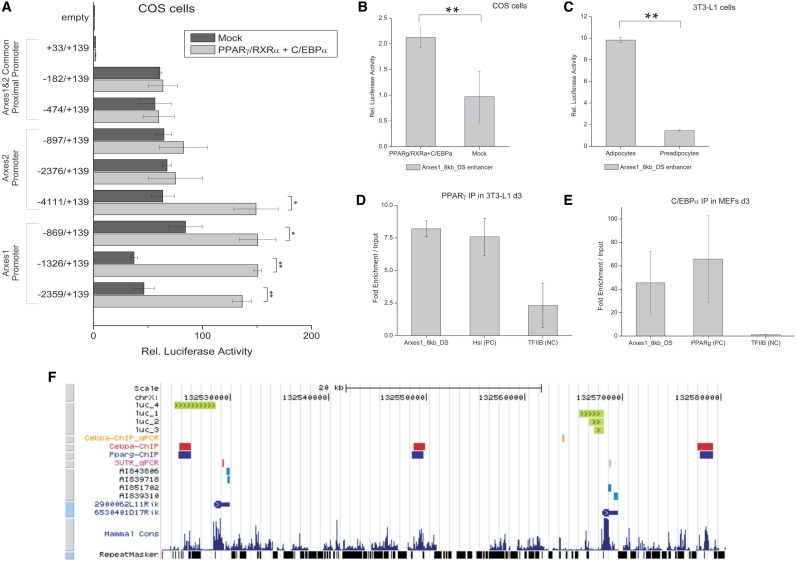
Figure 5.
Transactivation of Arxes promoters and enhancers by PPARγ and C/EBPα. (A) Indicated Arxes1 and Arxes2 promoter elements were cloned into a Luciferase reporter vector and cotransfected with either PPARγ, RXRα and C/EBPα expression vectors or empty vectors (mock) in COS cells. After 48 h, cells were lysed and assayed for luciferase activity using a luminometer. Data were normalized to Renilla luciferase readings to account for differences in transfection efficiencies. Finally, data were related to empty reporter vector activity. Data are represented as mean ± SEM from independent biological replicates (n = 2–3). Student’s t-test: *P < 0.05; **P < 0.01. (B) An enhancer region 8 kb downstream of the Arxes1 gene [Arxes1_8 kb_DS, identified by a recent genome-wide study (6)] was cloned in a luciferase reporter vector containing a minimal promoter, which was transfected and assayed as described above. Data are represented as mean ± SEM from three independent biological replicates. Student’s t-test: **P < 0.01. (C) The Arxes1_8 kb_DS enhancer region was electroporated in either preconfluent 3T3-L1 preadipocytes or mature 3T3-L1 adipocytes. Cells were lysed 48 h later and analyzed as described above. Data are represented as mean ± SEM from three different electroporation experiments. Student’s t-test: **P < 0.01. (D) ChIP was performed in d3 3T3-L1 cells using an anti-PPARγ antibody. PPARγ-bound immunoprecipitated DNA was quantified with qPCR using primers directed against the Arxes1_8 kb_DS enhancer region, Hsl promoter (published, positive control) and TFIIB promoter (as negative control). Measurements were normalized to signal of 18SrRNA and related to input DNA. Data are represented as mean ± SEM from two independent experiments. (E) ChIP-qPCR was performed in d3 MEFs with anti-C/EBPα antibody as described in Figure 1B. Primers target the Arxes1-8 kb-DS enhancer, PPARγ promoter (positive control) and the TFIIB promoter (negative control). Data are represented as mean ± SEM from two independent experiments. (F) Genomic overview of the Arxes loci in UCSC genome browser. (From top to bottom) Promoter regions showing a significant luciferase activity increase upon cotransfection with PPARγ, RXRα, and C/EBPα expression vectors (luc_1 to luc_4, compare to Figure 5A); C/EBPα ChIP-qPCR region from Figure 1B; C/EBPα and PPARγ ChIP–chip regions from Lefterova et al.; Amplicons from specific qPCR primers in 3′UTRs, used throughout this study; EST sequences represented on microarray; RefSeq database annotation (2900062L11Rik = Arxes2; 6530401D17Rik = Arxes2); Conservation and repeat masker track from UCSC genome browser.
The analysis of the predicted amino acid sequence of both Arxes suggest that they share a common architecture with Signal peptidase complex subunit 3 (Spcs3, NP_083977.1, Supplementary Figure S4), a known signal peptidase (28) catalyzing cotranslational cleavage of signal peptides from nascent proteins in the ER. Each of these three proteins contains a transmembrane segment (residues 11–32) and a PFAM domain SPC22 hit (residues 1–175). The Spcs3 locus can be found on chromosome 8 and the gene consists of five exons. On the nucleotide level, the Arxes CDSs are 79% identical to the Spcs3 CDS and on the amino acid level the Arxes and Spcs3 show 82% identity (see Table 1 for mutual alignments). These strong homologies suggest that the Arxes were integrated into their loci via retrotransposition of the Spcs3 mRNA. To investigate the retrotranspositon event in more detail we used a sliding window and sequence alignment approach to detect target site duplications [TSDs, short direct repeats flanking the sequences inserted by retrotransposition (23,29)] in the 5′ upstream and 3′ downstream regions of the sequences homologous between the Arxes loci and the Spcs3 mRNA. Indeed, this analysis yielded 7-nt TSDs directly 5′ and 3′ of the homologous region indicative of a retrotransposition event (Figure 2 and Supplementary Data file 1).
Table 1.
Multiple alignments of Arxes1, Arxes2 and Spcs3 promoters and transcripts
| Alignments | Promoter | 5′UTR | CDS | 3′UTR | Amino acid sequence |
|---|---|---|---|---|---|
| Spcs3 vs Arxes1 (%) | 1 | 12 | 79 | 23 | 82 |
| Spcs3 vs Arxes2 (%) | 3 | 18 | 79 | 7 | 82 |
| Arxes1 vs Arxes2 (%) | 85 | 99 | 100 | 5 | 100 |
Multiple alignments were performed by running ClustalW2 on the EBI server.
Numbers shown are percent identity at the nucleotide level (except the last column, which shows the amino acid identity of predicted protein sequences).
For the promoter alignment 1 kb upstream of the transcription start sites was considered.
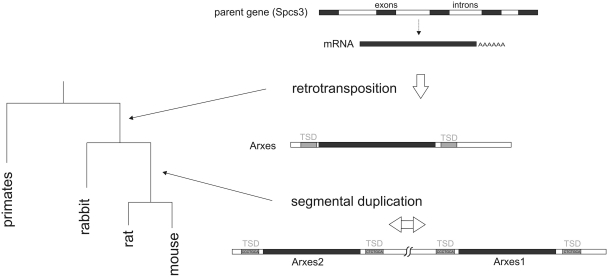
Figure 2.
Model of evolutionary history of Arxes (adipocyte-related X-chromosome expressed sequences) loci. Schematic of the ‘phylogeny’ of the Arxes genes based on bioinformatic analyses. Only one Arxes-like gene can be found on the rabbit x-chromosome while mouse and rat contain two paralog regions adjacent to each other on the x-chromosome. Arxes genes exhibit intact open reading frames. Target site duplication (TSD) sequences are given for mouse. A paralog stretch longer than the retrotransposed sequence (Arxes CDS including ∼800 bp upstream and 50 bp downstream) suggests a segmental duplication event.
Interestingly, the Arxes sequences were annotated as pseudogenes in the NCBI Build 36 (mm8, NR_003641 and NR_003642). Close inspection of the genomic features of the Arxes though revealed a host of indications that are atypical for nonfunctional pseudogenes: (i) Gene prediction algorithms (e.g. GeneScan) identified an open reading frame with no premature stop codons and no frameshift mutations, as usually found in pseudogenes; (ii) polyadenylation consensus sequences (ATTAAA) can be found in the 3′UTRs of both Arxes; (iii) a weak Kozak sequence (30) is situated upstream of the start codons; (iv) no repeat regions can be found within the Arxes CDSs; and (v) a scan of 5000 bp of upstream genomic sequence using PROMOTER2.0 (31) predicts promoter sites.
To clarify the evolutionary history of the Arxes, we applied BLAT of the mouse Arxes CDS against the genome versions of 29 organisms (Supplementary Data file 2). This analysis showed that both Arxes exist also on the X-chromosome of the rat genome, while on the X-chromosome of the rabbit only one homologous sequence could be found. However, in primates no Arxes homolog seems to exist. The homology of the two mouse Arxes regions extend ∼800 bp upstream and 50 bp downstream of the CDSs of Arxes1 and Arxes2, suggesting that one of the two paralogs arose by segmental duplication [defined as regions with a sequence similarity ≥90% and a length ≥1 bp (32)].
Hence, based on our analyses, we propose a model (Figure 2) of the Arxes as retrotransposed from the Spcs3 mRNA. This retrotransposition took place after the speciation of primates and glires (33) with an additional segmental duplication after the speciation of rabbit and mouse/rat. The resulting sequences appear to have contained an intact open reading frame.
Arxes1 and Arxes2 mRNA expression is highly upregulated during in vitro adipogenesis, in adipose tissue and by an antidiabetic agent
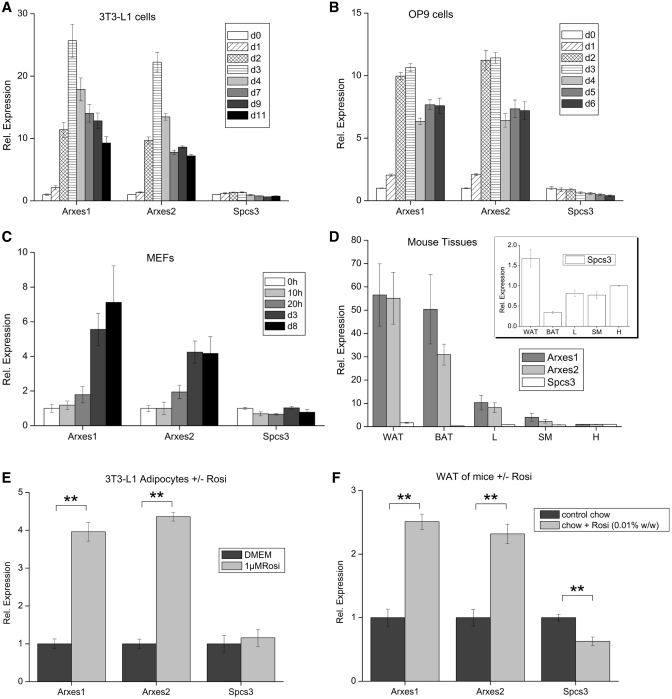
To distinguish mRNA expression of the two Arxes and the Spcs3, qPCR primers were designed in the highly distinct 3′ UTRs (Table 1, Table S2 and Figure 5F). With these specific primers, expression levels were measured in 3T3-L1 cells, OP9 cells and MEFs during adipogenic differentiation (Figure 3A–C), showing that the Arxes transcripts are strongly upregulated during adipogenesis. Their parental gene Spcs3, on the other hand, was slightly downregulated in these cell models, as was also observed for Spcs3 during differentiation of human cell culture models for adipogenesis (data not shown). The mRNA expression levels in metabolically relevant mouse tissues were then measured (Figure 3D). Arxes expressions in epididymal white (WAT) and brown (BAT) adipose tissue were up to 70-fold higher compared to expression in heart, while Spcs3 mRNA was differentially expressed to a much lower level in these tissues (inset of Figure 3D).
Figure 3.
mRNA expression patterns of Arxes1, Arxes2 and Spcs3 during in vitro adipogenesis, in mouse tissues and upon in vitro and in vivo rosiglitazone treatment. (A–C) Shown are qPCR measurements during the course of adipogenesis in three different cell models. Cells were induced with DMI cocktails 2 days post confluence (d0) as specified in ‘Materials and Methods’ section. Values are expressed relative to d0 measurements. Data are presented as mean ± SEM from two independent MEF isolations or two and three experiments for OP9 and 3T3-L1 cells, respectively. (D) Distribution of mRNA expression in metabolically relevant tissues in wild-type mice. Expression is shown relative to heart as fold-expression. Inset represents a scale-up of the Spcs3 expression values. Data are presented as mean ± SEM (n = 3). Abbreviations: WAT, white adipose tissue (epididymal); BAT, brown adipose tissue (interscapular); L, liver; SM, skeletal muscle; H. heart. (E) 3T3-L1 adipocytes were treated with 1 µM rosiglitazone at Day 7 of differentiation. RNA was harvested 24 h after rosiglitazone treatment or medium change (DMEM) and measured with qPCR using the indicated primers. Data are represented as mean ± SEM from three independent experiments. Student’s t-test: **P < 0.01. (F) Male mice were kept on a rosiglitazone-containing chow diet (0.01% w/w) or on normal chow for 7 weeks post-weaning. Prior to harvesting epididymal fat pads mice were fasted for ∼4 h to synchronize nutritional states. Data are represented as mean ± SEM (n = 5–6). Student’s t-test: **P < 0.01.
In age-matched female mice, the expression pattern among tissues was similar to their male counterparts (data not shown), suggesting that there is no sex-specific gene dosage effect. Addition of the PPARγ-agonist rosiglitazone to 3T3-L1 adipocytes (Figure 3E) or OP9 adipocytes (Supplementary Figure S6) increased the expression of Arxes mRNAs. Furthermore, feeding mice with a rosiglitazone-containing diet for 7 weeks elevated the Arxes1 and Arxes2 mRNA in epididymal fat pads when compared to mice on a normal diet (Figure 3F), whereas Spcs3 mRNA was downregulated by in vivo rosiglitazone treatment. Together, these results demonstrate that the Arxes mRNAs are highly upregulated during adipogenesis of several in vitro models as well as in adipose tissue (compared to other metabolic tissues) and that their mRNA levels increase massively upon rosiglitazone treatment, while their parental gene Spcs3 shows no upregulation in any of these conditions.
Protein evidence and subcellular localization of the Arxes
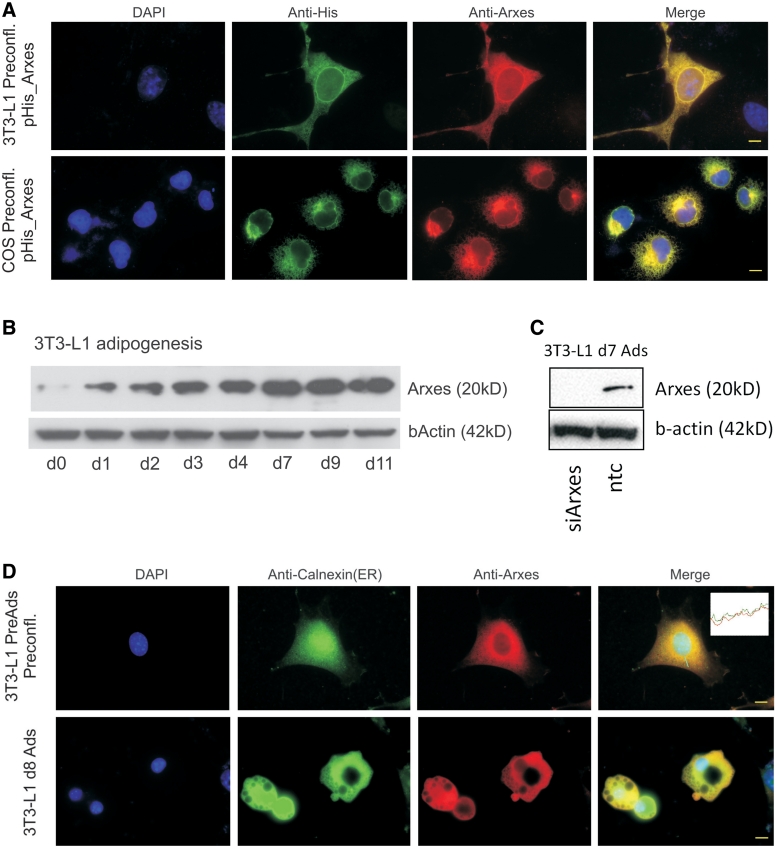
Processed pseudogenes are defined as genomic sequences retrotransposed from a parental mRNA, which no longer translate into functional proteins (34). To determine if the Arxes are nonfunctional, processed pseudogenes, we tested whether the Arxes yield a protein or if they exert their putative function as noncoding RNA as recently reported for other pseudogenes in mouse oocytes (35). Therefore, we established a polyclonal Arxes antibody directed against peptides that confer specificity over Spcs3 (Supplementary Figure S5A). To verify the specificity of this antibody we transiently expressed 6xHis-tagged Arxes in COS and 3T3-L1 cells. A strong overlap of the anti-His and the anti-Arxes staining could be shown with immunofluorescence microscopy in both cell types overexpressing His-tagged Arxes (Figure 4A).
Figure 4.
Protein evidence and localization of Arxes in the ER. (A) Shown are 3T3-L1 and COS cells overexpressing Arxes with a 6xHis-tag. Right-most panels show merge of all channels. Scale bars represent 10 µm. (B) Protein was harvested at indicated time points during 3T3-L1 differentiation and subjected to western blotting using the anti-Arxes antibody. β-Actin served as loading control. Increase in protein level is in concordance with Arxes mRNA measurements. (C) 3T3-L1 cells were transduced with silencing constructs (siArxes) or non-targeting control (ntc), and induced to undergo adipogenesis for 7 days with standard DMI treatment. Western blot analysis is performed on protein lysates to prove specificity of antibody. β-Actin served as loading control. (D) Endogenous Arxes protein location is shown in the red channel for preadipocytes and mature adipocytes (d8). The merged image shows a high degree of overlap with the ER-marker Calnexin, as also shown by the channel intensity profile in the inset. Scale bars represent 10 µm.
To rule out a cross-reactivity with Spcs3, western blot analysis on 3T3-L1 cells during adipogenesis was performed, which showed a steady increase of endogenous protein levels during differentiation (Figure 4B). This pattern correlates with the mRNA profile of Arxes1 and Arxes2, but not with that of Spcs3 (Figure 3A), thus indicating that the antibody is specific for the Arxes proteins. Further, knockdown of Arxes in 3T3-L1 cells, as described below, was used to prove the specificity of the antibody with western blotting (Figure 4C). Finally, we performed immunofluorescence microscopy to investigate the localization of endogenous Arxes in 3T3-L1 preadipocytes and mature adipocytes and found that the Arxes staining in both cases overlaps with the signal of the ER marker Calnexin (Figure 4D). Thus, we provide evidence that the Arxes transcripts are indeed translated into protein, which accumulates during adipogenesis and localizes to the ER in 3T3-L1 preadipocytes and adipocytes.
The Arxes are synergistically regulated by PPARγ and C/EBPα
The fact that the Arxes mRNAs are increased by the PPARγ agonist rosiglitazone and our initial identification of Arxes1 as potential C/EBPα target gene suggest that the Arxes might be regulated by both pivotal transcription factors of adipogenesis. In agreement with this, in silico analyses using the Genomatix software suite (36) identified potential PPARγ and C/EBP binding sites in both Arxes promoters (Supplementary Figure S7). According to these predictions, truncations of the Arxes proximal promoters, including the 5′UTR 139 bp downstream of the TSS, were cloned into Luciferase reporter vectors. Next, we cotransfected the Luciferase reporter vectors with PPARγ/RXRα and C/EBPα expression plasmids or empty vectors into COS cells. As depicted in Figure 5A, we found a >60-fold increase in Luciferase activity in promoter elements that contain the TSS (–182 to +139 bp) compared to downstream promoter elements (+33 to +139 bp) or to empty luciferase vectors. This increase, however, seems to be independent of whether PPARγ/RXRα and C/EBPα transcription factors are present and indicates a strong basal promoter activity mediated by elements in the region between −182 and +33 bp around the Arxes TSSs. Note that the first 689 bp upstream of the Arxes TSSs and their 5′UTRs are highly homologous (99% and 100%, respectively). However, a PPARγ/RXRα/C/EBPα-dependent increase in Luciferase activity could only be found further upstream (Figure 5A). For Arxes1, a significant increase was observed starting with promoter length of 869 bp upstream of the TSS. For Arxes2, around 4 kb upstream had to be cloned to elicit a significant transcription factor dependency (Figure 5A).
In addition to the proximal promoters, we investigated genomic sites around the Arxes that were identified to be bound by C/EBPα and PPARγ in a recently published genome-wide location study by Lefterova et al. (6). In the vicinity of the Arxes, three regions were identified by this study to be bound by both transcription factors (see genomic overview Figure 5F). One binding region upstream of Arxes2 (2900062L11Rik in Figure 5F) directly overlaps with the longest Arxes2 promoter fragment in Figure 5A, supporting our finding of significant increase of luciferase activity in a PPARγ/RXRα/C/EBPα-dependent manner. The two other regions—one about 20 kb upstream and one ∼8 kb downstream of the Arxes1 gene—were cloned into Luciferase reporter vectors containing a minimal promoter. Figure 5B shows a significant increase in reporter activity of the 8 kb downstream region after cotransfecting this reporter vector with PPARγ/RXRα and C/EBPα expression vectors into COS cells.
Compared to preadipocytes, luciferase activity increased nearly 7-fold when this construct was electroporated into 3T3-L1 adipocytes (which have high endogenous levels of PPARγ, RXRα, and C/EBPα; Figure 5C). However, the region ∼20 kb upstream of Arxes 1 did not show any significant difference in the tested experimental setups (Supplementary Figure S8). To verify binding of PPARγ to the Arxes1 8 kb downstream region (Arxes1_8 kb_DS) ChIP-qPCR was performed in 3T3-L1 cells at Day 3 of differentiation when Arxes mRNA levels are highest. Figure 5D shows an enrichment over input that is comparable with the positive control regions [known PPARγ binding site in the hormone-sensitive lipase promoter (37)]. The TFIIB promoter served as negative control. Also, the C/EBPα ChIP in MEFs was assayed with this primer pair specific for the Arxes1_8 kb_DS region and showed a strong enrichment over input (Figure 5E).
Collectively, our data suggest that the proximal promoters of Arxes1 and Arxes2 are functional and can be induced by the adipogenic master regulators, PPARγ and C/EBPα. Furthermore, an enhancer region 8 kb downstream of Arxes1 was shown to be strongly bound and activated both by transcription factors and in an adipogenic context.
Arxes-specific silencing abrogates adipogenesis and enhances osteoblastogenesis
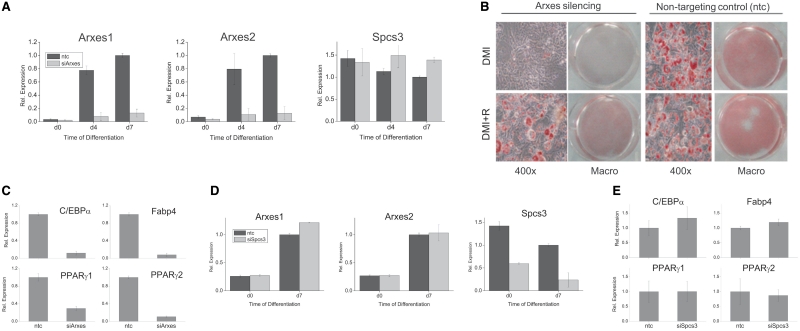
To investigate whether the Arxes are functional in adipogenesis we applied a loss-of-function approach using shRNA-based constructs that specifically and stably silence Arxes in 3T3-L1 cells (see Figure 6A and sequence in Supplementary Figure S5A). Intriguingly, when cells were transduced with lenti-viruses containing an Arxes-specific silencing construct (siArxes in Figure 6), adipogenesis was completely abolished, while cells infected with nontargeting control (ntc) shRNA differentiated normally after DMI treatment (see upper panel of Figure 6B for oil red O staining and Figure 6C for adipogenic markers at Day 7 of adipogenesis). However, if rosiglitazone is added to the differentiation cocktail the differentiation defect in silenced cells can be partly overcome (Figure 6B, lower panel of oil red O staining). In stark contrast to the Arxes silencing, specific (Figure 6D) and stable silencing of Spcs3 in 3T3-L1 cells did not impair adipogenesis or reduce adipogenic marker genes, as illustrated in Figure 6E for Day 7 of adipogenesis. These data suggest that an increase in transcript levels of the Arxes is required for the expression of adipogenic key regulators. Thus, the results of the silencing approach provide evidence that Arxes1 and Arxes2, but not their parental gene Spcs3, are required for adipogenesis, which suggests different functions of these evolutionary relatives. However, it should be noted that stable overexpression of Arxes proteins in 3T3-L1 cells did not influence adipogenesis, neither under normal adipogenic differentiation conditions (Supplementary Figure S9) nor when using suboptimal differentiation mixes (data not shown).
Figure 6.
Silencing of the Arxes abrogates adipogenesis, while Spcs3 knockdown does not change adipogenic marker gene expression. 3T3-L1 cells were transduced with silencing constructs (siArxes or siSpcs3) or nontargeting control (ntc), and induced to undergo adipogenesis with standard DMI treatment unless indicated otherwise. (A) qPCR mRNA measurement shows that Arxes knockdown using the siArxes silencing constructs is specific to the Arxes, while Spcs3 expression is not reduced. Data are presented as mean ± SEM from three independent transduction experiments. (B) Lipid droplets were visualized by oil red O staining of 7 days differentiated 3T3-L1 cells. Lower panel of the oil red O staining shows a partial rescue of the differentiation deficiency if 1 µM rosiglitazone is present during differentiation procedure. (C) Expression of marker genes of adipogenesis was measured with qPCR on Day 7 of adipogenesis in Arxes-silenced and control cells. Data are presented as mean ± SEM from three independent transduction experiments. (D) qPCR mRNA measurement shows that Spcs3 knockdown using the siSpcs3 silencing construct is specific to the Spcs3 while Arxes mRNA is not reduced. Data are presented as mean ± SEM from two independent transduction experiments. (E) Spcs3 knockdown does not influence adipogenic marker gene expression as measured with qPCR on Day 7 in DMI induced cells. Data are presented as mean ± SEM from two independent transduction experiments.
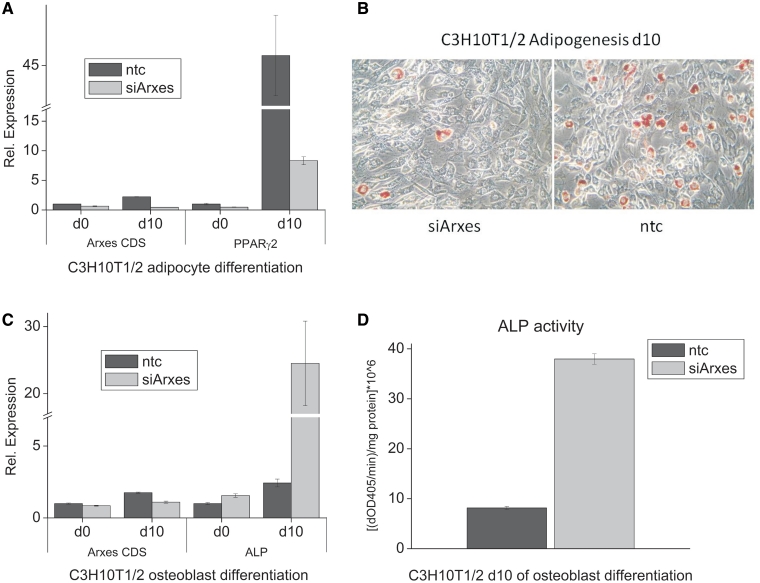
To evaluate whether Arxes are only necessary for the differentiation of preadipocytes to adipocytes or if they play a broader role in early developmental stages we used Arxes-specific silencing in mouse C3H10T1/2 cells. C3H10T1/2 is a pluripotent mesenchymal stem cell line that can be differentiated in a variety of cell types, including adipocytes and osteoblasts (13). Here, we stably silenced Arxes expression in C3H10T1/2 cells before we induced their differentiation into either adipogenic or osteoblastogenic lineages. Corroborating our findings in 3T3-L1 cells, Arxes-silenced C3H10T1/2 cells exhibit reduced adipogenesis when compared to ntc control cells, as assessed by oil red O staining (Figure 7B) and mRNA levels of adipogenic marker genes such as PPARγ2 (Figure 7A) and Fabp4 (data not shown). Interestingly, silencing of Arxes increased osteoblastogenic potential of C3H10T1/2 cells. This is evidenced by increased ALP mRNA expression (Figure 7C) and >4-fold elevated ALP activity (Figure 7D), a hallmark of osteoblast differentiation (13). Hence, our data suggest that Arxes play a role in lineage commitment of mesenchymal stem cells, because osteoblastogenesis is promoted while adipogenesis is repressed upon knockdown of Arxes in C3H10T1/2 cells.
Figure 7.
Silencing of the Arxes in C3H10T1/2 pluripotent stem cells reduces adipogenesis and enhances osteoblastogenesis. C3H10T1/2 cells were transduced with silencing constructs (siArxes) or nontargeting control (ntc), and induced to undergo adipogenesis or osteoblastogenesis for 10 days as described in ‘Material and Methods’. Results are representative of two independent replicates each performed in triplicates (mean ± SEM). (A) Expression of PPARγ was measured with qPCR before differentiation and on Day 10 of adipogenesis in Arxes-silenced and control cells. Arxes CDS primer pair recognizes mRNA levels of both Arxes as it is aimed at the coding sequence. (B) Oil red O staining shows reduced lipid droplet formation in Arxes-silenced cells at Day 10 of adipogenesis, which is in concordance to the results in 3T3-L1 cells. (C) qPCR measurements show that mRNA levels of ALP are increased in Arxes-silenced cells. (D) At Day 10 of osteoblastogenesis cells were homogenized and mixed with pNPP solution and kinetics were recorded for 3 min using a spectrophotometer at 405 nm. Measurements were normalized to protein contents.
DISCUSSION
In this study, we employed a combined computational/experimental approach and identified a set of previously unknown C/EBP target genes. We then further characterized one of the validated targets (Arxes1) and its paralog Arxes 2, both of which appeared to be retrotransposed genes of the signal peptidase Spcs3. Functional studies showed divergent expression, regulation, and function of the Arxes compared to their parental gene Spcs3. Moreover, several lines of evidence suggest that Arxes are novel key proteins in adipogenesis: (i) mRNAs of Arxes1 and Arxes2 were upregulated in cell models of adipogenesis and highly expressed in adipose tissue of mice, (ii) Arxes are synergistically regulated by C/EBPα and PPARγ, (iii) Arxes are translated and their gene products are localized in the endoplasmatic reticulum and (iv) silencing of Arxes1 and Arxes 2 blocked adipogenesis and enhanced osteoblastogenesis.
A major finding for fat cell biology is that upon silencing of the Arxes in 3T3-L1 cells differentiation is impaired, rendering the Arxes as essential players in the process of mouse fat cell differentiation. Moreover, this differentiation defect could be partly rescued by the addition of the PPARγ agonist rosiglitazone to the differentiation mix of the silenced cells. This may suggest that the Arxes function in pathways producing endogenous PPARγ ligands that can be bypassed by addition of synthetic ligands. With this, the Arxes join a line of recently described molecules that have been shown to be necessary for adipogenesis and that were proposed to be involved in PPARγ ligand production pathways, such as RetSat (7) and XOR (8).
Along this line of argumentation, the close relationship of the Arxes and Spcs3 raises the question of whether the Arxes function as signal peptidases and thereby in the secretory pathway. The subcelluar location of the Arxes proteins in the ER (Figure 4) supports this hypothesis. Spcs3 was shown to be a member of the ER signal peptidase complex comprised of five proteins in mammalian systems (38,39). Spc3p, the yeast homolog of mammalian Spcs3, has been shown to be essential for protein secretion in yeast, where upon silencing of Spc3p, precursor proteins accumulate in the ER, which is in turn incompatible with survival (40). However, secretion via the classical (adiponectin) and the nonclassical (visfatin) secretory pathway (41) was not changed 48 h after silencing of the Arxes in mature adipocytes (unpublished ELISA data). Also, silencing of Spcs3 did not impair adipogenic differentiation in 3T3-L1 cells. These data suggest that, although the Spcs3 protein and the Arxes proteins have a similar sequence architecture, the same hydrophobic pattern and the same predicted three-dimensional (3D) structure in the globular domain (amino acids 40–175), their function in adipogenesis is different. This difference is possibly due to many residues with polar side chains at homologous positions that are quite divergent in their physico-chemical properties (see alignment Supplementary Figure S4B). However, further studies are necessary to elucidate the pathway(s) in which the Arxes influence adipogenesis.
Another major finding is the identical transcriptional regulation of the Arxes despite their separation by a 40-kb genomic DNA stretch. The regulatory upstream regions of the Arxes are similar. Both Arxes contain a promoter around the TSS (–182/+33 relative to TSS, Figure 5A). Further upstream we could show PPARγ/C/EBPα-dependent induction of a reporter gene for both, the Arxes1 and Arxes2 promoter. Interestingly, there is an enhancer region situated 8 kb downstream of the 3′ end of Arxes1 to which C/EBPα and PPARγ bind. This region is capable of transactivating a reporter gene (Figure 5B and C). The mRNAs of the two Arxes genes were coregulated in all experimental circumstances shown. The strong homology of the first ∼800 bp of their promoters and the fact that both of the Arxes promoters are transactivated by PPARγ and C/EBPα may explain this coregulation. However, the contribution of the region 8 kb downstream of Arxes1 to the regulation of both Arxes gene remains elusive. Using the chromatin conformation capture (3C) method, the functionality and direct action of such long-range chromatin interactions were recently demonstrated for PPARγ -mediated activation of the Ucp2 and Ucp3 genes in 3T3-L1 adipocytes (42) and—on a genome-wide level—for human estrogen-receptor-α (43). Additional experiments using this 3C method could investigate a direct interaction by means of looping between this 8 kb downstream site and the Arxes promoters.
Finally, an intriguing result of this study is the specific genomic configuration of the Arxes. Arxes1 and Arxes2 are intronless genes and located adjacent to each other on the mouse X-chromosome. They show high sequence similarity in the regions immediately upstream of their TSSs and possess identical CDSs. The Arxes CDSs are highly homologous to the Spcs3 mRNA, expressed from a six-exon gene on the reverse strand on mouse chromosome 8. Further, our analyses identified a region syntenic to the mouse Arxes1–Arxes2 locus on the rat X-chromosome and a single Arxes-like sequence on the X-chromosome of the rabbit genome (Supplementary Data file 2), while there were no such homologous sequences in primate genomes. Based on this genomic setup, we propose that one of the Arxes genes was retrotransposed from Spcs3 mRNA after the speciation of primates and glires, followed by a segmental duplication event (32) after divergence between rabbit and mouse/rat (Figure 2). A similar ‘phylogeny’ was recently reported in a study on human GAPDH pseudogenes (44). The segmental duplication could account for the high sequence similarity in the ∼800 bp upstream of the mouse Arxes TSSs. This upstream promoter region was shown to be functional and is highly conserved in rat, but not in the rabbit Arxes locus. How the Arxes acquired a functional promoter after the retrotransposition remains elusive.
Deeper investigation of the retrotransposition event identified target site duplications (23,29) close to the 5′ and 3′ ends of the mouse and rat Arxes regions that are homologous to Spcs3 mRNA (Figure 2). If these TSDs indeed flank the retrotransposed sequences, this would present an explanation for the highly divergent 3′UTRs of Arxes1, Arxes2 and Spcs3 (Table 1). However, an L1-mediated retrotransposition event usually introduces a polyA stretch directly 5′ of the 3′ TSD (45) by inserting the entire mRNA of the parent gene. Such a polyA stretch cannot be found at the Arxes loci. We reason that the absence of a polyA stretch can be either due to sequence degeneration or because the Arxes arose by an L1-independent retrotransposition event.
Due to the high homology of Arxes CDSs and the Spcs3 mRNA sequence the Arxes loci were annotated as pseudogenes in the build 36 of the NCBI GenBank database. However, we show here that the Arxes are transcribed, regulated in adipogenic conditions and that they code for ER-located proteins, which are necessary for the process of in vitro fat development. Further, our experiments in mesenchymal stem cells suggest a role in lineage commitment. Hence, the Arxes defy the very definition of a (processed) pseudogene (34). Keeping in mind that there are about 20 000 predicted pseudogenes in each the mouse and the human genome (34), the results of our study provoke a reconsideration and reevaluation of these alleged evolutionary relics.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The GEN-AU projects BIN and GOLD from the Austrian Federal Ministry for Science and Research; SFB Project Lipotoxicity from the Austrian Science Fund. Funding for open access charge: Austrian Federal Ministry for Science and Research, GEN-AU project BIN.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Robert Rader for help with ArrayExpress, Florian Stöger, Nicole Golob, Jelena Kocic and Franziska Vogl for support in the lab, Daniel Schwartz for proofreading, Thomas Burkard for annotations, Gabriela Bindea for GO analysis and Fatima Sanchez-Cabo for statistical advice. Further, we thank Martina Lefterova for providing the ChIP–chip data set and Mitch Lazar for comments on the manuscript.
REFERENCES
- 1.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 2.Prokesch A, Hackl H, Hakim-Weber R, Bornstein SR, Trajanoski Z. Novel insights into adipogenesis from omics data. Curr. Med. Chem. 2009;16:2952–2964. doi: 10.2174/092986709788803132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lefterova MI, Lazar MA. New developments in adipogenesis. Trends Endocrinol. Metab. 2009;20:107–114. doi: 10.1016/j.tem.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Gray SL, Dalla NE, Vidal-Puig AJ. Mouse models of PPAR-gamma deficiency: dissecting PPAR-gamma's role in metabolic homoeostasis. Biochem. Soc. Trans. 2005;33:1053–1058. doi: 10.1042/BST0331053. [DOI] [PubMed] [Google Scholar]
- 5.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ, Jr, Liu XS, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schupp M, Lefterova MI, Janke J, Leitner K, Cristancho AG, Mullican SE, Qatanani M, Szwergold N, Steger DJ, Curtin JC, et al. Retinol saturase promotes adipogenesis and is downregulated in obesity. Proc. Natl Acad. Sci. USA. 2009;106:1105–1110. doi: 10.1073/pnas.0812065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung KJ, Tzameli I, Pissios P, Rovira I, Gavrilova O, Ohtsubo T, Chen Z, Finkel T, Flier JS, Friedman JM. Xanthine oxidoreductase is a regulator of adipogenesis and PPARgamma activity. Cell Metab. 2007;5:115–128. doi: 10.1016/j.cmet.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Christianson JL, Nicoloro S, Straubhaar J, Czech MP. Stearoyl-CoA desaturase 2 is required for peroxisome proliferator-activated receptor gamma expression and adipogenesis in cultured 3T3-L1 cells. J. Biol. Chem. 2008;283:2906–2916. doi: 10.1074/jbc.M705656200. [DOI] [PubMed] [Google Scholar]
- 10.Olive M, Williams SC, Dezan C, Johnson PF, Vinson C. Design of a C/EBP-specific, dominant-negative bZIP protein with both inhibitory and gain-of-function properties. J. Biol. Chem. 1996;271:2040–2047. doi: 10.1074/jbc.271.4.2040. [DOI] [PubMed] [Google Scholar]
- 11.Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolins NE, Quaynor BK, Skinner JR, Tzekov A, Park C, Choi K, Bickel PE. OP9 mouse stromal cells rapidly differentiate into adipocytes: characterization of a useful new model of adipogenesis. J. Lipid Res. 2006;47:450–460. doi: 10.1194/jlr.D500037-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Fan Q, Tang T, Zhang X, Dai K. The role of CCAAT/enhancer binding protein (C/EBP)-alpha in osteogenesis of C3H10T1/2 cells induced by BMP-2. J. Cell Mol. Med. 2009;13:2489–2505. doi: 10.1111/j.1582-4934.2008.00606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hackl H, Burkard TR, Sturn A, Rubio R, Schleiffer A, Tian S, Quackenbush J, Eisenhaber F, Trajanoski Z. Molecular processes during fat cell development revealed by gene expression profiling and functional annotation. Genome Biol. 2005;6:R108. doi: 10.1186/gb-2005-6-13-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maurer M, Molidor R, Sturn A, Hartler J, Hackl H, Stocker G, Prokesch A, Scheideler M, Trajanoski Z. MARS: microarray analysis, retrieval, and storage system. BMC Bioinformatics. 2005;6:101. doi: 10.1186/1471-2105-6-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 17.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 23.Lucier JF, Perreault J, Noel JF, Boire G, Perreault JP. RTAnalyzer: a web application for finding new retrotransposons and detecting L1 retrotransposition signatures. Nucleic Acids Res. 2007;35:W269–W274. doi: 10.1093/nar/gkm313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pabinger S, Thallinger GG, Snajder R, Eichhorn H, Rader R, Trajanoski Z. QPCR: application for real-time PCR data management and analysis. BMC Bioinformatics. 2009;10:268. doi: 10.1186/1471-2105-10-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J. Comput. Biol. 2005;12:1047–1064. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kliewer SA, Forman BM, Blumberg B, Ong ES, Borgmeyer U, Mangelsdorf DJ, Umesono K, Evans RM. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc. Natl Acad. Sci. USA. 1994;91:7355–7359. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bogner-Strauss JG, Prokesch A, Sanchez-Cabo F, Rieder D, Hackl H, Duszka K, Krogsdam A, Di Camillo B, Walenta E, Klatzer A, et al. Reconstruction of gene association network reveals a transmembrane protein required for adipogenesis and targeted by PPARgamma. Cell Mol. Life Sci. 2010;67:4049–4064. doi: 10.1007/s00018-010-0424-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newsome AL, McLean JW, Lively MO. Molecular cloning of a cDNA encoding the glycoprotein of hen oviduct microsomal signal peptidase. Biochem. J. 1992;282(Pt 2):447–452. doi: 10.1042/bj2820447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vanin EF. Processed pseudogenes: characteristics and evolution. Annu. Rev. Genet. 1985;19:253–272. doi: 10.1146/annurev.ge.19.120185.001345. [DOI] [PubMed] [Google Scholar]
- 30.Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knudsen S. Promoter2.0: for the recognition of PolII promoter sequences. Bioinformatics. 1999;15:356–361. doi: 10.1093/bioinformatics/15.5.356. [DOI] [PubMed] [Google Scholar]
- 32.Khurana E, Lam HY, Cheng C, Carriero N, Cayting P, Gerstein MB. Segmental duplications in the human genome reveal details of pseudogene formation. Nucleic Acids Res. 2010;38:6997–7007. doi: 10.1093/nar/gkq587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Margulies EH, Cooper GM, Asimenos G, Thomas DJ, Dewey CN, Siepel A, Birney E, Keefe D, Schwartz AS, Hou M, et al. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17:760–774. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng D, Gerstein MB. The ambiguous boundary between genes and pseudogenes: the dead rise up, or do they? Trends Genet. 2007;23:219–224. doi: 10.1016/j.tig.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 35.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scherf M, Klingenhoff A, Werner T. Highly specific localization of promoter regions in large genomic sequences by PromoterInspector: a novel context analysis approach. J. Mol. Biol. 2000;297:599–606. doi: 10.1006/jmbi.2000.3589. [DOI] [PubMed] [Google Scholar]
- 37.Yajima H, Kobayashi Y, Kanaya T, Horino Y. Identification of peroxisome-proliferator responsive element in the mouse HSL gene. Biochem. Biophys. Res. Commun. 2007;352:526–531. doi: 10.1016/j.bbrc.2006.11.054. [DOI] [PubMed] [Google Scholar]
- 38.Kalies KU, Hartmann E. Membrane topology of the 12- and the 25-kDa subunits of the mammalian signal peptidase complex. J. Biol. Chem. 1996;271:3925–3929. doi: 10.1074/jbc.271.7.3925. [DOI] [PubMed] [Google Scholar]
- 39.Evans EA, Gilmore R, Blobel G. Purification of microsomal signal peptidase as a complex. Proc. Natl Acad. Sci. USA. 1986;83:581–585. doi: 10.1073/pnas.83.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer HA, Hartmann E. The yeast SPC22/23 homolog Spc3p is essential for signal peptidase activity. J. Biol. Chem. 1997;272:13159–13164. doi: 10.1074/jbc.272.20.13159. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka M, Nozaki M, Fukuhara A, Segawa K, Aoki N, Matsuda M, Komuro R, Shimomura I. Visfatin is released from 3T3-L1 adipocytes via a non-classical pathway. Biochem. Biophys. Res. Commun. 2007;359:194–201. doi: 10.1016/j.bbrc.2007.05.096. [DOI] [PubMed] [Google Scholar]
- 42.Bugge A, Siersbaek M, Madsen MS, Gondor A, Rougier C, Mandrup S. A novel intronic peroxisome proliferator activated receptor (PPAR) {gamma} enhancer in the uncoupling protein (UCP) 3 gene as a regulator of both UCP2 and -3 expression in adipocytes. J. Biol. Chem. 2010;285:17310–17317. doi: 10.1074/jbc.M110.120584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu YJ, Zheng D, Balasubramanian S, Carriero N, Khurana E, Robilotto R, Gerstein MB. Comprehensive analysis of the pseudogenes of glycolytic enzymes in vertebrates: the anomalously high number of GAPDH pseudogenes highlights a recent burst of retrotrans-positional activity. BMC Genomics. 2009;10:480. doi: 10.1186/1471-2164-10-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.