Abstract
Voltage-dependent anion channels (VDACs) are critical regulators of outer mitochondrial membrane permeability in eukaryotic cells. VDACs have also been postulated to regulate cell death mechanisms. Erastin, a small molecule quinazolinone that is selectively lethal to tumor cells expressing mutant RAS, has previously been reported as a ligand for hVDAC2. While significant efforts have been made to elucidate the structure and function of hVDAC1, structural and functional characterization of hVDAC2 remains lacking. Here, we present an in vitro system that provides a platform for both functional and structural investigation of hVDAC2 and its small molecule modulator, erastin. Using this system, we found that erastin increases permeability of VDAC2 liposomes to NADH in a manner that requires the amino-terminal region of VDAC2. Furthermore, we confirmed that this VDAC2-lipsome sample is folded using solid-state NMR.
Voltage-dependent anion channel (VDAC) proteins are eukaryotic porins that provide the major route for metabolite trafficking across the mitochondrial outer membrane. VDACs are also involved in cell death and are therefore of great interest for their role in health and disease.(1) Humans possess three isoforms of VDAC channels; to date VDAC1, the major isoform, is the best characterized. Recently, the high-resolution structure of VDAC1 has been elucidated by both solution-state and solid-state NMR as well as X-ray crystallography,2,3 revealing a 19-stranded β-barrel fold with an N-terminal helical segment.
In contrast, biophysical and structural characterization of VDAC isoforms 2 and 3 is lacking. VDAC2 and 3 have distinct functions in cell death and serve as targets of the small molecule erastin, which triggers a previously unobserved form of tumor cell death(4) (Figure 1a and Figures S1 and S2 of the Supporting Information). Although VDAC2 is known to interact with and inhibit the pro-apoptotic protein Bak,(5) cell death mediated by erastin occurs by a nonapoptotic mechanism involving VDAC2 and VDAC3. A hallmark of erastin-induced cell death is the formation of reactive oxygen species, raising the possibility that erastin acts by affecting metabolite gating. Modulation of VDAC2 or VDAC3 metabolite gating activity may be a new approach to target disease, and knowledge of the erastin−VDAC2 interaction should improve our understanding of VDAC2 structure−function relationship.
Figure 1.
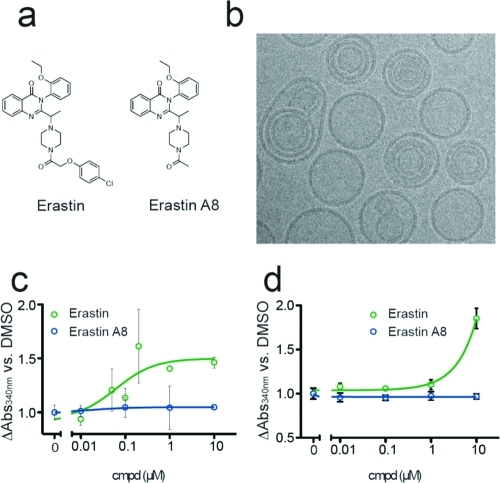
Effect of erastin and erastin A8 on NADH transport through hVDAC2. (a) Structure of erastin and its inactive analogue, erastin A8. (b) hVDAC2-containing liposomes visualized by CryoEM. (c) Relative NADH oxidation rates in wild-type VDAC2-containing liposomes, normalized to oxidation rates observed with DMSO treatment. (d) Relative NADH oxidation rates in N-terminal-truncated VDAC2-containing liposomes, normalized to oxidation rates observed with DMSO treatment.
To gain insight into the effects of erastin on hVDAC2, we developed an in vitro model of hVDAC2 that permits interrogation of structural and functional properties of purified hVDAC2. We purified recombinant His6-tagged hVDAC2 protein from inclusion bodies, followed by on-column refolding in the presence of detergents (see Supporting Information).(4) Similar to what has been reported for native VDAC,(6) this hVDAC2 protein reconstituted into the planar lipid membrane forms a typical VDAC single channel with characteristic gating behavior at high potentials (see Figure S3 of the Supporting Information).
Full-length hVDAC2 was incorporated into liposomes with near 100% efficiency (see Figure S4 of the Supporting Information) by mixing the protein with a lipid mixture of phosphatidylcholine and phosphatidylserine in a 4:1 mass ratio. Because previous studies have indicated the importance of cholesterol(7) in reconstituting VDACs in a functional conformation, we included 0.1 mg/mL cholesterol (see Supporting Information on exact precipitation procedure). The creation of liposomes was verified by CryoEM (Figure 1b and Figure S5 of the Supporting Information).
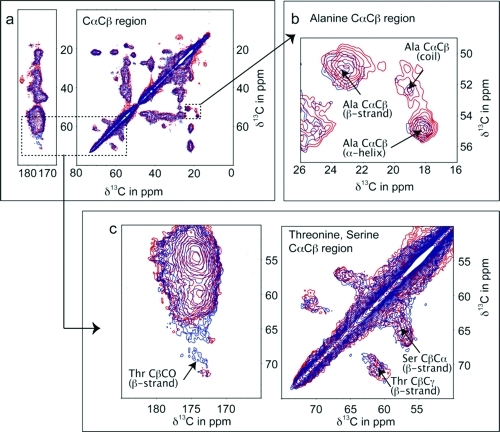
We sought to evaluate the structural integrity of precipitated hVDAC2 liposomes using solid-state NMR. In the past few years, this technique has emerged as a capable method for the investigation of integral membrane proteins8−10 and has recently been applied to structure determination of the N-terminal helix of VDAC1.(11) We prepared uniformly 13C-labeled hVDAC2 and found that hVDAC2 is folded under the conditions used for functional assays (see below and Supporting Information for sample preparation and additional spectra). A two-dimensional 13C−13C dipolar-assisted rotational resonance (DARR(10)) spectrum, which can be considered the “fingerprint” of the protein, differentiates between peaks arising from α-helical residues and those from the β-barrel, based on 13C chemical shift of the backbone and side-chain carbon atoms (Figure 2a). Resonances of Ala (Figure 2b), Thr and Ser (Figure 2c), with a strong structural chemical shift dependence of backbone resonances are highlighted; they all lie within expected chemical shift ranges for β-strands, in good correspondence with published structures of mVDAC1. The alanine resonances also indicate the presence of a helical environment. The strong resonances observed for aromatic residues also indicate the presence of a folded protein (see Supporting Information). On the basis of a comparison of the observed shift regions and structure-based statistics,(12) we conclude that hVDAC2 is in a β-barrel-like conformation, as expected from structural studies of VDAC1. An overlay of the erastin-treated and untreated samples reveals chemical shift changes, most notably in the β-strand threonine region, indicating that erastin may affect these residues by binding on the inside of the pore (see Figure S6 of the Supporting Information).
Figure 2.
The CαCβ and CO region of the 13C−13C DARR11 spectra for a uniformly 15N−13C-labeled sample of hVDAC2 shown in red contours, and in the presence of erastin in blue contours (hVDAC2e; 750 MHz AVANCE spectrometer with sample spinning at 14 kHz). Dashed lines represent spectral connectivities of 13Cα−13Cβ and 13CO backbone nuclei of residues with strong dependence on secondary structure, alanine (Cαstrand 50.86 ± 1.28 ppm, Cαhelix 54.86 ± 0.94 ppm, Cαcoil 52.67 ± 1.76 ppm and Cβstrand 21.72 ± 1.77 ppm, Cβhelix 18.27 ± 1.08 ppm, Cβcoil 19.03 ± 1.77 ppm), serine (Cαstrand 57.14 ± 1.11 ppm, Cβstrand 65.39 ± 1.48 ppm), and threonine (Cαstrand 56.28 ± 1.52 ppm, Cβstrand 70.82 ± 2.11 ppm). Common chemical shifts for backbone carbons for these residues based on the structural environment are given in parentheses.(9)
Pore size is a key indicator of correct folding in VDAC proteins. As an assessment of pore size for the liposome-incorporated hVDAC2, we monitored shrinkage and swelling of liposomes induced by osmolytes of varying sizes by light scattering.(17) In agreement with previous observations in liposomes containing mitochondrially isolated VDAC2, these experiments revealed that osmolytes smaller than 4 kDa can pass through the VDAC2 pore, while larger osmolytes do not induce reswelling (see Figure S8 of the Supporting Information).
Small molecule modulators of VDACs, such as erastin, may act by perturbing VDAC metabolite gating. To test this hypothesis, we developed an assay to monitor the passage of NADH through the VDAC2 pore. In cells, NADH serves as a key substrate and regulator of hVDAC2.13,14 To monitor NADH transport through the hVDAC2 pore, we introduced bovine lactate dehydrogenase (LDH) into liposomes by sonication (see Supporting Information). LDH serves to oxidize NADH entering the liposome, a process that is limited by VDAC2 permeability and which can be easily monitored by observing the decrease in absorption of NADH at 340 nm (see Figure S8 of the Supporting Information).
Liposomes containing hVDAC2 were subjected to the LDH-coupled gating assays and were found to have slower rates of NADH oxidation than samples containing LDH alone, while empty liposomes and liposomes containing LDH, but no VDAC2, showed little-to-no NADH oxidation (see Supporting Information). These data indicate that NADH enters the liposomes almost exclusively through the hVDAC2 pore.
Erastin was previously shown to bind hVDAC2 with a Kd value of 112 nM.(4) We evaluated the effect of erastin on NADH trafficking through hVDAC2. hVDAC2-liposome samples were incubated with varying concentrations of erastin or an inactive analogue, erastin A8, prior to NADH oxidation assays. By comparing the rates of NADH oxidation in DMSO-treated liposomes and compound-treated liposomes, we determined that erastin treatment increases VDAC2 permeability to NADH by 50.1 ± 5.1% with a mean EC50 = 52.6 ± 28.2 nM (Figure 1c). In contrast, erastin A8 (Figure 1a) failed to trigger a similar effect at any concentration tested.
We then investigated the possible involvement of the N-terminal loop in VDAC2 gating and erastin binding by expressing a truncated version of hVDAC2 lacking the first 20 amino acids (ΔhVDAC2). N-terminal truncations of VDACs have previously been described to be functional in lipid-bilayer gating experiments but show altered voltage-dependence and decreased initiation of cell death.(15) LDH-coupled gating assays with ΔhVDAC2 liposomes exhibited 30.6 ± 1.6% lower permeation rates than observed for the wild-type hVDAC2 (see Figure S9 of the Supporting Information). Erastin had no effect on NADH permeability in ΔhVDAC2 liposomes at sub-micromolar concentrations, suggesting involvement of the N-terminal region in mediating erastin’s effect on metabolite gating. At the highest concentration tested, 10 μM, a strong positive effect was observed on NADH permeability (Figure 1d). Consistent with these data is the observation that N-terminal mutants of Neurospora crassa VDAC have an increased preference for the cation selective “closed” state.(16) The N-terminal deletion of ΔhVDAC2 may similarly shift the channel into the closed state and cause a decreased basal rate of NADH permeation through the channel, thus amplifying an effect of high concentrations of erastin in our analysis. Whatever the mechanism, the high concentration makes it less relevant to erastin’s activity in cells.
In conclusion, we have created an in vitro functional system for monitoring metabolite gating of hVDAC proteins, a system that enables side-by-side structural and functional study of these proteins. Using this system, we determined that erastin affects metabolite gating of hVDAC2, leading to a better understanding of erastin’s mechanism of action. These studies should aid in understanding VDAC2 gating mechanisms and in developing novel antitumor agents.
Acknowledgments
We thank Tatiana Rostovtseva for single channel recordings of hVDAC2. We thank Ansgar Siemer (McDermott lab) and Boris Itin (NYSBC) for assistance with solid-state NMR. We thank Amanda Lefkowitz for assistance with cloning and mutagenesis. B.R.S. is an Early Career Scientist of the Howard Hughes Medical Institute and is supported by additional funding from the Arnold and Mabel Beckman Foundation, NYSTAR, and the National Institutes of Health (R01CA097061, R01GM085081, and RC2CA148308). A.E.M. is supported by funding from the National Institutes of Health (R01GM88724).
Supporting Information Available
General experimental details for the expression and purification of natural abundance and isotope-labeled hVDAC2, the cloning of ΔhVDAC2, protein precipitation, liposome swelling assays, LDH oxidation assays, and additional liposome swelling data are available free of charge via the Internet at http://pubs.acs.org.
Supplementary Material
References
- Rostovtseva T. K.; Tan W.; Colombini M. (2005) On the role of VDAC in apoptosis: fact and fiction. J. Bioenerg. Biomembr. 37, 129–142. [DOI] [PubMed] [Google Scholar]
- Hiller S.; Garces R. G.; Malia T. J.; Orekhov V. Y.; Colombini M.; Wagner G. (2008) Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science 321, 1206–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujwal R.; Cascio D.; Colletier J. P.; Faham S.; Zhang J.; Toro L.; Ping P.; Abramson J. (2008) The crystal structure of mouse VDAC1 at 2.3 Å resolution reveals mechanistic insights into metabolite gating. Proc. Natl. Acad. Sci. U. S. A. 105, 17742–17747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagoda N.; von Rechenberg M.; Zaganjor E.; Bauer A. J.; Yang W. S.; Fridman D. J.; Wolpaw A. J.; Smukste I.; Peltier J. M.; Boniface J. J.; Smith R.; Lessnick S. L.; Sahasrabudhe S.; Stockwell B. R. (2007) RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature 447, 864–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng E. H.; Sheiko T. V.; Fisher J. K.; Craigen W. J.; Korsmeyer S. J. (2003) VDAC2 inhibits BAK activation and mitochondrial apoptosis. Science 301, 513–517. [DOI] [PubMed] [Google Scholar]
- Rostovtseva T. K.; Kazemi N.; Weinrich M.; Bezrukov S. M. (2006) Voltage gating of VDAC is regulated by nonlamellar lipids of mitochondrial membranes. J. Biol. Chem. 281, 37496–37506. [DOI] [PubMed] [Google Scholar]
- Colombini M. (2009) The published 3D structure of the VDAC channel: native or not?. Trends Biochem. Sci. 34, 382–389. [DOI] [PubMed] [Google Scholar]
- Li Y.; Berthold D. A.; Gennis R. B.; Rienstra C. M. (2008) Chemical shift assignment of the transmembrane helices of DsbB, a 20-kDa integral membrane enzyme, by 3D magic-angle spinning NMR spectroscopy. Protein Sci. 17, 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M.; Krabben L.; Vinothkumar K. R.; Castellani F.; van Rossum B. J.; Kuhlbrandt W.; Oschkinat H. (2005) Solid-state magic-angle spinning NMR of outer-membrane protein G from Escherichia coli. ChemBioChem 6, 1679–1684. [DOI] [PubMed] [Google Scholar]
- Takegoshi K.; Nakamura S.; Terao T. (2001) C-13-H-1 dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 344, 631–637. [DOI] [PubMed] [Google Scholar]
- Schneider R.; Etzkorn M.; Giller K.; Daebel K.; Eisfeld J.; Zweckstetter M.; Griesinger C.; Becker S.; Lange A. (2010) The native conformation of the human VDAC1 N terminus. Angew. Chem., Int. Ed. 49, 1882–1885. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Jardetzky O. (2002) Probability-based protein secondary structure identification using combined NMR chemical-shift data. Protein Sci. 11, 852–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.; Decker W.; Sampson M. J.; Craigen W. J.; Colombini M. (1999) Mouse VDAC isoforms expressed in yeast: channel properties and their roles in mitochondrial outer membrane permeability. J. Membr Biol. 170, 89–102. [DOI] [PubMed] [Google Scholar]
- Zizi M.; Forte M.; Blachly-Dyson E.; Colombini M. (1994) NADH regulates the gating of VDAC, the mitochondrial outer membrane channel. J. Biol. Chem. 269, 1614–1616. [PubMed] [Google Scholar]
- Lee A. C.; Zizi M.; Colombini M. (1994) Beta-NADH decreases the permeability of the mitochondrial outer membrane to ADP by a factor of 6. J. Biol. Chem. 269, 30974–30980. [PubMed] [Google Scholar]
- Abu-Hamad S.; Arbel N.; Calo D.; Arzoine L.; Israelson A.; Keinan N.; Ben-Romano R.; Friedman O.; Shoshan-Barmatz V. (2009) The VDAC1 N-terminus is essential both for apoptosis and the protective effect of anti-apoptotic proteins. J. Cell Sci. 122, 1906–1916. [DOI] [PubMed] [Google Scholar]
- Popp B.; Court D. A.; Benz R.; Neupert W.; Lill R. (1996) The role of the N and C termini of recombinant Neurospora mitochondrial porin in channel formation and voltage-dependent gating. J. Biol. Chem. 271, 13593–13599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.