Abstract
LigD 3′-phosphoesterase (PE) is a component of the bacterial NHEJ apparatus that performs 3′-end-healing reactions at DNA breaks. The tertiary structure, active site and substrate specificity of bacterial PE are unique vis–à-vis other end-healing enzymes. PE homologs are present in archaea, but their properties are uncharted. Here, we demonstrate the end-healing activities of two archaeal PEs—Candidatus Korarchaeum cryptofilum PE (CkoPE; 117 amino acids) and Methanosarcina barkeri PE (MbaPE; 151 amino acids)—and we report their atomic structures at 1.1 and 2.1 Å, respectively. Archaeal PEs are minimized versions of bacterial PE, consisting of an eight-stranded β barrel and a 310 helix. Their active sites are located in a crescent-shaped groove on the barrel’s outer surface, wherein two histidines and an aspartate coordinate manganese in an octahedral complex that includes two waters and a phosphate anion. The phosphate is in turn coordinated by arginine and histidine side chains. The conservation of active site architecture in bacterial and archaeal PEs, and the concordant effects of active site mutations, underscore a common catalytic mechanism, entailing transition state stabilization by manganese and the phosphate-binding arginine and histidine. Our results fortify the proposal that PEs comprise a DNA repair superfamily distributed widely among taxa.
INTRODUCTION
DNA ligase D (LigD) is the central agent of the bacterial non-homologous end-joining (NHEJ) pathway of DNA double-strand break (DSB) repair (1). LigD is a single polypeptide composed of three autonomous domains: an ATP-dependent ligase (LIG), a polymerase (POL), and a 3′-phosphoesterase (PE). The POL domain incorporates dNMP/rNMPs at DSB ends and gaps prior to strand sealing by the LIG domain (2–6) and is largely responsible for the mutagenic quality of bacterial NHEJ in vivo (7). The PE domain provides a 3′-end-healing function, whereby it hydrolyzes 3′-phosphates from DSB ends to yield a 3′-OH that can serve as a primer for polymerase extension or as a strand donor for ligation (8). PE also trims short 3′-ribonucleotide tracts (produced by POL) to generate the 3′-monoribonucleotide ends that are the preferred substrates for sealing by bacterial NHEJ ligases (8,9).
The bacterial LigD PE domains catalyze two sequential manganese-dependent end-healing steps at a DNA primer-template containing a 3′-teminal diribonucleotide (Figure 2C). First, the 3′-terminal nucleoside is removed by a phosphodiesterase activity to yield a primer strand with a ribonucleoside 3′-PO4 terminus. Then, the 3′-PO4 is hydrolyzed by a phosphomonoesterase activity to a 3′-OH. Mutational analyses identified six conserved side chains that were essential for the monoesterase and diesterase activities and thus candidates to comprise their common active site (8,10–12). We recently determined the 1.9 Å crystal structure of the active phosphodiesterase core of Pseudomonas aeruginosa PE (PaePE) spanning amino acids 30–187 (13). PaePE has a novel fold in which an eight-stranded β barrel with a hydrophobic interior supports a crescent-shaped hydrophilic active site on its outer surface (Figure 1A). The six essential side chains coordinate manganese and a sulfate mimetic of the scissile phosphate (13). The PaePE active site is unlike that of any known DNA/RNA end-healing enzymes or any known phosphotransferases. These distinctive features, and the detection of stand-alone PE homologs in many bacterial, archaeal and eukaryal proteomes, suggested that PEs comprise a new and widely distributed DNA end-healing superfamily.
Figure 2.
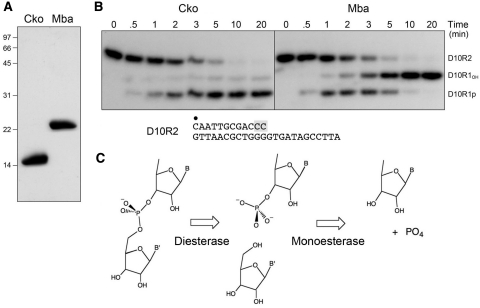
Phosphoesterase activity of recombinant CkoPE and MbaPE. (A) Purification. Aliquots (7 µg) of purified recombinant CkoPE and MbaPE were analyzed by SDS–PAGE. The Coomassie Blue-stained gel is shown. The positions and sizes (kDa) of marker polypeptides are indicated on the left. (B) Phosphoesterase reaction mixtures (90 µl) containing 50 mm Tris–HCl (pH 8.0) for CkoPE or 50 mM MES (pH 6.5) for MbaPE, 5 mM DTT, 0.5 mM MnCl2, 4.5 pmol 32P-labeled D10R2 primer-template substrate, and 8 µg of PE protein were incubated at 45°C. Aliquots (10 µl) were withdrawn at the times specified and quenched immediately with EDTA/formamide. The time 0 sample was withdrawn prior to adding PE proteins. The products were resolved by electrophoresis through an 18% polyacrylamide gel containing 7 M urea in TBE (90 mM Tris–borate, 2.5 mM EDTA) and then visualized by autoradiography. The D10R2 primer-template substrate is shown at bottom with the 5′-32P-label denoted by filled circle and the 3′-diribonucleotide shaded gray. (C) Two-step 3′-end-healing activity. The reaction mechanism entails an initial phosphodiesterase step that incises the 3′-diribonucleotide linkage of the primer-template to generate a monoribonucleotide-3′-phosphate-terminated primer strand and release a monoribonucleoside. The phosphomonoesterase activity then releases inorganic phosphate and leaves a 3′-monoribonucleotide primer-template end-product.
Figure 1.
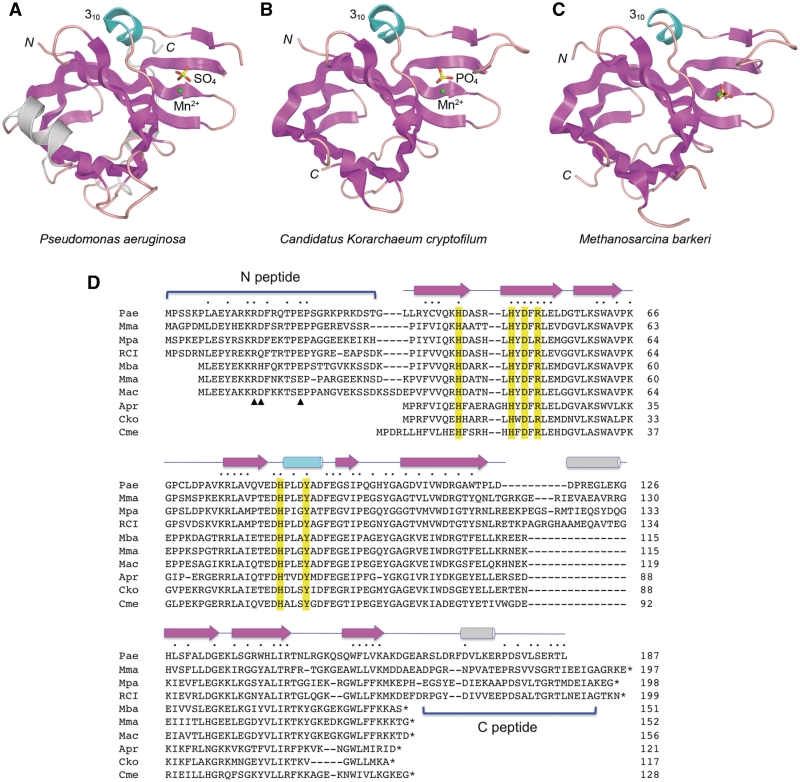
Tertiary and primary structures of bacterial and archaeal PEs. The tertiary structure of the P. aeruginosa LigD PE domain (13) is shown in (A). The structures of Candidatus Korarchaeum cryptofilum PE and M. barkeri PE reported herein are shown in (B) and (C) respectively. The three structures were superimposed and then offset laterally. The amino (N) and carboxyl (C) termini are labeled. The conserved PE folds are shown with β strands colored magenta and the 310 helix colored cyan. Structural elements unique to the Pseudomonas PE are colored gray. The active sites are demarcated by a manganese cation (green sphere) and a nearby sulfate or phosphate anion (stick model). (D) The amino-acid sequence of P. aeruginosa (Pae) PE is aligned to the homologous polypeptides encoded by M. marisnigri (Mma), M. paludicola (Mpa), uncultured methanogenic archaeon RC-I (RCI), M. barkeri (Mba), Methanosarcina mazei (Mma), M. acetivorans (Mac), A. profundus (Apr), Canidatus Korarchaeum cryptofilum (Cko) and Candidatus Methanoregula boonei (Cme). Positions of amino-acid side chain identity/similarity are denoted by filled circle above the aligned sequences. Gaps in the alignment are indicated by dash. Conserved PE active site residues are highlighted in yellow. The secondary structure elements of the PaePE fold (strands depicted as arrows and helices as cylinders) are shown above the sequences. The N- and C-peptides that are variously present or missing from the archaeal PEs are demarcated by brackets. Three conserved residues in the N-peptide that are important for 3′-phosphatase activity in PaePE are denoted by arrowheads below the aligned N-peptides.
Here, we test this idea by studying the activities and structures of exemplary archaeal PE proteins. An alignment of the primary structures of PaePE and its homologs from nine archaeal taxa reveals three distinct subgroups of archaeal PEs (Figure 1D). The ‘minimized’ PEs of Archaeoglobus profundus (121 amino acids), Candidatus Korarchaeum cryptofilum (117 amino acids), and Candidatus Methanoregula boonei (128 amino acids) appear to conserve the PaePE β-barrel and active site constituents, but they lack counterparts of the N-terminal (N) peptide and C-terminal (C) peptide segments of bacterial PE (Figure 1D). In contrast, the PEs of Methanoculleus marisnigri (197 amino acids), Methanocella paludicola (198 amino acids), and uncultured methanogen RC-I (199 amino acids) appear to be bonafide full-length homologs of PaePE. Between these extremes are the PEs of Methanosarcina mazei (152 amino acids), Methanosarcina acetivorans (156 amino acids) and Methanosarcina barkeri (151 amino acids), which retain the N-peptide, but not the C-peptide (Figure 1D). The N-peptide segment of PaePE is sensitive to limited proteolysis (10), suggesting that it is disordered in solution. Deletions of the N-peptide in PaePE selectively diminish 3′-phosphomonoesterase activity without affecting the 3′-phosphodiesterase (10).
Thus, we suspected that: (i) the six archaeal PE proteins that do have conserved counterparts of the N-peptide might possess 3′-phosphodiesterase and monoesterase activities à la the bacterial LigD PE domains; and (ii) the three minimal archaeal PEs lacking the N-peptide might be proficient as a 3′-phosphodiesterase, but have weaker monoesterase activity, similar to the N-terminal deletants of PaePE. Alternatively, the archaeal PE proteins might be dedicated to phosphohydrolase reactions having nothing to do with polynucleotide end-healing. To evaluate these scenarios, we produced and characterized the PE homologs from Candidatus Korarchaeum cryptofilum (hereafter named CkoPE) and M. barkeri (MbaPE) and confirmed their end-healing functions in vitro. We also determined the atomic structures of CkoPE and MbaPE at 1.1 and 2.1 Å resolution, respectively (Figure 1B and C). Concordant mutational effects highlight conservation of the active sites of archaeal and bacterial PEs.
MATERIALS AND METHODS
Recombinant CkoPE and MbaPE proteins
The DNAs encoding CkoPE and MbaPE were synthesized by GenScript USA Inc. to optimize codon usage in Escherichia coli and introduce a BamHI site 5′ of the start codon and a HindIII site 3′ of the stop codon. The BamHI–HindIII fragments were inserted into pET28b-His10Smt3 to generate pET28b-His10Smt3-CkoPE and pET28b-His10Smt3-MbaPE plasmids, respectively. The inserts were sequenced completely to exclude the acquisition of unwanted changes during synthesis or cloning. The expression plasmids were transformed into E. coli BL21(DE3). Cultures (2 l) of E. coli BL21(DE3)/pET28b-His10Smt3-PE derived from single transformants were grown at 37°C in Luria–Bertani medium containing 0.06 mg/ml kanamycin until the A600 reached 0.6. The cultures were adjusted to 0.5 mM isopropyl-ß-d-thiogalactopyranoside and then incubated at 17°C for 15 h. Cells were harvested by centrifugation and the pellets stored at −80°C. All subsequent procedures were performed at 4°C. Thawed bacteria were resuspended in 50 ml of lysis buffer (50 mM Tris–HCl, pH 7.5, 1.5 M NaCl, 200 mM Li2SO4, 10% glycerol, 15 mM imidazole). Lysozyme and Triton X-100 were added to final concentrations of 50 µg/ml and 0.1%, respectively. The lysates were sonicated to reduce viscosity and insoluble material was removed by centrifugation. The supernatants were applied to 6-ml columns of Ni2+-nitrilotriacetic acid-agarose (Qiagen) that had been equilibrated with lysis buffer. The columns were washed with 50 ml of lysis buffer and then eluted stepwise with 12-ml aliquots of buffer A (50 mM Tris–HCl, pH 7.5, 0.2 M NaCl, 10% glycerol) containing 50, 100 and 500 mM imidazole. The polypeptide compositions of the fractions were monitored by SDS–PAGE. The CkoPE and MbaPE proteins were recovered predominantly in the 500 mM imidazole eluate fraction. The 500 mM imidazole eluate fractions containing Smt3-CkoPE or MbaPE were digested with the Smt3-specific protease Ulp1 at a 500:1 ratio of PE:Ulp1 during overnight dialysis in buffer A. The released His10Smt3 tag was removed by passing the digest over a second Ni2+-nitrilotriacetic acid-agarose column. The tag-free CkoPE and MbaPE proteins were recovered in the flow-through fractions (which contained 40 mg CkoPE and 43 mg MbaPE, respectively). The protein concentrations were determined by using the Bio-Rad dye reagent with bovine serum albumin as the standard. The recombinant proteins were stored at −80°C.
Crystallization and X-ray diffraction
The tag-free CkoPE and MbaPE proteins were purified further by gel filtration through a 120-ml Superdex-200 column pre-equilibrated in 20 mM Tris–HCl, pH 8.0, 350 mM NaCl, 1 mM DTT. The peak CkoPE and MbaPE fractions were concentrated by centrifugal ultrafiltration to 9.7 and 8.5 mg/ml respectively in 10 mM Tris–HCl, pH 8.0, 100 mM NaCl, 1 mM DTT. The protein solutions were stored at −80°C. Crystallization was performed by sitting-drop vapor diffusion at 22°C. A solution of 0.7 mM CkoPE and 1.8 mM MnCl2 was mixed 1:1 with and equilibrated against a reservoir solution of 0.2 M Na2HPO4 and 20% PEG-3350. CkoPE crystals grew overnight. A solution of 0.5 mM MbaPE and 2 mM MnCl2 was mixed 1:1 with and equilibrated against a reservoir solution of 0.18 M MgCl2, 2.5% 1,4-dioxane, 32% PEG-4000, 0.1 M Tris–HCl (pH 7.5). Crystals of MbaPE appeared within 1 week. The CkoPE and MbaPE crystals were frozen in liquid nitrogen directly from their respective mother liquors.
Diffraction data were collected at NSLS beamline X25 with an ADXV-Q315 CCD detector. Data from a single CkoPE crystal was collected in three passes using three different detector-to-crystal distances and three different X-ray dosage schemes to give a total of 556° of oscillation data. These data were merged to give a dataset that was 96.5% complete to a limiting resolution of 1.1 Å with no signs of radiation damage. A single MbaPE crystal was used to collect a 180° sweep of oscillation data that was 97.5% complete to a limiting resolution of 2.1 Å. All data were indexed and integrated in HKL2000 and merged in SCALEPACK (14). CkoPE and MbaPE crystallized in space groups P21 and P212121, respectively, with one protomer in their asymmetric units. Diffraction statistics are compiled in Table 1.
Table 1.
Crystallographic data and refinement statistics
| MbaPE | CkoPE | |
|---|---|---|
| Space group | P212121 | P21 |
| Unit cell dimensions @ 130K | a = 30.37 Å, b = 60.17 Å, c = 76.48 Å | a = 27.77 Å, b = 58.46 Å, c = 33.18 Å, β=102.25° |
| Radiation source | NSLS–X25 | NSLS–X25 |
| Wavelength (Å) | 1.1 | 0.95 |
| Crystallographic data quality | ||
| Resolution (Å) | 30.0–2.1 (2.18–2.1) | 30.0–1.1 (1.12–1.1) |
| Rsym | 12.2% (46.5%) I > −σI | 3.8% (37%) I > −3σI |
| Unique reflectionsa | 8606 (843) | 40 543 (1902) |
| Mean redundancy | 4.9 (4.4) | 5.1 (2.3) |
| Completeness (%) | 97.5 (97.2) | 96.5 (90.8) |
| Mean I/σI | 11.12 (2.7) | 42.0 (2.1) |
| Phasing statistics | ||
| Phasing method | Phenix AutoMR | Phenix AutoMR |
| Resolution (Å) | 30.0–3.0 | 30.0–1.8 |
| Log-likelihood gain / FOM | 215.6 / 0.33 | 74.45 / 0.46 |
| Refinement statistics | ||
| Resolution (Å) | 30.0–2.1 (2.23–2.1) | 30.0–1.1 (1.14–1.1) |
| Rfree/Rworkb (%) for F > 0 | 25.7/20.3 (28.8/24.2) | 16.5/13.2 (20.6/18.8) |
| Estimated coordinate error | 0.26 Å | 0.1 Å |
| Model Statistics | ||
| RMS deviation for bonds lengths | 0.005 Å | 0.01 Å |
| RMS deviation for bonds angles | 0.95° | 1.41° |
| Ramachandran outliers | 0 | 0 |
| B-factors (Åb) Wilson/Overall | 20.6/23.1 | 10.9/18.2 |
| Mean anisotropy | 0.47 | |
| Protein | 22.5 | 15.6 |
| Heteroatoms, Solvent | 29.6 | 27.4 |
| Model Contents | ||
| Amino acid residues | 126 | 118 |
| Alternate conformations | 0 | 51 |
| Heteroatoms | 1 PO4, 1 Mn | 1 PO4, 1 Mn, 1 PEG |
| Waters | 62 | 215 |
| PDB ID | 3P43 | 3P4H |
Standard definitions are used for all of the parameters. Figures in parentheses refer to data in the highest resolution bin. The data collection statistics come from SCALEPACK. The refinement and geometric statistics come from PHENIX.
aI+ and I− were treated as equivalent observations throughout.
bRfree sets for cross validation consisted of 5% of data selected at random.
Structure determination
Phases and models of the CkoPE and MbaPE structures were obtained via automated molecular replacement and model rebuilding in PHENIX (15). The search model for solving the CkoPE structure was derived from a polypeptide fragment of the PaePE structure domain (PDB ID 3N9B) truncated at residue 160 and then converted to polyalanine. (The CkoPE structure was subsequently used as a search model to solve the MbaPE structure.) After manual rebuilding in COOT (16), all models were refined in PHENIX using standard procedures. Full anisotropic atomic displacement parameters were refined for CkoPE. The refined models of both structures show excellent geometry with no Ramachandran outliers and no large Fo–Fc difference Fourier peaks. The MbaPE structure refined to R/Rfree values of 20.3/25.7% while the CkoPE refined to final R/Rfree values of 13.2/16.5%. The refinement statistics are compiled in Table 1.
PE mutants
Alanine mutations were introduced into the pET28b-His10Smt3-CkoPE and pET28b-His10Smt3-MbaPE plasmids by two-stage overlap extension PCR. The PE open reading frames in the mutated plasmids were sequenced completely to exclude the acquisition of unwanted changes during PCR and cloning. The CkoPE-Ala and MbaPE-Ala proteins were produced in E. coli and purified in parallel with wild-type CkoPE and MbaPE through the second Ni-agarose step as described above.
RESULTS
End-healing activities of recombinant archaeal PE proteins
The CkoPE and MbaPE proteins were produced in E. coli as His10Smt3 fusions and then isolated from soluble bacterial extracts by Ni-affinity chromatography. The tags were removed by treatment with the Smt3-specific protease Ulp1 and the tag-free CkoPE and MbaPE proteins were separated from His10Smt3 by a second Ni-affinity step, during which the PEs were recovered in the flow-through. SDS–PAGE analysis revealed that the preparations were highly enriched with respect to the CkoPE and MbaPE polypeptides, which migrated at ∼15 and ∼23 kDa, respectively (Figure 2A).
The PE proteins were reacted with a 5′-32P-labeled D10R2 primer-template composed of a 12-mer primer strand with two terminal ribonucleotides and a 24-mer DNA template strand (Figure 2B). MbaPE converted the input labeled strand to two more rapidly migrating products, corresponding to D10R1p and D10R1, respectively (Figure 2B), that migrated identically to the D10R1p and D10R1 products generated by PaePE (not shown). A kinetic analysis of the MbaPE reaction in enzyme excess showed the transient appearance of D10R1p at early times (Figure 2B, 0.5–3 min) and its subsequent conversion to D10R1 (Figure 2B, 5–20 min), suggesting that MbaPE removes the terminal ribonucleotide in a two-step pathway similar to that described for PaePE (Figure 2C).
In contrast, CkoPE yielded D10R1p as the predominant product even in enzyme excess (Figure 2B). The kinetic profile indicated that nearly all of the D10R2 substrate was converted to D1R1p in 10 min, but relatively little of this material was processed to D10R1 between 10 and 20 min (Figure 2B). Thus, CkoPE has a relatively weak 3′-phosphomonoesterase activity, similar to PaePE that lacks the N-peptide (10).
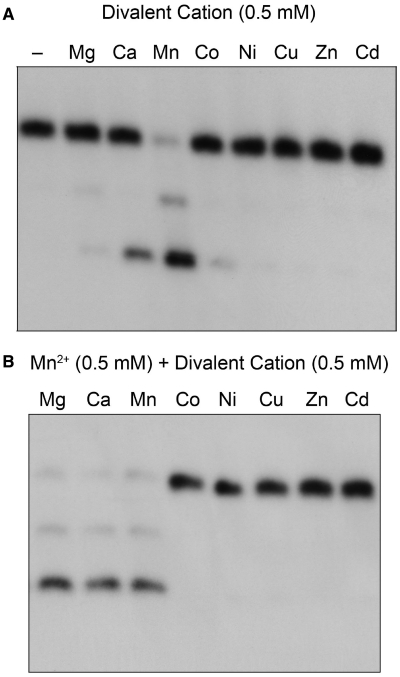
The ribonucleoside resection reaction of CkoPE was strictly dependent on a divalent cation cofactor (Figure 3A). The metal requirement was satisfied by 0.5 mM manganese, which triggered processing of 86% of the input substrate (Figure 3A). In contrast, magnesium was ineffective, either at 0.5 mM (Figure 3), or at any of several higher concentrations tested (1, 2, 5 or 10 mM; not shown). Calcium (0.5 mM) supported processing of 23% of the input substrate (Figure 3). Other metals at 0.5 mM concentration were either very weakly active (2.6% product formation with cobalt) or inactive (no detectable product formed with nickel, copper, zinc and cadmium) (Figure 3A). Additional insights to the metal specificity of CkoPE were provided by mixing experiments, in which reaction solutions containing 0.5 mM manganese were supplemented with 0.5 mM of each of the series of divalent cations (Figure 3B). Cobalt, nickel, zinc, copper and cadmium abolished PE activity in the presence of manganese, suggesting that these metals might out-complete manganese for a putative metal-binding site on the enzyme, wherein engaged they are unable to support reaction chemistry. In contrast, magnesium had no such deleterious effect in combination with manganese, suggesting that magnesium does not bind effectively to the CkoPE active site.
Figure 3.
Divalent cation specificity of CkoPE. (A) Reaction mixtures (10 µl) containing 50 mM Tris–HCl (pH 8.0), 0.5 pmol 32P-labeled D10R2 primer-template, 15 pmol CkoPE, and either no divalent metal (−) or 0.5 mM of the indicated divalent cation were incubated for 10 min at 45°C. (All metals were chloride salts except for zinc sulfate.) The reactions were quenched with EDTA/formamide. The extents of end processing (%) supported by the various metals were quantified by scanning the gel with a phosphorimager and calculated as [D10R1p + D10R1]/[D10R2 + D10R1p + D10R1] × 100. The values were as follows, with each datum being the average of three independent experiments ±SEM: manganese 86.4 ± 0.5; calcium 22.9 ± 1.6; cobalt 2.6 ± 1.7; magnesium 0.7 ± 0.7. Activity with the other metals was <0.5%. (B) Reaction mixtures (10 µl) containing 50 mM Tris–HCl (pH 8.0), 0.5 pmol 32P-labeled D10R2 primer-template, 15 pmol CkoPE, 0.5 mM MnCl2 and 0.5 mM of the indicated divalent cation were incubated for 10 min at 45°C.
Crystallization of CkoPE and structure determination
Crystallization trials were performed with a solution of 0.7 mM CkoPE and 1.8 mM Mn2+ by sitting drop vapor diffusion against various commercial precipitant solutions. Crystals grew overnight in the presence of PEG and sodium phosphate. Diffraction data at 1.1 Å resolution were collected for a native crystal in space group P21. Initial phases were obtained by molecular replacement using an all-alanine version of a trimmed PaePE fold as a search model. The crystal contained one CkoPE protomer in the asymmetric unit. The structural model, comprising the entire CkoPE polypeptide (amino acids 1–117), was refined at 1.1 Å resolution with R/Rfree values of 0.132/0.165 and excellent geometry (Table 1).
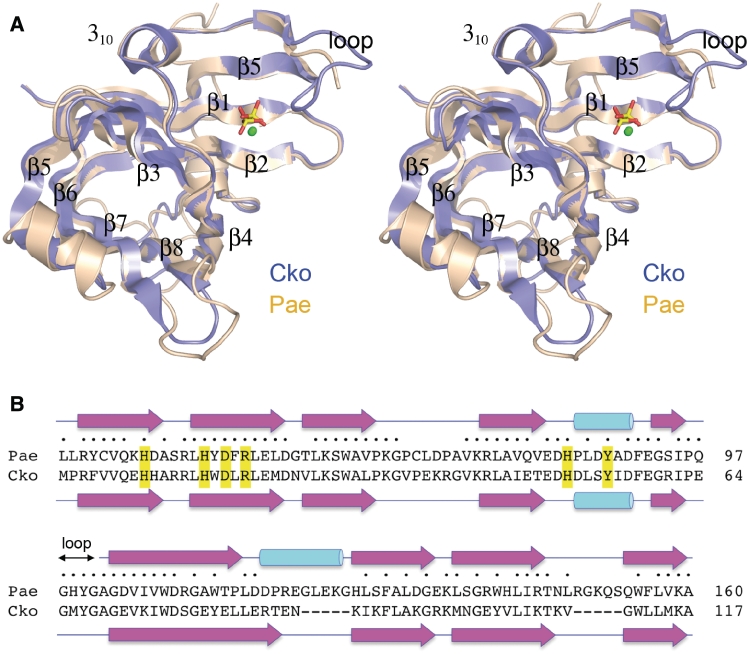
CkoPE consists of a central eight-stranded anti-parallel β barrel and a 310 helix (Figure 1B). Figure 4A shows a superposition of CkoPE (blue) and PaePE (beige). The folds align in DALI (17) with an rmsd of 1.3 Å at 116 Cα positions (Z-score 18.6). The PaePE and CkoPE secondary structure elements are displayed in Figure 4B above and below their aligned amino-acid sequences, respectively. The archaeal protein lacks the α-helix situated between strands β5 and β6 of PaePE and, as expected, CkoPE has no counterpart of the C-terminal segment of PaePE that includes a surface α-helix. The CkoPE β barrel has a narrow aperture filled with hydrophobic residues that are conserved in PaePE.
Figure 4.
Comparison of bacterial and archaeal PE structures. (A) A stereo view of the superimposed CkoPE (blue) and PaePE (beige) structures is shown. The PE fold comprises an eight-stranded anti-parallel β barrel flanked by a 310 helix. The β strands of the barrel are numbered according to their order in the primary structure. The active sites are located in a crescent-shaped groove on the outer surface of the β-barrels, and are demarcated by protein-bound manganese (green spheres) and sulfate or phosphate (stick model) ligands. (B) Structure-based alignment of the PaePE and CkoPE amino-acid sequences, with positions of side-chain identity/similarity indicated by filled circle. The secondary structure elements of the PaePE and CkoPE folds are shown above and below the amino-acid sequences, with strands depicted as arrows and helices as cylinders. The figure highlights a disordered surface loop flanking the active site in PaePE (amino acids 98–101), the counterpart of which was visible in the CkoPE structure (amino acids 65–68).
CkoPE active site and catalytic mechanism
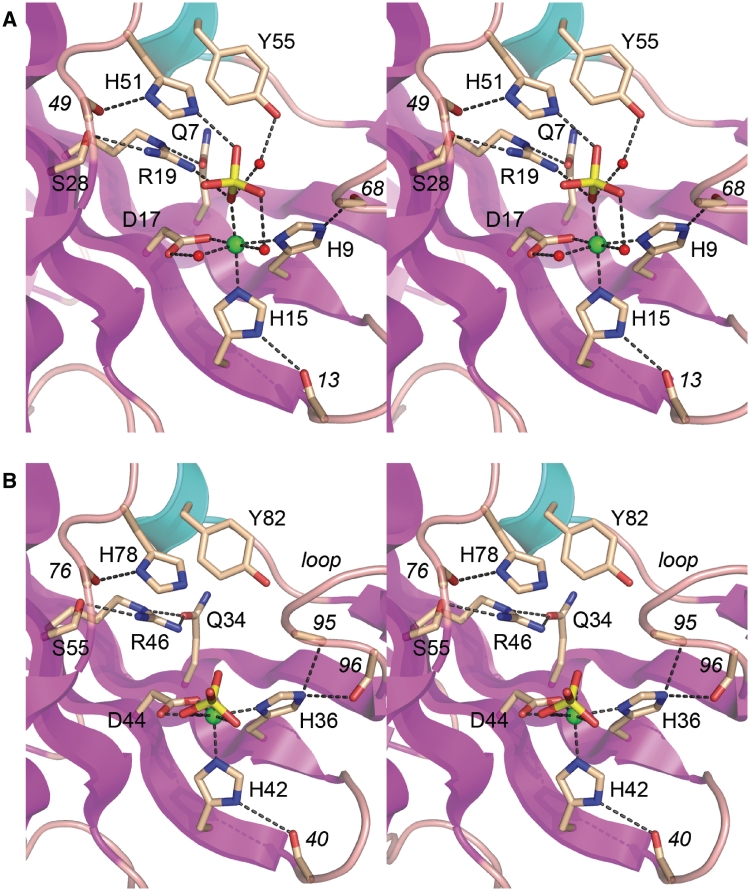
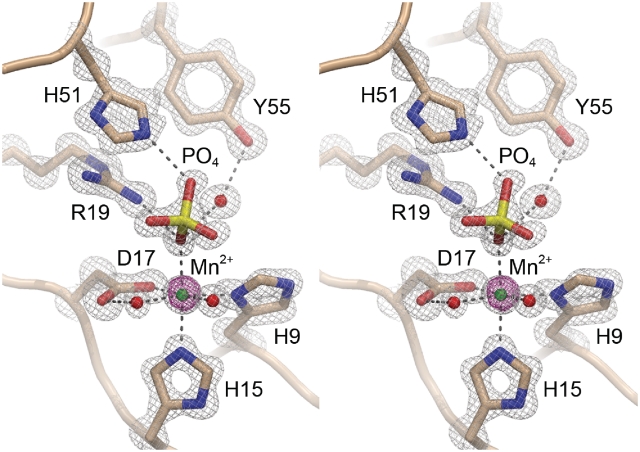
The CkoPE β barrel supports a crescent-shaped hydrophilic active site on its outer surface that binds the essential manganese cofactor and a phosphate anion that likely mimics the scissile phosphate (Figure 1B). The PaePE active site contains manganese and a sulfate anion at the equivalent positions (Figures 1A and 4A). The CkoPE active site architecture and pertinent atomic interactions are depicted in detail in Figures 5A and 6. The electron density map in Figure 6 highlights the octahedral coordination complex about the manganese ion, which is rendered as a green sphere surrounded by its 15 σ anomalous difference density. The six metal ligands are His9-Nδ, His15-Nε, Asp17-Oε, a phosphate oxygen and two waters (Figure 5A). A likely catalytic role of the metal ion is to aid in substrate binding and then stabilize the developing negative charge on the scissile phosphate in the presumptive associative transition state. The structure suggests an explanation for the requirement of CkoPE for manganese as the metal cofactor, and its inability to utilize magnesium, insofar as the reliance on His9 and His15 nitrogens as metal ligands might favor ‘soft’ metal interactions in contrast to the ‘hard’ oxygen-based contacts preferred by magnesium (Supplementary Table S1). In addition to binding manganese, His9 and His15 appear to stabilize the active site architecture, via hydrogen bond donation: from His9-Nε to the main chain carbonyl of Gly68 and from His15-Nδ to the Arg13 main chain carbonyl (Figure 5A). The interactions of His9 and His15 with the main chain also promote suitable tautomerization states on the metal-binding nitrogens of the respective imidazole rings of His9 and His15.
Figure 5.
CkoPE and MbaPE active sites. Stereo views of the active sites of CkoPE (A) and MbaPE (B). Selected amino-acid side chains and main chain atoms are rendered as stick models with beige carbons. Manganese and waters are depicted as green and red spheres, respectively. A phosphate anion (a putative mimic of the scissile phosphate) is shown as a stick model. Interatomic contacts are indicated by dashed lines.
Figure 6.
CkoPE active site electron density. Stereo view of a 1.1 Å resolution 2Fo−Fc density map of the CkoPE active site contoured at 2σ (gray mesh) for the amino acids, waters, and phosphate anion or 6σ (gray mesh) for the manganese cation. The magenta mesh is the anomalous difference density for the manganese ion contoured at 15σ.
The phosphate anion is bound in the active site via: (i) direct contacts from phosphate oxygens to the metal and one of its coordinated waters; (ii) the Arg19 and His51 side chains; and (iii) a water-bridged contact with the Tyr55 hydroxyl (Figure 5A). We predict that Arg19 and His51 promote catalysis via transition state stabilization. They also maintain the proper active site conformation via additional atomic contacts. For example, Arg19 donates hydrogen bonds to Ser28-Oγ and Gln7-Oε and thereby tethers three of the β strands that line the concave active site (Figure 5A). His51 donates a hydrogen bond from Nδ to the Glu49 main chain carbonyl while His51 Nε coordinates the phosphate (Figure 5A), implying that both imidazole nitrogens are protonated and that His51 is positively charged.
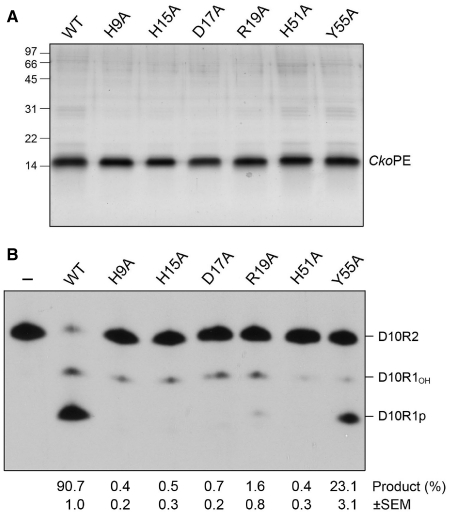
To test the proposed catalytic mechanism for CkoPE, we introduced single alanine substitutions in lieu of active site residues His9, His15, Asp17, Arg19, His51 and Tyr55. The recombinant proteins were produced in E. coli as His10Smt3 fusions and then purified from soluble extracts in parallel with wild-type CkoPE by affinity chromatography, tag removal, and recovery of the tag-free proteins (Figure 7A). Equivalent amounts of CkoPE proteins were assayed for end-healing activity on the 5′-32P-labeled D10R2 primer-template substrate. An exemplary product analysis by PAGE is shown in Figure 7B and the extents of end processing are indicated below the lanes, whereby each datum is the average of three independent experiments (±SEM). Whereas >90% of the substrate reacted with WT CkoPE, mutations H9A, H15A and D17A in the metal-binding site reduced substrate consumption to <1% (Figure 7B). The R19A and H51A changes in the phosphate-binding site exerted similarly severe effects on CkoPE activity (1.6 and <1% product formation, respectively). In contrast, Y55A retained 3′-phosphodiesterase activity, to an extent of 23% conversion of substrate to product (Figure 7A). These results are concordant with the impact of alanine mutations at the equivalent active site constituents of PaePE, especially the low residual activity of the tyrosine-to-alanine mutant (8,10).
Figure 7.
Effects of alanine mutations in the CkoPE active site. (A) Aliquots (2 µg) of the Ni-agarose preparations of wild-type CkoPE and the indicated Ala mutants were analyzed by SDS–PAGE. Polypeptides were visualized by staining with Coomassie Blue dye. The positions, and sizes (kDa) of marker polypeptides are indicated on the left. (B) Phosphoesterase reaction mixtures (10 µl) containing 50 mM Tris–HCl (pH 8.0), 5 mM DTT, 0.5 mM MnCl2, 0.5 pmol 32P-labeled D10R2 primer-template, and 13 pmol of wild-type (WT) or mutant CkoPE as specified were incubated for 10 min at 45°C. Enzyme was omitted from control reaction mixtures (−). An exemplary product analysis by PAGE and autoradiography is shown. The extents of end processing (quantified by scanning the gel with a phosphorimager) are indicated below the lanes; each datum is the average of three independent experiments (±SEM).
Crystal structure of MbaPE
We undertook to crystallize full-length MbaPE in an attempt to visualize the N-peptide segment that confers vigorous 3′-phosphomonoesterase activity. A mixture of MbaPE and manganese yielded crystals by sitting drop vapor diffusion against a precipitant solution containing MgCl2, dioxane and PEG. Diffraction data at 2.1 Å resolution were collected for a native crystal in space group P212121. The crystal contained one MbaPE protomer in the asymmetric unit. The MbaPE structure was solved by molecular replacement. Like its homologs, MbaPE consists of an eight-stranded anti-parallel β barrel and a 310 helix (Figure 1C). The modeled MbaPE polypeptide initiated at residue Asp27 and extended to the C-terminus at Ser151, punctuated by a single amino-acid gap at residue Lys140 that is located at the tip of a surface loop connecting the β7 and β8 strands of the barrel fold. Unfortunately for us, there was no interpretable electron density for the MbaPE N-peptide, implying that it is disordered. This disorder resonates with the protease-sensitivity of the equivalent N-peptide segment of PaePE (10). The MbaPE and PaePE folds aligned in DALI with an rmsd of 1.4 Å at 121 Cα positions (Z-score 19.7). MbaPE and CkoPE aligned with an rmsd of 1.0 Å at 117 Cα positions (Z-score 22.2).
The active site of MbaPE is similar to that of CkoPE. Putative MbaPE catalytic residues His36, His42, Asp44, Arg46, His78 and Tyr82 (Figure 5B) superimpose on their CkoPE counterparts (Figure 5A). The divalent cation in the MbaPE active site overlaps the metal ion in CkoPE (∼0.6 Å offset of the metal centers when the two proteins are aligned globally). Whereas MbaPE His36 and His42 coordinate the metal in the same manner as do CkoPE His9 and His15, there are differences elsewhere in the metal coordination complex in MbaPE versus CkoPE. The MbaPE metal coordination complex is not octahedral as it is in CkoPE (or PaePE) and it also lacks the two waters seen in the CkoPE and PaePE structures. In MbaPE, the metal interacts with both of the carboxylate oxygens of Asp44 (rather than one Asp oxygen and one water as in CkoPE).
Most pertinent is that the MbaPE active site includes a tetrahedral shaped density modeled as a phosphate in a site that is significantly displaced compared to CkoPE (by 2.7 Å at the phosphorus atom) and consequently closer to the metal ion. Indeed, the MbaPE metal coordinates three of the four phosphate oxygens at distances of 2.7–2.8 Å (Figure 5B and Supplementary Figure S1). [We were initially uncertain of the source of the phosphate anion modeled in the MbaPE active site, given that phosphate was not an explicit ingredient of the crystallization buffer. Assaying the buffer components via the malachite green method revealed that the precipitant solution in the well contained 50 µM Pi. Testing the individual stock solutions used to prepare the precipitant buffer showed that the 2M MgCl2 stock contained 0.4 mM Pi.] The sequelae of the displacement of the phosphate in MbaPE are that; (i) there are no contacts between the phosphate and MbaPE residues Arg46 and His78 (the counterparts of the CkoPE phosphate-binding residues Arg19 and His51); and (ii) the MbaPE surface loop 91AGYEGA96 at the lateral edge of the concave active site moves inward compared to the equivalent 65GMYGAG70 loop in CkoPE (Figure 5B). The more ‘closed’ conformation of this loop in MbaPE, which is stabilized by hydrogen bonds from His36 Nε to the Gly95 and Ala96 main-chain carbonyls (Figure 5B), would appear to occlude the putative DNA binding site overlying the metal and the scissile phosphate (13). These several deviations in metal coordination, phosphate position, and loop position suggest that the MbaPE structure represents a catalytically inactive conformation.
We considered the possibility that the manganese included in the protein sample might have been displaced from the MbaPE active site by the large excess of magnesium in the crystallization buffer. This appeared not to be the case, insofar as there was an anomalous difference peak overlying the metal ion (Supplementary Figure S1). Thus, we modeled the bound metal as manganese, not magnesium.
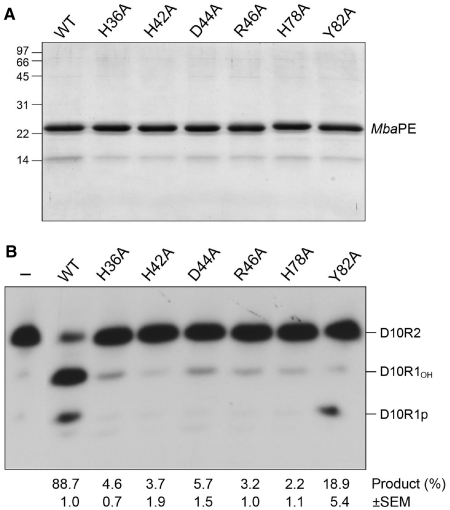
To test the active site requirements for catalysis by MbaPE, we introduced alanine changes at His36, His42, Asp44, Arg46, His78 and Tyr82 (Figure 8A) and tested the Ala-mutants for 3′-end-healing activity in parallel with wild-type MbaPE (Figure 8B). Mutations H36A, H42A, D44A, R46A and H78A suppressed MbaPE activity (to between 2 and 6% product formation compared to 89% for wild-type). In contrast, Y82A retained 3′-phosphodiesterase activity, to an extent of 18% conversion of substrate to product, which was predominantly D10R1p (Figure 8B). The corresponding tyrosine-to-alanine mutant in PaePE had a similar impact on product distribution during reaction with the D10R2 substrate, reflecting a more stringent requirement for the tyrosine in the 3′-phosphomonoesterase reaction than in the 3′-phosphodiesterase step (10). The tyrosine hydroxyl of PaePE and CkoPE coordinate a phosphate-bound water in the PE active site that is proposed to be the immediate nucleophile in the 3′-phosphomonoesterase reaction (13). Although the active site contacts are deviant in the MbaPE structure, the concordant mutational results for bacterial and archaeal PEs suggest a conserved mechanism of 3′-end processing.
Figure 8.
Effects of alanine mutations in the MbaPE active site. (A) Aliquots (2 µg) of the Ni-agarose preparations of wild-type MbaPE and the indicated Ala mutants were analyzed by SDS–PAGE. Polypeptides were visualized by staining with Coomassie Blue dye. The positions and sizes (kDa) of marker polypeptides are indicated on the left. (B) Phosphoesterase reaction mixtures (10 µl) containing 50 mM MES (pH 6.5), 5 mM DTT, 0.5 mM MnCl2, 0.5 pmol 32P-labeled D10R2 primer-template and 4 pmol of wild-type (WT) or mutant MbaPE as specified were incubated for 10 min at 45°C. Enzyme was omitted from control reaction mixtures (–). An exemplary product analysis by PAGE and autoradiography is shown. The extents of end processing are indicated below the lanes; each datum is the average of three independent experiments (±SEM).
DISCUSSION
The present study consolidates our earlier proposal (13) that the LigD PE domain defines a novel DNA end-healing enzyme superfamily distributed widely among taxa. The structures of two exemplary stand-alone archaeal PE proteins were determined and, as anticipated from primary structure conservation, they adopt the distinctive β-barrel fold and active site first seen in the structure of Pseudomonas LigD PE (13). A key finding here was that the archaeal PE proteins catalyzed the same set of manganese-dependent 3′ end-healing reactions (ribonucleoside resection and 3′-phosphate removal) as do the PE domains of Pseudomonas, Agrobacterium and Mycobacterium (8,12), implying that archaeal PEs are engaged in pathways of DNA repair. Yet, the nature of the hypothetical pathway to which archaeal PEs contribute must be different from that of the bacterial LigD-associated PE domains, insofar as none of the archaea that have a PE enzyme encode a homolog of Ku, the essential agent, along with LigD, of the bacterial NHEJ system (7). Moreover, none of the archaeal PE genes are clustered on their respective chromosomes with other DNA repair genes. The paucity of information regarding DNA repair in the archaea that have PE enzymes precludes us from speculating further on what these PEs do and with what other proteins they might collaborate in vivo.
The strong conservation of active site architecture in bacterial and archaeal PEs underscores a common catalytic mechanism for the phosphodiesterase step via transition state stabilization by the manganese and arginine and histidine side chains. It is likely that the metal ion is necessary to attain a proper conformation of the concave active site groove on the PE surface, insofar as a metal was critical to attain diffracting crystals of PaePE, CkoPE and MbaPE. Specificity for manganese over magnesium is a shared feature of archaeal and bacterial PEs (8,12) that we suggest is attributable to the soft metal contacts with histidine nitrogens at two of the six positions of the octahedral coordination complex. To support this point, we attempted to alter the metal specificity of PaePE—i.e. to render it active in magnesium—by introducing aspartate or glutamate (as hard metal ligands) in place of one or both of the metal-binding histidines. All combinations of Asp/Glu changes were tested and the results were uniform: the introduction of acidic residues eliminated manganese-dependent activity and failed to elicit any gain-of-function in magnesium (H.Z., our unpublished data).
The present characterization of MbaPE and CkoPE fortifies the case that the N-peptide segment is needed for a vigorous DNA 3′-phosphomonoesterase function to follow the 3′-phosphodiesterase step of the PE end-processing pathway. Previous studies identified three individual residues in the PaePE N-peptide (Arg14, Asp15 and Glu21) that are critical for the 3′-phosphomonoesterase reaction, but not for the 3′-phosphodiesterase (11). These amino acids are conserved in all six archaeal PEs that have an N-peptide (Figure 1D), implying a shared mechanism for enhancement of 3′-phosphatase activity by the N-peptide. Crystallization of PaePE was achieved only after deleting amino acids 1–17 (a chymotrypsin-sensitive segment); moreover the PaePE model had virtually no electron density for the peptide segment upstream of the first β strand of the core PE fold. Here, we did grow crystals of full-length MbaPE containing the N-peptide, but there was no interpretable electron density for this segment of the protein. The apparent theme is that the N-peptide is inherently disordered, even when manganese is present in the active site. We propose a conformational change that imparts structure to the N-peptide that is triggered by the DNA primer-template substrate. The effect of that structural transition might be to mobilize the three essential residues in the N-peptide to participate in binding the 3′-phosphate strand or in promoting the 3′-phosphohydrolase reaction.
We infer another conformational switch related to DNA binding that involves the interstrand loop at the outer edge of the concave active site groove to which DNA must bind. This loop is flexible in its position (e.g. varying in the CkoPE and MbaPE structures) and is partially disordered in the PaePE structure (13) (Figure 4A). We suggest that the loop reflects away from the active site to permit ingress of the DNA primer-template and then closes around the nucleic acid to help clamp it in place over the active site.
Elucidating the existence and nature of these two proposed conformational steps will obviously depend on capturing a crystal structure of PE proteins bound to DNA substrates in the presence of manganese (to ensure a proper active site conformation). This is a non-trivial matter, insofar as the inclusion of manganese with DNA substrate will trigger catalysis of 3′-end processing. Based on the available structures of PE•Mn2+•PO4(SO4) complexes, it seems unlikely that a PE active site mutant (with a defective metal coordination complex or anion-binding site) would be effective in trapping a substrate complex. Alternative strategies could include crystallizing PE with: (i) chemically modified nucleic acid substrates that resist attack by PE; (ii) alternative metals that bind in the active site, but do not support catalysis; or (iii) a transition state mimetic.
ACCESSION NUMBERS
PDB 3P43, 3P4H.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US Department of Energy (Offices of Biological and Environmental Research and Basic Energy Sciences to NSLS beamline); National Institutes of Health (grant GM63611, National Center for Research Resources). Funding for open access charge: National Institutes of Health (GM63611).
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to NSLS staff members Niel Whalen and Alex Soares for their help. S.S. is an American Cancer Society Research Professor.
REFERENCES
- 1.Shuman S, Glickman MS. Bacterial DNA repair by non-homologous end joining. Nat. Rev. Microbiol. 2007;5:852–861. doi: 10.1038/nrmicro1768. [DOI] [PubMed] [Google Scholar]
- 2.Zhu H, Shuman S. A primer-dependent polymerase function of Pseudomonas aeruginosa ATP-dependent DNA ligase (LigD) J. Biol. Chem. 2005;280:418–427. doi: 10.1074/jbc.M410110200. [DOI] [PubMed] [Google Scholar]
- 3.Zhu H, Nandakumar J, Aniukwu J, Wang LK, Glickman MS, Lima CD, Shuman S. Atomic structure and NHEJ function of the polymerase component of bacterial DNA ligase D. Proc. Natl Acad. Sci. USA. 2006;103:1711–1716. doi: 10.1073/pnas.0509083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu H, Shuman S. Gap filling activities of Pseudomonas LigD polymerase and functional interactions of LigD with the DNA end-binding Ku protein. J. Biol. Chem. 2010;285:4815–4825. doi: 10.1074/jbc.M109.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pitcher RS, Brissett NC, Picher AJ, Andrade P, Juarez R, Thompson D, Fox GC, Blanco L, Doherty AJ. Structure and function of a mycobacterial NHEJ DNA repair polymerase. J. Mol. Biol. 2007;366:391–405. doi: 10.1016/j.jmb.2006.10.046. [DOI] [PubMed] [Google Scholar]
- 6.Brissett NC, Pitcher RS, Juarez R, Picher AJ, Green AJ, Dafforn TR, Fox GC, Blanco L, Doherty AJ. Structure of a NHEJ polymerase-mediated synaptic complex. Science. 2007;318:456–459. doi: 10.1126/science.1145112. [DOI] [PubMed] [Google Scholar]
- 7.Aniukwu J, Glickman MS, Shuman S. The pathways and outcomes of mycobacterial NHEJ depend on the structure of the broken DNA ends. Genes Dev. 2008;22:512–527. doi: 10.1101/gad.1631908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu H, Shuman S. Novel 3′-ribonuclease and 3′-phosphatase activities of the bacterial non-homologous end-joining protein, DNA ligase D. J. Biol. Chem. 2005;280:25973–25981. doi: 10.1074/jbc.M504002200. [DOI] [PubMed] [Google Scholar]
- 9.Zhu H, Shuman S. Bacterial nonhomologous end joining ligases preferentially seal breaks with a 3′-OH monoribonucleotide. J. Biol. Chem. 2008;283:8331–8339. doi: 10.1074/jbc.M705476200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu H, Wang LK, Shuman S. Essential constituents of the 3′-phosphoesterase domain of bacterial DNA ligase D, a nonhomologous end-joining enzyme. J. Biol. Chem. 2005;280:33707–33715. doi: 10.1074/jbc.M506838200. [DOI] [PubMed] [Google Scholar]
- 11.Zhu H, Shuman S. Substrate specificity and structure-function analysis of the 3′-phosphoesterase component of the bacterial NHEJ protein, DNA Ligase D. J. Biol. Chem. 2006;281:13873–13881. doi: 10.1074/jbc.M600055200. [DOI] [PubMed] [Google Scholar]
- 12.Zhu H, Shuman S. Characterization of Agrobacterium tumefaciens DNA ligases C and D. Nucleic Acids Res. 2007;35:3631–3645. doi: 10.1093/nar/gkm145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nair PA, Smith P, Shuman S. Structure of bacterial LigD 3′-phosphoesterase unveils a DNA repair superfamily. Proc. Natl Acad. Sci. USA. 2010;107:12822–12827. doi: 10.1073/pnas.1005830107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 15.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 16.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 17.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:1780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.