Abstract
The cell’s ability to sense and respond to specific stimuli is a complex system derived from precisely regulated protein-protein interactions. Some of these protein-protein interactions are mediated by the recognition of linear peptide motifs by protein modular domains. BRCT (BRCA1 C-terminal) domains and their linear motif counterparts, which contain phosphoserines, are one such pair-wise interaction system that seems to have evolved to serve as a surveillance system to monitor threats to the cell’s genetic integrity. Evidence indicates that BRCT domains found in tandem can cooperate to provide sequence-specific binding of phosphorylated peptides as is the case for the breast and ovarian cancer susceptibility gene BRCA1 and the PAX transcription factor–interacting protein PAXIP1. Particular interest has been paid to tandem BRCT domains as “readers” of signaling events in the form of phosphorylated serine moieties induced by the activation of DNA damage response kinases ATM, ATR, and DNA-PK. However, given the diversity of tandem BRCT-containing proteins, questions remain as to the origin and evolution of this domain. Here, we discuss emerging views of the origin and evolving roles of tandem BRCT domain repeats in the DNA damage response.
Keywords: BRCT, DNA damage, networks, cancer, systems biology, modular domains
The DNA Damage Response
During the life of a cell, its DNA is constantly subjected to damage. This damage can be from external sources, such as chemical carcinogens and ionizing radiation, or from internal sources such as errors in replication.1 Cells have evolved a network of interconnecting pathways to promptly fix these errors and prevent illegitimate changes to DNA from being propagated to daughter cells. This network can be collectively called the DNA damage response (DDR) and includes a wide array of overlapping systems that recognize abnormal DNA structures such as single- and double-stranded breaks, topological distortions of the double helix, or DNA adducts.2,3 An important feature of this network in eukaryotes is the coordination of DNA repair with a tight control of cell cycle progression.4,5 In addition, cells in metazoan organisms will undergo apoptosis as a fail-safe mechanism when they cannot properly fix the damage.3 Defects in this network can lead to a series of diseases, in particular, cancer.6,7
Many proteins involved in DNA repair are found in all superkingdoms (Archaea, Bacteria, Eukaryota), but one particular feature found primarily in eukaryotes is the integration of DNA repair processes with cell cycle check points and apoptosis.8 Indeed, a problem that has captured the attention of many researchers is how organisms have evolved more complex signaling networks during the course of evolution.9
A Modular Domain View of the DDR
Signaling events are normally mediated by a system of interacting partners, which utilize the recognition of linear motifs, usually present in unstructured regions of proteins, and by modular domains.10 Even a cursory analysis of the DDR reveals that the system is driven by a series of phosphorylations and ubiquitinations and that there is an overrepresentation of 3 particular protein domains: BRCTs, FHA, and 14-3-3.11
Modular domains that can be found in combination with a large number of other modular domains are sometimes called promiscuous.12,13 This is the case of the BRCT and FHA domains, while the 14-3-3 domain can be considered a lonely domain in the sense that it does not occur with other domains.9 Here, we will focus our discussion on the BRCT domains.
BRCT Domains
Elements of the DNA repair machinery typically exist as large macromolecular complexes that incorporate a diverse array of proteins required for the faithful detection and resolution of the lesion. Outside of the obvious need for DNA binding components of the complex, adaptor proteins (and their domains) are prevalent and mediate the specific protein-protein interactions required for the organization of the complexes. Present in many of the known scaffold proteins involved in the DDR, the BRCT domain is an important component of adaptor proteins involved in DNA repair.
Originally identified in the C-terminal region of the breast and ovarian cancer susceptibility protein BRCA1, known to be truncated or mutated in cases of hereditary breast and ovarian cancer,14 BRCT domains have been found in a number of different proteins in species ranging from bacteria to humans.15,16 Although it contains no intrinsic enzymatic activity, the BRCT domain is essential for orchestrating a variety of enzymatic reactions involved in repair and check point regulation at sites of DNA damage.17-22 The domain itself is relatively small, consisting of approximately 90 to 100 amino acids.14 Concordantly, most if not all of the proteins that have been found to contain BRCT domains are intimately connected to the regulation of DNA integrity.15,16
BRCT domains occur in proteins as single (e.g., PARP1) or multiple units (e.g., TOPBP1). Interestingly, in proteins in which they occur as multiple units, BRCTs tend to be organized as pairs of closely juxtaposed units (e.g., BRCA1). Domains that occur as single isolated units function in a variety of homodimerization or heterodimerization interactions, leading to the formation of protein complexes that play important roles in DNA damage signaling, repair, and coordination of cell cycle check points.23 Tandem BRCT domains can be found in BRCA1 (Fig. 1), LIG4, TOPBP1, PAXIP1 (PTIP), ECT2, NBN, MCPH1, TP53BP1, XRCC1, BARD1, MDC1, and ANKRD32 and seem to function as one structural unit. Although not all of these proteins have yet been systematically analyzed, several of them have been shown to recognize phosphorylated peptides.24-26 Interestingly, the phosphopeptide recognition pocket of the BRCA1 tandem BRCTs was independently identified by mutation analysis in breast cancer, highlighting the importance of this function to tumor suppression.27 It is not uncommon for these 2 subclasses of BRCT domains (single or tandem) to occur in the same protein; such is the case for TOPBP1 and MCPH1.
Figure 1.
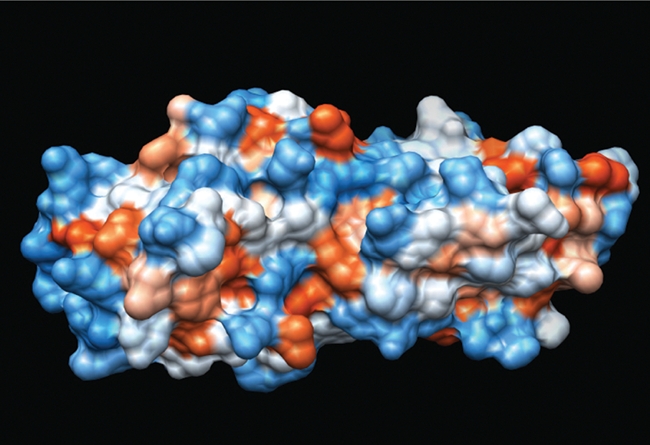
Tandem BRCT domains from BRCA1. Space-filling structure derived from 1JNX.29
The BRCT domain presents as a globular α/β fold composed of a 4-stranded central β sheet flanked by 3 α helices (α1 and α3 on one side with α2 on the opposite face).28,29 There is also a strong conservation of the amino acid tryptophan in helix α3 in most, but not all, BRCT domains.15 When present as a tandem pair, the individual domains fold in a head-to-tail fashion connected by a linker region that can vary in structure and composition.29-33 Interestingly, the BRCT domain includes a NAD(P)-binding Rossmann fold,34 a protein architecture commonly found in several different superfamilies35 and in ancient proteins.36
While the phosphorylated serine is normally recognized by a groove in one of the BRCT units, the specificity is conferred by the amino acid located in the +3 position of the phosphopeptide (with the serine being the 0 position).19,25,33,37-40 In BRCA1, the ability to recognize phosphopeptides is mediated by conserved residues S1655 and K1702 in the first BRCT domain that allow hydrogen bonding to the pS/T residue. A hydrophobic pocket composed of F1704, M1775, and L1839 in the second BRCT domain of BRCA1, which form the core of the fold, provides the specificity for phenylalanine at the +3 position.25,37,38,40 Indeed, this type of “2-knob” interaction has been shown to be essential for BRCA1 binding to DDR proteins CtIP, BACH1, and ABRAXAS as well as others including MDC1 binding to the C-terminal tail of γH2AX and PAXIP1 binding to phosphorylated TP53BP1.25,33,37,38,40-42 However, because of the diversity in the proteins in which tandem BRCT domains exist and the specific functions of each one, it is not surprising that the phosphopeptides they bind differ as well. For instance, it was found that the MDC1 tandem domain prefers tyrosine at the +3 position.33,39 Differences in the sequence of amino acids around the pS/T residue allow for tight regulation of the interactions, which likely is further fine tuned by spatial and temporal regulation of specific kinases and phosphatases.
Importantly, not all interactions between tandem BRCT domains and their partners are mediated by phosphorylation events. The example best characterized by comprehensive structural analysis is that of TP53BP1 binding to the tumor suppressor p53, which occurs on the face opposite that of the hydrophobic cleft of the tandem BRCT domains of TP53BP1.30,32 As these domains exist as globular domains, the conserved phosphopeptide binding pockets only occupy a small portion of the available surface area of the domains. This clearly leaves room for a number of other potential surface interactions to occur. Therefore, caution should be made in only classifying these domains as phosphopeptide binding.
Evolution of BRCT Domains
The tandem BRCT domain organization has undergone significant evolutionary expansion from prokaryotes to higher order metazoans (Fig. 2). Although the specific mechanisms that fostered this expansion remain speculative, interesting correlations can be made between the phyletic distribution of tandem BRCT proteins and aspects of DNA repair and cell cycle check points in diverse lineages.
Figure 2.
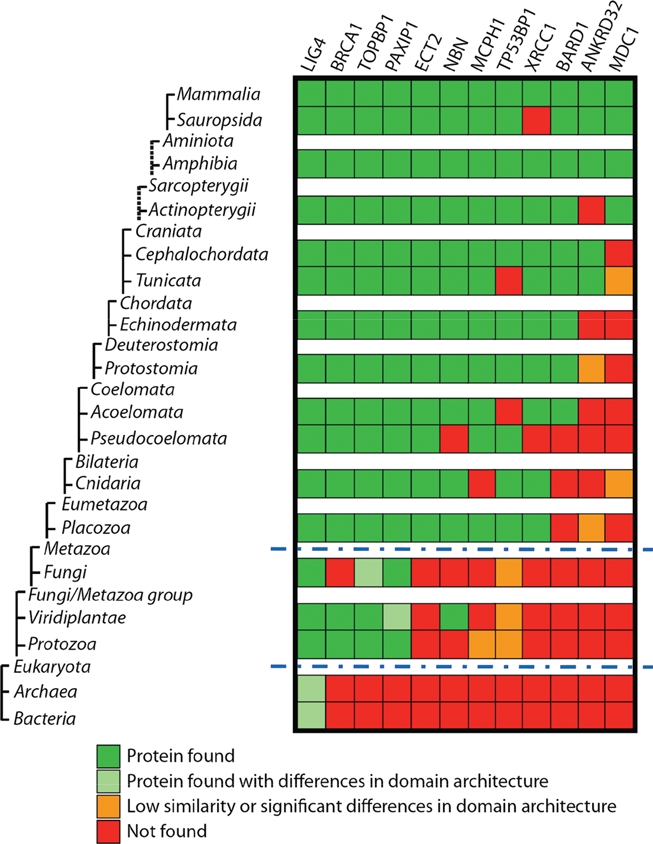
Taxonomic distribution of tandem BRCT-containing proteins. Distant orthologs of human tandem BRCT-containing proteins were identified in different taxa and curated individually. Orthologs were retrieved using human tandem BRCT-containing proteins (including LIG4, BRCA1, TOPBP1, PAXIP1, ECT2, NBN, MCPH1, TP53BP1, XRCC1, BARD1, MDC1, and ANKRD32) to perform saturated60 and reverse61 blast searches on a nonredundant database (NCBI), allowing for small changes in conserved domain architecture. Common blast (NCBI) and conserved domain analyses were performed locally using a HMMER and blast manager software (FAT [functional analysis tool] developed by our group) and PFAM database.62 All results were manually curated. Taxonomy tree organization was based on NCBI taxonomy, although some taxa were omitted (a complete list of taxa used can be provided upon request) and do not show evolutionary distances. Dashed lines on the tree show indirect taxa links. *Only taxa Alveolata (txid 33630), Amoebozoa (txid 554915), and Euglenozoa (txid 33682) were used for Protozoa division. Blue dashed lines indicate major transitions to eukaryotes and to metazoans.
The minimal complement of BRCT-containing proteins in the human genome is 24 (Mesquita and Monteiro, unpublished results). Of those proteins, 12 present with more than one BRCT unit (Fig. 2). We hypothesize that these 12 proteins might therefore make up the core of the phosphoserine recognition system during the DDR.
One of the earliest analyses of DDR-associated domain architecture by Aravind et al. did not find any instances of the BRCT domain in Archaea, although it was evident in the carboxy terminus of bacterial DNA ligases.8 Additionally, the presence of the BRCT domain in Trypanosomes (which is not considered part of the early radiation of eukaryotes) prompted speculation that horizontal gene transfer from prokaryotes could be the mechanism by which eukaryotes acquired BRCT domains.8 However, instances of BRCT domains have since been found in Archaea.43 A search of currently deposited sequences also identifies the presence of a BRCT domain in proteins with similarity to the human LIG4 in several Archaea genera (e.g., Halorhabdus, Haloarcula, Natronomonas, Haloferax, Halorubrum, and Haloquadratum). The presence of BRCT domain in all 3 superkingdoms suggests that vertical descent of this domain from the last common ancestor, or cenancestor, could explain its phyletic distribution. In addition, 2 “transition points,” the eukaryotic/prokaryotic division and the emergence of the metazoans, can be identified (Fig. 2).
Interestingly, SH2 domains are present almost exclusively in metazoans, suggesting that its origin is close to the emergence of multicellularity.25 Although the tandem BRCT domain clearly has roots in single-cell organisms, multicellularity requires a more complex and tightly controlled protection of DNA integrity as a genetic insult to a single cell can be detrimental to the whole organism. As organisms expand in complexity, so too does the number of tandem BRCT domain–containing proteins. This could be attributed to the cell’s penchant for highly redundant systems that provide added insurances of successful completion of vital cellular processes such as chromatin regulation, DNA damage repair, and cell cycle check points.
It is likely that the first occurrence of the tandem BRCT domain came from the duplication of a single domain either by genetic duplication or domain shuffling between 2 different genes. Hints of a possible origin of tandem BRCT domains can be extrapolated from the existence of an apparent third, less defined, subclass of BRCT domains found in RFC1-, PARP1-, and NAD+-dependent bacterial DNA ligases. These BRCTs are defined by replacement of the highly conserved tryptophan residue found in helix α3 as well as several other specific structural alterations.44 Most striking is that some of the individual domains within this subclass display a capacity to bind DNA mediated by the BRCT domain in which binding is mediated by the phosphate binding pocket and an N-terminally located helix outside the BRCT domain.44-46 Compellingly, in the case of RFC1 binding to the 5′-phosphate of DNA, there is excellent conservation of both the 3-dimensional structure and the chemical nature of the phosphate binding site found in other BRCT domains. Furthermore, there is conservation between the DNA binding residues of RFC1 and bacterial NAD+-dependent ligases, and mutation of these residues in the ligases severely affects their ability to bind DNA, suggesting conservation in the mode of DNA binding.44,46 These studies suggest that the BRCT domain could have originated as a DNA binding motif through recognition of the 5′-phosphate moiety and the binding to phosphorylated peptides may have developed later.
To probe further into this problem, we generated a tree comparing BRCTs (not the full proteins) from 1) the E. coli ligase, 2) all Archaea BRCTs (all of them from DNA ligases), and 3) all 24 BRCT-containing human proteins (Fig. 3). Surprisingly, the RFC1 BRCT clustered with the Archaea BRCTs, followed by DBF4 and PARP1. These results strengthen the notion raised in the previous paragraph, although a more comprehensive and exhaustive analysis is needed to fully investigate these issues. Importantly, the presence of the BRCT domain in Archaea does not necessarily exclude the possibility of horizontal gene transfer as an evolutionary process that could have contributed to the expansion of this domain.
Figure 3.
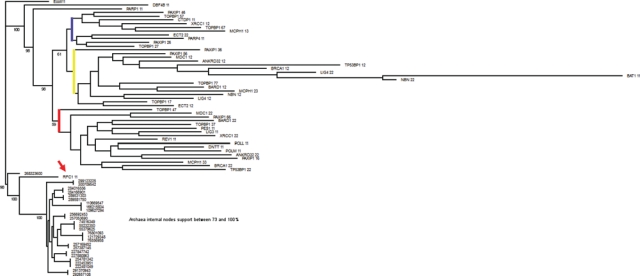
RFC1 BRCT clusters with Archaea BRCTs. RFC1 BRCT is indicated by an arrow. Blue, yellow, and red lines indicate major branches of BRCT domains. Tree comparing single BRCT units from E. coli ligase BRCT, all BRCTs from Archaea (all derive from DNA ligases), and BRCTs from all 24 human BRCT proteins. BRCTs were aligned with PRALINE. Bootstrap created 1,000 replicates using the partial jackknife method where 1% positions were randomly omitted (SEQBOOT, PHYLIP package). Distance matrix calculations were performed using PMB matrix (BLOCKS conserved domain-based matrix) and software PROTDIST (PHYLIP package). Trees were calculated for each replicate using E. coli as a root and software neighbor. The consensus tree and percentage support were calculated using software consensus (PHYLIP package) and visualized with TREEVIEW and edited with GIMP to include percentage support (numbers included at branching points). Numbers appended to protein names identify BRCT domains. For example, PAXIP1 36 represents the third BRCT repeat out of 6 BRCTs contained in PAXIP1. Archaea BRCTs are identified by protein gi numbers.
Of all human tandem BRCT proteins, DNA ligase IV (LIG4) contains the earliest identifiable ortholog found in all 3 superkingdoms, although found as a single unit in Archaea and bacteria (Fig. 2). In early prokaryotes and Archaea, where there is less pressure to maintain genomic fidelity and error-prone repair could aid in adaptability of the species, the error-prone NHEJ pathway appears to be the first to utilize the tandem BRCT domain. LIG4 complexes with XRCC4 at sites of double-stranded breaks through recruitment by DNA-PK and KU70/KU86 and completes the final step in repair by ligating the 2 strands of DNA.
Interestingly, in the case of the tandem BRCT domain of LIG4, it is not the domains themselves but rather the linker between the domains that mediate the important interaction with XRCC4.18 Indeed, this mode of interaction is conserved in other instances of the tandem such as the phospho-independent binding of the TP53BP1 tandem BRCT to p53, which utilizes the linker region between the BRCT domains for binding.30,32 However, the linker region does not always directly bind to the interaction partner. Indeed, each of the BRCT domains found in the tandems of both BRCA1 and MDC1 confers sequence binding specificity for phosphopeptides.38 Despite their conservation in structure and composition, it is obvious that their scaffolding properties are utilized in a diverse fashion within the BRCT family. It is also important to note that BRCT domains have also been identified in plants47 and a BRCT-related fold has been observed in the T antigen helicase domain of polyoma viruses.48
This apparent plasticity in binding functionality is likely to be a driving force in the utilization of these domains as adaptor proteins in the DNA damage response. Furthermore, it is tempting to speculate that combination of the most ancient singleton BRCT domains with other non-BRCT regions (or with other duplicated BRCT units) led to the emergence of new binding interfaces not present in the single domains alone that could confer binding selectivity and specificity. This ability to specifically bind new proteins may have provided the basis for the expansion of this motif as a tandem domain in a single protein.
Perhaps more perplexing is how these conserved domains with diverse modes of mediating interactions do not appear to have branched out into other cellular processes, especially those that also involve serine and threonine kinase signaling. Perhaps the most non-DNA damage use of the BRCT domain has been described in ECT2 control of cleavage furrow formation.49 However, since this process involves coordination of cytokinesis with chromosome segregation, it is evident that this BRCT domain functions in maintaining genome fidelity. Considering the FHA and 14-3-3 scaffolding domains involved in the DDR, which participate in a number of diverse cellular pathways, BRCT domains have been relatively confined to the DDR. This could possibly be explained by evolutionary determinants that prevent its utilization in other processes or constraints on recognizing consensus regions that are preferred by kinases involved in the DDR.
Conclusion
Recently, approaches to understand the origin and evolution of cellular signal transduction mechanisms have yielded important insights when “writers” (kinases), “erasers” (phosphatases), and “readers” (modular domains) are analyzed as a 3-component signaling system.9,50-52 These approaches have focused on a well-characterized system involving protein tyrosine kinases, tyrosine phosphatases, and SH2 domain–containing proteins that has a prominent role in growth factor receptor signaling. However, only in the last 15 years has a clearer picture of the DNA damage response network begun to be delineated.53 Presently, we have accumulated significant knowledge on the kinases involved in signaling proximal to DNA damage sites such as ATM, ATR, and DNA-PK,54-56 and the main effector kinases CHK1 and CHK2,57,58 but our knowledge of other kinases and phosphatases involved in DNA damage signaling is still rudimentary.59
Just like the Roman Praetorian Guard, BRCT domains function as elite body guards keeping close attention on the integrity of the emperor (the cell’s DNA). Also, just like the Praetorian Guard that eventually had its role expanded from the battlefield to politics, BRCT domains’ functions seem to have evolved from participating in DNA repair to include negotiating cell cycle check points. Here, we presented a rough analysis of the occurrence of BRCT domains in diverse lineages. This analysis suggests that the RFC1 BRCT might be the functional prototype of tandem BRCT domains. However, this analysis also highlights several limitations in our understanding and suggests several questions that need to be answered to illuminate the role and evolution of the DNA damage response.
Acknowledgments
The authors thank Hanna Silva Condelo for help with data mining, Yun Liu for help with the figures, and J.N. Mark Glover and Rachel Karchin for helpful suggestions. This article is dedicated to the memory of Saburo Hanafusa.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the authorship and/or publication of this article.
This work was supported by the National Institutes of Health [grant nos. CA148112, CA116167, and CA11809] and by the US Army Medical Research and Materiel Command, National Functional Genomics Center project [grant no. W81XWH-08-2-0101]. The opinions, interpretations, conclusions, and recommendations are those of the author(s) and are not necessarily endorsed by the US Army. N.T.W. is supported by a Florida Breast Cancer Foundation fellowship.
References
- 1. Friedberg EC. DNA damage and repair. Nature. 2003;421:436-40 [DOI] [PubMed] [Google Scholar]
- 2. Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39-85 [DOI] [PubMed] [Google Scholar]
- 3. Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316-23 [DOI] [PubMed] [Google Scholar]
- 5. Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433-9 [DOI] [PubMed] [Google Scholar]
- 6. Bartkova J, Rezaei N, Liontos M, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633-7 [DOI] [PubMed] [Google Scholar]
- 7. Bartkova J, Horejsi Z, Koed K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864-70 [DOI] [PubMed] [Google Scholar]
- 8. Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jin J, Xie X, Chen C, et al. Eukaryotic protein domains as functional units of cellular evolution. Sci Signal. 2009;2:ra76. [DOI] [PubMed] [Google Scholar]
- 10. Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445-52 [DOI] [PubMed] [Google Scholar]
- 11. Mohammad DH, Yaffe MB. 14-3-3 proteins, FHA domains and BRCT domains in the DNA damage response. DNA Repair (Amst). 2009;8:1009-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285:751-3 [DOI] [PubMed] [Google Scholar]
- 13. Basu MK, Carmel L, Rogozin IB, Koonin EV. Evolution of protein domain promiscuity in eukaryotes. Genome Res. 2008;18:449-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koonin EV, Altschul SF, Bork P. BRCA1 protein products: functional motifs. Nat Genet. 1996;13:266-8 [DOI] [PubMed] [Google Scholar]
- 15. Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68-76 [PubMed] [Google Scholar]
- 16. Callebaut I, Mornon JP. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Letters. 1997;400:25-30 [DOI] [PubMed] [Google Scholar]
- 17. Cantor SB, Bell DW, Ganesan S, et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149-60 [DOI] [PubMed] [Google Scholar]
- 18. Dore AS, Furnham N, Davies OR, et al. Structure of an Xrcc4-DNA ligase IV yeast ortholog complex reveals a novel BRCT interaction mode. DNA Repair (Amst). 2006;5:362-8 [DOI] [PubMed] [Google Scholar]
- 19. Glover JN, Williams RS, Lee MS. Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem Sci. 2004;29:579-85 [DOI] [PubMed] [Google Scholar]
- 20. Goldberg M, Stucki M, Falck J, et al. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952-6 [DOI] [PubMed] [Google Scholar]
- 21. Kim JE, Billadeau DD, Chen J. The tandem BRCT domains of Ect2 are required for both negative and positive regulation of Ect2 in cytokinesis. J Biol Chem. 2005;280:5733-9 [DOI] [PubMed] [Google Scholar]
- 22. Lou Z, Minter-Dykhouse K, Franco S, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187-200 [DOI] [PubMed] [Google Scholar]
- 23. Dulic A, Bates PA, Zhang X, et al. BRCT domain interactions in the heterodimeric DNA repair protein XRCC1-DNA ligase III. Biochemistry. 2001;40:5906-13 [DOI] [PubMed] [Google Scholar]
- 24. Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636-9 [DOI] [PubMed] [Google Scholar]
- 25. Williams RS, Lee MS, Hau DD, Glover JN. Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nat Struct Mol Biol. 2004;11:519-25 [DOI] [PubMed] [Google Scholar]
- 26. Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639-42 [DOI] [PubMed] [Google Scholar]
- 27. Mirkovic N, Marti-Renom MA, Weber BL, Sali A, Monteiro AN. Structure-based assessment of missense mutations in human BRCA1: implications for breast and ovarian cancer predisposition. Cancer Res. 2004;64:3790-7 [DOI] [PubMed] [Google Scholar]
- 28. Shiozaki EN, Gu L, Yan N, Shi Y. Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: implications for signaling. Mol Cell. 2004;14:405-12 [DOI] [PubMed] [Google Scholar]
- 29. Williams RS, Green R, Glover JN. Crystal structure of the BRCT repeat region from the breast cancer-associated protein BRCA1. Nat Struct Biol. 2001;8:838-42 [DOI] [PubMed] [Google Scholar]
- 30. Derbyshire DJ, Basu BP, Serpell LC, et al. Crystal structure of human 53BP1 BRCT domains bound to p53 tumour suppressor. EMBO J. 2002;21:3863-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Edwards RA, Lee MS, Tsutakawa SE, Williams RS, Tainer JA, Glover JN. The BARD1 C-terminal domain structure and interactions with polyadenylation factor CstF-50. Biochemistry. 2008;47:11446-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Joo WS, Jeffrey PD, Cantor SB, Finnin MS, Livingston DM, Pavletich NP. Structure of the 53BP1 BRCT region bound to p53 and its comparison to the Brca1 BRCT structure. Genes Dev. 2002;16:583-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee MS, Edwards RA, Thede GL, Glover JN. Structure of the BRCT repeat domain of MDC1 and its specificity for the free COOH-terminal end of the gamma-H2AX histone tail. J Biol Chem. 2005;280:32053-6 [DOI] [PubMed] [Google Scholar]
- 34. Rao ST, Rossmann MG. Comparison of super-secondary structures in proteins. J Mol Biol. 1973;76:241-56 [DOI] [PubMed] [Google Scholar]
- 35. Orengo CA, Thornton JM. Protein families and their evolution-a structural perspective. Annu Rev Biochem. 2005;74:867-900 [DOI] [PubMed] [Google Scholar]
- 36. Ma BG, Chen L, Ji HF, et al. Characters of very ancient proteins. Biochem Biophys Res Commun. 2008;366:607-11 [DOI] [PubMed] [Google Scholar]
- 37. Botuyan MV, Nomine Y, Yu X, et al. Structural basis of BACH1 phosphopeptide recognition by BRCA1 tandem BRCT domains. Structure (Camb). 2004;12:1137-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Clapperton JA, Manke IA, Lowery DM, et al. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat Struct Mol Biol. 2004;11:512-8 [DOI] [PubMed] [Google Scholar]
- 39. Rodriguez M, Yu X, Chen J, Songyang Z. Phosphopeptide binding specificities of BRCA1 COOH-terminal (BRCT) domains. J Biol Chem. 2003;278:52914-8 [DOI] [PubMed] [Google Scholar]
- 40. Shiozaki EN, Gu L, Yan N, Shi Y. Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: implications for signaling. Mol Cell. 2004;14:405-12 [DOI] [PubMed] [Google Scholar]
- 41. Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213-26 [DOI] [PubMed] [Google Scholar]
- 42. Munoz IM, Jowsey PA, Toth R, Rouse J. Phospho-epitope binding by the BRCT domains of hPTIP controls multiple aspects of the cellular response to DNA damage. Nucleic Acids Res. 2007;35:5312-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao A, Gray FC, MacNeill SA. ATP- and NAD+-dependent DNA ligases share an essential function in the halophilic archaeon Haloferax volcanii. Mol Microbiol. 2006;59:743-52 [DOI] [PubMed] [Google Scholar]
- 44. Kobayashi M, Ab E, Bonvin AM, Siegal G. Structure of the DNA-bound BRCA1 C-terminal region from human replication factor C p140 and model of the protein-DNA complex. J Biol Chem. 2010;285:10087-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wilkinson A, Smith A, Bullard D, et al. Analysis of ligation and DNA binding by Escherichia coli DNA ligase (LigA). Biochim Biophys Acta. 2005;1749:113-22 [DOI] [PubMed] [Google Scholar]
- 46. Feng H, Parker JM, Lu J, Cao W. Effects of deletion and site-directed mutations on ligation steps of NAD+-dependent DNA ligase: a biochemical analysis of BRCA1 C-terminal domain. Biochemistry. 2004;43:12648-59 [DOI] [PubMed] [Google Scholar]
- 47. Singh SK, Choudhury SR, Roy S, Sengupta DN. Sequential, structural, and phylogenetic study of BRCT module in plants. J Biomol Struct Dyn. 2008;26:235-45 [DOI] [PubMed] [Google Scholar]
- 48. Kumar A, Joo WS, Meinke G, Moine S, Naumova EN, Bullock PA. Evidence for a structural relationship between BRCT domains and the helicase domains of the replication initiators encoded by the Polyomaviridae and Papillomaviridae families of DNA tumor viruses. J Virol. 2008;82:8849-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wolfe BA, Takaki T, Petronczki M, Glotzer M. Polo-like kinase 1 directs assembly of the HsCyk-4 RhoGAP/Ect2 RhoGEF complex to initiate cleavage furrow formation. PLoS Biol. 2009;7:e1000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pincus D, Letunic I, Bork P, Lim WA. Evolution of the phospho-tyrosine signaling machinery in premetazoan lineages. Proc Natl Acad Sci U S A. 2008;105:9680-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mayer BJ. Clues to the evolution of complex signaling machinery. Proc Natl Acad Sci U S A. 2008;105:9453-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Manning G, Young SL, Miller WT, Zhai Y. The protist, Monosiga brevicollis, has a tyrosine kinase signaling network more elaborate and diverse than found in any known metazoan. Proc Natl Acad Sci U S A. 2008;105:9674-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739-45 [DOI] [PubMed] [Google Scholar]
- 54. Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Matsuoka S, Ballif BA, Smogorzewska A, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160-6 [DOI] [PubMed] [Google Scholar]
- 56. Meek K, Dang V, Lees-Miller SP. DNA-PK: the means to justify the ends? Adv Immunol. 2008;99:33-58 [DOI] [PubMed] [Google Scholar]
- 57. Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421-9 [DOI] [PubMed] [Google Scholar]
- 58. Bartek J, Falck J, Lukas J. CHK2 kinase: a busy messenger. Nat Rev Mol Cell Biol. 2001;2:877-86 [DOI] [PubMed] [Google Scholar]
- 59. Freeman AK, Monteiro AN. Phosphatases in the cellular response to DNA damage. Cell Commun Signal. 2010;8:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li W, Pio F, Pawlowski K, Godzik A. Saturated BLAST: an automated multiple intermediate sequence search used to detect distant homology. Bioinformatics. 2000;16:1105-10 [DOI] [PubMed] [Google Scholar]
- 61. Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997; 278:631-7 [DOI] [PubMed] [Google Scholar]
- 62. Finn RD, Mistry J, Tate J, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211-22 [DOI] [PMC free article] [PubMed] [Google Scholar]


