Abstract
The domain Archaea has historically been divided into two phyla, the Crenarchaeota and Euryarchaeota. Although regarded as members of the Crenarchaeota based on small subunit rRNA phylogeny, environmental genomics and efforts for cultivation have recently revealed two novel phyla/divisions in the Archaea; the ‘Thaumarchaeota’ and ‘Korarchaeota’. Here, we show the genome sequence of Candidatus ‘Caldiarchaeum subterraneum’ that represents an uncultivated crenarchaeotic group. A composite genome was reconstructed from a metagenomic library previously prepared from a microbial mat at a geothermal water stream of a sub-surface gold mine. The genome was found to be clearly distinct from those of the known phyla/divisions, Crenarchaeota (hyperthermophiles), Euryarchaeota, Thaumarchaeota and Korarchaeota. The unique traits suggest that this crenarchaeotic group can be considered as a novel archaeal phylum/division. Moreover, C. subterraneum harbors an ubiquitin-like protein modifier system consisting of Ub, E1, E2 and small Zn RING finger family protein with structural motifs specific to eukaryotic system proteins, a system clearly distinct from the prokaryote-type system recently identified in Haloferax and Mycobacterium. The presence of such a eukaryote-type system is unprecedented in prokaryotes, and indicates that a prototype of the eukaryotic protein modifier system is present in the Archaea.
INTRODUCTION
The Archaea have long been presumed to consist of two phyla, the Crenarchaeota and Euryarchaeota. However, it has been established that diverse uncultivated lineages of Archaea inhabit every niche on this planet (1). Recent metagenomic analyses have revealed that two previously uncultivated Archaea, the group I marine crenarchaeote Candidatus (Ca.) ‘Cenarchaeum symbiosum’ and the hyperthermophilic deeply branching Ca. ‘Korarchaeum cryptofilum’, harbor both Crenarchaeota- and Euryarchaeota-specific genomic traits (2–5). Based on their unique phylogenetic positions and distinct genomic features, it has been proposed that C. symbiosum represents a novel phylum/division ‘Thaumarchaeota’ (4). The unique genomic features of K. cryptofilum also support the proposal of ‘Korarchaeota’ whose phylogenetic position had been discussed only based on SSU rRNA gene phylogenetic analysis (5). The proposal of ‘Thaumarchaeota’ has further been supported by the genome sequences of the marine archaeon Ca. ‘Nitrosopumilus maritimus’ and the moderately thermophilic archaeon Ca. ‘Nitrososphaera gargensis’ (6–9). On the other hand, the phylum ‘Nanoarchaeota’, represented by the obligate symbiont Ca. ‘Nanoarchaeum equitans’, has been proposed based on SSU rRNA gene phylogeny (10), but a later study using its genomic information suggested that the archaeal group is a fast evolving group within the Euryarchaeota (11).
Proteasome-mediated protein degradation coupled with protein modification with ubiquitin (Ub) is one of the hallmarks of eukaryotes (12). In eukaryotes, proteasome-mediated proteolysis is regulated by the Ub system, which is responsible for the conjugation of Ub to target proteins via the function of Ub-activating (E1), Ub-conjugating (E2) and Ub-protein ligating (E3) enzymes (12). Ub, E1 and E2 are members of distinct protein superfamilies that include structurally related proteins termed Ub-like (Ubl), E1-like (E1l) and E2-like (E2l) proteins, respectively. Although only distantly related to their eukaryotic counterparts, Ubl, E1l and E2l proteins are present in prokaryotes (13–15). For simplicity, based on primary structure, we will refer to these proteins as the ‘prokaryote-type’ Ubl, E1l and E2l proteins. In prokaryotes, some of the prokaryote-type Ubls and E1ls are responsible for sulfur incorporation in the biosynthesis of thiamine, molybdenum/tungstate cofactors and siderophores, while functions of other prokaryote-type proteins remain obscure (13,15). Recently, two proteasome-mediated proteolysis systems utilizing prokaryote-type proteins have been identified; the prokaryotic Ub-like protein (Pup)-proteasome system in Mycobacterium tuberculosis and the Ub-like small archaeal modifier proteins (SAMPs)-proteasome system in the halophilic archaeon Haloferax volcanii (16–18). In the Haloferax system, two prokaryote-type Ubls of the ThiS/MoaD family, which generally had been presumed to contribute in thiamine and molybdenum/tungstate cofactor biosynthesis together with prokaryote-type E1ls, have been shown to be involved in protein degradation via protein conjugation in the absence of E2/E3 homologs (16,18). These studies provided the first evidence that Ub–proteasome protein degradation occurs in Archaea and Bacteria. As these systems utilize prokaryote-type components, it is of increasing interest whether the origin of the eukaryote-type system resides in the prokaryotes.
The Hot Water Crenarchaeotic Group I (HWCGI) comprises putative thermophiles that have been detected in high-temperature environments such as terrestrial surface and subsurface hot springs, and deep sea hydrothermal environments, but have not yet been cultivated (7,19–22). The phylogroup is known to occupy a relatively deep position within crenarchaeotic lineages but distinct from hyperthermophilic Crenarchaeota or Thaumarchaeota in SSU rRNA gene phylogenetic analyses (7,21,22). From a geothermal water stream in a subsurface gold mine, we previously found unusual mat formation dominated by uncultured crenarchaeotic lineages including members of HWCGI, and constructed a metagenomic library to elucidate the physiology and genomic traits of these crenarchaeotes (21). Here, we present a composite genome sequence of a member of HWCGI, Ca. ‘Caldiarchaeum subterraneum’, from the metagenomic library, and its unique genomic features that are distinct from previously reported archaeal genomes. In particular, the genome has revealed the presence of a eukaryote-type protein modifier system, a trait that had been believed to be inherent in Eucarya. The C. subterraneum genome harbors unique features that are distinct from previously reported archaeal genomes. The genome set provides clear insight into the biology of the novel deeply branching crenarchaeotic lineage, as well as the evolution of Archaea especially in the lineages which include the HWCGI, hyperthermophilic Crenarchaeota, Thaumarchaeota and Korarchaeota.
MATERIALS AND METHODS
Sampling, sample preparation and fosmid library construction
Sampling, DNA isolation and fosmid library construction have been previously described (21). The microbial mat community, in which HWCGI dominated, was taken from a geothermal water stream located at a depth of 320 m from the ground surface from a subsurface mine in Japan. High-molecular DNA up to 50 kb was extracted from microbial mat formation, and fosmid library using pCC1FOS (EPICENTRE, Madison, WI, USA) vector was constructed. Resulting totally 5280 fosmid clones were stored as glycerol stock in 96-well microtiter dishes at −80°C.
Screening for archaeal genome fragments encoding SSU rRNA gene
Genome fragments encoding archaeal SSU rRNA genes in the metagenomic library were reexamined by dot-blot hybridization with a digoxigenin-labeled DNA probe and anti digoxigenin antibody coupled to alkaline phosphatase using a DNA labeling and detection kit (Roche, Basel, Switzerland). SSU rRNA genes amplified from the genome fragments 10-H-8 (HWCGI (C. subterraneum); AB201309) and 45-H-12 [HWCGIII (Nitrosocaldus sp.); AB201308] obtained previously (7,21) were used as DNA probes. Archaeal SSU rRNA genes in the fosmids acquired by the dot-blot hybridization were amplified by PCR using primers A21F and U1492R (23,24) and directly sequenced from both strands.
Sequencing and enrichments of archaeal genome fragments, and annotation
All fosmid clones in the metagenomic library were extracted from E. coli culture, and paired-end sequences of each cloned genomic fragment were sequenced using Big Dye ver. 3.1 sequencing kit (Applied Biosystems, Foster City, CA, USA) in accordance with the manufacturer’s recommendations by an ABI3730 DNA sequencer (Applied Biosystems). The end-sequences from cloned genomic fragments were analyzed by BLAST algorithm targeted to NCBI/EMBL/DDBJ database. On the other hand, as a part of metagenomic assessment for the whole microbial community (Takami et al., unpublished data), 151 fosmid clones; 15 clones encoding SSU rRNA gene and 136 clones were randomly selected and sequenced by the whole-genome random-sequencing method described previously using ABI 3730 and the MegaBase 1000 (GE Healthcare, Piscataway, NJ, USA) (25,26).
Fifty-two fosmid clones encoding putative archaeal genome fragments were grouped into four individual pools containing equal weight of 13 fosmids. Each fosmid pool was analyzed in a half plate of the 454 DNA Genome Sequencer 20 (GS20) (Roche) at Takara Bio Inc. (Otsu, Japan). Large contigs obtained by 454 pyrosequencing were analyzed using BLAST algorithm targeted to genomic fragments encoding archaeal SSU rRNA genes reported previously (21), complete sequences of 151 fosmid clones analyzed by Sanger method (Takami et al., unpublished data) and end-sequences of the genome fragments in the metagenomic library. Based on the homology search using BLAST, large scaffolds containing large contigs from 454 sequencing, complete fosmid clone sequences and fosmid-end sequences were manually constructed. In the second round of 454 sequencing, a total of 80 fosmids involving genome fragments extending previously sequenced regions and putative archaeal genome fragments were separated into four groups each containing 20 fosmids. The 20 fosmids in each group were analyzed in a half plate of the 454 GS20. Large contigs obtained from a total of four runs of GS20 were analyzed by BLAST targeting fosmid sequences analyzed by Sanger sequencing and fosmid end-sequences from the metagenomic library. A single large scaffold was manually constructed. Gap-regions in the scaffold were amplified by PCR with appropriate fosmids as templates, and the amplified fragments were analyzed using an ABI 3130xl DNA sequencer. Assembly in overlapping regions and gap regions was accomplished with Sequencher ver. 4.7 software (Gene Codes Corp, Ann Arbor, MI, USA). Finally, the large circular scaffold was constructed by the fosmid clone 10-H-8 (AB201309) reported previously (21), and JFF001_H02 (AP011633), JFF004_H08 (AP011650), JFF011_H10 (AP011675), JFF016_D08 (AP011689), JFF022_F09 (AP011708), JFF029_E04 (AP011723), JFF029_F10 (AP011724), JFF030_F06 (AP011727), JFF037_B02 (AP011745), JFF040_C01 (AP011751), JFF055_C09 (AP011796) analyzed by Sanger method (Takami et al., unpublished data), and JFF001_G10 (AP011862), JFF002_G05 (AP011850), JFF004_B03 (AP011868), JFF005_B08 (AP011872), JFF008_E07 (AP011864), JFF009_A08 (AP011867), JFF009_F01 (AP011875), JFF009_F10 (AP011844), JFF011_A11 (AP011858, AP011859), JFF012_C01 (AP011870), JFF013_A09 (AP011845), JFF015_C06 (AP011842), JFF015_C07 (AP011830), JFF015_E11 (AP011831), JFF017_C01 (AP011851), JFF021_E09 (AP011873), JFF021_G03 (AP011856), JFF022_C07 (AP011838), JFF025_E12 (AP011827), JFF027_H06 (AP011834), JFF028_A01 (AP011854), JFF028_A10 (AP011876), JFF028_E01 (AP011852), JFF029_A12 (AP011865), JFF029_F08 (AP011836), JFF030_C12 (AP011869), JFF030_H11 (AP011855), JFF031_B05 (AP011861), JFF032_D08 (AP011843), JFF033_A05 (AP011857), JFF033_F07 (AP011840), JFF033_G03 (AP011849), JFF034_A01 (AP011853), JFF035_A09 (AP011828), JFF035_E02 (AP011848), JFF036_A12 (AP011839), JFF036_E03 (AP011833), JFF036_H04 (AP011837), JFF039_F10 (AP011846), JFF040_F12 (AP011871), JFF042_C08 (AP011829), JFF049_D05 (AP011863), JFF050_B05 (AP011866), JFF051_A09 (AP011832), JFF051_C10 (AP011826), JFF052_D03 (AP011874), JFF052_E01 (AP011841), JFF052_H05 (AP011847), JFF053_A03 (AP011860) and JFF055_E04 (AP011835) analyzed by the GS20 in this study. Numbers in parentheses following each fosmid clone are accession numbers in DDBJ/EMBL/GenBank database.
The predicted ORFs were initially defined by Glimmer program (http://www.cbcb.umd.edu/software/glimmer/), and putative functions for predicted ORFs were identified by comparing against all non-redundant (NR) sequences deposited in the NCBI database using BLASTP (27). Truncated ORFs and frame shifts found in the initial BLASTP search were confirmed by re-sequencing by the Sanger method. Clusters of Orthologous Groups (COGs) (28), archaeal Clusters of Orthologous Groups (arCOGs) (29) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) (30) databases were used for further functional information. For the comparison of genome core genes, publically available archaeal genome sequences in the arCOG database were used, and arCOGs in K. cryptofilum were referred to from Elkins et al. (5). Assignments of arCOGs for C. subterraneum and N. maritimus were performed under the following condition; the BLAST E-value threshold was set at 10−3, and the homologous region covers >70% of the hit sequences in arCOGs. Proteins that were putatively separated or fused compared to those in the databaes were manually concatenated or divided, and reexamined. Forty-six tRNA genes were identified by using tRNAscan-SE (31) with Archaea-specific search mode and SPLITSX (32) with the following parameters: –p 0.55 –f 0 –h 3. Clusters of regularly interspaced repeats (CRISPR) were identified using the CRISPR Finder (33).
Phylogenetic analyses
The small and large subunit rRNA gene alignments were constructed by ARB software (34). Then, concatenated alignments were constructed using only unambiguously aligned region for phylogenetic analysis. The maximum likelihood tree was computed by using the program package PhyML with HKY85 (35). The support values for the internal nodes were estimated from 100 bootstrap replicates. Protein sequences; RNAP subunits, ribosomal proteins, D-type DNA polymerase (DNAP) small and large subunits and elongation factor II (EFII) were aligned by using CLUSTAL W 1.8 program (36), and ambiguous regions were automatically trimmed according to Gblocks (37,38). Two concatenated alignments were constructed for the phylogenetic analyses of ribosomal proteins (L10, L10e, L11, L13, L14, L15, L15e, L18e, L19e, L2, L22, L3, L30, L44e, L4e, L5, L6, L7Ae, S10, S11, S13, S19, S19e, S2, S27e, S3, S3Ae, S4, S4e, S5, S6e, S7, S8, S8e, S9, S17, S17e, L1, L18, L24, L31e, L32e, S12, S15, L23) and RNAP subunits (RpoA′, RpoA′′, RpoB′, RpoB′′, RpoD, RpoE′, RpoH and RpoK), and concatenated (SSU+LSU) DNAP. Maximum likelihood trees were constructed using the program package RAxML with WAG+I+G (39). The support values for the internal nodes were estimated from 200 bootstrap replicates. Almost full length of ef2 sequence from the Nitrosocaldus sp. (HWCGIII) was obtained by PCR amplification from the DNA assemblage. A primer set (5′-AATNGCNCAYGTNGAYCAYGGMAARAC-3′, and 5′-GTCTCWGMTGCAGGTATCTC-3′) for the amplification of ef2 was constructed based on DNA alignments of ef2 from crenarchaeal lineages including partial ef2 sequence from the Nitrosocaldus sp. (HWCGIII) (31-F-01; GI 106364417) that were obtained from the metagenomic fosmid library used in this study.
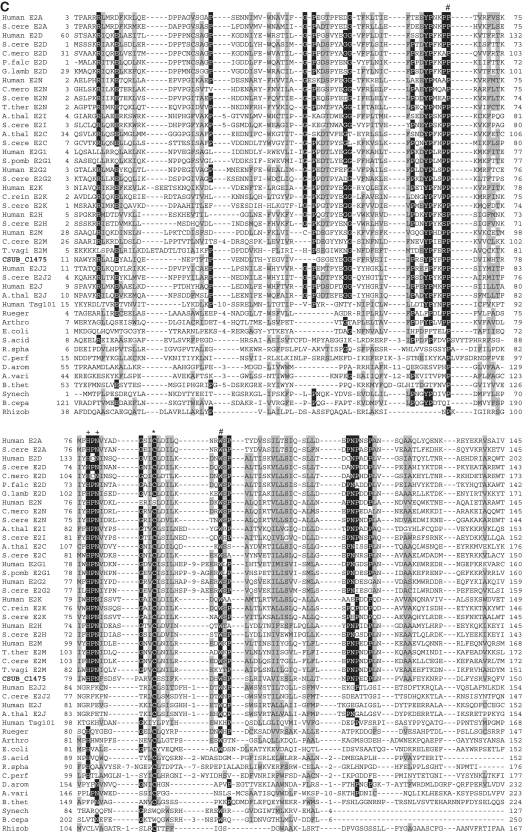
Alignments of Ub-like protein family, E1-like protein family, E2-like protein family and JAMM protease family shown in Figure 2 were constructed by ClustalX (40) and edited manually based on the previously reported secondary structures of each protein family (13–15,41–44).
Figure 2.
Sequence alignments of Ub, E1, E2 (super-) and JAMM family proteins. (A) Sequence alignments of eukaryotic and archaeal Ub superfamily proteins; proteins from Saccharomyces cerevisiae; S.cere Smt3 (6320718) and S.cere Rpl40 (6322043), from human; Human sumo2 (54792071), Human sumo1 (54792065), Human NEDD8 (5453760) and Human Ufm1 (7705300), from Cyanidioschyzon merolae; C.mero smt3 (CME004C), C.mero ubl (CML042C) and C.mero Rps27 (CMN125C), from Tetrahymena thermophila; T.ther ubl1 (229594936) and T.ther ubl2 (118367859), from Cryptosporidium parvum; C. parv ubl1 (126654302), C.parv Rps27 (66357428) and C.parv ubl2 (66363058), from Giardia lamblia; G.lamb sumo (159114790), G.lamb Epl40 (159108136), G.lamb ub1 (159112981), G.lamb ub2 (159111413), from Trypanosoma brucei; T.bruc ub (72387960) and T.bruc ubl (72387818), from C. subterraneum; eukaryote-type Ubl (CSUB_C1474) and prokaryote-type Ubls (ThiS/MoaD) (CSUB_C0525, CSUB_C0702, CSUB_C1012, CSUB_C1603), from H. volcanii; SAMPs, HVO_0202 (302595884) and HVO_2619 (302595883), from Bacillus subtilis; B.sub ThiS (CAB13025), from Streptomyces avermitilis; S.aver ThiS (BAC73805), from Nitrosomonas europaea; N.euro ThiS (CAD84196), from Escherichia coli; E.coli MoaB (AAN79339), from Pyrococcus furiosus; P.furi MoaB (1VJK_A), from Methanosarcina acetivorans; M.acet MoaB (AAM05120), from Aromatoleum aromaticum; A.arom NrfH (CAI07579) and from Pseudomonas syringae; P.syri NrfH (AAY39230). Asterisks indicate the C-terminal Gly-Gly motif. (B) Sequence alignments of adenylation and catalytic cysteine domains in E1 superfamily proteins; proteins from human; Human E1L (23510338), Human sumoE1 (60594167), Human UBA1 (23510338), Human UBA2 (4885649), Human UBA3 (38045942), Human UBA5 (13376212), Human ATG7 (119584500) and Human MOCS3 (7657339), from Schizosaccharomyces pombe; S.pomb E1L (162312305) and S.pomb UBA3 (19113852), from S. cerevisiae; S.cere Aos1 (6325438), S.cere UBA1 (6322639), S.cere UBA2 (6320598), S.cere ATG7 (6321965), S.cere UBA4 (6321903) and S.cere YgdLl (6322825), from T. thermophila; T.ther E1L (118383519), T.ther E1B (118351055), T.ther UBA4 (118351953) and T.ther YgdLl (118400480), from Trypanosoma cruzi; T.cruz E1 (71411317), from Plasmodium yoelii; P. yoel UBA2 (82595829) and P.uoel MoeB (83315401), from Trichomonas vaginalis; T.vagi APG7 (123446747), from C. subterraneum; E1l (CSUB_C1476) and MoeB (CSUB_C1135), from H. volacanii; HVO_0558 (292654724), Cupriavidus metallidurans; C.meta ThiF (4039868), from Clostridium perfringens; C.perf (86559649), from Shewanella sp. ANA3; S.ANA3 (117676291), from Rhizobium etli; R.etli (86359719), from Anabaena variabilis; A.vari (ABA25158), from Polaromonas naphthalenivorans; P.naph (121605347), from Nostoc sp. PCC7120; Nostoc (BAB77147), from Xanthomonas axonopodis; X.axon MoeB (21242767), from E. coli; E.coli MoeB (1JW9_B) from C. symbiosum; C.symb ThiF (ABK78649), from P. furiosus; P.furi MoeB (18977661), from Geobacillus kaustophilus; G.kaus MoeBl (56419161), Desulfuromonas acetoxidans; D.acet ThiF (95930339), from Desulfovibrio desulfuricans; D.desu ThiF (78357502), from Bacteroides thetaiotaomicron; B.thet (29349047), from M. tuberculosis; M.tube Rv (15609475), from Cytophaga hutchinsonii; C.hutc (110639176), and from Bacillus thuringiensis; B.thur (110639176). Asterisks and plus indicate adenylation active sites and thiolating cysteine, respectively. Mg2+ chelating motifs (CxxC) are shown by octothorpes. (C) Alignment of E2 superfamily proteins; proteins from human; Human E2A (32967280), Human E2D (5454146), Human E2N (61175265), Human E2G1 (13489085), Human E2G2 (29893557), Human E2K (163660385), Human E2H (4507783), Human E2M (4507791), Human E2J2 (37577124), Human E2J (37577122) and Human Tsg101 (5454140), from Arabidopsis thaliana; A.thal E2I (15230881), A.thal E2C (18403097) and A.thal E2J (18401338), from Chlamydomonas reinhardtii; C.rein E2K (159463008), from C. merolae; C.mero E2D (CMB015C) and C.mero E2N (CMR010C), from Plasmodium falciparum; P.fal E2D (124805463), from S. cerevisiae; S.cere E2A (6321380), S.cere E2D (6319556), S.cere E2N (6320297), S.cere E2I (6320139), S.cere E2C (6324915), S.cere E2G2 (6323664), S.cere E2K (6320382), S.cere E2H (6579192), S.cere E2M (6323337) and S.cere E2J2 (6320947), from S. pombe; S.pomb E2G1 (6323664), from T. thermophila; T.ther E2M (118382495), from T. vaginalis; T.vagi E2M (123484378), from G. lamblia; G. lamb E2D (159111264), from C. subterraneum; CSUB_C1475, from Ruegeria sp; Rueger (22726448), from Arthrobacter sp.; Arthro (A0AW81), from E. coli; E.coli (37927532), from Syntrophus aciditrophicus; S.acid (85859492), from Rhodobacter sphaeroides; R.spha (77387013), from Clostridium perfringens; C.perf (86559649), from Dechloromonas aromatica; D.arom (71847775), from Anabaena variabilis; A.vari (75705484), from Bacteroides thetaiotaomicron; B.thet (29339960), from Synechocystis sp. PCC6803; Synech (38423903), from Burkholderia cepacia; B.cepa (A4JA91), and from Rhizobium sp. NGR234; Rhizob (2496664). Astetisk and octothorpes indicate catalytic cysteine residue and residues forming a conserved stabilizing contact in E2 from eukaryotes, respectively. Flap histidine and asparagine residues are shown by plus. Identical and similar amino acids are shaded in black and gray, respectively. (D) Sequence alignment of JAMM family proteins; proteins from human; Human COPS5 (12654695) and Human PSMD14 (5031981), from A. thaliana; A.thal CSN5A (15219970), from S. cerevisiae; S. cere RPN11 (14318526), from T. brucei; T.bruc RPN11 (18463065) and T.bruc SCN5 (72393165), from G. lamblia; G.lamb RPN11 (159114272), from S. pombe; S.pomb AMSHP (19115685), from C. subterraneum; CSUB_C1473, from Archaeoglobus flugidus; A.flugi JAB (11499780), from Pyrococcus horikoshii; P.hori JAB (3257912), from Pseudomonas aeruginosa; P.aeru JAB (15597298), from Pyrobaculum aerophilum; Py.aer JAB (18313041), from E. coli; E.coli RadC (15801143), from B. subtilis; B.subt RadC (16079856), from M. acetivorans; M.acet RadC (20090827), from Thermotoga maritima; T.mari RadC (15644305), from Aquifex aeolicus; A.aeol (2984019); from Deinococcus radiodurans; D.radi (15805429), from Pseudomonas putida; P.puti (84994017), from Salinibacter rubber; S.rubb (83814538), from M. tuberculosis; M.tube (13880984), from Nocardia farcinica; N.farc (54014564), from Wolinella succinogenes; W.succ, and from Geobacter metallireducens; G.meta. Asterisks indicate the JAMM motif residues. Identical and similar amino acids are shaded in black and gray, respectively.
RESULTS
Archaeal diversity within the metagenomic library
As a result of dot blot hybridization and previous PCR screening, a total of 21 and three fosmids-encoding SSU rRNA genes of HWCGI and HWCGIII (Ca. ‘Nitrosocaldus’ sp.; SSU rRNA gene similarity between ammonia oxidizing thaumarchaeon Ca. ‘Nitrosocaldus yellowstonii’ (21) and the HWCGIII sequences in the metagenomic library [AB201308] was 95%) lineages, respectively, were obtained from the metagenomic library. Among the 21 fosmids-harboring HWCGI SSU rRNA genes, 19 SSU rRNA gene sequences belonged to ribotype I represented by the SSU rRNA gene included in the fosmid clone 10-H-08, while the other two sequences constituted another single ribotype. Here, we named the predominant HWCGI archaeon represented by the 10-H-08 SSU rRNA gene ribotype as Ca. ‘C. subterraneum’ (Caldiarchaeum type I) (‘calidus’ and ‘subterraneum’ meaning hot and underground, respectively) and the other minor HWCGI population as ‘Caldiarchaeum type II’. Similarity between the two ribotypes of Caldiarchaeum SSU rRNA gene sequences was 96.6%. Sixteen of the C. subterraneum SSU rRNA genes, each harbored two introns. Three orthologous sequences with 99% similarity were observed among the 16 sequences of the first intron, while five sequences with 95–99% similarity were found for the second intron. No diversity was present among all exon SSU rRNA gene sequences in the C. subterraneum SSU rRNA.
Reconstruction of a composite genome
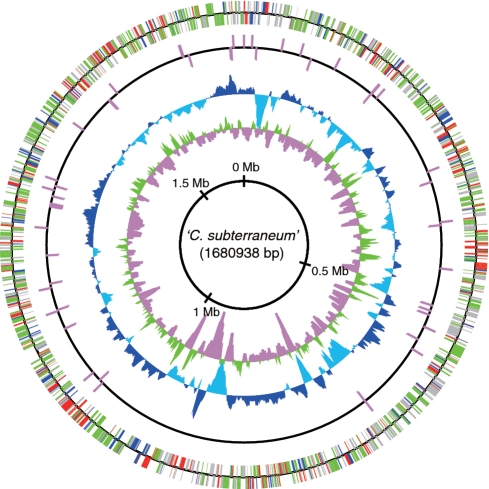
In order to investigate the genomic properties of the metagenomic library, paired- or one-end sequences of the genome fragments were obtained from 3375 fosmid clones, and 151 fosmids (136 randomly selected fosmids and 15 fosmids encoding SSU rRNA gene) were analyzed by Sanger method (Takami et al., unpublished data). Among a total 5965 end-sequences from these cloned fragments, 883 end-sequences (∼13.5 % of total end-sequences) displayed highest similarity with sequences derived from Archaea. Among these ‘archaeal’ sequences, fosmids were selected for 454 sequencing based on the following two criteria: (i) the presence of paired-ends sequences predicted to encode open reading frames (ORFs) most similar to archaeal sequences; or (ii) the presence of ORFs in either end encoding homologues of archaeal translation, transcription or replication genes. Large contigs obtained by initial 454 sequencing of the 52 fosmids were manually assembled with the sequences from the 151 fosmids described above, two genome fragments-encoding archaeal SSU rRNA genes obtained previously (21) and the end-sequences of all fosmids, followed by a BLAST search. In this step, a scaffold of >1 Mb including the C. subterraneum SSU rRNA gene was assembled, but we did not find a large scaffold with other archaeal SSU rRNA genes. For the second round of 454 sequencing, 80 fosmids that met the following criteria were further analyzed: (i) linkage with the scaffold including the C. subterraneum SSU rRNA gene sequence; (ii) presence of paired-ends predicted to encode ORFs most similar to archaeal sequences; and (iii) presence of ORFs in either end showing high similarity with archaeal sequences. After the second 454 sequencing, large contigs obtained from 454 sequencing, fosmids analyzed by Sanger method and end-sequences were manually assembled and subjected to BLAST search. As a result, a circular scaffold including complete sequences of 12 fosmid clones analyzed by Sanger sequencing was obtained. The similarities of overlapping regions were generally >99%. Afterwards, gap-regions were obtained by PCR with appropriate fosmid clones as templates, and the amplified fragments were sequenced by Sanger method. Finally, a composite circular genome sequence of C. subterraneum (1 680 938 bp) was assembled from a set of 62 complete or partial fosmid sequences (Figure 1). We also obtained 28 complete or partial fosmid sequences derived from C. subterraneum, and 10 of them completely overlapped with the composite circular genome. However, 18 sequences harbored distinct insertion (a total of 68 kb)/deletion regions compared to the composite circular genome, or consisted of two genomic regions distantly located on the composite circular genome. The similarities of these regions with the circular genome were >99%. The genomic heterogeneity is likely the result of recombination or rearrangement within a species because we could not obtain any evidence of inter-species genomic recombination in the distinct insertion regions.
Figure 1.
Circular representation of the C. subterraneum composite genome. From the inside, the first and second circles show the GC skew (values >0 or <0 are indicated in green and pink, respectively) and the G+C percent content (values greater or smaller than the average percentage in the overall chromosome are shown in blue and sky blue, respectively) in a 10-kb window with 100-bp step, respectively. The third and fourth circles show the presence of RNAs (rRNA and tRNA); CDSs aligned in the clock-wise and counterclock-wise directions are indicated in the upper and lower sides of the circle, respectively. Colors of CDSs indicate their functional categories; red for information storage and processing, green for metabolism, blue for cellular processes and signaling, and gray for poorly characterized function.
General features
The G + C content of the genome from C. subterraneum is 51.6%. A single rRNA gene set is identified but rRNA genes do not form an operon structure in the composite genome. Forty-five tRNAs were identified. A total of 1730 predicted ORFs were detected. Among these, 1054 of the predicted protein-encoding sequences (CDSs) could be assigned a function, 352 of the CDSs could be identified as hypothetical conserved proteins and the remaining 324 CDSs did not show significant similarity to any of the amino acid sequences in the protein databases (Supplementary Table S1).
Mobile genes
The genome contains three genes encoding transposases of the IS6 family and one of the IS4 family. Both of these transposase families, originally found in Bacteria, are distributed only in the Euryarchaeota and not in the Crenarchaeota within the archaeal domain (45). Four clustered regularly interspaced short palindromic repeats (CRISPR) and one CRISPR-related gene cluster, presumed to provide resistance against virus infection, are present (46). The genome encodes one prophage-like gene cluster.
DNA replication, repair, cell cycle
Caldiarchaeum subterraneum carries three orc1/cdc6 orthologues and a single minichromosome maintenance protein. The genome encodes multiple DNA-dependent DNA polymerases including two family B type enzymes; the BII type found only in crenarchaeal lineages (47) and the inactivated type (48), and both the small and large subunits of a D-type enzyme (Table 1). Genes for the large and small subunits of replication factor C form a gene cluster. Single genes each encoding the small subunit of primase, sliding clamp (PCNA), ATP dependent ligase, RNase HII, flap endonuclease (FEN1) and ERCC4-like helicase, are present. Genes for one truncated and two complete large subunits of primase are found. Unlike the Hef protein found in P. furiosus that consists of ERCC4-like helicase (COG1111) and XPF protein domains (ERCC4-type nuclease), which is the case in most of the euryarchaeotes, both domains are located separately on the genome of C. subterraneum as observed in Thaumarchaeota and a minority of euryarchaeotes (8,49,50). The ERCC4-like helicase domain (COG1111) is absent from the genome of Korarchaeota (8). Both topoisomerase IA and IB were found in C. subterraneum as in the case of Thaumarchaeota (8) (Table 1). One reverse gyrase gene, which had been considered a genomic signature for hyperthermophiles, but now also detected in thermophiles, is observed (51–53). Genes for chromatin-associated proteins, two Alba and one histone, are present. The archaeon possesses genes for euryarchaeal chromosome segregation proteins including SMC family ATPase, chromosome segregation and condensation protein B and kleisin family Rec8/ScpA/Scc1-like protein (chromosome segregation and condensation protein A) in a single, operon-like structure. The genome harbors one gene for the cell division protein FtsZ. Among the newly identified crenarchaeal cell division proteins CdvA, CdvB and CdvC that have been identified in Thaumarchaeota and hyperthermophilic Crenarchaeota (with the exception of the Thermoproteales), CdvB and CdvC are present but a gene for CdvA is absent in C. subterraneum (8,54).
Table 1.
Distribution patterns of representative components for DNA replication/repair, cell division, translation and transcription among Crenarchaeota, Euryarchaeota, Thaumarchaeota, Korarchaeota and C. subterraneum
| C. subterraneum | Crenarchaeota | Euryarchaeota | Thaumarchaeota | Korarchaeota | |
|---|---|---|---|---|---|
| Major DNA polymerasesa | BII, D | BI, BII | BI, D | BII, D | BI, BII, D |
| Chromosome segregation ATPase | + | − | + | + | + |
| ERCC4 like helicase (COG01111) | + | − | + | − | |
| Topoisomerase I | IA, IB | IA | IA | IAb, IB | IA |
| FtsZ | + | − | + | + | + |
| Hisotne | + | −c | + | + | + |
| RNA polymerase RpoA | fusion | split | split | fusion | fusion |
| RNA polymerase RpoB | fusion | split | split/fusiond | fusion | fusion |
| RNA polymerase RPB8 | − | + | − | − | + |
| Ribosomal protein S25, S26, S30 | + | + | − | + | + |
| Ribosomal protein L14e, 34e | + | + | + (some) | − | + |
| Ribosomal protein L13e | − | + | − | (+)e | + |
| Ribosomal protein LXa | − | + | + (most) | − | − |
| Ribosomal protein L39e | − | + | + | + | − |
+, present; −, absent.
aCharacterization of DNA polymerase is based on Ref. (47).
bOnly C-terminal domain is found in C. symbiosum and N. maritimus.
cOnly found in Thermofilum pendens and Caldivirga maquilingensis.
dFusion form is observed in Thermococcales and Thermoplasmatales.
eOnly found in N. gargensis.
The genome contains genes for double-strand-break repair, direct repair, base excision repair and nucleotide excision repair including photolyase and family Y DNA polymerase, which have previously been found only in Sulfolobales among the hyperthermophilic crenarchaeotes (55,56). However, XPB helicase for excision repair, mismatch detection proteins MutS and MutL, mismatch glycosylase MIG and bacterial nucleotide excision repair protein UvrABC are absent.
Translation and transcription
Forty-six tRNAs corresponding to all 61 sense codons and one initiator codon can be identified. Thirteen tRNAs are predicted to be intron-containing tRNAs and three out of the 13 harbor multiple introns (tRNALeu UAA, tRNAGln CUG, tRNAThr GGU). The introns are located not only at anticodon loop regions (canonical position) but also various non-canonical positions (D-arm, V-arm and T-arm), as observed in other crenarchaeal species (57,58). The BHB structure, a well-known motif of archaeal tRNA splicing, is found at exon–intron junctions of tRNA and the corresponding heterotetrameric splicing endonuclease can be identified. Aminoacyl tRNA synthetases for all of the amino acids are encoded in the genome except for the enzyme for glutaminyl tRNA synthesis, however, glutaminyl tRNA formation is likely dependent on heterodimeric glutamyl-tRNA amidotransferase (GatD and GatE). A selenocysteine incorporation system is lacking, resembling other genomes from crenarchaeal lineages (59).
The archaeal DNA-dependent RNA polymerase in C. subterraneum lacks the orthologue of the eukaryotic subunit RPB8 found in the hyperthermophilic crenarchaeotes and Korarchaeota (5,60), and possesses all other subunits found in the Archaea. RpoA is not fragmented as in eukaryotes, and is similar to those of Thaumarchaeota and Korarchaeota (Table 1). An ortholog of the eukaryotic RNA polymerase III subunit RPC34 is also found in C. subterraneum as in the hyperthermophilic Crenarchaeota, Thaumarchaeota and some of the Euryarchaeota but not the Korarchaeota (61). Archaeal homologs related to transcriptional initiation such as transcription factor B (TFB), TATA-binding protein (TBP) and transcription factor E (TFE) are present.
A complete set of 28 archaeal SSU ribosomal proteins are present, including S25e, S26e and S30e, that are absent in the Euryarchaeota (4,8,62) (Table 1). A total of 34 LSU ribosomal proteins are present. Although L39e is conserved in the Euryarchaeota and hyperthermophilic Crenarchaeota, L39e, along with L13e, L35ae, L38e, L41e and LXa (L20a/L18s), was not present on the genome. The absence of L13e is a euryarchaeal feature, and that of L35ae and LXa (L20a/L18a) is common to the Thaumarchaeota and Korarchaeota (4,5,8,62). The lack of L39e has also been noted in the Korarchaeota (4). We observed that L14e and L34e, which are not conserved in the Thaumarchaeota, are present on the C. subterraneum genome (Table 1).
Energy metabolism
The predicted gene set suggests the potential of chemolithotrophic growth in C. subterraneum using hydrogen or carbon monoxide as an electron donor, and oxygen, nitrate or nitrite as an electron acceptor. One Ni–Fe NADP-reducing hydrogenase and one potential aerobic type carbon monoxide dehydrogenase were detected. However, the hydrogenase is phylogenetically similar to those of heterotrophic organisms and potential aerobic type carbon monoxide dehydrogenase lacks biochemical evidence (21). In the respiratory chain, one set of complex II (succinate dehydrogenase), an incomplete complex I (NADH dehydrogenase), cytochrome b, rieske protein, heme-copper terminal oxidase, membrane-bound nitrate reductase and periplasmic nitrite reductase are each present. Genes for cytochrome b, rieske protein and potential cytochrome c are distributed separately on the genome. The subunit II of heme-copper terminal oxidase harbors copper-binding motif residues that are signatures of cytochrome c oxidase but not quinone oxidase (63).
Central metabolism
An almost complete Emden-Meyerhof pathway and complete tricarboxylic acid (TCA) cycle are present, but phosphofructokinase that is necessary for glycolysis is missing. ATP citrate lyase and its alternatives such as citryl-CoA synthetase and citryl-CoA lyase (64) are also lacking in the genome. Therefore, the reductive TCA cycle most likely does not function. Genes encoding enzymes for the Calvin–Benson cycle and reductive acetyl-CoA pathway are also not observed. Recently, two carbon assimilation pathways; the 3-hydroxypropionate/4-hydroxybutyrate cycle and the dicarboxylate/4-hydroxybutyrate cycle have been recognized in crenarchaeal lineages. The two cycles utilize distinct carbon dioxide/bicarbonate-fixing pathways to convert acetyl-CoA to succinyl-CoA, but share a common route in converting the succinyl-CoA to two acetyl-CoA molecules (65–69). Enzymes responsible for the conversion of acetyl-CoA and bicarbonate into succinyl-CoA in the 3-hydroxypropionate/4-hydroxybutyrate cycle, methylmalonyl-CoA epimerase, methylmalonyl-CoA mutase and biotin carboxylase and L-chain subunit of acetyl-CoA carboxylase (65,69), are not found in C. subterraneum . In contrast, all enzymes converting acetyl-CoA into succinyl-CoA by fixing carbon dioxide and bicarbonate in the dicarboxylate/4-hydroxybutyrate cycle are present. Intriguingly however, although all other enzymes necessary for the regeneration of acetyl-CoA from succinyl-CoA are present, the gene for 4-hydroxybutyryl-CoA dehydratase cannot be found on the genome.
The organism does not have the non-oxidative pentose phosphate pathway that is required for standard pentose/nucleic acid biosynthesis. However, three alternative pathways that replace the non-oxidative pentose phosphate pathway can be identified; the ribulose monophosphate (RuMP) pathway that converts fructose 6-phosphate to ribulose 5-phosphate (70,71), the archaeal 2-deoxyribose 5-phophate aldolase (DERA) pathway that can produce deoxyribose 1-phosphate from glyceraldehyde 3-phosphate and acetaldehyde (72), and the 6-deoxy-5-ketofructose-1-phosphate (DEFP) pathway that supplies 3-dehydroquinate (73,74).
Protein folding and heat shock proteins
The genome possesses gene sets of heat shock proteins such as sHsp, Hsp60, Hsp70 and HtpX. Homologues of Hsp70 related proteins such as DnaJ, DnaK and GrpE have only been found in mesophilic euryarchaeotes and the Thaumarchaeota among the Archaea (75). Genes for NAC protein, prefoldin, FKBP-type peptidyl-prolyl cis-trans isomerase and thioredoxin are present, but those for Lon and Clp protease are absent.
Ub-like protein modifier system
Among the various unique traits of C. subterraneum, an unparalleled finding is the presence of a potential protein-degradation pathway consisting of a eukaryote-type Ub conjugation system associated with proteasome and AAA+ family ATPase. As mentioned above, the structural features of the components of this system clearly distinguishes this system from the prokaryote-type systems recently identified in the Archaea and Bacteria. In the H. volcanii SAMPs-proteasome system, two of five prokaryote-type Ubls (ThiS/MoaD) identified in this haloarchaeon have been shown to conjugate with proteins and function as SAMPs, and conjugation between the SAMPs and a prokaryote-type E1l (MoeB) has been observed (18). C. subterraneum possesses four prokaryote-type Ubl (ThiS/MoaD) genes (CSUB_C0702, CSUB_C1012, CSUB_C0525 and CSUB_C1603) along with a molybdenum cofactor/tungstate cofactor biosynthesis pathway including a single prokaryote-type E1l (MoeB) gene (CSUB_C1135). These genes may be involved in a prokaryote-type protein modifier system similar to that found in H. volcanii (Figure 2A). Interestingly however, two of the prokaryote-type Ubls (CSUB_C0702 and CSUB_C1603) in C. subterraneum have 89 and 12 additional residues following the C-terminal Gly-Gly motif, in contrast to most archaeal prokaryote-type Ubl (MoaD) sequences which terminate after the Gly-Gly sequence (13) (Figure 2A).
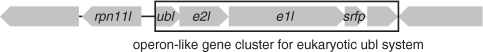
In addition to these homologues, the C. subterraneum genome harbors an operon-like gene cluster encoding homologues of eukaryote-type Ubl, E1l and E2l (CSUB_C1474, CSUB_C1476 and CSUB_C1475, respectively), suggesting the presence of an unprecedented eukaryote-type Ubl system (Figures 2 and 3). Furthermore, while an apparent homologue of E3 is absent in the genome, a gene for a small Zn finger protein (CSUB_C1477) containing a RING finger motif (C-X2-C-X11-C-X2-C-X4-H-X2-C-X10-C-X2-C) that mediates the Ub ligase activity of RING-type E3s (76) is also found in the same operon-like gene cluster (Figure 3). Moreover, a gene for RPN11-like protein (RPN11l) (CSUB_C1473), which is the homologue of eukaryotic 26 S proteasome regulatory subunit constituting a part of the proteasome lid sub-complex that catalyzes de-ubiquitination of captured substrates (77,78), is juxtaposed to the operon-like structure in the reverse strand (Figures 2D and 3). The Ubl, E1l and E2l harbor the key residues necessary for their respective functions, and are much more similar to their eukaryotic counterparts than to the prokaryote-type proteins (Figure 2). Ubl found in C. subterraneum shares >30–35% identity with the eukaryotic Ub-ribosomal fusion proteins and Ub B, and harbors the Gly–Gly motif found at the C-terminal region of eukaryotic Ub/Ubl (Figure 2A). As nine residues follow the Gly-Gly motif in the C. subterraneum Ubl, this suggests that this organism possesses a post-translational modification system, generally presumed to be a trait of the eukaryotic Ub/Ubl system (79). The C. subterraneum E1l retains the second-catalytic-cysteine domain involved in Ub-E1 interaction and the adenylation domains found in eukaryote-type E1s (UBA2, UBA3) (80,81) (Figure 2B). The significant eukaryote-type feature in the C. subterraneum E1l is the presence of two insertion helices (Asp197–Ser208 and Ile224–Leu239) between the Ub-E1 interaction domain and second Mg2+-chelating domain, which are found only in eukaryote-type E1s such as UBA1, UBA2, UBA3 and Aos1 (15) (Figure 2B). The JAMM (JAB1/MPN/Mov34 metalloenzyme) motif is a highly conserved motif found in various metal proteases from all three domains of life (82). The motif is known to be essential for the de-ubiquitination of captured substrate by RPN11 to facilitate their degradation, and is conserved in the RPN11l found in C. subterraneum (83). The C. subterraneum protein also possesses a C-terminal extension that forms sheet structures, which is a specific characteristic of the eukaryotic RPN11 proteins associated with the proteasome, and not found in archaeal and bacterial JAMM proteins (84) (Figure 2D). However the C. subterraneum protein seemingly lacks the central region of the eukaryotic RPN11, consisting of ∼55 residues and including one helix.
Figure 3.
The gene cluster of the Ub-like protein modifier system in C. subterraneum. CDSs without gene annotation encode hypothetical proteins. CDSs; rpn11l (CSUB_C1473), ubl (CSUB_C1474), e2l (CSUB_C1475), e1l (CSUB_C1476) and srfp (CSUB_C1477) encode eukaryotic RPN11, Ubl, E2l and E1l and small RING finger protein, respectively.
Phylogenetic analyses
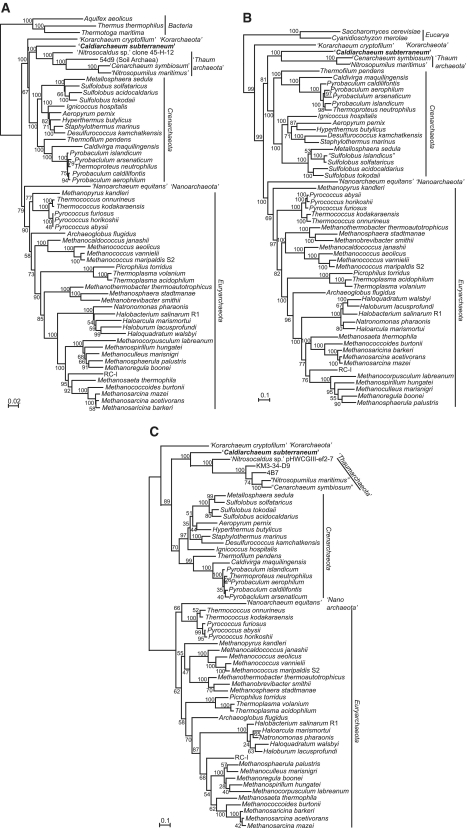
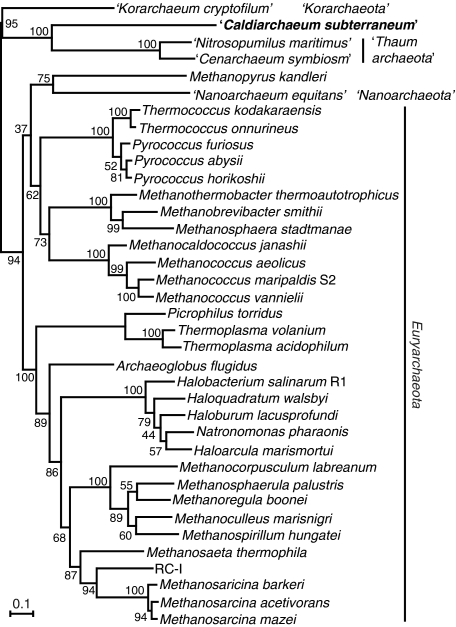
In order to confirm the phylogenetic position of HWCGI, we used the genomic information of C. subterraneum along with those from other archaeal complete genome sequences and environmental genome fragments to perform phylogenetic analyses based on (i) concatenated SSU+LSU rRNA genes; (ii) concatenated ribosomal proteins and RNA polymerase subunits; and (iii) translation elongation factor 2 (EFII) (Figure 4). Taken together, all of these phylogenetic analyses demonstrate that C. subterraneum forms a robust cluster with the Thaumarchaeota, and is distinct from the hyperthermophilic Crenarchaeota. The Korarchaeota is placed in a deeply branching lineage with affinity to the crenarchaeal cluster in the trees of SSU+LSU rRNA genes and EFII, and occupies the deepest position of the Archaea in the tree based on concatenated r-proteins+RNAP subunits sequence. Most orders in the Euryarchaeota are sturdily recovered in all of these trees (Figure 4). The phylogenetic positions of C. subterraneum based on these multiple gene phylogenetic analyses are consistent with those suggested from previously reported phylogenetic trees including environmental SSU rRNA gene sequences (7,21,22; Supplementary Figure S1). The results appear to conflict with the deep branching of Thaumarchaeota as a sister group of all other Archaea, and the potential of a mesophilic last archaeal common ancestor (4,8,9).
Figure 4.
Phylogenetic analyses of Archaea including C. subterraneum. (A) Maximum likelihood phylogenetic tree of concatenated (SSU+LSU) rRNA genes using 3063 identical nucleotide positions. Bacterial sequences were used as out-group. Numbers indicate bootstrap values from 100 replications. (B) Maximum likelihood phylogenetic tree of concatenated universally conserved 45 ribosomal proteins and nine RNA polymerase subunits using aligned identical 5993 amino acid residues. Eukaryotic sequences were used as out-group. Numbers indicate bootstrap values (%) from 200 replications. (C) Maximum likelihood phylogenetic tree made from archaeal translation EF2 proteins based on 590 identical residues. Numbers indicate bootstrap values (%) from 200 replications.
Furthermore, in order to examine the origin of the ‘euryarchaeal genes’ in the novel creanarchaeal lineages, we performed phylogenetic analyses targeting DNAP, which is a signature of Euryarchaeota (47) (Table 1). The phylogenetic tree of concatenated SSU+LSU D-type DNAP presents a robust cluster of crenarchaeal lineages that can be considered as a sister group of the enzymes from Euryarchaeota (Figure 5). When the cluster of crenarchaeal sequences was placed as an outgroup of the euryarchaeal sequences, the tree topology does not contradict with the phylogenetic analyses for rRNA genes, r-proteins+RNAP subunits and EFII (Figures 4 and 5). It can thus be concluded that the D-type DNAPs in the novel crenarchaeal lineages were vertically inherited from the last common archaeal ancestor and did not originate in euryarchaeotes.
Figure 5.
Maximum likelihood phylogenetic tree of concatenated (SSU+LSU) DNAP. Number of identical amino acid residues used were 829. Numbers indicate bootstrap values (%) from 200 replications.
Genome core
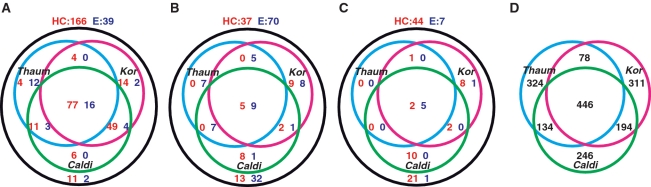
In order to compare the gene complement among the novel crenarchaeal lineages, C. subterraneum, Thaumarchaeota and Korarchaeota, and to investigate the differences between C. subterraneum and hyperthermophilic Crenarchaeota, the numbers of arCOGs in these crenarchaeal lineages that are in common with the genome core genes of Euryarchaeota (E) and hyperthermophilic Crenarchaeota (HC) were examined (Figure 6, Supplementary Table S2). The CDSs in C. subterraneum were tentatively assigned to arCOGs based on BLASTP analysis (<e−3) targeting the arCOG database (29). In this study, genome cores were defined as follows: (i) genes defined in an arCOG that are represented in all sequenced genomes of one division, but are missing in at least some organisms of the other division (5); (ii) genes present in more than two-thirds of the genomes from one division and absent in the other division (5); and (iii) genes that are present in at least one representative of each order of one division, but are absent from all genomes in the other division (4). When examining the presence of euryarchaeotic or crenarchaeotic genome core genes based on definition (I), C. subterraneum (HC:E = 80%:59%) and Korarchaeota (HC:E = 81%:22%) apparently show higher affinity with hyperthermophilic Crenarchaeota than Euryarchaeota, while Thaumarchaeota (HC:E = 58%:79%) had a more euryarchaeotic genomic feature (Figure 6, Supplementary Table S2). When the numbers of genome core genes defined by (II) and (III) were compared, we found that all three lineages shared similar euryarchaeotic features, but Thaumarchaeota exhibited fewer crenarchaeal features among the three. With definition (III), we found that C. subterraneum and Korarchaeota demonstrate significant euryarchaeotic features (Figure 6; Supplementary Table S2). Interestingly, only a small number of HC and E genome core genes defined by (II) and (III) are shared among the three novel crenarchaeal lineages (Figure 6; Supplementary Table S2). In addition, we summarized the numbers of shared arCOGs among the novel crenarchaeal lineages in order to examine genomic affinity (Figure 6). The numbers of arCOGs shared in two lineages but lost in the other probably reflect the relative affinity among the three lineages. As a result, while a total of 446 arCOGs was shared among the three lineages, K. cryptofilum and C. subterraneum showed higher affinity (194 arCOGs shared) to one another compared to the affinity between C. subterraneum and Thaumarchaeota (134 arCOGs shared), and K. cryptofilum and Thaumarchaeota (78 arCOGs shared).
Figure 6.
Venn diagrams presenting number of arCOGs among crenarchaeotic lineages; Caldiarchaeum, Korarchaeum and Thaumarchaeota. (A, B and C) Venn diagrams presenting number of arCOGs represents genome core genes of hyperthermophilic Crenarchaeota (HC: red) and Euryarchaeota (E: blue) in the genomes of the novel crenarchaeal lineages; Caliarchaeum subterraneum (Caldi), Thaumarchaeota (Thaum) and K. cryptofilum (Kor). A total of 11 hyperthermophilic-crenarchaeal and 27 euryarchaeal genomes in arCOG database were used in this analysis. (A) Genes that are represented in all sequenced genome used in arCOG from the represented division, but that are missing in at least some organisms of the other division. (B) Genes present in more than two-thirds of the genomes from one division and absent in the other division. (C) Genes that are present in at least one representative of each order of one division, but are absent from all genomes in the other division. (D) A Venn diagram presenting number of arCOGs shared among three crenarchaeotic lineages; Caldiarchaeum, Korarchaeum and Thaumarchaeota.
DISCUSSION
Genomic coherence and assembly
The high similarities of overlapping regions and the presence of potential insertion/deletion regions indicate that the composite genome sequence of C. subterraneum was successfully assembled from individual, closely related sympatric donor genotypes. The metagenomic library contains DNA from two uncultivated crenarchaeotic lineages; the HWCGI and HWCGIII (Ca. ‘Nitrosocaldus’ sp.). The HWCGIII populations were thought to be more abundant than the HWCGI populations in the metagenomic library based on PCR dependent screening for archaeal SSU rRNA genes (21). However, the dot-blot screening in this study indicates that the number of genome fragments encoding SSU rRNA gene from the HWCGI is seven times as much as those from the HWCGIII, and the result is consistent with the successful genome assemblage of C. subterraneum. Among the 19 C. subterraneum SSU rRNA genes found in the metagenomic library, we observed the co-existence of intron coding or non-coding SSU rRNA genes within one ribotype population. The finding suggests the occurrence of intron transfer events among the C. subterraneum populations associated with double-strand break by intron-coding homing endonuclease and homologous recombination for double strand break repair (85).
Metabolism and ecology
The bacterial communities of microbial mat formation in the geothermal water stream in the subsurface gold mine are dominated by hydrogen-, ammonia- or nitrite-oxidizing chemolithoautotrophs and methanotrophs while hetetrotrophs represent minor populations (86). Considering the high abundance of C. subterraneum and bacterial chemolithoautotrophs and methanotophs in the microbial ecosystem, the archaeon also likely displays chemolithoautotrophic metabolism. In fact, the presence of hydrogen up-take hydrogenase and aerobic carbon monoxide dehydrogenase implies the capability of chemolithotrophic metabolism in C. subterraneum. However, we cannot assert the metabolism because of several uncertainties in the function of these enzymes as described above. On the other hand, dicarboxylate/4-hydroxybutyrate cycle is the most likely carbon assimilation pathway though one key enzyme, 4-hydroxybutyryl-CoA dehydratase, is missing. This resembles the situation of Pyrobaculum arsenaticum, which is known to exhibit autotrophic growth with the dicarboxylate/4-hydroxybutyrate cycle but does not harbor a 4-hydroxybutyryl-CoA dehydratase gene on its genome (68). Non-homologous enzymes, such as members of other dehydratase groups, which are present on the composite genome, may be used as an alternative to support the function of the dicarboxylate/4-hydroxybutyrate cycle.
The HWCGI has been detected from terrestrial and subsurface hot springs, and, recently, dominance of the group in anaerobic hot hydrothermal sediments was reported (7, 19–22). In such hot anaerobic environments, the most probable metabolism is anaerobic hydrogen oxidation dependent chemolithoautotrophy coupled with sulfur or sulfate reduction (22). Judging from the genome sequence, this does not seem to be the case in C. subterraneum. Consequently, the HWCGI is expected to be driven by a versatile energy metabolism as in the case of hyperthermophilic crenarchaeotes (87), and the composite genome of C. subterraneum probably does not represent all of the diverse energy metabolisms of the HWCGI.
In the unique archaeal genome, we found genomic signatures of potential hyperthermphilic life such as the presence of reverse gyrase and the relatively high G+C content of the SSU rRNA gene (21,51). On the other hand, we also observed the presence of DnaJ, DnaK and GrpE genes, reported only in the mesophilic and thermophilic, but not hyperthermophilic, archaea. The microbial mat formation studied here derives from a geothermal water stream with a temperature of 70°C, and other HWCGI SSU rRNA gene sequences have been detected from hot water (70°C, 72°C and 92°C) (7,20), hot spring sediments (74°C) (19,88) and hydrothermal sediments (from 35°C to 60°C) (22). Genes for reverse gyrase have recently been found from genomes of thermophilic bacteria (52,53), and it has been clarified that the gene is not necessarily a prerequisite for hyperthermophilic life (89). Taking all of these factors into account, the HWCGI including C. subterraneum can be considered to be thermophilic, but their optimum growth temperatures are most likely lower than those of hyperthermophilic crenarchaeotes. Considering the potential growth temperatures of C. subterraneum, the Nitrosocaldales, the most deeply branching thaumarchaeal group (74°C) (7) and mesophilic thaumarchaeotes, and the branch lengths of C. subterraneum and thaumarchaeal sequences in the phylogenetic tree (Figure 4 and Supplementary Figure S1), the HWCGI and Thaumarchaeota most likely evolved from a (hyper-)thermophilic common ancestor in the course of adapting to lower temperature environments.
Evolutionary considerations
Differences in replicative functions, transcription and translation are one of the major criteria for phylum level characterization in the domain Archaea (90). The overall mechanisms of DNA replication/repair and cell division in C. subterraneum are more typical of the Euryarchaeota whereas the ribosomal proteins of this archaeon are shared more with crenarchaeotic lineages than with euryarchaeotes (Table 1). We also examined the number of arCOGs present on the genomes of C. subterraneum, Thaumarchaeota and K. cryptofilum that correspond to genome core genes of the Euryarchaeota and hyperthermophilic Crenarchaeota. These comparisons, along with the number of shared arCOGs among the novel crenarchaeal lineages, were used to clarify the affinity between C. subterraneum and other archeal phyla/divisions (Figure 6; Supplementary Table S2). The results indicate that (i) C. subterraneum is distinct from hyperthermophilic Crenarchaeota; (ii) Thaumarchaeota differs from C. subterraneum and K. cryptofilum with its significant euryarchaeotic features; and (iii) C. subterraneum shares more genes with K. cryptofilum than Thaumarchaeota. Moreover, judging from phylogenetic topology, indications of horizontal gene transfer (HGT) were not observed in most of the other euryarchaeal proteins in the crenarchaeal lineages that we examined, a typical case represented in the phylogenetic tree of D-type DNAPs (Figure 5). Taking all of these observations into consideration, we conclude that the complexity in the genomic core structures of the archaeal domain is mostly attributed to a combination of inheritance from an archaeal common ancestor and gene loss events, and that HGT events are not a major factor.
Considering the unique genomic features of C. subterraneum among the crenarchaeal lineages described above (C. subterraneum, the hyperthermophilic Crenarchaeota, Thaumarchaeota and Korarchaeota), the HWCGI occupies a position that can be considered an independent candidatus division among these lineages. On the other hand, phylogenetic trees suggest a close relationship between the Thaumarchaeota and C. subterraneum with high-bootstrap values (Figure 4), also raising the possibility that HWCGI represented by C. subterraneum is a deeply branching group in the Thaumarchaeota. Although conclusions will have to await further data accumulation, we would like to note several points that seem difficult to explain with the latter interpretation. At least two uncultivated crenarchaeal groups; Miscellaneous Crenarchaeotic Group (MCG) and Deep Sea Archaeal Group (DSAG) [also known as the Marine Benthic Group B (MBGB), whose phylogenetic position is still under debate] have been recognized (91) (Supplementary Figure S1). Although the HWCGI and Thaumarchaeota appear to be closely related in the phylogenetic trees shown in Figure 4, the inclusion of the MCG and DSAG sequences in the phylogenetic analysis based on SSU rRNA genes may influence the topology between the HWCGI and Thaumarchaeota. In addition, the genomes of Thaumarchaeota present more euryarchaeotic and less hyperthermophilic crenarchaeotic features than that of C. subterraneum as described above. It is difficult to explain, without considering the occurrence of HGT, that a deeply branching group conserves more crenarchaeotic features while a related group with longer branches within the same phylum/division shares more euryarchaeotic features. Therefore, there is the possibility that the HWCGI can be proposed as a novel division among the crenarchaeal lineages as ‘Aigarchaeota’ (from the Greek αυγη ‘aigi’, meaning dawn and aurora for the intermediate features of hyperthermophilic and mesophilic life during the evolution of the crenarchaeal lineage). However, the current analyses are based on the comparison of one HWCGI genome, one korarchaeal genome and two complete and two partial thaumarchaeal genomes. Thus, we cannot rule out the possibilities of the HWCGI as members of the Crenarchaeota or Thaumarchaeota. The classification of Archaea described in this study may have to be reconsidered in the light of future genomic analyses.
The genome of C. subterraneum also represents several eukaryotic features that have not observed in most of the previously known archaeal lineages. One such feature could be the presence of a type I DNA topoisomerase IB (TopoIB) family that has been found only in the Thaumarchaeota in the domain Archaea (8,92). The gene in C. subterraneum forms a clade with the Thaumarchaeota as a sister group of the eukaryotic cluster, and the phylogenetic topology supports the hypothesis presented by Brochier-Armanet et al. (92) that TopoIB was present in the last common ancestor of the Archaea and Eucarya, and lost in the Euryarchaeota and hyperthermophilic Crenarchaeota.
A striking eukaryotic feature of C. subterraneum is the presence of a potential protein degradation pathway that utilizes an Ub conjugation system. Although the possibility of the C. subterraneum Ubl gene cluster originating in eukaryotes was of concern, the structure of the gene cluster rules out the potential of HGT from eukaryotes. Most importantly, the gene cluster consists of five genes, which are partially overlapped (Figure 3), strongly indicating that this cluster is transcribed as an operon, a signature of prokaryotes. In addition, genes encoding prokaryote-type Ubl, E1l, E2l and JAMM proteins usually constitute fusion genes and/or form operon-like structures. The gene order of prokaryote-type Ubl, E2l and E1l genes in these operon-like gene clusters is highly conserved in the bacterial and archaeal genomes, and is also maintained in the eukaryote-type Ubl, E2l and E1l genes in C. subterraneum. No eukaryotic genome has ever been found to encode the protein modifier system in the form of a gene cluster, and it is highly unlikely that individual components derived by HGT from eukaryotes afterwards reorganize to form operon-like gene clusters. Furthermore, the gene for RPN11l is located adjacent to this gene cluster (Figure 3). The operon-like structure, the conserved prokaryotic gene organization, and the high similarity of the individual components to their eukaryotic counterparts strongly indicate that the eukaryote-type Ubl, E1l, E2l and adjacent RPN11l found in C. subterraneum had already evolved before the divergence between Eucarya and Archaea. The presence of the gene encoding the small Zn RING finger protein in this operon-like gene cluster raises the possibilities that a progenitor of RING-type E3, previously unidentified in prokaryotes, also occurred in the last common ancestor of Eucarya and Archaea. The only other possibility is that HGT occurred from an ancestral eukaryote still retaining prokaryotic gene organization. Such unexpected distributions of eukaryote-specific genes in particular archaeal groups have also been recently identified in cell division and vesicle-formation mechanisms, and these findings suggest a more complex gene composition in the genome of the last common ancestor of Eucarya and Archaea than those found in the genomes of individual modern Archaea (93).
As genes encoding the components of the Haloferax SAMPylation system, such as MoeB (prokaryote-type E1l) and MoaE, are present as single copies on various archaeal genomes (18,94), these genes might exhibit dual roles in both protein degradation and molybdenum/tungstate cofactor biosynthesis. C. subterraneum harbors both the molybdenum/tungstate cofactor biosynthesis systems in addition to the eukaryote-type Ub-like protein modifier system. The unique presence of the eukaryote-type Ub-like system in C. subterraneum and its absence in other Archaea are intriguing. As the Haloferax SAMPylation has been suggested to function in proteasome-dependent protein degradation, the eukaryote-type Ubl, E1l, E2l and RPN11l found in C. subterraneum might have been functionally replaced by the proteins for molybdenum cofactor/tungstate cofactor biosynthesis, allowing the gene loss of the eukaryotic system in most of the presently known archaeal lineages.
The composite genome of C. subterraneum provides further strong evidence that variations of the genome core in the domain Archaea are the result of a combination of vertically inherited ancient features and gene loss events rather than HGT. Furthermore, the genome provides novel insight into the evolutional relationship between Archaea and Eucarya, especially in the Ub–proteasome system. It is well recognized that many lineages of uncultivated Archaea exist on our planet that have yet to be examined. Future multidisciplinary studies combining cultivation, metagenomic or single-cell genomic analyses targeting these unexplored archaeal lineages will surely provide new perspective toward the understanding of the early evolution of life, especially in the Archaea and Eucarya.
ACCESSION NUMBERS
Sequences obtained or used in this study have been deposited in the DDBJ/EMBL/GenBank database under the accession numbers described below. A composite circular genome of C. subterraneum; BA000048. Complete or partial fosmid sequences from of C. subterraneum; AP011633, AP011650, AP011675, AP011689, AP011708, AP011723, AP011724, AP011727, AP011745, AP011751, AP011796 and AP011826-AP011902. Sequences and quality scores from pyrosequencing runs; DRP000160. Fosmid-end sequences of the metagenomic library; AG993735–AG999698. Fosmid sequences encoding representative intron-coding SSU rRNA genes from Caldiarchaeum type I (C. subterraneum); AP011786 and AP011878. Caldiarchaeum type II SSU rRNA gene sequence identified from the metagenomic library; AB566230. Partial ef2 sequence from the Nitrosocaldus sp. (HWCGIII), pHWCGIII-ef2-7; AB543518. Sequences from C. subterraneum are also publically accessible from our ExtremoBase web site (http://www.jamstec.go.jp/gbrowser/cgi-bin/top.cgi).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Education, Culture, Sports, Science & Technology of Japan (Grant-in-Aid No. 18681030 to T.N., in part); Ministry of Education, Culture, Sports, Science & Technology of Japan (Grant-in-Aid No. 20310124 to H.T., in part); Ministry of Education, Culture, Sports, Science & Technology of Japan (Grant-in-Aid No. 22370066 to A.K., in part); Scientific Research on Priority Areas ‘Comprehensive Genomics’ (Grant-in-Aid to M.H., in part); Research fund at the Institute for Fermentation, Osaka, Japan (Grant-in-Aid to A.K., in part); Research funds at the Yamagata Prefectural Government and Tsuruoka City in Japan (Grant-in-Aid to A.K., in part). Funding for open access charge: Japan Agency for Marine-Earth Science & Technology (JAMSTEC).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Matsui and M. Hirai (JAMSTEC) for technical supports in bioinformatics and gene cloning, respectively, and K. Furuya, C. Yoshino (University of Tokyo), A. Yamashita, A. Nakazawa and N. Ito (Kitasato University) for technical assistance in fosmid end-sequencing. We also thank Y. Watanabe (University of Tokyo) and Y. Ishino (Kyushu University) for discussion.
REFERENCES
- 1.Schleper C. Diversity of uncultivated Archaea: perspectives from microbial ecology and metagenomics. In: Garrett RA, Klenk H-P, editors. Archaea; Evolution, Physiology, and Molecular Biology. Oxford: Blackwell Publishing; 2007. pp. 39–50. [Google Scholar]
- 2.Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y, Sugahara J, Preston C, de la Torre J, Richardson PM, DeLong EF. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc. Natl Acad. Sci. USA. 2006;103:18296–18301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM, DeLong EF. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 2006;4:e95. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 5.Elkins JG, Podar M, Graham DE, Makarova KS, Wolf Y, Randau L, Hedlund BP, Brochier-Armanet C, Kunin V, Anderson I, et al. A korarchaeal genome reveals insights into the evolution of the Archaea. Proc. Natl Acad. Sci. USA. 2008;105:8102–8107. doi: 10.1073/pnas.0801980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 7.de la Torre JR, Walker CB, Ingalls AE, Könneke M, Stahl DA. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ. Microbiol. 2008;10:810–818. doi: 10.1111/j.1462-2920.2007.01506.x. [DOI] [PubMed] [Google Scholar]
- 8.Spang A, Hatzenpichler R, Brochier-Armanet C, Rattei T, Tischler P, Spieck E, Streit W, Stahl DA, Wagner M, Schleper C. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 2010;18:331–340. doi: 10.1016/j.tim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, Brochier-Armanet C, Chain PS, Chan PP, Gollabgir A, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc. Natl Acad. Sci. USA. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC, Stetter KO. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature. 2002;417:63–67. doi: 10.1038/417063a. [DOI] [PubMed] [Google Scholar]
- 11.Brochier C, Gribaldo S, Zivanovic Y, Confalonieri F, Forterre P. Nanoarchaea: representatives of a novel archaeal phylum or a fast-evolving euryarchaeal lineage related to Thermococcales? Genome Biol. 2005;6:R42. doi: 10.1186/gb-2005-6-5-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iyer LM, Burroughs AM, Aravind L. The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like beta-grasp domains. Genome Biol. 2006;7:R60. doi: 10.1186/gb-2006-7-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burroughs AM, Jaffee M, Iyer LM, Aravind L. Anatomy of the E2 ligase fold: implications for enzymology and evolution of ubiquitin/Ub-like protein conjugation. J. Struct. Biol. 2008;162:205–218. doi: 10.1016/j.jsb.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burroughs AM, Iyer LM, Aravind L. Natural history of the E1-like superfamily: implication for adenylation, sulfur transfer, and ubiquitin conjugation. Proteins. 2009;75:895–910. doi: 10.1002/prot.22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322:1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darwin KH, Hofmann K. SAMPyling proteins in archaea. Trends Biochem. Sci. 2010;35:348–351. doi: 10.1016/j.tibs.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humbard MA, Miranda HV, Lim J-M, Krause DJ, Pritz JR, Zhou G, Chen S, Wells L, Maupin-Furlow JA. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature. 2010;463:54–62. doi: 10.1038/nature08659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barns SM, Delwiche CF, Palmer JD, Pace NR. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc. Natl Acad. Sci. USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marteinsson VT, Hauksdóttir S, Hobel CFV, Kristmannsdóttir H, Hreggvidsson GO, Kristjánsson JK. Phylogenetic diversity analysis of subterranean hot springs in Iceland. Appl. Environ. Microbiol. 2001;67:4242–4248. doi: 10.1128/AEM.67.9.4242-4248.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nunoura T, Hirayama H, Takami H, Oida H, Nishi S, Shimamura S, Suzuki Y, Inagaki F, Takai K, Nealson KH. Genetic and functional properties of uncultivated thermophilic crenarchaeotes from a subsurface gold mine as revealed by analysis of genome fragments. Environ. Microbiol. 2005;7:1967–1984. doi: 10.1111/j.1462-2920.2005.00881.x. [DOI] [PubMed] [Google Scholar]
- 22.Nunoura T, Oida H, Nakaseama M, Kosaka A, Ohkubo S, Kikuchi T, Kazama H, Tanabe SH, Nakamura K, Kinoshita M, et al. Archaeal diversity and distribution along thermal and geochemical gradients in hydrothermal sediments at the Yonaguni Knoll IV hydrothermal field in the Southern Okinawa Trough. Appl. Environ. Microbiol. 2010;76:1198–1211. doi: 10.1128/AEM.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lane DJ. 16S-23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic Acid Techniques in Bacterial Systematics. New York: John Wiley and Sons; 1985. pp. 115–175. [Google Scholar]
- 24.DeLong EF. Archaea in coastal marine environments. Proc. Natl Acad. Sci. USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM. Wholegenome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 26.Takami H, Nakasone K, Takaki Y, Maeno G, Sasaki R, Masui N, Fuji F, Hirama C, Nakamura Y, Ogasawara N, et al. Complete genome of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilils. Nucleic Acids Res. 2000;28:4317–4331. doi: 10.1093/nar/28.21.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makarova KS, Sorokin AV, Novichkov PS, Wolf YI, Koonin EV. Clusters of orthologous genes for 41 archaeal genomes and implications for evolutionary genomics of archaea. Biol. Direct. 2007;2:33. doi: 10.1186/1745-6150-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugahara J, Yachie N, Arakawa K, Tomita M. In silico screening of archaeal tRNA-encoding genes having multiple introns with bulge-helix-bulge splicing motifs. RNA. 2007;13:671–681. doi: 10.1261/rna.309507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar H, Buchner A, Lai T, Steppi S, Jobb G, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 36.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 38.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 39.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 40.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 41.Hochstrasser M. Evolution and function of ubiquitin-like protein-conjugation systems. Nat. Cell Biol. 2000;2:E153–157. doi: 10.1038/35019643. [DOI] [PubMed] [Google Scholar]
- 42.Lake MW, Wuebbens MM, Rajagopalan KV, Schindelin H. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature. 2001;414:325–329. doi: 10.1038/35104586. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, Hu W, Cai S, Lee B, Song J, Chen Y. The intrinsic affinity between E2 and the Cys domain of E1 in ubiquitin-like modifications. Mol. Cell. 2007;27:228–237. doi: 10.1016/j.molcel.2007.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee I, Schindelin H. Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell. 2008;134:268–278. doi: 10.1016/j.cell.2008.05.046. [DOI] [PubMed] [Google Scholar]
- 45.Filée J, Siguier P, Chandler M. Insertion sequence diversity in archaea. Microbiol. Mol. Biol. Rev. 2007;71:121–157. doi: 10.1128/MMBR.00031-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sorek R, Kunin V, Hugenholtz P. CRISPR- a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 47.Cann IK, Ishino Y. Archaeal DNA replication: identifying the pieces to solve a puzzle. Genetics. 1999;152:1249–1267. doi: 10.1093/genetics/152.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rogozin IB, Makarova KS, Pavlov YI, Koonin EV. A highly conserved family of inactivated archaeal B family DNA polymerases. Biol. Direct. 2008;3:32. doi: 10.1186/1745-6150-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Komori K, Fujikane R, Shinagawa H, Ishino Y. Novel endonuclease in Archaea cleaving DNA with various branched structure. Genes Genet. Syst. 2002;77:227–241. doi: 10.1266/ggs.77.227. [DOI] [PubMed] [Google Scholar]
- 50.Roberts JA, Bell SD, White MF. An archaeal XPF repair endonuclease dependent on a heterotrimeric PCNA. Mol. Microbiol. 2003;48:361–371. doi: 10.1046/j.1365-2958.2003.03444.x. [DOI] [PubMed] [Google Scholar]
- 51.Forterre P. A hot story from comparative genomics: reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 2002;18:236–237. doi: 10.1016/s0168-9525(02)02650-1. [DOI] [PubMed] [Google Scholar]
- 52.Brochier-Armanet C, Forterre P. Widespread distribution of archaeal reverse gyrase in thermophilic bacteria suggests a complex history of vertical inheritance and lateral gene transfers. Archaea. 2007;2:83–93. doi: 10.1155/2006/582916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Campbell BJ, Smith JL, Hanson TE, Klotz MG, Stein LY, Lee CK, Wu D, Robinson JM, Khouri HM, Eisen JA. Adaptations to submarine hydrothermal environments exemplified by the genome of Nautilia profundicola. PLoS Genet. 2009;5:e1000362. doi: 10.1371/journal.pgen.1000362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lindås AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R. A unique cell division machinery in the Archaea. Proc. Natl Acad. Sci. USA. 2008;105:18942–18946. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White MF. DNA repair. In: Garrett RA, Klenk H-P, editors. Archaea; Evolution, Physiology, and Molecular Biology. Oxford: Blackwell Publishing; 2006. pp. 171–184. [Google Scholar]
- 56.Nohmi T, Yamada M, Gruz P. DNA repair and DNA damage tolerance in archaeal bacteria: extreme environments and genome integrity. In: Blum P, editor. Norfolk: New models for prokaryotic biology, Casier Academic Press; 2008. pp. 147–169. [Google Scholar]
- 57.Sugahara J, Kikuta K, Fujishima K, Yachie N, Tomita M, Kanai A. Comprehensive analysis of archaeal tRNA genes reveals rapid increase of tRNA introns in the order Thermoproteales. Mol. Biol. Evol. 2008;25:2709–2716. doi: 10.1093/molbev/msn216. [DOI] [PubMed] [Google Scholar]
- 58.Sugahara J, Fujishima K, Morita K, Tomita M, Kanai A. Disrupted tRNA gene diversity and possible evolutionary scenarios. J. Mol. Evol. 2009;69:497–504. doi: 10.1007/s00239-009-9294-6. [DOI] [PubMed] [Google Scholar]
- 59.Stock T, Rother M. Selenoproteins in Archaea and Gram-positive bacteria. Biochim. Biophys. Acta. 2009;1790:1520–1532. doi: 10.1016/j.bbagen.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 60.Koonin EV, Makarova KS, Elkins JG. Orthologs of the small RPB8 subunit of the eukaryotic RNA polymerases are conserved in hyperthermophilic Crenarchaeota and ‘‘Korarchaeota’’. Biol. Direct. 2007;2:38. doi: 10.1186/1745-6150-2-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blombach F, Makarova KS, Marrero J, Siebers B, Koonin EV, van der Oost J. Identification of an ortholog of the eukaryotic RNA polymerase III subunit RPC34 in Crenarchaeota and Thaumarchaeota suggests specialization of RNA polymerases for coding and non-coding RNAs in Archaea. Biol. Direct. 2009;4:39. doi: 10.1186/1745-6150-4-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lecompte O, Ripp R, Thierry J-C, Moras D, Poch O. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res. 2002;30:5382–5390. doi: 10.1093/nar/gkf693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kelly M, Lappalainen P, Talbo G, Haltia T, Van der Oost J, Saraste M. Two cysteines, two histidines, and one methionine are ligands of a binuclear purple copper center. J. Biol. Chem. 1993;268:16781–16787. [PubMed] [Google Scholar]
- 64.Hügler M, Huber H, Molyneaux SJ, Vetriani C, Sievert SM. Autotrophic CO2 fixation via the reductive tricarboxylic acid cycle in different lineages within the phylum Aquificae: evidence for two ways of citrate cleavage. Environ. Microbiol. 2007;9:81–92. doi: 10.1111/j.1462-2920.2006.01118.x. [DOI] [PubMed] [Google Scholar]
- 65.Berg IA, Kockelkorn D, Buckel W, Fuchs G. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science. 2007;318:1782–1786. doi: 10.1126/science.1149976. [DOI] [PubMed] [Google Scholar]
- 66.Huber H, Gallenberger M, Jahn U, Eylert E, Berg IA, Kockelkorn D, Eisenreich W, Fuchs G. A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic archaeum Ignicoccus hospitalis. Proc. Natl Acad. Sci. USA. 2008;105:7851–7856. doi: 10.1073/pnas.0801043105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kockelkorn D, Fuchs G. Malonic semialdehyde reductase, succinic semialdehyde reductase, and succinyl-coenzyme A reductase from Metallosphaera sedula: enzymes of the autotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle in Sulfolobales. J. Bacteriol. 2009;191:6352–6562. doi: 10.1128/JB.00794-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ramos-Vera WH, Berg IA, Fuchs G. Autotrophic carbon dioxide assimilation in Thermoproteales revisited. J. Bacteriol. 2009;191:4286–4297. doi: 10.1128/JB.00145-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Teufel R, Kung JW, Kockelkorn D, Alber BE, Fuchs G. 3-hydroxypropionyl-coenzyme A dehydratase and acryloyl-coenzyme A reductase, enzymes of the autotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle in the Sulfolobales. J. Bacteriol. 2009;191:4572–4581. doi: 10.1128/JB.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grochowski LL, Xu H, White RH. Ribose-5-phosphate biosynthesis in Methanocaldococcus jannaschii occurs in the absence of a pentose-phosphate pathway. J. Bacteriol. 2005;187:7382–7389. doi: 10.1128/JB.187.21.7382-7389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Orita I, Sato T, Yurimoto H, Kato N, Atomi H, Imanaka T, Sakai Y. The ribulose monophosphate pathway substitutes for the missing pentose phosphate pathway in the archaeon Thermococcus kodakaraensis. J. Bacteriol. 2006;188:4698–4704. doi: 10.1128/JB.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rashid N, Imanaka H, Fukui T, Atomi H, Imanaka T. Presence of a novel phosphopentomutase and a 2-deoxyribose 5-phosphate aldolase reveals a metabolic link between pentoses and central carbon metabolism in the hyperthermophilic archaeon Thermococcus kodakaraensis. J. Bacteriol. 2004;186:4185–4191. doi: 10.1128/JB.186.13.4185-4191.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tumbula DL, Teng Q, Bartlett MG, Whitman WB. Ribose biosynthesis and evidence for an alternative first step in the common aromatic amino acid pathway in Methanococcus maripaludis. J. Bacteriol. 1997;179:6010–6013. doi: 10.1128/jb.179.19.6010-6013.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.White RH. L-Aspartate semialdehyde and a 6-deoxy-5-ketohexose 1-phosphate are the precursors to the aromatic amino acids in Methanocaldococcus jannaschii. Biochemistry. 2004;43:7618–7627. doi: 10.1021/bi0495127. [DOI] [PubMed] [Google Scholar]
- 75.Meereis F, Kaufmann M. Extension of the COG and arCOG databases by amino acid and nucleotide sequences. BMC Bioinformatics. 2008;9:479. doi: 10.1186/1471-2105-9-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Joazeiro CA, Weissman AM. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 77.Verma R, Aravind L, Oania R, McDonald WH, YatesIII JR, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 78.Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell. Biol. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- 79.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 80.Walden H, Podgorski MS, Schulman BA. Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature. 2003;422:330–334. doi: 10.1038/nature01456. [DOI] [PubMed] [Google Scholar]
- 81.Lois LM, Lima CD. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 2005;24:439–451. doi: 10.1038/sj.emboj.7600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 83.Maytal-Kivity V, Reis N, Hofmann K, Glickman MH. MPN?, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002;3:28–39. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ambroggio XI, Rees DC, Deshaies RJ. JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2004;2:E2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jurica MS, Stoddard BL. Homing endonucleases: structure, function and evolution. Cell. Mol. Life Sci. 1999;55:1304–1326. doi: 10.1007/s000180050372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hirayama H, Takai K, Inagaki F, Yamato Y, Suzuki M, Nealson KH, Horikoshi K. Bacterial community shift along a subsurface geothermal water stream in a Japanese gold mine. Extremophiles. 2005;9:169–184. doi: 10.1007/s00792-005-0433-8. [DOI] [PubMed] [Google Scholar]
- 87.Huber R, Huber H, Stetter KO. Towards the ecology of hyperthermophiles: biotopes, new isolation strategies and novel metabolic properties. FEMS Microbiol. Rev. 2000;24:615–623. doi: 10.1111/j.1574-6976.2000.tb00562.x. [DOI] [PubMed] [Google Scholar]
- 88.Barns SM, Fundyga RE, Jeffries MW, Pace NR. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl Acad. Sci. USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Atomi H, Matsumi R, Imanaka T. Reverse gyrase is not a prerequisite for hyperthermophilic life. J. Bacteriol. 2004;186:4829–4833. doi: 10.1128/JB.186.14.4829-4833.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bernander R. Chromosome replication, nucleoid segregation and cell division in archaea. Trends Microbiol. 2000;8:278–283. doi: 10.1016/s0966-842x(00)01760-1. [DOI] [PubMed] [Google Scholar]
- 91.Teske A, Sørensen KB. Uncultured archaea in deep marine subsurface sediments: have we caught them all? ISME J. 2008;2:3–18. doi: 10.1038/ismej.2007.90. [DOI] [PubMed] [Google Scholar]
- 92.Brochier-Armanet C, Gribaldo S, Forterre P. A DNA topoisomerase IB in Thaumarchaeota testifies for the presence of this enzyme in the last common ancestor of Archaea and Eucarya. Biol. Direct. 2008;3:5. doi: 10.1186/1745-6150-3-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Makarova KS, Yutin N, Bell SD, Koonin EV. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat. Rev. Microbiol. 2010;8:731–741. doi: 10.1038/nrmicro2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hartman AL, Norais C, Badger JH, Delmas S, Haldenby S, Madupu R, Robinson J, Khouri H, Ren Q, Lowe TM, et al. The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS One. 2010;5:e9605. doi: 10.1371/journal.pone.0009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.